Background: Phospholipase C (PLC) isozymes are increasingly attractive therapeutic targets; however, pharmacological modulators are lacking.
Results: A facile fluorescent high-throughput screen was developed and used to identify small molecule inhibitors of PLC activity.
Conclusion: The new assay is robust and suitable for the rapid discovery of novel PLC modulators.
Significance: This new methodology eliminates the major roadblock hampering the discovery of small molecule PLC inhibitors.
Keywords: High-throughput Screening (HTS); Inositol 1,4,5-Trisphosphate; Phosphatidylinositol; Phospholipase C; Small Molecules
Abstract
Phospholipase C (PLC) isozymes are important signaling molecules, but few small molecule modulators are available to pharmacologically regulate their function. With the goal of developing a general approach for identification of novel PLC inhibitors, we developed a high-throughput assay based on the fluorogenic substrate reporter WH-15. The assay is highly sensitive and reproducible: screening a chemical library of 6280 compounds identified three novel PLC inhibitors that exhibited potent activities in two separate assay formats with purified PLC isozymes in vitro. Two of the three inhibitors also inhibited G protein-coupled receptor-stimulated PLC activity in intact cell systems. These results demonstrate the power of the high-throughput assay for screening large collections of small molecules to identify novel PLC modulators. Potent and selective modulators of PLCs will ultimately be useful for dissecting the roles of PLCs in cellular processes, as well as provide lead compounds for the development of drugs to treat diseases arising from aberrant phospholipase activity.
Introduction
Extracellular stimuli, including hormones, growth factors, and neurotransmitters, promote activation of phospholipase C (PLC)2 isozymes and cleavage of the membrane lipid phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) into the classical second messengers, diacylglycerol and inositol 1,4,5-trisphosphate (1). These second messengers coordinately control numerous signaling cascades through the mobilization of intracellular Ca2+ stores and the activation of protein kinase C. Aberrant regulation of PLCs contributes to diverse human diseases such as cancer (2–4), cardiovascular diseases (5, 6), and neuropathic pain (7). Consequently, small molecule PLC inhibitors will be valuable pharmacological tools to dissect the roles of PLCs in development and disease and could potentially serve as candidates for drug development.
Few small molecule PLC inhibitors have been reported, among which U73122 is the most widely used agent (8, 9). However, U73122 has been reported to have pleiotropic effects on cellular processes. Because most experiments with U73122 monitored events secondary to activation of PLCs, e.g. intracellular calcium release, it seems increasingly likely that U73122 does not faithfully report direct inhibition of these isozymes. For example, it was shown that U73122 prevented calcium release by directly inhibiting various Ca2+ pumps (10–13). In addition, the maleimide group within U73122 makes it highly reactive such that most U73122 modifies membrane components and does not enter cells (14). Internalized U73122 reacts with and inhibits a variety of unrelated enzymes, including key enzymes regulating lipid metabolism: phosphatidylinositol-4-phosphate kinase and 5-lipoxygenase (15, 16). U73122 has also been reported to sequester the PLC substrate PtdIns(4,5)P2 (17) and even activate the phospholipase activity of purified PLCs (18). Thus, U73122 is a singularly poor reagent to probe signaling by PLC isozymes. Similarly, small peptides previously used to inhibit PLC enzymes also suffer from indirect effects, as well as from limited bioavailability. Thus, there is overwhelming evidence that the current repertoire of small molecules used to inhibit PLCs do so indirectly and can generate effects that are mistakenly attributed to PLCs. Clearly, a substantial need exists to develop small molecules that directly and selectively modulate PLC isozymes.
Current assays of the phospholipase activity of PLCs rely upon quantification of radioactive inositol phosphates derived from the hydrolysis of radiolabeled PtdIns(4,5)P2. These assays are not readily amenable to high-throughput screens. Although several fluorogenic reporters have been tested to monitor continuously the phospholipase activity of PLCs, they have significant drawbacks, including limited applicability, availability, and reproducibility. For example, fluorescent substrates typically used to study bacterial PLCs are expected to be poorly hydrolyzed by mammalian PLCs (19–23), which have more stringent substrate requirements, including an absolute need for a 4′-phosphate on the inositol ring (24) that is absent from these compounds and some more recently described reporters (25). A second-generation fluorescein derivative of phosphatidylinositol 4-phosphate has been reported to be a fluorescent substrate of PLCδ1 (26); however, it is not commercially available and has not been used in subsequent reports to monitor mammalian PLC activity. Furthermore, this compound is likely to be a poor substrate for mammalian PLCs because it lacks an acyl chain shown to be necessary for efficient hydrolysis by these enzymes (27), a common flaw for most fluorescent substrates reported for mammalian PLCs. More recently, PLCδ1 was shown to efficiently hydrolyze phosphorothiolate analogues of PtdIns(4,5)P2 (28). However, product detection requires a coupled secondary assay that would introduce unnecessary artifacts during high-throughput screens.
We recently developed WH-15, a robust fluorescent reporter useful for directly monitoring the phospholipase activity of mammalian PLCs (29). Here, we used WH-15 to develop a high-throughput PLC assay and verified its utility by identifying three new PLC inhibitors.
EXPERIMENTAL PROCEDURES
Screening of the LOPAC1280 Collection
Chemical compounds (1 mm in 1 μl of dimethyl sulfoxide (DMSO)) were added to assay buffer (19 μl) containing 50 mm HEPES (pH 7.2), 70 mm KCl, 3 mm CaCl2, 3 mm EGTA, 2 mm DTT, and 0.04 mg/ml fatty acid-free BSA. The resulting stock solutions (2 μl) were then added to each well of a PerkinElmer ProxiPlateTM-384 Plus F black plate that contained purified PLCδ1 (4 ng) in assay buffer (4 μl). The mixture was incubated at room temperature for 10 min, and the fluorogenic reporter WH-15 (30 μm) in assay buffer (4 μl) was added to initiate the reaction. After incubation at room temperature for 1 h, 5 μl of stop solution (0.2 m EGTA in H2O (pH 10.2)) was added, and fluorescence was recorded on a PerkinElmer Wallac EnVision 2103 multilabel reader with an excitation wavelength of 355 nm (bandwidth of 10 nm) and an emission wavelength of 535 nm (bandwidth of 10 nm).
Quantification of PLC Inhibition in the Fluorescence-based Assay
Similar to the procedure described above, 2 μl of small molecule inhibitors (10 mm) in DMSO were diluted with assay buffer (78 μl) to make 250 μm stock solutions, which were subsequently serially diluted at a 1:3 ratio with assay buffer containing 2.5% DMSO. Inhibitors (4 μl) at the indicated concentrations were incubated with PLCδ1 (0.5 ng) in assay buffer (2 μl) in a PerkinElmer ProxiPlateTM-384 Plus F black plate at room temperature for 15 min before WH-15 (30 μm, 4 μl) was added to initiate the reaction. The final assay mixtures contained various concentrations of inhibitors (100, 33.3, 11.1, 3.70, 1.23, 0.411, 0.137, 0.046, 0.015, or 0.005 μm), PLCδ1 (0.5 ng), WH-15 (12 μm), 1% DMSO, HEPES (50 mm, pH 7.2), KCl (70 mm), CaCl2 (3 mm), EGTA (3 mm), DTT (2 mm), cholate (0.5%), and fatty acid-free BSA (0.1 mg/ml). DMSO was used instead of inhibitors as a control. Fluorescence was recorded every 5 min as described above, and phospholipase activity was quantified as the ratio of fluorescence intensity in the presence and absence (DMSO only) of inhibitor.
Quantification of Phospholipase Activity of Purified PLC Enzymes
Detergent mixed micelles containing 50 μm PtdIns(4,5)P2 and ∼10,000 cpm of [3H]PtdIns(4,5)P2 per assay were generated by combining lipids in a borosilicate glass tube and drying under a stream of nitrogen. The dried lipids were resuspended by sonication in assay buffer, and assays were initiated by the addition of PLC dissolved in assay buffer. Activity assays were run for 10 min in 60 μl and stopped with trichloroacetic acid (200 μl) and 10 mg/ml BSA (100 μl). The reactions were centrifuged at 5000 × g for 10 min, and the supernatant containing soluble [3H]inositol 1,4,5-trisphosphate was measured by scintillation counting.
Quantification of [3H]Inositol Phosphate Accumulation in Cells
HEK293A cells were maintained in high-glucose DMEM containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in an atmosphere of 95% air and 5% CO2. Forty-eight hours prior to measurements, cells were seeded at a density of ∼20,000 cells/well into a 96-well culture dish coated with 0.01% poly-l-lysine. The culture medium was changed to inositol-free DMEM containing 1 μCi/well myo-[2-3H]inositol (American Radiolabeled Chemicals) ∼24 h after plating, and metabolic labeling was allowed to proceed for 12–18 h. One hour prior to measurements, the medium was changed to 20 mm HEPES-buffered Hanks' balanced salt solution with various concentrations of the PLC inhibitors. After 1 h, cells were challenged with 25 μl of a 5-fold solution of carbachol in 20 mm HEPES (pH 7.2) containing LiCl (50 mm) at 37 °C for 30 min. Incubations were terminated by aspiration of the medium and addition of ice-cold formic acid (50 mm), followed by neutralization with NH4OH (150 mm) after cell lysis. [3H]Inositol phosphates were isolated and quantified using Dowex chromatography. Experiments using 1321N1 human astrocytoma cells stably expressing the human P2Y6 receptor were carried out similarly, except the cells were maintained with 5% fetal bovine serum, and uridine diphosphate (100 μm) was used as the agonist.
cAMP Accumulation and Quantification of ATP Levels
cAMP accumulation in P2Y6-1321N1 cells was quantified as described previously (30). Briefly, cells in 24-well plates were preincubated with [3H]adenine (1 μCi) for 2 h in serum-free DMEM. This medium was then aspirated, and various concentrations of aurintricarboxylic acid (ATA), 3013, or 3017 were added in 25 mm HEPES-buffered Hanks' balanced salt solution. After a 30-min incubation at 37 °C, a solution of 200 μm 3-isobutyl-1-methylxanthine, 10 μm forskolin, and 10 μm isoproterenol was added, and the incubation was continued for an additional 15 min. Incubations were terminated by aspiration of the medium and addition of 5% trichloroacetic acid. [3H]cAMP was isolated by sequential Dowex and alumina chromatography and quantified by liquid scintillation counting as described previously (31). [3H]ATP was isolated by Dowex chromatography and quantified by liquid scintillation counting as described previously (32).
Thermal Shift Assay
Melting temperatures were determined by monitoring binding of the dye SYPRO Orange (Invitrogen) to versions of PLCδ1 during thermal denaturation. PLCδ1 (1 μm) or PLCδ1(E341A) (1 μm) was incubated with ATA in buffer containing 100 mm HEPES (pH 7.5), 70 mm KCl, 3 mm EGTA, 10 mm CaCl2, 2 mm DTT, 1% (v/v) DMSO, and 1:1000 SYPRO Orange. To determine melting temperatures in the absence of Ca2+, the buffer consisted of 100 mm HEPES (pH 7.5), 70 mm KCl, 10 mm EGTA, 2 mm DTT, 1% (v/v) DMSO, and 1:1000 SYPRO Orange. All reactions were performed in triplicate in a volume of 20 μl in 384-well PCR plates (Genesee Scientific). Fluorescence was monitored with an Applied Biosystems 7900HT fast real-time PCR system, using the filter sets for NEDTM, as the temperature was increased from 25 to 95 °C at a ramp rate of 3%.
RESULTS AND DISCUSSION
Assay Development and Pilot Screen
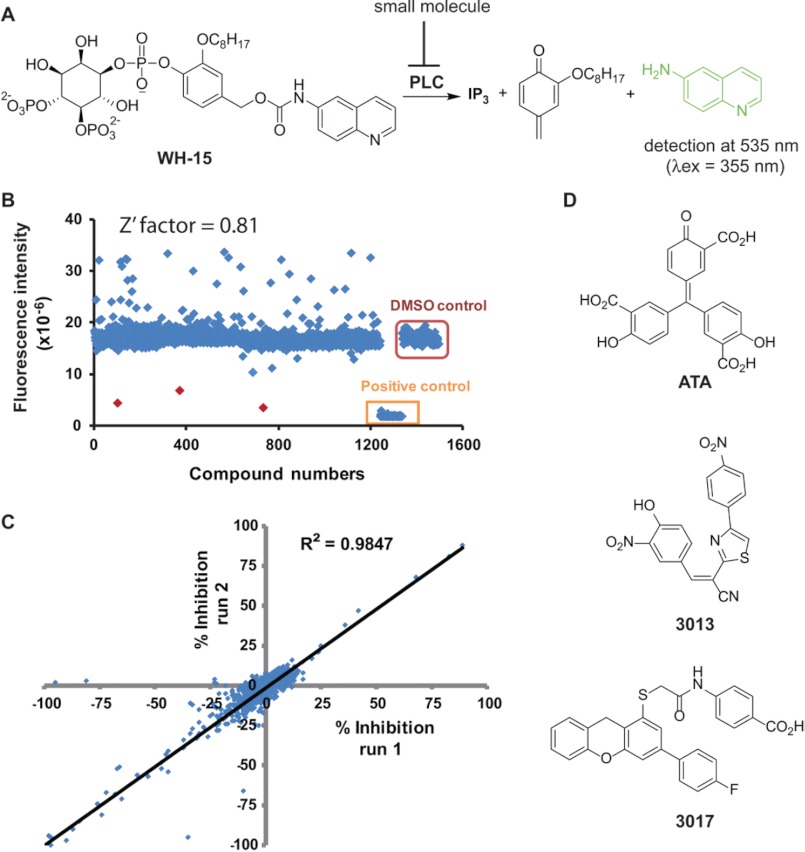
The hydrolysis of WH-15 by PLC isozymes generates up to a 20-fold increase in fluorescence. Thus, we determined whether WH-15 and PLCδ1 could be used to implement a prototype assay suitable for high-throughput screens using 384-well microtiter plates (Fig. 1A). Several parameters were assessed, including (i) order of reagent addition; (ii) concentrations of WH-15, DMSO, and PLCδ1; (iii) reaction time; and (iv) potential stop conditions.
FIGURE 1.
High-throughput screen for small molecule PLC inhibitors. A, fluorescence-based high-throughput assay of PLC activity. Upon PLC action, WH-15 is cleaved to inositol 1,4,5-trisphosphate (IP3), a quinomethide derivative, and 6-aminoquinoline. When excited at 355 nm and detected at 535 nm, WH-15 is non-fluorescent, whereas 6-aminoquinoline is highly fluorescent. B, scatter plot of fluorescence changes after incubation of PLCδ1 and WH-15 with individual compounds of the LOPAC1280 collection. The Z′-factor for the screen is 0.81, with a hit rate of 0.23% for >50% inhibition relative to DMSO. C, screen reproducibility. The correlation coefficient for two parallel screens of the LOPAC1280 library is 0.98. D, chemical structures of representative hits.
For instance, the capacity of PLCδ1 to hydrolyze WH-15 was unaffected by up to 5% DMSO (data not shown), indicating that residual DMSO in the final assay conditions and carried over from the compound stocks will not affect assay performance. Furthermore, the hydrolysis of 10 μm WH-15 by as little as 4 ng of wild-type PLCδ1 was sufficient to generate an ∼15-fold increase in fluorescence over the course of 1 h (data not shown). In contrast, in the absence of PLCδ1 or in the presence of a catalytically inactive form (E341A) of the isozyme, no increase in fluorescence occurred over the same time. WH-15 is stable for up to 24 h at room temperature over a pH range of 7–10 (data not shown). Differences in fluorescence upon incubation of WH-15 with either wild-type or catalytically inactive PLCδ1 in multiple wells of a 384-well microtiter plate were used to calculate an initial Z′-factor of 0.9, indicating that this format is highly reproducible for high-throughput screening (data not shown). Because PLCδ1, PLCβ2, and PLCγ1 hydrolyze WH-15 with similar rates (29), we expect this assay format to be applicable to all PLC isozymes with only minor variations. Finally, the addition of EGTA at a final concentration of 60 mm completely terminated the phospholipase activity of PLCδ1, presumably through chelation with Ca2+ normally required with the catalytic site. Efficient termination of reaction conditions provides for flexibility in plate handling.
Using the optimized assay conditions, we screened in duplicate the 1280 compounds composing the Library of Pharmacologically Active Compounds LOPAC1280 (Fig. 1B). Two microliters of a 50 μm stock of each compound was incubated with 4 ng of PLC (4 μl) for 10 min prior to incubation with 10 μm WH-15 in a final volume of 10 μl. The correlation coefficient for the parallel runs was 0.98, with an average Z′-factor of 0.81 for all plates (DMSO versus EGTA) (Fig. 1C). The hit rate was 0.23% for compounds (10 μm) exhibiting >50% inhibition relative to identical reactions containing DMSO alone. Although this format exhibits excellent characteristics for a high-throughput screen, we noted identification of a disproportionate number of putative activators relative to inhibitors of PLCδ1 (Fig. 1B). The majority of these putative activators most likely represent false hits due to high intrinsic fluorescence. This contention was confirmed by incorporating a secondary counterscreen using catalytically inactive PLCδ1. Overall, the high signal-to-noise ratio and reproducibility of the assay should ensure the successful identification of both activators and inhibitors of PLCs.
As an interesting aside, U73122, which is included in LOPAC1280 and is widely accepted as an inhibitor of PLCs, was not flagged as an inhibitor of PLCδ1 in this screen. This result is consistent with literature reports showing that U73122 does not inhibit a panel of PLCs, including PLCδ1 (18). The success of the pilot screen prompted us also to screen an in-house collection of 5000 compounds with diverse scaffolds. The three most promising hit compounds from both screens (Fig. 1D) inhibited >80% of the phospholipase activity of PLCδ1 at 10 μm and were further assessed for inhibitory potential in additional complementary assays.
Inhibitory Profiles of Hit Compounds with Purified Enzymes
An unknown fraction of the hits identified from high-throughput screens are expected to consist of nonspecific modulators. For instance, a ubiquitous class of “promiscuous” inhibitors tends to self-aggregate, leading to nonspecific adsorption of proteins and concomitant inhibition (33–35). The addition of low amounts (0.01–0.1%, w/v) of various detergents tends to disrupt these aggregates and prevent nonspecific inhibition (36). Unfortunately, the addition of detergents also tends to increase artifacts in high-throughput screens due to increased difficulties in reproducible liquid handling, such as bubbles and varying menisci arising from lowered surface tension. In an effort to minimize potential difficulties in handling the low volumes required for 384-well plates, the high-throughput assays described above were designed without detergents.
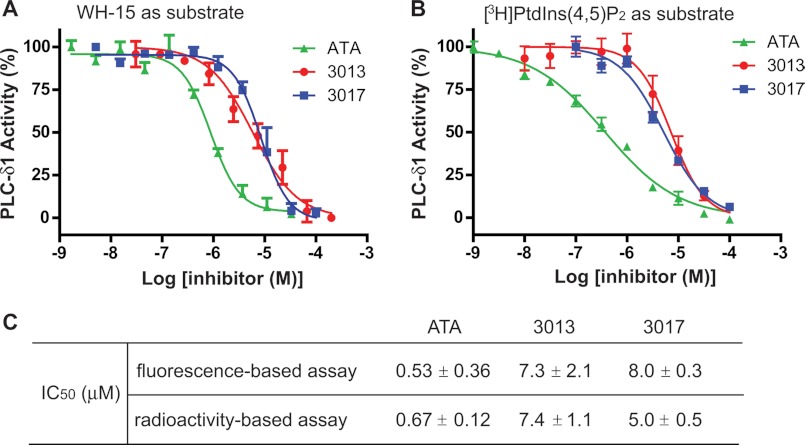
We thus retested the three identified hits (ATA, 3013, and 3017) to generate concentration effect curves for each molecule in the presence of 0.5% cholate. This concentration of cholate is traditionally used to solubilize PtdIns(4,5)P2 in detergent mixed micelles for presentation to PLCs and does not diminish phospholipase activity relative to the presentation of PtdIns(4,5)P2 in lipid vesicles (37). Accordingly, the inhibitors at various concentrations were incubated with PLCδ1 in the presence of 0.5% cholate at room temperature for 15 min before WH-15 was added to initiate the reactions. A plot of the percentage modulation of activity as a function of compound concentration was used to calculate the potency of each inhibitor (Fig. 2A). The half-maximal inhibitory concentrations (IC50) for ATA, 3013, and 3017 were 0.53 ± 0.36, 7.3 ± 2.1, and 8.0 ± 0.3 μm, respectively (Fig. 2C).
FIGURE 2.
Concentration-dependent inhibition of phospholipase activity of PLCδ1 by inhibitors. A, plot of PLC activity in the presence of increasing concentrations of ATA, 3013, or 3017 in the fluorescence-based assays. Purified full-length PLCδ1 was incubated with inhibitors at the indicated concentrations for 15 min before WH-15 was added to initiate the enzymatic reactions. Phospholipase activity was calculated as described under “Experimental Procedures.” The experiments were carried out in triplicate. Error bars are S.D. B, plot of PLC activity in the presence of increasing concentrations of ATA, 3013, or 3017 in the radioactivity-based assays. Purified full-length PLCδ1 was incubated with inhibitors at the indicated concentrations for 15 min before [3H]PtdIns(4,5)P2 was added to initiate the enzymatic reactions. Phospholipase activity was calculated as described under “Experimental Procedures.” The experiments were carried out in triplicate. Error bars are S.D. C, The IC50 values for ATA, 3013, and 3017.
The above analyses monitored fluorescence change derived from hydrolysis of WH-15 by PLCs. However, compounds that either are intrinsically fluorescent or affect the fluorescent properties of WH-15 will likely produce confounding results. To prevent mistaken interpretations based on monitoring the fluorescence of WH-15, we also measured the effects of hit compounds on the enzymatic activity of purified PLC proteins using [3H]PtdIns(4,5)P2 solubilized in cholate as described in the literature (Fig. 2B). In this traditional assay, the IC50 values for ATA, 3013, and 3017 were 0.67 ± 0.12, 7.4 ± 1.1, and 5.0 ± 0.5 μm, respectively (Fig. 2C). Although the slopes of the IC50 plots are slightly different (possibly due to different forms of presentation of WH-15 and PtdIns(4,5)P2 in mixed micelles), these values are essentially the same as those obtained from the fluorescence-based assay, indicating that ATA, 3013, and 3017 are direct inhibitors of PLC isozymes.
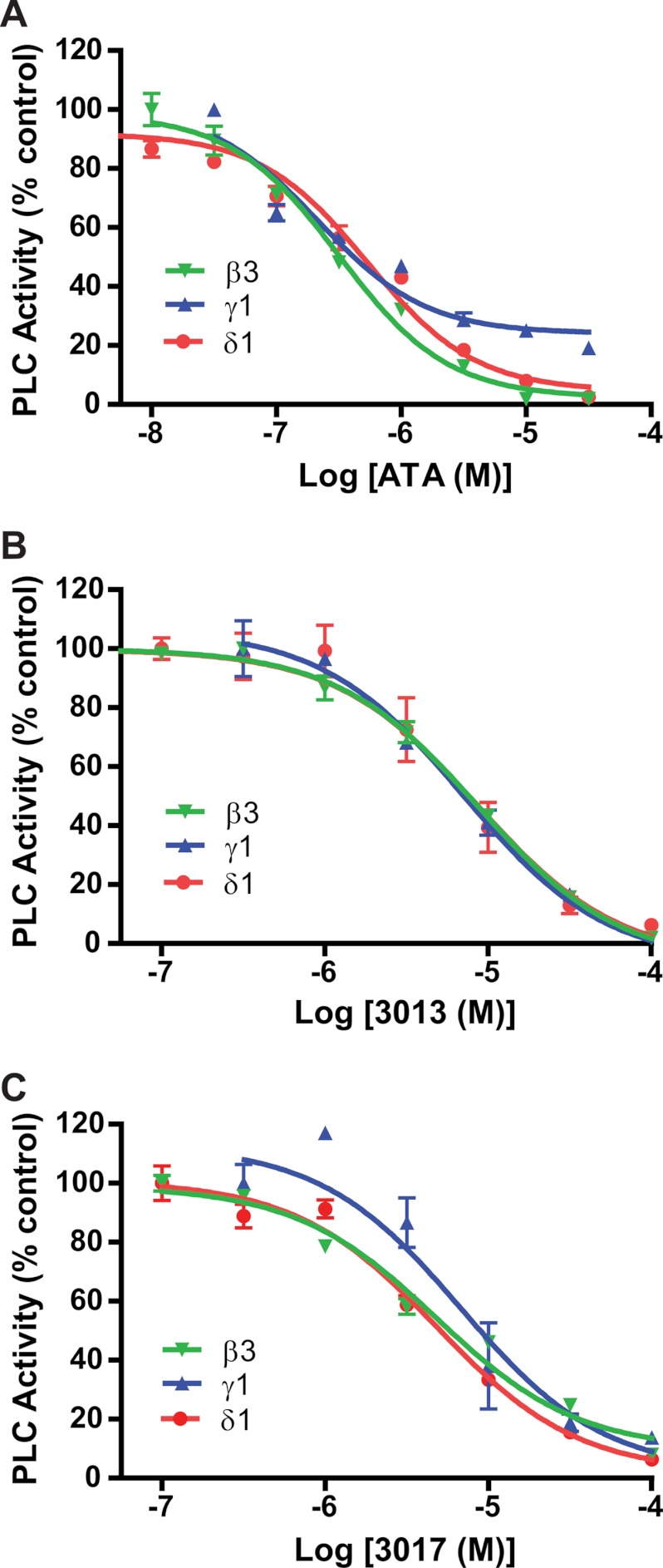
Next, we tested whether the inhibitors are selective among PLC isoforms. Under initial velocity conditions, the concentration-dependent inhibition of the phospholipase activity of PLCδ1, PLCγ1, and PLCβ3 by ATA (Fig. 3A), 3013 (Fig. 3B), or 3017 (Fig. 3C) was measured. Each of the three inhibitors exhibited similar effects on the different PLC isoforms, suggesting that they are general inhibitors of PLCs. These results also are consistent with the idea (but do not prove) that the inhibitors bind to the active site because PLC isozymes share a conserved binding pocket and mechanism of catalysis.
FIGURE 3.
Inhibitory effects of ATA, 3013, and 3017 on different PLC isoforms. The phospholipase activities of purified PLCδ1, PLCγ1, and PLCβ3 in the presence of increasing concentrations of ATA (A), 3013 (B), or 3017 (C) were measured and plotted as described in the legend to Fig. 2.
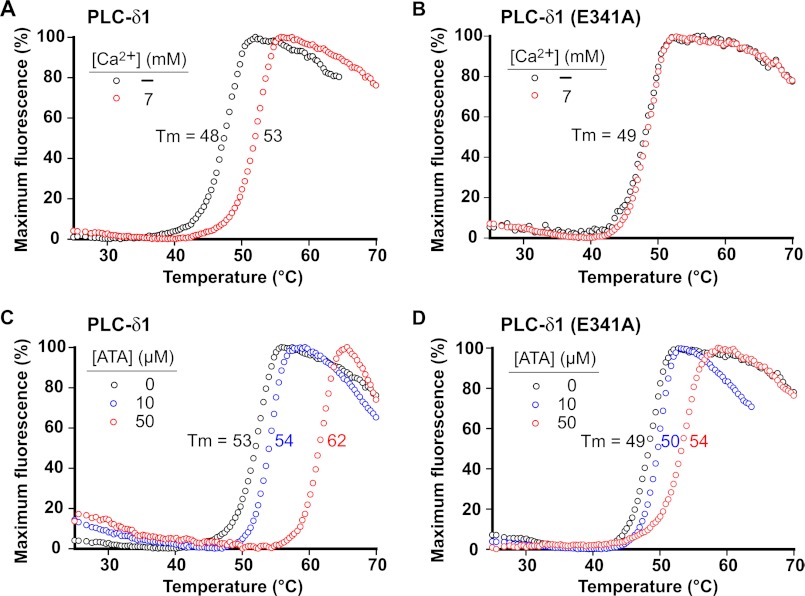
A fluorescence-based thermal shift assay was applied to confirm that the inhibitors directly bind to PLCδ1. Complex formation of a small molecule with PLCδ1 should stabilize the native state of the protein, leading to enhanced thermal stability. In the active site of PLCs, Ca2+ binds to both the enzyme and the substrate to facilitate catalysis. Consequently, we used Ca2+ as a positive control for the assay. Indeed, the melting temperature (Tm) (27) of wild-type PLCδ1 was 5 °C higher in the presence of a high concentration of Ca2+ than when Ca2+ was depleted (Fig. 4A). In contrast, the Tm of PLCδ1(E341A), a mutant form that is unable to properly ligate Ca2+ in its active site due to a single mutation, did not change irrespective of the Ca2+ concentrations (Fig. 4B). Increasing concentrations of ATA were added, and Tm values were quantified similarly. In the presence of 50 μm ATA, the Tm of wild-type PLCδ1 was increased by ∼9 °C (Fig. 4C), whereas that of PLCδ1(E341A) was increased by only ∼5 °C (Fig. 4D). These results suggest that ATA directly binds within the active site of PLCδ1. The other two hits, 3013 and 3017, were not sufficiently soluble under the required solution conditions for the assay.
FIGURE 4.
PLCδ1 is stabilized by ATA in thermal shift assay. Shown are plots of the fluorescence changes of PLCδ1 (A) and PLCδ1(E341A) (B) as the temperature was increased in the presence or absence of Ca2+ (7 mm). With the concentration of Ca2+ at 7 mm, the fluorescence changes of PLCδ1 (C) and PLCδ1(E341A) (D) as the temperature was increased in the presence of increasing concentrations of ATA were then recorded. The mean Tm was calculated from a single experiment in which each condition was assayed in triplicate as described under “Experimental Procedures.”
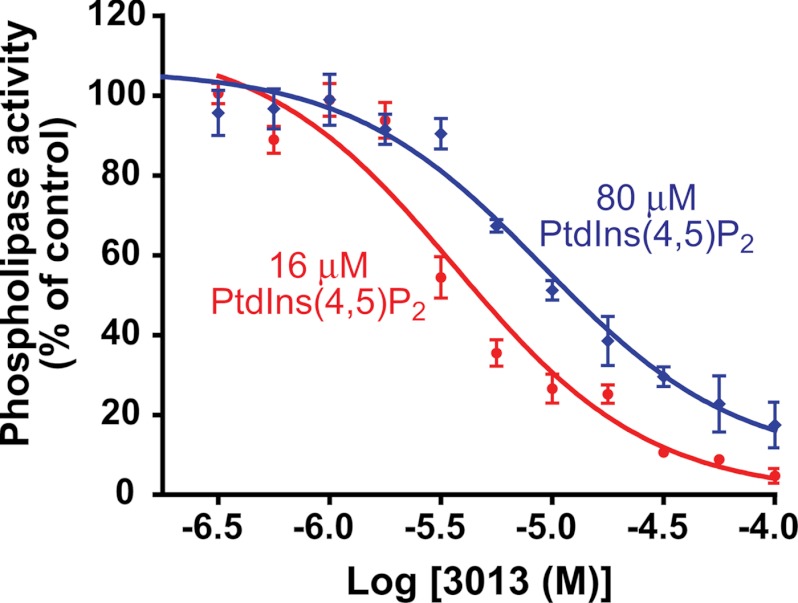
Finally, concentration-dependent inhibition of phospholipase activity of PLCδ1 by 3013 was quantified in experiments in which different concentrations of PtdIns(4,5)P2 were used (Fig. 5). The IC50 values of 3013 were 3.7 and 9.1 μm in the presence of PtdIns(4,5)P2 at 16 and 80 μm, respectively. These data also are consistent with the idea that 3013 binds in the lipase active site and inhibits activity at least in part by a competitive mechanism.
FIGURE 5.
Concentration-dependent inhibition of PLCδ1 by 3013 at two different concentrations of PtdIns(4,5)P2. Purified PLCδ1 was incubated with the indicated concentrations of 3013 in the presence of two different concentrations (16 and 80 μm) of the PtdIns(4,5)P2 substrate. Phospholipase activity was calculated as described under “Experimental Procedures.”
Inhibitory Potential of Hit Compounds in Cellular Assays
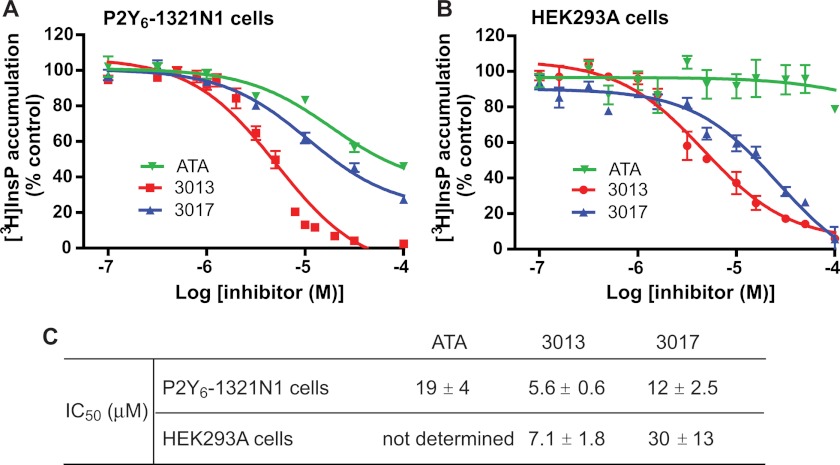
One critical feature of a useful chemical probe is its cell permeability and interaction with its molecular target in intact cells. Consequently, we also assessed the three inhibitors in two cell-based assays. First, we investigated whether cellular PLCs were inhibited by ATA, 3013, or 3017 using previously published protocols to measure phospholipase activity (38, 39). Briefly, 1321N1 human astrocytoma cells stably expressing the UDP-activated P2Y6 receptor were metabolically prelabeled with myo-[2-3H]inositol, and UDP-stimulated [3H]inositol phosphate accumulation was quantified in the presence of various concentrations of ATA, 3013, or 3017 (Fig. 6A). Similarly, carbachol-stimulated [3H]inositol phosphate accumulation was quantified in HEK293A cells in the presence of various concentrations of inhibitors (Fig. 6B). Both 3013 and 3017 caused concentration-dependent inhibition of agonist-stimulated responses in the 1321N1 and HEK293A cell test systems. The potency of ATA in these assays was lower than that of 3013 or 3017, likely due to low cell permeability.
FIGURE 6.
Concentration-dependent inhibition of receptor-promoted inositol lipid signaling by ATA, 3013, or 3017. A, P2Y6-1321N1 cells metabolically prelabeled with [3H]inositol were incubated with increasing concentrations of ATA, 3013, or 3017 for 1 h before stimulation with UDP (100 μm). The cells were lysed, and the radioactivity of the lysate was quantified as described under “Experimental Procedures.” The experiment was repeated three times. B, as described for A, HEK293A cells were metabolically prelabeled with [3H]inositol and treated with increasing concentrations of the inhibitors before incubation with carbachol (10 μm) and quantification of radioactivity. C, the IC50 values for ATA, 3013, and 3017.
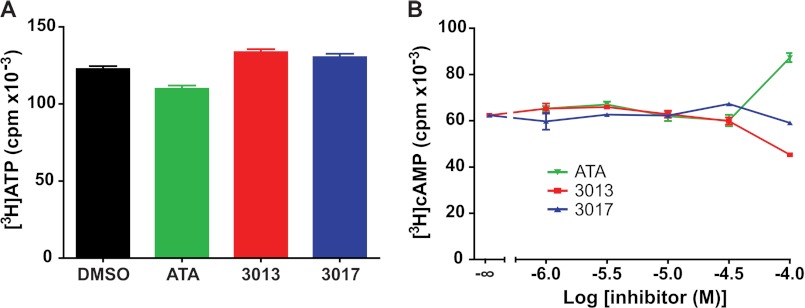
To rule out the possibility that these molecules cause a nonspecific loss of cell viability, their effect on ATP levels was determined. Thus, P2Y6-1321N1 cells were preincubated with [3H]adenine for 2 h, and these metabolically labeled cells were then incubated with 100 μm ATA, 100 μm 3013, or 100 μm 3017 for an additional 30 min. [3H]ATP levels were quantified as described under “Experimental Procedures,” and as illustrated in Fig. 7A, no effect of ATA, 3013, or 3017 on ATP concentration was observed. We also examined whether these phospholipase inhibitors exhibited inhibitory effects on another G protein-regulated effector enzyme. Preincubation of P2Y6-1321N1 cells with a broad range of concentrations of ATA, 3013, or 3017 had no effect on adenylyl cyclase-dependent cAMP accumulation in the presence of forskolin plus the β-adrenergic receptor agonist isoproterenol (Fig. 7B). We conclude from these studies that the inhibitory effects of ATA, 3013, and 3017 on receptor-stimulated inositol lipid signaling are not due to nonspecific effects that diminish cell viability or generally interfere with G protein-mediated signaling cascades.
FIGURE 7.
ATA, 3013, and 3017 do not change cellular levels of basal ATP or (forskolin + isoproterenol)-stimulated cAMP levels. A, ATP levels in the presence of ATA, 3013, and 3017. ATP pools of P2Y6-1321N1 cells were metabolically labeled by preincubation with [3H]adenine for 3 h. ATA (100 μm), 3013 (100 μm), or 3017 (100 μm) was then added, the incubation was terminated after 30 min, and [3H]ATP levels were quantified as described under “Experimental Procedures.” The results are means ± S.E. and are representative of results obtained in two independent experiments. B, (forskolin + isoproterenol)-stimulated cAMP accumulation in the presence of ATA, 3013, or 3017. [3H]Adenine-prelabeled P2Y6-1321N1 cells were incubated with the indicated concentrations of ATA, 3013, or 3017 for 30 min prior to a 10-min challenge with 10 μm forskolin and 10 μm isoproterenol. Accumulation of [3H]cAMP was quantified as described under “Experimental Procedures.” The results are means ± S.E. and are representative of results obtained in two independent experiments. Basal cpm accumulation (data not shown) was ∼400 cpm for each condition and was not changed by the addition of ATA, 3013, or 3017 at concentrations up to 100 μm.
In summary, we have developed a robust fluorogenic high-throughput assay of PLC activity and used it to screen a collection of 6280 compounds. Three new PLC inhibitors were identified and verified in assays with purified enzymes or in cells. Given the small set of compounds that have been screened and the preliminary nature of the studies on the hit compounds, the current three inhibitors might not be optimal to inhibit cellular PLC activity due to poor cell permeability, insufficient potency, or unaccounted nonspecific effects. Indeed, ATA has been shown to inhibit platelet aggregation (40), modulate glucocorticoid receptor-mediated signaling (41), and promote survival and regeneration of retinal ganglion cells (42). In addition, ATA inhibits the activity of multiple enzymes such as DNA topoisomerase II (43), the cytosine deaminase APOBEC3G (44), and the kinase c-Met (45). Thus, it is unlikely that ATA could serve as an effective chemical probe to study PLC-regulated cellular processes. Although there are no reports of the biological activity of 3013 and 3017 in the literature, their moderate potency and solubility should be reasons for caution when using these two inhibitors to modulate the phospholipase activity of PLCs. Despite these limitations, this work provides a complete set of screening protocols that are suitable for high-throughput screening to identify PLC-selective modulators. These modulators would then be exceptionally useful for dissecting signaling cascades controlled by PLCs. Abnormal signaling through PLC isozymes is implicated in a multitude of diseases, including numerous cancers (2, 3, 46–51), atherosclerosis and cardiac failure (5, 52), and schizophrenia and epilepsy (7, 53). Thus, this work also has the potential to provide new and exciting avenues to understand and treat human diseases.
Acknowledgments
We thank William Janzen and Emily Hull-Ryde (University of North Carolina) for help in running the pilot screen.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM098894 (to Q. Z. and J. S.), GM57391 (to J. S. and T. K. H.), and GM38213 (to T. K. H.).
- PLC
- phospholipase C
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- DMSO
- dimethyl sulfoxide
- ATA
- aurintricarboxylic acid.
REFERENCES
- 1. Harden T. K., Sondek J. (2006) Regulation of phospholipase C isozymes by Ras superfamily GTPases. Annu. Rev. Pharmacol. Toxicol. 46, 355–379 [DOI] [PubMed] [Google Scholar]
- 2. Sala G., Dituri F., Raimondi C., Previdi S., Maffucci T., Mazzoletti M., Rossi C., Iezzi M., Lattanzio R., Piantelli M., Iacobelli S., Broggini M., Falasca M. (2008) Phospholipase Cγ1 is required for metastasis development and progression. Cancer Res. 68, 10187–10196 [DOI] [PubMed] [Google Scholar]
- 3. Shepard C. R., Kassis J., Whaley D. L., Kim H. G., Wells A. (2007) PLCγ contributes to metastasis of in situ-occurring mammary and prostate tumors. Oncogene 26, 3020–3026 [DOI] [PubMed] [Google Scholar]
- 4. Bertagnolo V., Benedusi M., Brugnoli F., Lanuti P., Marchisio M., Querzoli P., Capitani S. (2007) Phospholipase Cβ2 promotes mitosis and migration of human breast cancer-derived cells. Carcinogenesis 28, 1638–1645 [DOI] [PubMed] [Google Scholar]
- 5. Zhang L., Malik S., Kelley G. G., Kapiloff M. S., Smrcka A. V. (2011) Phospholipase Cϵ scaffolds to muscle-specific A kinase anchoring protein (mAKAPβ) and integrates multiple hypertrophic stimuli in cardiac myocytes. J. Biol. Chem. 286, 23012–23021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woodcock E. A., Grubb D. R., Iliades P. (2010) Potential treatment of cardiac hypertrophy and heart failure by inhibiting the sarcolemmal binding of phospholipase Cβ1b. Curr. Drug Targets 11, 1032–1040 [DOI] [PubMed] [Google Scholar]
- 7. Kurian M. A., Meyer E., Vassallo G., Morgan N. V., Prakash N., Pasha S., Hai N. A., Shuib S., Rahman F., Wassmer E., Cross J. H., O'Callaghan F. J., Osborne J. P., Scheffer I. E., Gissen P., Maher E. R. (2010) Phospholipase Cβ1 deficiency is associated with early-onset epileptic encephalopathy. Brain 133, 2964–2970 [DOI] [PubMed] [Google Scholar]
- 8. Bleasdale J. E., Thakur N. R., Gremban R. S., Bundy G. L., Fitzpatrick F. A., Smith R. J., Bunting S. (1990) Selective inhibition of receptor-coupled phospholipase C-dependent processes in human platelets and polymorphonuclear neutrophils. J. Pharmacol. Exp. Ther. 255, 756–768 [PubMed] [Google Scholar]
- 9. Bala G. A., Thakur N. R., Bleasdale J. E. (1990) Characterization of the major phosphoinositide-specific phospholipase C of human amnion. Biol. Reprod. 43, 704–711 [DOI] [PubMed] [Google Scholar]
- 10. Hollywood M. A., Sergeant G. P., Thornbury K. D., McHale N. G. (2010) The PI-PLC inhibitor U-73122 is a potent inhibitor of the SERCA pump in smooth muscle. Br. J. Pharmacol. 160, 1293–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pulcinelli F. M., Gresele P., Bonuglia M., Gazzaniga P. P. (1998) Evidence for separate effects of U73122 on phospholipase C and calcium channels in human platelets. Biochem. Pharmacol. 56, 1481–1484 [DOI] [PubMed] [Google Scholar]
- 12. Berven L. A., Barritt G. J. (1995) Evidence obtained using single hepatocytes for inhibition by the phospholipase C inhibitor U73122 of store-operated Ca2+ inflow. Biochem. Pharmacol. 49, 1373–1379 [DOI] [PubMed] [Google Scholar]
- 13. Wang J. P. (1996) U-73122, an aminosteroid phospholipase C inhibitor, may also block Ca2+ influx through phospholipase C-independent mechanism in neutrophil activation. Naunyn-Schmiedebergs Arch. Pharmacol. 353, 599–605 [DOI] [PubMed] [Google Scholar]
- 14. Wilsher N. E., Court W. J., Ruddle R., Newbatt Y. M., Aherne W., Sheldrake P. W., Jones N. P., Katan M., Eccles S. A., Raynaud F. I. (2007) The phosphoinositide-specific phospholipase C inhibitor U73122 (1-(6-((17β-3-methoxyestra-1,3,5(10)-trien-17-yl)amino)hexyl)-1H-pyrrole-2,5-dione) spontaneously forms conjugates with common components of cell culture medium. Drug Metab. Dispos. 35, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 15. Burgdorf C., Schäfer U., Richardt G., Kurz T. (2010) U73122, an aminosteroid phospholipase C inhibitor, is a potent inhibitor of cardiac phospholipase D by a PIP2-dependent mechanism. J. Cardiovasc. Pharmacol. 55, 555–559 [DOI] [PubMed] [Google Scholar]
- 16. Feisst C., Albert D., Steinhilber D., Werz O. (2005) The aminosteroid phospholipase C antagonist U-73122 (1-[6-[[17-β-3-methoxyestra-1,3,5(10)-trien-17-yl]amino]hexyl]-1H-pyrrole-2,5-dione) potently inhibits human 5-lipoxygenase in vivo and in vitro. Mol. Pharmacol. 67, 1751–1757 [DOI] [PubMed] [Google Scholar]
- 17. Vickers J. D. (1993) U73122 affects the equilibria between the phosphoinositides as well as phospholipase C activity in unstimulated and thrombin-stimulated human and rabbit platelets. J. Pharmacol. Exp. Ther. 266, 1156–1163 [PubMed] [Google Scholar]
- 18. Klein R. R., Bourdon D. M., Costales C. L., Wagner C. D., White W. L., Williams J. D., Hicks S. N., Sondek J., Thakker D. R. (2011) Direct activation of human phospholipase C by its well known inhibitor U73122. J. Biol. Chem. 286, 12407–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zaikova T. O., Rukavishnikov A. V., Birrell G. B., Griffith O. H., Keana J. F. (2001) Synthesis of fluorogenic substrates for continuous assay of phosphatidylinositol-specific phospholipase C. Bioconjug. Chem. 12, 307–313 [DOI] [PubMed] [Google Scholar]
- 20. Scholze H., Stütz H., Paltauf F., Hermetter A. (1999) Fluorescent inhibitors for the qualitative and quantitative analysis of lipolytic enzymes. Anal. Biochem. 276, 72–80 [DOI] [PubMed] [Google Scholar]
- 21. Hendrickson E. K., Johnson J. L., Hendrickson H. S. (1991) A fluorescent substrate for the assay of phosphatidylinositol-specific phospholipase C: 4-(1-pyreno)butylphosphoryl-1-myo-inositol. Bioorg. Med. Chem. Lett. 1, 619–662 [Google Scholar]
- 22. Rukavishnikov A. V., Smith M. P., Birrell G. B., Keana J. F., Griffith O. H. (1998) Synthesis of a new fluorogenic substrate for the assay of phosphoinositide-specific phospholipase C. Tetrahedron Lett. 39, 6637–6640 [DOI] [PubMed] [Google Scholar]
- 23. Shashidhar M. S., Volwerk J. J., Keana J. F., Griffith O. H. (1991) A fluorescent substrate for the continuous assay of phosphatidylinositol-specific phospholipase C: synthesis and application of 2-naphthyl-myo-inositol 1-phosphate. Anal. Biochem. 198, 10–14 [DOI] [PubMed] [Google Scholar]
- 24. Heinz D. W., Essen L. O., Williams R. L. (1998) Structural and mechanistic comparison of prokaryotic and eukaryotic phosphoinositide-specific phospholipases C. J. Mol. Biol. 275, 635–650 [DOI] [PubMed] [Google Scholar]
- 25. Rose T. M., Prestwich G. D. (2006) Synthesis and evaluation of fluorogenic substrates for phospholipase D and phospholipase C. Org. Lett. 8, 2575–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rukavishnikov A. V., Zaikova T. O., Birrell G. B., Keana J. F., Griffith O. H. (1999) Synthesis of a new fluorogenic substrate for the continuous assay of mammalian phosphoinositide-specific phospholipase C. Bioorg. Med. Chem. Lett. 9, 1133–1136 [DOI] [PubMed] [Google Scholar]
- 27. Rebecchi M. J., Eberhardt R., Delaney T., Ali S., Bittman R. (1993) Hydrolysis of short acyl chain inositol lipids by phospholipase Cδ1. J. Biol. Chem. 268, 1735–1741 [PubMed] [Google Scholar]
- 28. Liu Y., Mihai C., Kubiak R. J., Rebecchi M., Bruzik K. S. (2007) Phosphorothiolate analogues of phosphatidylinositols as assay substrates for phospholipase C. ChemBioChem 8, 1430–1439 [DOI] [PubMed] [Google Scholar]
- 29. Huang W., Hicks S. N., Sondek J., Zhang Q. (2011) A fluorogenic, small molecule reporter for mammalian phospholipase C isozymes. ACS Chem. Biol. 6, 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carter R. L., Fricks I. P., Barrett M. O., Burianek L. E., Zhou Y., Ko H., Das A., Jacobson K. A., Lazarowski E. R., Harden T. K. (2009) Quantification of Gi-mediated inhibition of adenylyl cyclase activity reveals that UDP is a potent agonist of the human P2Y14 receptor. Mol. Pharmacol. 76, 1341–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harden T. K., Scheer A. G., Smith M. M. (1982) Differential modification of the interaction of cardiac muscarinic cholinergic and beta-adrenergic receptors with a guanine nucleotide binding component(s). Mol. Pharmacol. 21, 570–580 [PubMed] [Google Scholar]
- 32. Salomon Y., Londos C., Rodbell M. (1974) A highly sensitive adenylate cyclase assay. Anal. Biochem. 58, 541–548 [DOI] [PubMed] [Google Scholar]
- 33. Feng B. Y., Shoichet B. K. (2006) A detergent-based assay for the detection of promiscuous inhibitors. Nat. Protoc. 1, 550–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McGovern S. L., Helfand B. T., Feng B., Shoichet B. K. (2003) A specific mechanism of nonspecific inhibition. J. Med. Chem. 46, 4265–4272 [DOI] [PubMed] [Google Scholar]
- 35. Feng B. Y., Shelat A., Doman T. N., Guy R. K., Shoichet B. K. (2005) High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 1, 146–148 [DOI] [PubMed] [Google Scholar]
- 36. Feng B. Y., Simeonov A., Jadhav A., Babaoglu K., Inglese J., Shoichet B. K., Austin C. P. (2007) A high-throughput screen for aggregation-based inhibition in a large compound library. J. Med. Chem. 50, 2385–2390 [DOI] [PubMed] [Google Scholar]
- 37. Hicks S. N., Jezyk M. R., Gershburg S., Seifert J. P., Harden T. K., Sondek J. (2008) General and versatile autoinhibition of PLC isozymes. Mol. Cell 31, 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bourdon D. M., Wing M. R., Edwards E. B., Sondek J., Harden T. K. (2006) Quantification of isozyme-specific activation of phospholipase Cϵ2 by Rac GTPases and phospholipase Cϵ by Rho GTPases in an intact cell assay system. Methods Enzymol. 406, 489–499 [DOI] [PubMed] [Google Scholar]
- 39. Bembenek M. E., Jain S., Prack A., Li P., Chee L., Cao W., Spurling H., Roy R., Fish S., Rokas M., Parsons T., Meyers R. (2003) Development of a high-throughput assay for two inositol-specific phospholipase Cs using a scintillation proximity format. Assay Drug Dev. Technol. 1, 435–443 [DOI] [PubMed] [Google Scholar]
- 40. Owens M. R., Holme S. (1996) Aurin tricarboxylic acid inhibits adhesion of platelets to subendothelium. Thrombosis Res. 81, 177–185 [DOI] [PubMed] [Google Scholar]
- 41. Gerber A. N., Masuno K., Diamond M. I. (2009) Discovery of selective glucocorticoid receptor modulators by multiplexed reporter screening. Proc. Natl. Acad. Sci. U.S.A. 106, 4929–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heiduschka P., Thanos S. (2000) Aurintricarboxylic acid promotes survival and regeneration of axotomised retinal ganglion cells in vivo. Neuropharmacology 39, 889–902 [DOI] [PubMed] [Google Scholar]
- 43. Benchokroun Y., Couprie J., Larsen A. K. (1995) Aurintricarboxylic acid, a putative inhibitor of apoptosis, is a potent inhibitor of DNA topoisomerase II in vitro and in Chinese hamster fibrosarcoma cells. Biochemical Pharmacol. 49, 305–313 [DOI] [PubMed] [Google Scholar]
- 44. Li M., Shandilya S. M., Carpenter M. A., Rathore A., Brown W. L., Perkins A. L., Harki D. A., Solberg J., Hook D. J., Pandey K. K., Parniak M. A., Johnson J. R., Krogan N. J., Somasundaran M., Ali A., Schiffer C. A., Harris R. S. (2012) First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS Chem. Biol. 7, 506–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Milanovic M., Radtke S., Peel N., Howell M., Carrière V., Joffre C., Kermorgant S., Parker P. J. (2012) Anomalous inhibition of c-Met by the kinesin inhibitor aurintricarboxylic acid. Int. J. Cancer 130, 1060–1070 [DOI] [PubMed] [Google Scholar]
- 46. Abnet C. C., Freedman N. D., Hu N., Wang Z., Yu K., Shu X. O., Yuan J. M., Zheng W., Dawsey S. M., Dong L. M., Lee M. P., Ding T., Qiao Y. L., Gao Y. T., Koh W. P., Xiang Y. B., Tang Z. Z., Fan J. H., Wang C., Wheeler W., Gail M. H., Yeager M., Yuenger J., Hutchinson A., Jacobs K. B., Giffen C. A., Burdett L., Fraumeni J. F., Jr., Tucker M. A., Chow W. H., Goldstein A. M., Chanock S. J., Taylor P. R. (2010) A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat. Genet. 42, 764–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lo Vasco V. R., Calabrese G., Manzoli L., Palka G., Spadano A., Morizio E., Guanciali-Franchi P., Fantasia D., Cocco L. (2004) Inositide-specific phospholipase Cβ1 gene deletion in the progression of myelodysplastic syndrome to acute myeloid leukemia. Leukemia 18, 1122–1126 [DOI] [PubMed] [Google Scholar]
- 48. Li M., Edamatsu H., Kitazawa R., Kitazawa S., Kataoka T. (2009) Phospholipase Cϵ promotes intestinal tumorigenesis of ApcMin/+ mice through augmentation of inflammation and angiogenesis. Carcinogenesis 30, 1424–1432 [DOI] [PubMed] [Google Scholar]
- 49. Kassis J., Moellinger J., Lo H., Greenberg N. M., Kim H. G., Wells A. (1999) A role for phospholipase Cγ-mediated signaling in tumor cell invasion. Clin. Cancer Res. 5, 2251–2260 [PubMed] [Google Scholar]
- 50. Follo M. Y., Finelli C., Clissa C., Mongiorgi S., Bosi C., Martinelli G., Baccarani M., Manzoli L., Martelli A. M., Cocco L. (2009) Phosphoinositide-phospholipase Cβ1 mono-allelic deletion is associated with myelodysplastic syndromes evolution into acute myeloid leukemia. J. Clin. Oncol. 27, 782–790 [DOI] [PubMed] [Google Scholar]
- 51. Bai Y., Edamatsu H., Maeda S., Saito H., Suzuki N., Satoh T., Kataoka T. (2004) Crucial role of phospholipase Cϵ in chemical carcinogen-induced skin tumor development. Cancer Res. 64, 8808–8810 [DOI] [PubMed] [Google Scholar]
- 52. Wang Z., Liu B., Wang P., Dong X., Fernandez-Hernando C., Li Z., Hla T., Li Z., Claffey K., Smith J. D., Wu D. (2008) Phospholipase Cβ3 deficiency leads to macrophage hypersensitivity to apoptotic induction and reduction of atherosclerosis in mice. J. Clin. Invest. 118, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McOmish C. E., Burrows E. L., Howard M., Hannan A. J. (2008) PLCβ1 knockout mice as a model of disrupted cortical development and plasticity: behavioral endophenotypes and dysregulation of RGS4 gene expression. Hippocampus 18, 824–834 [DOI] [PubMed] [Google Scholar]