Abstract
Angiogenesis requires coordinated dynamic regulation of multiple phenotypic behaviors of endothelial cells in response to environmental cues. Multi-scale computational models of angiogenesis can be useful for analyzing effects of cell behaviors on the tissue level outcome, but these models require more intensive experimental studies dedicated to determining the required quantitative “rules” for cell-level phenotypic responses across a landscape of pro- and anti-angiogenic stimuli in order to ascertain how changes in these single cell responses lead to emerging multi-cellular behavior such as sprout formation. Here we employ single-cell microscopy to ascertain phenotypic behaviors of more than 800 human microvascular endothelial cells under various combinational angiogenic (VEGF) and angiostatic (PF4) cytokine treatments, analyzing their dynamic behavioral transitions among sessile, migratory, proliferative, and apoptotic states. We find that an endothelial cell population clusters into an identifiable set of a few distinct phenotypic state transition patterns (clusters) that is consistent across all cytokine conditions. Varying the cytokine conditions, such as VEGF and PF4 combinations here, modulates the proportion of the population following a particular pattern (referred to as phenotypic cluster weights) without altering the transition dynamics within the patterns. We then map the phenotypic cluster weights to quantified population level sprout densities using a multi-variate regression approach, and identify linear combinations of the phenotypic cluster weights that associate with greater or lesser sprout density across the various treatment conditions. VEGF-dominant cytokine combinations yielding high sprout densities are characterized by high proliferative and low apoptotic cluster weights, whereas PF4-dominant conditions yielding low sprout densities are characterized by low proliferative and high apoptotic cluster weights. Migratory cluster weights show only mild association with sprout density outcomes under the VEGF/PF4 conditions and the sprout formation characteristics explored here.
Keywords: angiogenesis, cell tracking, population heterogeneity, Markov models
Introduction
Angiogenesis, the generation of new blood vessel branches from existing vessels, arises from activation of an endothelial cell population constituting a previously quiescent vessel by angiogenic cytokines such as vascular endothelial cell growth factor (VEGF). This process involves stimulation of cells among this population to become proliferative and migratory. A few cells adopt an invasive ‘tip cell’ phenotype, whose penetration of the extracellular matrix (ECM) lead to formation of extending sprouts in which following cells (‘stalk cells’) proliferate and migrate1,2. Given a presumably homogeneous starting population, an interesting question is how do individual cells carry out the various kinds of phenotypic behaviors in coordinated manner leading to successful formation of new vessel branches. This question is further complicated by the presence of angiostatic cytokines, such as platelet factor 4 (PF4), which can inhibit sprout formation even when VEGF is present3,4. The net outcome of potential angiogenic sprouting situations for a given tissue environment comprising both angiogenic and angiostatic factors results from an integration of the stimuli’s effects on the various endothelial cell phenotypic behaviors, including not only proliferation and migration but also apoptotic death. Understanding how various molecular and cellular properties may influence the outcome is important for envisioned therapeutic manipulation, such as for anti-cancer treatments or for tissue regeneration strategies5.
Because of the daunting complexity of this multi-factorial system – i.e., multiple phenotypic behaviors governed by multiple environmental cues – computational approaches have been developed by many laboratories with the goal of helping to predict sprouting angiogenic outcomes across diverse landscape of potential circumstances; an excellent review has been provided by Peirce6. These models take into account the differential responses of individual cells in a population using a variety of alternative approaches. For continuum models7,8, a population is described in terms of cell number distribution and its evolution is governed by deterministic or probabilistic differential equations, allowing calculation of how cell number evolves in space and time. Lattice models9,10 generalize a population as individual cells occupying discrete “lattice sites” in a geometrically organized space; the evolution of this population arises from functions governing cell behavior in terms of site properties. In agent-based models, including hybrid continuum/discrete variants, cells are treated as autonomous entities whose behaviors are governed by rules for their phenotypic states11,12. In recent years, hybrid agent-based modeling has gained popularity as it allows simulation of changing cellular and extracellular microenvironment in response to cell-cell and cell-ECM interactions over the course of sprout initiation and extension into ECM9,11,13.
The power of computational models for complex multi-cellular systems such as angiogenesis is critically dependent on having a strong foundation in experimental characterization of the phenotypic behavioral responses of endothelial cells to various angiogenic and angiostatic stimuli that may be encountered in their environment. This characterization needs to possess a number of complex features, including: {a} a probabilistic nature, since individual cells may exhibit different phenotypic behaviors in a given environment; {b} a dynamic nature, since any given cell may exhibit different phenotypic behaviors over the course of time; {c} a contingent nature, since these behaviors will likely be influenced by environmental conditions such as cytokine stimuli. Our work here aims to contribute a helpful advance in this basic experimental foundation.
In particular, our goal is to demonstrate a useful framework by which quantitative data concerning phenotypic behaviors of individual endothelial cells within a population can be characterized across diverse angiogenic and angiostatic conditions in a manner that can be related to overall population behavior such as sprout formation. We have designed and implemented an experimental and analytical methods to determine stochastic transition rate parameters among the four key phenotypic behaviors involved in angiogenic sprouting: proliferation, migration, apoptosis, and a state representing quiescent/sessile (i.e., not proliferating, dying, or migrating). Focusing on the endothelial cell behavior prior to the sprout initiation, we establish a method to identify phenotypic cell behavioral state from time-lapse live-cell microscopic imaging, showing that the basic phenotypic states can be inferred from cell morphology and movement parameters. Using this state identification tool, we characterize the state transition dynamics of more than 800 individual human microvascular endothelial cells (hMVECs) over 24–30 hours across combinations of angiogenic and angiostatic cytokines (VEGF and PF4, respectively) within their respective angiogenesis modulating concentration ranges (Fig S1b,c,e). We find that the transitions among the phenotypic states are consistent with the conditional independence and the memory-less properties of a continuous-time Markov (CTM) process. Thus, a CTM framework provides a valid model of dynamic phenotypic state transitions, with the transition rates estimable from single-cell trajectories.
We discover that the hMVEC population comprises several subpopulations, which cluster with respect to distinct state transition patterns that remain categorically consistent across the landscape of cytokine conditions. VEGF and PF4 treatments alter the hMVEC population behavior by changing the proportions of cells distributed within these state transition patterns. We quantify these proportions in terms of ‘phenotypic cluster weights’, and find that the VEGF-/PF4-induced changes in phenotypic cluster weights correlate successful with population sprout density across all treatment conditions.
Results
Human microvascular endothelial cells can generate angiogenic sprouts from confluent monolayer on Collagen I gel
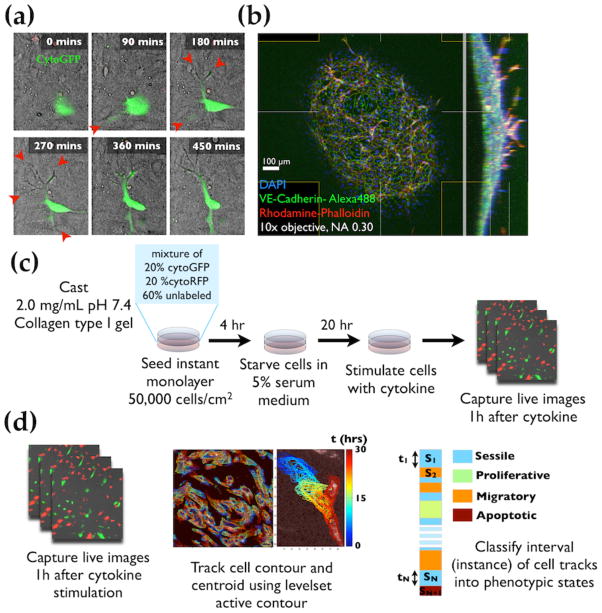
With the longer-term goal of constructing an integrative model encompassing multiple types of experimental data, we select an experimental platform that: {a} is amenable to angiogenic sprout formation; {b} allows the observation of single cells through the course of their phenotypic state transitions; and {c} permits interrogation of the intracellular signaling through which angiogenic cytokines’ effect can be measured. The collagen gel invasion assay (endothelial monolayer on gel as reported in14,15) meets all these criteria. Using this experimental setup, we can modulate the extent to which sprout initiation occurs by treatments with the potent angiogenic cytokine VEGF from a monolayer of human microvascular endothelial cells (hMVECs) (Fig 1a,b). The sprouting responses in Collagen gel invasion assay are comparable to those in in microfluidic devices under the same angiogenic VEGF and angiostatic PF4 cytokine combinations (Fig S1b,c,e). To understand how combinations of cytokines affect the hMVEC population at individual cell level, we used videomicroscopy to track cells over the course of 24–30 hours and ascertained their phenotypic patterns by image analysis. To follow individual cells in fluorescent time-lapse images, a mixture of cytoplasmic GFP-labeled, RFP-labeled, and unlabeled hMVECs were seeded at a 1:1:3 ratio. This mixed cell population was allowed to adhere on 2.0 mg/mL Collagen I gel for 4 hours in endothelial growth medium and then incubated with cytokine-free, 5% fetal bovine serum (FBS) supplemented medium for 20 hours, completing a 24-hour period after seeding. The purpose of FBS incubation is to allow the intrinsic intracellular signaling to subside to basal level without significantly synchronizing the cells to non-mitotic state. The cells are then stimulated with cytokines and immediately imaged in a live-cell imaging chamber with regulated temperature, humidity, and CO2 levels (Fig 1c). Cell contours were detected using a modified level set active contour algorithm16, with cell centroids computed as the center of mass of the detected contour points (Fig 1d). From these individual-cell contours and centroid tracks, we determined the phenotypic state of each time point using morphology- and movement-related features as discussed in the following section.
Fig 1.
Angiogenic human microvascular endothelial cells (hMVECs) cultured on collagen type I gel are capable of initiating angiogenic sprout. (a) Angiogenic protrusions of hMVECs in the collagen gel observed as shadow (red arrowheads) in phase contrast time-lapse images. Time indicated is in minutes. (b) Wide-field confocal image of hMVEC angiogenic sprouts showing 100 – 200 μm invasions. hMVECs were stained with DAPI, VE-cadherin, and Phalloidin to visualize nuclei, cell-cell junctions, and actin cytoskeleton respectively. (c) Experimental setup for live cell imaging experiments. First, a mixture of GFP-labeled, RFP-labeled, and unlabeled hMVECs were seeded at an optimized seeding density (50,000 cells/cm2) to yield a confluent monolayer after 24 hours of seeding. Cells are incubated for 20 hrs under 5% fetal bovine serum (FBS) supplemented cytokine-free base medium, then stimulated with angiogenic/angiostatic cytokines. After cytokine stimulation, cells are imaged over a 24 – 30 hour period. (d) Image analysis procedure. Cell contours and centroids at each time point were detected by a level set active contour algorithm. The detected cell contour and centroid trajectories were classified into different phenotypic states: sessile, proliferative, migration, and apoptotic.
Individual endothelial cells exhibit four identifiable phenotypic behaviors: Sessile, Proliferative, Migratory, and Apoptotic
Time-lapse imaging and tracking of hMVEC contours and centroids revealed four distinct major phenotypes exhibited by any given cell at any given time-point: proliferative, apoptotic, migratory, and sessile. These phenotypes are identifiable based on cell morphology and dynamics of transient motility features (Fig 2a). Proliferative and apoptotic phenotypes are characterized by doubling/splitting and disappearance/shrinkage of existing contour respectively and can be detected as changes in contour topology. Migratory phenotype is characterized by productive and persistent translocation of the centroid, whereas sessile phenotype is characterized by erratic and unproductive centroid variation. A single-cell track is obtained from live-cell imaging as a temporal series of snapshots of the cell contour and centroid properties; we refer to individual snapshot as an ‘instance’ in the track. To assign a phenotypic state to each instance of a given track, we used a hybrid approach outlined in Figure 2b. First, we determined whether the contour topology changes (splits, or collapses), with respect to the previous time-step within the instance being state assigned. If a contour instance splits or collapses, we assigned the instance to be in proliferative or apoptotic state respectively. If the contour topology does not change, the instance is subjected to further classification into either migratory or sessile states.
Fig 2.
Timelapse images of hMVECs reveal four major phenotypes: proliferative, apoptotic, migratory, and sessile. (a) Example contours outlining hMVEC in sessile, proliferative, migratory, and apoptotic states as detected by level set active contour. Proliferative instances are characterized by contour splitting, while apoptotic instances by contour collapsing. Scale bars are 20 μm. (b) Semi-hierarchical scheme for state classification. Contours are first classified based on whether there is a change in contour topology. If topology is conserved, they are further classified into sessile or migratory instances based on contour morphology and motility-related features. (c) Hierarchical clustering reveals two clusters within the subset of non-proliferative, non-apoptotic instances. The main discriminatory features between the two subsets are the mean and the variance of VACFs over 1 – 6 hour track interval. Sessile instances (blue) are characterized by low μVACF, high σ2VACF while migratory instances (orange) are characterized by high μVACF, low σ2VACF. (d) PCA projection of the motile vs. sessile instances as classified by an optimized ensemble of sessile vs. motile base classifiers (details in Supplementary Method). (e) Example instances of single cell contour and centroid tracks labeled to show the sessile vs. motile state identification results.
Based on the above description of migratory vs. sessile states, we asked whether a single motility feature is sufficient as a discriminatory criterion. To undertake migratory vs. sessile classification in a more informed yet unbiased manner, we computed morphological and motility related features of instances of centroid tracks (Table 1). Among these features, we included cell step size and velocity autocorrelation function (VACFN), which are one-interval equivalents of speed and directional persistence of cell migration respectively. Hierarchical clustering analysis in the feature space reveals two distinct subsets of instances (Fig 2c). Notably, the first subset (orange cluster) is characterized by high mean VACFN and low variance of VACFN, fitting the migratory description. (Here N denotes the number of time interval over which mean and variance of VACF are computed). Conversely, the other subset is characterized by low mean VACFN and high variance of VACFN (aqua cluster) that follows the sessile description. Three-dimensional principle component embedding of the instances shows separation of these two clusters (Fig 2d), demonstrating that multiple features are significantly useful as determinants of migratory vs. sessile instances (Fig 2d).
Table 1.
Types and descriptions of morphological and motility related features used in classifying of instances of cell tracks into sessile vs. migratory states. Morphological features were computed based on the detected contour points of individual contour instance. Motility related features were computed based on the detected centroid across N intervals around the instance being feature computed. Step size is the one time step equivalent of cell speed and velocity autocorrelation function is the one time step equivalent of directional persistence.
| Features | Notation | Type | Definition |
|---|---|---|---|
| step size | d | motility | centroid-to-centroid distance in subsequent time step |
| mean velocity autocorrelation function across N time step | μVACFN | motility | sample mean of dot products of N adjacent unit velocity vectors |
| variance velocity autocorrelation function across N time steps | σ2VACFN | motility | sample variance of dot products of N adjacent unit velocity vectors |
| cell size | A | morphological | area enclosed by cell contour |
| cell elongation | ℓ | morphological | ratio between cell major and minor axes |
| cell orientation | θ | morphological | dot product of unit instantaneous velocity vector and unit major axis |
Our single cell trajectory dataset comprises more than 500,000 cell track instances to be classified into either sessile or motile state. To meet the computational challenge of classifying these instances, we drew non-overlapping subsamples of cell track instances that make up about 2% of all instances and performed initial hierarchical clustering analysis. In all of the subsamples, hierarchical clustering yields two well separated clusters (example shown in Fig 2d) characterized by differences in mean and variance VACFN. We used these cluster assigned subsamples (of cell track instances) as labeled data to train, test, and cross validate an ensemble of base classifiers using the Adaptive Boosting algorithm17,18. The validated ensemble classifier achieved the desired classification task as seen from the images of cell centroid tracks and the sessile vs. migratory state prediction (Fig 2e). In the instances in which cells exhibit productive locomotion, the instances are labeled migratory (shown as orange centroid track and contours), while those in which cell contours or centroids fluctuate erratically the instances are labeled sessile (shown as aqua centroid track and contours). These results demonstrated that our state annotation approach yields satisfactory classification results using an unbiased method.
Individual-cell phenotypic state transition patterns statistically follow one-step dependent continuous time Markov chain (CTMC) dynamics
Applying the state classification method described above, we converted the time-series snapshots of each cell track into a sequence of states with corresponding waiting time prior to each transition; we refer to this sequence as a ‘single-cell state trajectory’ (illustration in Table 2). The transition dynamics of these single cell trajectories are well characterized by continuous time Markov process based on two criteria. First, the waiting time distributions of the state transition in the data are well estimated by an exponential distribution (Fig S3a). Second, the state transitions are consistent with the conditional independence properties of a one step dependence continuous time Markov chain19 (Fig S3b,c,d).
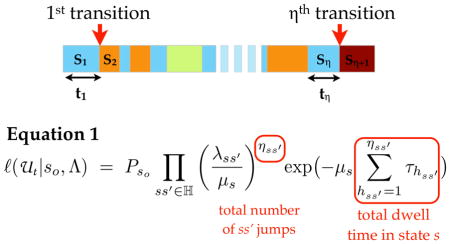
Table 2.
A hypothetical state trajectory with η state transitions. States are color-labeled. According to the continuous time Markov (CTM) model, the likelihood of a particular transition rate parameter set Λ given the observed state trajectory U is a product of the likelihoods of all individual transitions observed in the trajectory, and the marginal probability of the first state So (PSo). After an arithmetic manipulation, the likelihood can be written as in Equation 1 (details in Supplementary Modeling Approaches 2.1). The likelihood expression contains two types of experimentally obtainable variables. The first is the total number of transitions of certain type ss′ (ηss′). The second set is total dwell time in a certain state s (Σhτh). Since these variables are involved in determining the likelihood, they are used as state transition dynamic features for classifying single cell state trajectories.
| |||
|---|---|---|---|
| Feature | Notation | Definition | |
| frequency of ss′ type jumps (trajectory length normalized number of ss′ type jumps) | fss′ |
|
|
| frequency of jumps of all types (trajectory length normalized number of any jumps) | f tot |
|
|
| total dwell time in state s | τtot | Σhs τhs | |
| mean dwell time in state s | τs |
|
|
| variance dwell time in state s | σ2τs |
|
|
An advantage of modeling single cell trajectories in terms of a continuous time Markov chain (CTMC) is that the parameter estimation problem based on likelihood function can be solved analytically. In a CTMC, the probability at which a cell transitions from a state s to another state s′ after some time t depends on the relative rates to s′ compared to the rates to other states s″ reachable from s (SI Modeling Approaches 2.1). Since individual state transitions in CTMC are independent, the likelihood of a single cell trajectory (as a sequence of state transitions and corresponding waiting time) is a product of likelihood of all individual transitions (illustration in Table 2). From this likelihood of single cell trajectories (expression in Table 2), we can determine the set of transition rate parameter values most consistent with the observed single cell trajectories by either a maximum likelihood estimation (MLE) or Bayesian inference (BI). In either case, we rely on the same likelihood distribution of the phenotypic transition rates given the observed single cell trajectories. For MLE, we solved for the rate parameter sets that maximize the likelihood distribution function whereas for BI we weighted the likelihood distribution by a conjugate prior and renormalized the resulting distribution.
By combining automatic phenotypic state identification from single-cell data and the parameter estimation procedure, we have established a method that enables determination of the phenotypic state transition rates consistent with agent-based modeling. Our rate parameter estimation methodology consists of three main aspects. First is the contour tracking method that maps time-lapse images to sets of contour points outlining individual cells. Second is the automated state annotation based on features derived from the images, the detected contour points, and the centroids. Third is parameter estimation method based on CTMC. We now proceed to the application of our method to a particular biological system: quantitative analysis of how cytokine-modulated individual-cell phenotypic behavioral state transition patterns may govern changes in population-level sprouting.
VEGF and PF4 differentially influence hMVEC dynamic phenotypic state transitions by altering the distribution of cells among diverse behavioral subpopulations
With our analysis methodology in hand, we proceeded to examine the phenotypic state transition dynamics of hMVECs treated with vascular endothelial growth factor (VEGF) and platelet factor 4 (PF4) -- opposing angiogenesis modulators that are co-released from activated platelets during the onset of inflammation20,21. The cytokine conditions selected for this study (0 – 80 ng/mL VEGF and 0 – 500 ng/mL PF4 in the background of VEGF) are physiologically relevant for angiogenesis under acute inflammation conditions and effectively modulate sprouting angiogenesis in vitro (Fig S1) and in vivo3,22,23. Within these concentration ranges, we found that VEGF dose dependently induces sprouting in collagen gel invasion assay and in microfluidic device sprouting assay. On the other hand, PF4 at the higher end of this range suppresses VEGF-induced sprouting.
Figure 3 shows the single-cell state trajectories for each of the cytokine treatment conditions selected from the VEGF and PF4 sprouting dose response curve, with approximately 100 cells observed in each condition. Some differences among the sets of trajectories seem readily apparent, such as more migratory states for the VEGF-treated cells, but the quantitative comparisons of both kinds of states and transitions between them are not obvious from inspection. Thus, we need to bring the next layer of computational analysis to bear on these data, in order to elucidate the cytokine influences.
Fig 3.
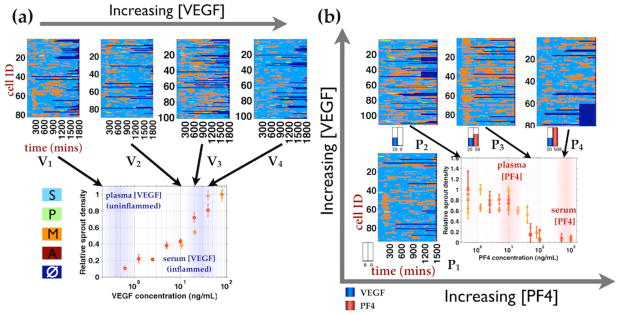
Phenotypic state labeled cell trajectories of hMVEC treated with physiologically relevant VEGF and PF4 concentration ranges. Single cell state trajectories are plotted over time along the rows and phenotypic states are colored labeled as follow: Sessile(S) - aqua; Migratory (M) - orange; Proliferative (P) - pale green; Apoptotic (A) - dark red, and No data (□) - dark blue. The corresponding dose response curves of hMVEC sprout densities under VEGF and PF4 treatments are shown underneath the single state cell trajectories plots. (a) hMVECs were treated with VEGF concentrations within the transition region of hMVEC sprouting dose response curve in collagen gel invasion assay (fig S2), which is the same setup as the imaging experiment (Fig 1). V1, …, V4 = 0, 10, 20, 40 ng/mL VEGF. (b) hMVECs were treated with PF4 concentrations spanning with transition region of PF4 dose response curve in the background of physiological VEGF concentration (20 ng/mL). P1 = no cytokine, P2, …, P4 = 0, 50, 500 ng/mL PF4 in the background of 20 ng/mL VEGF.
We considered two kinds of conceptual models to account for the individual-cell phenotypic state transitions as they are distributed across a cell population (Fig 4). A Uniform Population Model (UPM) posits that all cells in the population intrinsically possess identical potential to adopt different phenotypic states, such that under a cytokine combination, the population state transition rates are described by a single transition rate parameter set. In contrast, a Diverse Population Model (DPM) posits that endothelial cells within angiogenic population are heterogeneous in their state transition dynamics such that, under a cytokine combination, it cannot be described by a single parameter set. One or the other kind of model might prove superior with respect to capturing the features of our experimental data, although it is possible that both kinds of models can do so satisfactorily. If the UPM is more consistent with the data, the dependence of phenotypic transitions on context conditions is due to a uniform population of cells exhibiting transition probabilities that are as a whole modulated by context (angiogenic/angiostatic cytokine treatment). If, on the other hand, the DPM is more consistent, the context dependence is better explained as due to cell subpopulations exhibiting transition probabilities invariant with respect to treatment but with the treatment modulating the proportion of cells in each subpopulation.
Fig 4.

Conceptual model of angiogenic population. (a) Uniform population model posits that endothelial population is homogeneous in state transition dynamics and the population is unimodally distributed in transition rate parameter space. Under control condition, the population assumes a unimodal distribution in state transition rate parameters (top). Treatments with cytokines (shown as black arrows) cause individual cells to respond in a similar manner and the population distribution to shift unidirectionally (bottom left and right shown as results of two different stimulations). (b) Diverse population model posits that endothelial population is heterogeneous in state transition dynamics. Under control condition, the population consists of multiple subpopulations characterized by different transition rate parameters (top). Treatments with cytokines cause changes in the fractions of cell within subpopulations without shifting subpopulations centers (bottom left and right).
To address this question, we applied our parameter estimation method in two ways. First, under the UPM, we performed parameter estimation on the all state trajectories within each condition separately and refer to the rates estimates obtained via this approach as condition-based estimates (λ(cond)). Alternatively, under the DPM, we first cluster the state trajectories based on relevant transition dynamic descriptors (listed in Table 2). As we observed subpopulations of distinct state transition dynamics, we subsequently performed parameter estimation on the single-cell trajectories within each cluster separately. The rate estimates obtained via this approach is referred to as phenotypic program-cluster based estimates (λ(clust)).
For this set of cytokine treatments, the UPM-derived condition-based estimates are shown in Figure 5a-b. Some differences can be seen between the phenotypic state transition rates for different cytokine treatment conditions. Following the λ(cond) in the VEGF dose conditions (first to fourth bars in Fig 5c subplots), VEGF treatments increase the S-to-P transition rate, consistent with previous findings that VEGF promotes proliferation24. VEGF also appears to increase S-to-M while decrease S-to-A transition rates. However, the changes in these two transition types are non-monotonic (first to fourth bars in Fig 5c S-to-M and S-to-A subplots). Going from VEGF-treated to combination with PF4 (second to forth bars in Fig 5d subplots), the S-to-P and P-to-S transition rates directionally decrease as PF4 concentration increases, while S-to-A transition rates directionally increases. Other transition rates do not change directionally. These results suggest that while the majority of the cytokine-induced changes may be consistent with the current understanding of VEGF and PF4 roles, these changes are difficult to interpret with the UPM framework.
Fig 5.
Maximum likelihood estimates (MLEs) of the condition based transition rates under the physiologically relevant cytokine conditions. Under the uniform population model’s assumptions, MLEs of the phenotypic transition rates were computed from the cell trajectories under each treatment condition separately. (a and b) The resulting transition rate matrices exhibits slight difference across increasing VEGF (V1 - V4) and PF4 concentration (P1 - P4). (c and d) Alternative way of visualizing the rate estimates of the same transition types across cytokine conditions. (c) Rate estimates of the same starting and ending states are presented in the same subplot as a function of increasing VEGF (V1 - V4). (d) Rate estimates for the same starting and end staets shown as a function of PF4 concentrations in the background of constant VEGF (P1 - P4).
To pursue the alternative DPM-derived cluster-based estimates, we sought clustering features that will resolve the differential state transition dynamics of single cell trajectories within the population pool. In the likelihood expression of a cell trajectory (equation in Table 2), there are two sets of key variables determining the state transition dynamics: {a} fss′ - the (trajectory length) normalized frequencies of the transition from s to s′ states; and {b} Στs - the total dwell or waiting time in a particular state s for that trajectory. fss′ describes the average frequency at which ss′ state transition occurs, while Στs the total s state dwell time is inversely proportional to the rate at which an agent escapes state s. Intuitively, these parameters are relevant descriptors of state transition dynamics and can be used as clustering features.
Hierarchical clustering of single cell trajectories based on fss′ and Στs reveals three to five identifiable clusters (Fig 6a). Notably, the clustering pattern and the cluster assignment correspond well to the different cellular phenotypes in which cells dwell in the most. Clusters 1 and 2 consist exclusively of single-cell trajectories that transition through proliferative and apoptotic states and are referred to as proliferative (P cluster) and apoptotic (A cluster) respectively. Clusters 3, 4, and 5 contain trajectories that traverse only through sessile and migratory states and are more related. Trajectories in Cluster 3 are characterized by relatively longer dwell time in sessile and Cluster 4 by relatively longer dwell time migratory states and are referred to as sessile (S cluster) and migratory (M cluster) respectively. Cluster 5 is highlighted by an exceptionally high rate of back-and-forth shifting between sessile and migratory states, so is referred to as switching (Sw cluster). It is conceivable that the M and Sw clusters could be combined into a single cohort, but because they clearly exhibit different quantitative properties with respect to their individual-cell trajectories we held it important to consider their contributions separately.
Fig 6.
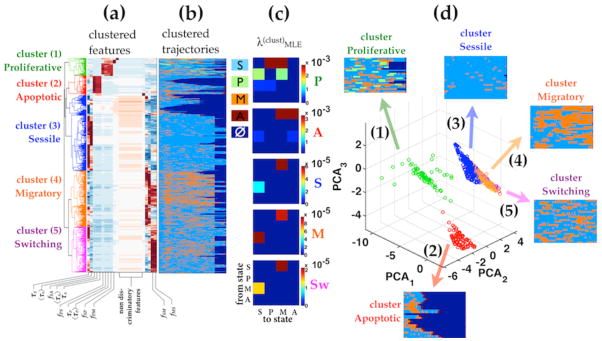
Hierarchical clustering of all cell trajectories in all cytokine conditions reveals 3–5 identifiable clusters. Hierarchical clustering was performed based on the trajectory features described in Table 2. (a) Dendrogram of the clustered features and (b)the corresponding clustered single cell trajectories. Clusters color labels are consistent from (a – d). (c) The corresponding transition rate MLEs computed from the single cell trajectories in each cluster separately λ(clust). The transition rates matrices in sessile, migratory and switching clusters are qualitatively similar. Under DPM, the effect of cytokine treatment can described as changes in the fraction of cells adopting different dynamic phenotypic transition patterns (also referred to as cluster weights). (d) Principle component projections of cell trajectories shows the separation of the endothelial subpopulations. The apoptotic and proliferative clusters (1 and 2) are well separated in state transition dynamics from each other and from the sessile, migratory, and switching clusters. Sessile, migratory and switching clusters (3, 4, 5) consist of cells that transition between sessile vs. motile states and are only differentiated by the frequencies of transition and the dwell times within the sessile vs. motile states.
The transition dynamics of trajectories in each of these clusters is determined by the phenotypic program-based transition rate values (Fig 6c), characterizing the probabilities for each subpopulation in quantitative manner. The cluster based transition rate MLE matrices here permit explicit appreciation of the underlying structure of the key phenotypic transitions, which may not be easily discerned from the trajectories. For instance, A cluster exhibits the largest rate constants for transition into A state, but predominantly from S state and M state. For another, P cluster shows a large rate constant for transition into P state but only from S state (Fig 6c). The same hierarchical clustering analysis performed on single-cell trajectories of different treatment conditions yield similar clustering patterns (Fig S4), suggesting that the hMVEC population is heterogeneous with respect to angiogenesis-related phenotypic behaviors and can adopt one of a few distinct state transition patterns. Projection in principle component subspace (Fig 6d) shows clear contrast between the A cluster and the P cluster and a separate aggregate of S, M, and Sw clusters. Interestingly, the quantitative relationship for transitioning between S and M clusters is significantly different from that between S and Sw clusters, indicating an underlying distinction among these three exists with respect to their influence by the cytokine treatments.
To examine the cytokine effects within the DPM framework, we computed the phenotypic cluster weight coefficients of each treatment condition with respect to proportion of cells in each behavioral subpopulation (Fig 7a,b). In the no cytokine control condition (first bars in Fig 7a,b), the proliferative (green), and apoptotic (red) cluster weights are small compared to that of switching, migratory, and sessile clusters. With VEGF (second to fourth bars in Fig 7a), the proliferative cluster weight (wP) increases while the apoptotic cluster weight (wA) decreases as a function of VEGF doses. Upon adding PF4 from 0 to 500 ng/mL under constant 20 ng/mL VEGF concentration (second to fourth bars in Fig 7b), wP decreases from 0.16 to 0.03, while wA increases from 0.04 to 0.10. Compared to the corresponding conditions with VEGF, the conditions with no VEGF have higher wA (0.186 for 0 ng/mL VEGF to 0.037 for 40 ng/mL VEGF) and lower wP (from 0.096 at 0 ng/mL VEGF to 0.192 at 40 ng/mL VEGF).
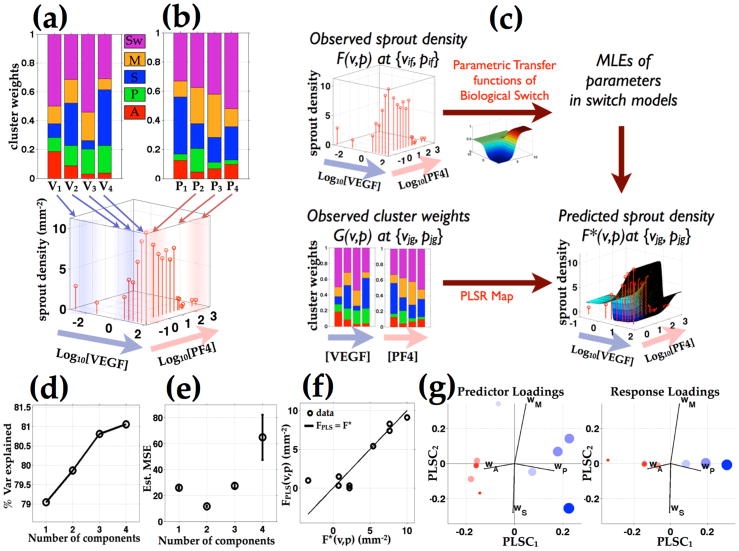
Fig 7.
Proliferative and apoptotic cluster weight axes predicts the VEGF and PF4 modulated population level sprout densities. (a and b) Phenotypic cluster weights and sprout densities vary as functions of input VEGF and PF4 concentrations. (c) A quantitative mapping between the observed phenotypic cluster weights and sprout densities are determined using parametric transfer functions of biological switch and partial least square regression (PLSR). (d) Percent variance explained plot as a function of number of components shows that one component PLS model captures almost 80% of the variance in model prediction. Increasing model components from 2 to 4 components achieves additional 2% of the percent variance explained. (e) Mean square prediction error plot shows that two component PLS model achieves the smallest MSE. (f) The sprout density predicted by the two component PLS model (FPLS(v,p)) is plotted against those estimated from the parametric switch model (F*(v,p)), based on 3D confocal images of sprout densities at varying input VEGF and PF4 combinations. The two component PLS model produces a relatively accurate prediction with most of the prediction points fall close to F*(v,p) = FPLS(v,p) line. (g) The PLS component projections of model predictors and response are scaled by the predicted sprout densities and color-coded by the level of VEGF vs. PF4 relatively their respective EC50 and IC50 values in sprout densities.
To determine whether the changes in cluster weights are statistically significant, we estimated the distribution of weight coefficients by bootstrapping. We drew 1000 random samplings with replacement of 50 trajectories from the pool of all single-cell trajectories. For each of these samplings, we assigned clusters based on relative similarities of the sampled trajectories to the cluster prototypes (cluster means in the feature space) in Supplementary Figure 5 and computed the cluster weight coefficients. We performed pair-wise comparisons of the resulting cluster weight coefficient distributions across different cytokine conditions using non-parametric Kolmogorov-Smirnov (KS) tests with the null hypothesis that the two samplings are drawn from the same distributions. For 134 of the 140 pair-wise comparisons, the weight coefficient differences are statistically significant to the 0.05 level (Fig S5a–c).
hMVEC dynamic phenotypic cluster weights are predictive of population-level sprout density response
Finally, in order to connect individual-cell behaviors with cell-population outcomes, we sought to determine if the cytokine-induced modulation of phenotypic cluster weights might be predictive of overall sprout density across the landscape of treatment conditions. Establishment of this association would further support the notion that VEGF and PF4 influence population-level sprout formation at least partly by governing the quantitative proportion of cells exhibiting these dynamic transition behaviors rather than giving rise to new classes of behavior. To address this question, we acquired and quantitatively analyzed additional sprout density data by confocal imaging; accordingly, we generated measurements of the number of angiogenic sprouts moving into the underlying gel per unit cell monolayer area. Then we devised and employed a method -- outlined in Figure 7c -- to elucidate a potential quantitative mapping between sprout density and phenotypic cluster weights over the full set of VEGF and PF4 concentrations.
To construct this mapping between the two data sets, we fitted the observed sprout density data to parametric models of biological switches as functions of VEGF and PF4, and for each model, estimated an optimized model parameter set. Among variants of the parametric models, each with corresponding, the five parameter hyperbolic tangent (Tanh) switch model made the closest prediction to the observed sprout densities (quantified as the smallest Kullback-Liebler divergence between the model prediction and the observed sprout density data (Fig S6)). Using the optimized Tanh switch model, we estimated the sprout densities at the VEGF and PF4 combinations corresponding to those in phenotypic cluster weight dataset - thus creating consistent data sets from which quantitative mapping can be drawn. We used partial least-square regression (PLSR) to simplify the dimensions of the input phenotypic cluster weights and the output sprout density datasets by projecting them onto alternative bases on which the redundancy and co-linearity within the input dataset were eliminated. To determine an optimal number of model dimensions, we examined the percent variance explained and mean square prediction error (MSE) with increasing numbers of PLS components. The 1-component PLSR model explains about 79% of the variance in the predicted sprout density; models with increasing number of PLS components from 2 to 4 only capture an additional few percent of the variance (Fig 7d). Importantly, the 2-component version achieves the smallest MSE (Fig 7e), so we chose the this model to analyze the relationship between phenotypic cluster weights and sprout density. To assess the PLSR mapping performance, we plotted the predictions of the 2-component model (FPLS(v,p)) against the corresponding estimates by the Tanh switch equation (F*(v,p)) (Fig 7f). The model predictions fall close to the identity line (FPLS(v,p) = F*(v,p)), affirming that the PLS mapping based on phenotypic cluster weights captures the majority of trends in VEGF- and PF4-modulated sprout density across all the angiogenic and angiostatic treatment combinations.
As a complementary means to visualize model performance, we plotted the PLSR projections of the model predictors (phenotypic cluster weights in each cytokine combination) and model predictions (sprout density in each cytokine combination). To denote the different inputs (cytokine combinations) and outputs (sprout densities), we scaled the projection markers by sprout density and color-coded them by the relative VEGF and PF4 concentrations (Blue: VEGF dominant relative to EC50, Red: PF4 dominant relative to IC50 for sprout density response respectively). A few important features can be seen in the PLS projections. First, the markers are segregated by size and color along the first component (PLSC1) (Fig 7g). The relatively large (high sprout density) markers are mostly blue (VEGF dominant) while the smaller ones are mostly red (PF4 dominant). The co-segregation of the marker color with size (VEGF-dominance with high relative sprout density) is consistent with biological understanding that VEGF and PF4 play the opposite roles in sprout formation. In addition, the first component (PLSC1) axis is composed of large coefficients for wA and wP but small coefficients for wM and wS, indicating that the proportions of the population in the apoptotic and proliferative clusters are the strongest predictors of sprout density. Notably, most of the large blue markers show up on the positive PLSC1 side (with which wP axis is strongly positively correlated) while the small red ones lie mostly on the negative PLSC1 (with which wA axis is strongly positively correlated), indicating that high wP is associated with the VEGF dominant, high sprout density conditions, while high wA is associated with the PF4 dominant, low sprout density conditions (Fig 7g).
DISCUSSION
An important challenge in developing reliable, and even predictive, approaches to modulate multi-cellular processes in biomedical applications, such as promoting angiogenesis for tissue regeneration or diminishing it for anti-cancer therapies, is the need for design principles that offer guidelines for how molecular-level interventions will alter tissue-level properties via cell-level behavioral processes. The past decade has witnessed advances in multi-scale (often agent-based) computational modeling of angiogenesis aimed toward this objective, generating theoretical predictions and insights concerning the molecular-to-cell and cell-to-tissue relationships6. A consequent need is foundational quantitative experimental information at the cell level, across a diverse landscape of conditions that might be encountered in physiological contexts, to aid in model construction and constraint. We have designed and implemented an experimental measurement and data analysis method that quantifies multiple phenotypic behavioral responses of individual endothelial cells to various concentration combinations of an angiogenic cytokine (VEGF) and an angiostatic cytokine (PF4) that yield noticeably different cell-level phenotypic behaviors along with noticeably different population-level sprout formation. Our method consists of three main procedures: cell contour detection, automated state annotation, and rate maximum likelihood estimation based on continuous time Markov process (Fig 1–2).
To address the effect of opposing inflammatory cytokines VEGF and PF4 on phenotypic state transition dynamics, we have considered two conceptual models to account for variations in phenotypic transition dynamics across cell population: uniform population model (UPM) and dynamic population model (DPM) (Fig 4). We have shown that under DPM, the endothelial subpopulations are distinct and well-separated in identifiable behaviors (Fig 6a) consistent across all cytokine treatment conditions (Fig S4). These behaviors are characterized not by a single time-point measurements of a traditional phenotype (e.g., proliferation, death, migration), but instead by dynamic patterns for more-or-less predominance of one or more of these traditional phenotypes followed over a 24-to-30 hour period. That is, any particular cell can exhibit multiple traditional phenotypes during a sustained time period, and our analysis indicates that its behavior is most usefully represented by its dynamic pattern of multiple behaviors rather than requiring only one behavior be specified. The dynamic patterns, however, can be clustered into a small number of identifiable categories, which we have terms as follows (Fig 6): Apoptotic (A), Proliferative (P), Sessile (S), Migratory (M), and Switching (Sw). The latter two show qualitative similarities in shifts between Sessile and Migratory, but the Switching category is quantitatively separable in exhibiting a much greater frequency of shifting. Because the same five categories are identifiable regardless of cytokine treatment, we learn that the cytokines influence cell population behavior by quantitatively influencing the proportion of cells following the various categorical dynamic behaviors.
Importantly, the population-level outcome across all the angiogenic and angiostatic treatments can be accounted for by the quantitative proportions of cells in these five dynamic behavioral categories, within the framework of a multi-variate regression model. We are confident that alternative models can be employed to pursue this kind of cell-to-population “multi-scale” association, but it is valuable to have proven it in concept here using PLSR methodology. Thus, our work illustrates how the behaviors of single cell in a heterogeneous population can be quantitatively described and reduced empirically to a simpler computational mapping that relates to population biological response.
Agent-based models of angiogenesis in the literature (including previous contributions from our laboratory12,13 typically assume that the vascular endothelial cells can be described in terms of a uniform population model such that cells in the population follow the same rules and any given treatment condition modulates the phenotypic behaviors according to an identical relationship for all cells. We have found that the phenotypic behavior of a human microvascular endothelial cell population is heterogeneous – and, most remarkably, that this heterogeneity can be mapped to a small number of distinct patterns based on phenotypic state transition dynamics. This heterogeneity may reflect cell-to-cell variability in genetic, epigenetic regulation and protein expression. Alternatively, we speculate that the observed heterogeneity may serve as diverse potential phenotypic programs among which endothelial cell can adopt, especially if such heterogeneity can be directionally altered by relevant cytokine stimuli. In this study, we demonstrate examples supporting cases of such directional alterations by potent angiogenic cytokine VEGF and PF4 -- as VEGF induces higher fraction of the population to adopt the transition pattern consistent with a proliferative program and lower fraction to adopt an apoptotic program while angiostatic cytokine PF4 exerts the opposite directional effect on an endothelial cell population.
Taken together, our findings offer a novel perspective in understanding the phenotypic behaviors of endothelial cell population as a unique consistent set of dynamic transition cassettes across angiogenic and angiostatic cytokine stimuli. In this light, the influence of the angiogenic and angiostatic cytokines can be usefully characterized as the directional changes in cell population proportions within the various dynamic transition programs. By quantitatively summarizing the cytokine induced changes in the population proportions adopting different state transition patterns as phenotypic cluster weights, we have identified the single cell behavioral basis that can be used to predict the effect of cytokine on population-level sprout density. As endothelial cells must take on multiple roles in various physiological and pathological contexts (e.g., apart from playing central roles in sprouting angiogenesis, they act as selective barrier for transport of molecules and immune cells between blood and tissues, help regulate blood fluidity, and participate in inflammatory responses). In each of these roles, endothelial cells need to flexibly exhibit different phenotypic behaviors25,26 so individual cells in a homogeneous endothelial population have to be able to switch their responses accordingly. Our findings suggest that in microvascular endothelial cells, at least transiently, the population may achieve such diverse phenotypic requirement by permitting subpopulation behavioral dynamics to be quantitatively modulated by their environmental stimuli. This may turn out to represent a more widely general principle across a broad spectrum of tissue types.
Methods
Live imaging of hMVECs
Human adult microvascular endothelial cells (hMVECs) of dermal origin (purchased from Lonza at passage 4; Cat No. CC-2543) were maintained in culture according to manufacturers recommendations. Cells were passage once then stably infected with GFP and RFP plasmid following a standard retroviral infection protocol. For all live cell imaging experiments, GFP-labeled and RFP-labeled hMVECs were used at second passage after infection, while unlabeled hMVECs were used after three passages after the cells were received. hMVECs were mixed at 1:1:3 ratio in complete medium to allow visualization of single cell when seeded at instant monolayer density (50000 cells/cm2) to allow sprout initiation on 1 mm thick 2.0 mg/mL Collagen I gel (BD Biosciences; Cat No. 356236). At 4 hours after seeding, adhered hMVECs were replaced with 5% FBS no cytokine medium. At 24 hours after seeding, hMVECs on gel were stimulated with VEGF and PF4 (Peprotech; Cat No. 100-20 and 300-16 respectively). The cell were imaged upon stimulation using Cellomic array scan microscope. The chosen imaging frequency (15 min interval) does not appear to drastically affect cell behavior (Supplementary Data Set 1).
Collagen gel invasion assay
Type I rat tail collagen (BD Bioscience; Cat.No. 354236) diluted in 0.02 N Acetic acid to 2.0 mg/mL at pH 7.4 was prepared on ice and was immediately casted in glass bottom 24-well multi-well plate (MatTek Corp P24G-0-13-F) and allowed to solidify at 37°C for 30 minutes in a humidified environment. After gel solidification, cell culture medium was added to the plate to prevent the gel from desiccation. After at least 1 hour of medium incubation with collagen gel, hMVECs or HUVECs were seeded on the gel at instant monolayer density (50,000 cells/cm2). At this seeding density, HUVECs and hMVECs consistently formed a confluent monolayer on collagen gel at 18 hours after seeding (Fig 1a,b and Fig S1d). To test the effect of cytokines on sprouting, cytokine containing medium was introduced at 24 hours after seeding. The conditioned media were refreshed every 24 hours until the end of experiment (72 hours after seeding, unless indicated otherwise).
Sprout density quantification
To build the sprout density dataset, hMVECs were seeded on Collagen I gel (as in single cell tracking experiment) and treated with varying VEGF and PF4 combinations for 72 hours. The samples were fixed at 72 hours and stained with DAPI(Sigma D8417) and Rhodamine Phalloidin (Molecular Probes R415) for sprout visualization. Phase contrast and fluorescent images of the fixed and stained samples were acquired using Olympus FV1000 confocal microscope through a 20x long working distance air objective (LMPLFL 20x, Olympus USA). In each input cytokine condition, 12–16 non-overlapping confocal imaging voxels of approximately 1350×1350×100–200 μm3 were acquired and reconstructed in 3D using Imaris software package (Bitplane Inc). The sprout density is determined by the number of sprouts in each voxel were counted divided by the monolayer area.
Contour and centroid tracking by level set active contour
Fluorescent live cell images were enhanced by median and entropy filtering algorithms using the built-in scripts in the MATLAB R2011a image processing toolbox (MathWorks). Preprocessed image were binarized (thresholded based on intensity histogram to partition intensity levels into that of the foreground and the background) and the boundary of the preprocessed image was used as an initial contour. Contours were detected by a level set active contour algorithm of Chan-Vese16 implemented in MATLAB and applied to filtered, unbinarized images. The level set routine was adopted from Yue Wu’s contribution on publicly accessible MathWorks’ file exchange website (http://www.mathworks.com/matlabcentral/fileexchange/23445) and the level set’s objective function was slightly modified so the algorithm works well for our images. For time series analysis, the first image in the series was contour detected as described. Then, the subsequent image was initialized with the optimal contour of an immediately previous image in the series. All single cell tracks in the data set are validated manually to ensure accurate tracking.
State annotation and determination of the length of proliferative and apoptotic state interval
In this study, the proliferative state is defined with respect to the time-point at which the cell contour splits from one to two (tsplit). We note that, for most ‘proliferative’ trajectories, the mother cells slow down prior to splitting (pre-splitting phase) and the daughter cells pull apart antipodally such that their velocity vectors are anti-correlated for some time before they become uncorrelated (post-splitting phase). We use these phases to define proliferative interval of individual trajectories. For apoptosis states, cells are characterized by continual progressive shrinkage of contours which eventually lead to contour disappearance, causing the characteristic lost contour track. This series of events are flagged and manually validated at the final step of tracking.
Additional methods
Additional details for all experimental methods are available in SI Text descriptions, including semi-supervised sessile vs. motile state classification, CTMC model of single cell state trajectories, comparisons of condition based and cluster based rate estimations, and parametric modeling of sprout density response.
Supplementary Material
Acknowledgments
The authors express gratitude to Shannon Alford and Ta Hang for reagents and advice on multiple aspects of the project, Waleed Farahat and Levi Wood for advice on stochastic modeling, and Ioannis Zervantonakis and Aprotim Mazumder for help with imaging. This work was supported by NSF grant EFRI-0735007, NIH grant R01-GM081336, and NIH grant R01-EB010246.
Abbreviations
- ECM
extracellular matrix
- hMVECs
human microvascular endothelial cells
- MLE
maximum likelihood estimate
- PF4
platelet factor 4
- VACF
velocity autocorrelation function
- VEGF
vascular endothelial growth factor
- CTM
continuous time Markov
- CTMC
continuous time Markov chain
- UPM
uniform population model
- DPM
diverse population model
References
- 1.Adams R, Alitalo K. Nat Rev Mol Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- 2.Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, Betsholtz C. J Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vandercappellen J, Liekens S, Bronckaers A, Noppen S, Ronsse I, Dillen C, Belleri M, Mitola S, Proost P, Presta M, Struyf S, Van Damme J. Molecular Cancer Research. 2010;8:322–334. doi: 10.1158/1541-7786.MCR-09-0176. [DOI] [PubMed] [Google Scholar]
- 4.Aidoudi S, Bikfalvi A. Thromb Haemost. 2010;104:941–948. doi: 10.1160/TH10-03-0193. [DOI] [PubMed] [Google Scholar]
- 5.Ferrara N, Kerbel RS. Nat Cell Biol. 2005;438:967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 6.Peirce SM. Microcirculation. 2008;15:739–751. doi: 10.1080/10739680802220331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine HA, Tucker AL, Nilsen-Hamilton M. Growth Factors. 2002;20:155–175. doi: 10.1080/0897719031000084355. [DOI] [PubMed] [Google Scholar]
- 8.McDougall SR, Anderson ARA, Chaplain MAJ. Journal of Theoretical Biology. 2006;241:564–589. doi: 10.1016/j.jtbi.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Bauer AL, Jackson TL, Jiang Y. Biophysical Journal. 2007;92:3105–3121. doi: 10.1529/biophysj.106.101501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer AL, Jackson TL, Jiang Y. PLoS Computational Biology. 2009;5:e1000445. doi: 10.1371/journal.pcbi.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milde F, Bergdorf M, Koumoutsakos P. Biophysical Journal. 2008;95:3146–3160. doi: 10.1529/biophysj.107.124511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das A, Lauffenburger D, Asada H, Kamm RD. Philos Transact A Math Phys Eng Sci. 2010;368:2937–2960. doi: 10.1098/rsta.2010.0085. [DOI] [PubMed] [Google Scholar]
- 13.Wood L, Kamm R, Asada H. The International Journal of Robotics Research. 2011;30:659–677. [Google Scholar]
- 14.Yamamura N, Sudo R, Ikeda M, Tanishita K. Tissue Engineering. 2007;13:1443–1453. doi: 10.1089/ten.2006.0333. [DOI] [PubMed] [Google Scholar]
- 15.Bayless KJ, Kwak H-I, Su S-C. Nature Protocols. 2009;4:1888–1898. doi: 10.1038/nprot.2009.221. [DOI] [PubMed] [Google Scholar]
- 16.Vese L. International Journal of Computer Vision. 2002 [Google Scholar]
- 17.Schapire R. Machine learning. 1999 [Google Scholar]
- 18.Rätsch G, Mika S, Schölkopf B. IEEE Transactions on …. 2002 doi: 10.1109/72.914517. [DOI] [PubMed] [Google Scholar]
- 19.Norris JR. Markov chains. Cambridge Univ Pr; 1998. [Google Scholar]
- 20.Italiano JE, Richardson JL, Patel-Hett S, Battinelli E, Zaslavsky A, Short S, Ryeom S, Folkman J, Klement GL. Blood. 2007;111:1227–1233. doi: 10.1182/blood-2007-09-113837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatterjee M, Huang Z, Zhang W, Jiang L, Hultenby K, Zhu L, Hu H, Nilsson GP, Li N. Blood. 2011;117:3907–3911. doi: 10.1182/blood-2010-12-327007. [DOI] [PubMed] [Google Scholar]
- 22.Lasagni L, Francalanci M, Annunziato F, Lazzeri E, Giannini S, Cosmi L, Sagrinati C, Mazzinghi B, Orlando C, Maggi E, Marra F, Romagnani S, Serio M, Romagnani P. Journal of Experimental Medicine. 2003;197:1537–1549. doi: 10.1084/jem.20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Struyf S. Circulation Research. 2004;95:855–857. doi: 10.1161/01.RES.0000146674.38319.07. [DOI] [PubMed] [Google Scholar]
- 24.Hutchings H. The FASEB Journal. 2003 doi: 10.1096/fj.02-0691fje. [DOI] [PubMed] [Google Scholar]
- 25.Sumpio BE, Riley JT, Dardik A. Int J Biochem Cell Biol. 2002;34:1508–1512. doi: 10.1016/s1357-2725(02)00075-4. [DOI] [PubMed] [Google Scholar]
- 26.Michiels C. J Cell Physiol. 2003;196:430–443. doi: 10.1002/jcp.10333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.