Abstract
The Bantu languages are widely distributed throughout sub-Saharan Africa. Genetic research supports linguists and historians who argue that migration played an important role in the spread of this language family, but the genetic data also indicates a more complex process involving substantial gene flow with resident populations. In order to understand the Bantu expansion process in east Africa, mtDNA hypervariable region I variation in 352 individuals from the Taita and Mijikenda ethnic groups was analyzed, and we evaluated the interactions that took place between the Bantu- and non-Bantu-speaking populations in east Africa. The Taita and Mijikenda are Bantu-speaking agropastoralists from southeastern Kenya, at least some of whose ancestors probably migrated into the area as part of Bantu migrations that began around 3,000 BCE. Our analyses indicate that they show some distinctive differences that reflect their unique cultural histories. The Taita are genetically more diverse than the Mijikenda with larger estimates of genetic diversity. The Taita cluster with other east African groups, having high frequencies of haplogroups from that region, while the Mijikenda have high frequencies of central African haplogroups and cluster more closely with central African Bantu-speaking groups. The non-Bantu speakers who lived in southeastern Kenya before Bantu speaking groups arrived were at least partially incorporated into what are now Bantu-speaking Taita groups. In contrast, gene flow from non-Bantu speakers into the Mijikenda was more limited. These results suggest a more complex demographic history where the nature of Bantu and non-Bantu interactions varied throughout the area.
Keywords: Demographic history, Africa, Gene flow
The Bantu languages are spoken across a broad geographic area that encompasses 27 countries in sub-Saharan Africa (Nurse and Philippson, 2003). They are the most commonly spoken languages in Africa, with over 240 million speakers, or one third of Africans. Given the wide geographic distribution and large number of speakers of the approximately 500 Bantu languages, linguists have been puzzled by the lack of linguistic diversity and its position as a small subgroup on one of the branches of the much larger Niger-Congo languages phylogenetic tree (Nurse and Philippson, 2003; Schadeberg, 2003; Bostoen, 2007).
The Bantu languages were recognized as a part of the Niger-Congo language phylum in the mid-1800s but an arbitrary division of them into separate groups for study created a perception that the Bantu family was a self-contained unit (Schadeberg, 2003). Greenberg (1963; 1972) dispelled this misconception with his phylogeny of Niger-Congo languages, and Greenberg and other linguists proposed that the Bantu language family arose in southeastern Nigeria or central Cameroon around 2000 to 3000 BCE (Vansina, 1995; Blench, 2006) and spread out of the rainforests around 500 to 1000 BCE (Nurse and Philippson, 2003). For many years, an emphasis on an east-west separation (“early split” model) has been the foundation of many reconstructions (Ehret, 2000; 2001; 2002).
Accurate phylogeny reconstruction has proven difficult because the Bantu languages are adjacent to closely related Niger-Congo languages in the core area. Lexicostatistical analyses are often criticized for their heavy dependence on the 100-word Swadesh list and have been unable to resolve many of the internal issues. Holden (2002) and others have argued that the western languages are paraphyletic, while Bastin et al. (1999) found little support for a widely accepted monophyletic eastern group. Many Bantu linguists (Nurse and Philippson, 2003; Bostoen, 2007) consider lexicostatistics a short-cut or first approximation method and call for more in-depth and nuanced comparative linguistic studies. Consequently, a growing number of Bantu specialists (Vansina, 1995; Ehret, 2001; Nurse and Philippson, 2003; Blench, 2006; Holden and Gray, 2006) now argue that the strict migration model of Bantu-speaking people, as originally proposed by historical linguists, is overly simplistic and that language shifts and other forms of cultural diffusion played important roles in the spread of Bantu languages as well.
In the last half century, archaeological data became more important as historians and archaeologists linked the linguistic data to archaeological cultures whose geographic distributions mirrored current Bantu language distribution and appeared in the archaeological record at dates similar to those derived by glottochronology for various Bantu language branches. Phillipson (1974; 1975; 1976; 1977; 2005) developed a two-stream model that fit Greenberg’s (1963) and Guthrie’s (1962) west-east division of Bantu languages that corresponds to eastern and western flows of the Bantu migration. He proposed that proto-Bantu was initially spoken by people living in western Cameroon around 1000 BCE who made stone tools, kept domesticated goats and possibly cultivated some root crops. A group of these Bantu speakers dispersed across the northern fringes of the equatorial forests, interacting with farmers and acquiring sheep, cattle and domesticated crops such as sorghum from them. These groups formed Phillipson’s ‘eastern stream’ and were represented in the archaeological record by the Chifumbaze complex. Its distribution throughout eastern and southern Africa matches the area where East Bantu languages are spoken today. Robertson and Bradley (2000) dispute the existence of a cohesive Chifumbaze complex, however, citing the lack of standardization in ceramic analyses and emphasizing local continuity in ceramic sequences instead of a replacement scenario.
Genetic studies generally support a migration model of people bringing their Bantu language and culture with them as they colonized new territories (Excoffier et al., 1987; Cavalli-Sforza et al., 1994; Pereira et al., 2001; Salas et al., 2002; Wood et al., 2005). Although some recent studies (Castrì et al., 2009; Montano et al., 2011; Sikora et al., 2011) have reported greater genetic heterogeneity than had been previously observed, the Bantu-speaking populations today tend to be genetically homogeneous (Excoffier et al., 1987; Cavalli-Sforza et al., 1994; Salas et al., 2002; Berniell-Lee et al., 2009; Alves et al., 2011; Schlebusch et al., 2012). Researchers have identified 1) both mitochondrial DNA (mtDNA) and Y chromosome haplogroups that are shared in high frequencies in Bantu speaking groups from western central Africa to southeastern Africa and 2) reduced genetic diversity toward southern periphery of Bantu expansion (Pereira et al., 2002; Salas et al., 2002; Castrì et al., 2009; de Filippo et al., 2011; de Filippo et al., 2012). de Filippo et al. (2012) find little evidence for an ‘early-split’ model, however.
We analyzed the Hypervariable Region I (HVRI) in the control region of the mtDNA of men belonging to two east African Bantu-speaking populations, the Taita and Mijikenda, to assess the extent of interaction among Bantu speaking groups and their neighbors as they settled in east Africa. Recent linguistic, archaeological, and genetic research is yielding a more nuanced portrait of Bantu expansion where both migration and cultural exchange play a role (Eggert, 2005; de Filippo et al., 2012), and the expansion process was likely to have been complicated in east Africa where speakers of all four major African language families reside (Niger-Congo, Nilo-Saharan, Afro-Asiatic, and Khoisan), often living in close proximity to one another. Sampling density in this region is still relatively low in genetics studies given the demographic, ethnic, and linguistic diversity, but the genetic studies undertaken to date (Watson et al., 1996; Semino et al., 2002; Cruciani et al., 2004; Kivisild et al., 2004; Hassan et al., 2008; Tishkoff et al., 2009), indicate that the region is genetically diverse as well.
Samples and methods
Subjects
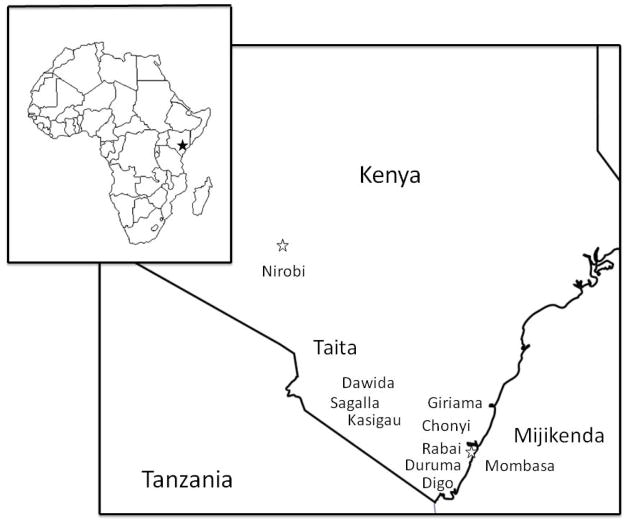
The Taita and Mijikenda are two Bantu-speaking ethnic groups who live in southeastern Kenya (Fig. 1). The Taita and the Mijikenda languages belong to the Northeast Bantu language group (Hinnesbusch et al., 1981). The Taita languages are most closely related to the Chaga languages spoken in Tanzania, south of Mt. Kilimanjaro and form the Kilimanjaro Bantu group (Philippson and Montlahuc, 2003). The Taita are agropastoralists who number about 213,000 today (Bravman, 1998). They live in the Taita hills located about 150 km west of the port city of Mombasa and about 150 km east of Mt. Kilimanjaro. The Taita are composed of three subgroups named for each of the Taita hills they occupy; the Davida, the Sagalla and the Kasigau. The Taita speak two different Bantu languages which both show evidence of borrowing from Cushitic languages (Ehret and Nurse, 1981). The five dialects of Davida are spoken throughout the Taita hills, while Sagalla is spoken only on Sagalla. The Mijikenda language is closely related to the Swahili that belongs to the Sabaki language cluster (Hinnesbusch et al., 1981). The Mijikenda reside in the southeastern coastal region of Kenya, in and around Mombasa (Spear, 1981). The Mijikenda are composed of nine tribes (Chonyi, Digo, Duruma, Giriama, Jibana, Kambe, Kauma, Rabai, and Ribe) and have a population size of 1,208,000. Linguists separate the Mijikenda languages into two groups. Digo is given its own designation as southern Mijikenda, while the remaining eight languages are grouped together in a northern Mijikenda cluster (Maho, 2003). According their oral history, the Taita, Mijikenda, and other Bantu-speaking groups in the area, including the Pokomo and Swahili, left their mythological ancestral land, Singwaya, located in somewhere northeastern Kenya or southern Somalia around the 16th century, and settled in the coastal region of southeastern Kenya and northeastern Tanzania (Spear, 1974; 1977; 1981). Historians, such as Willis (1993) and Bravman (1998), have argued that the Taita and Mijikenda ethnic designation are relatively recent phenomena, emerging from colonial political and economic processes.
Figure 1.
Map of southeastern Kenya showing geographical location of the Taita and Mijikenda.
MtDNA analyses
Swab samples were collected from 352 men from all three Taita subgroups and five of the nine Mijikenda groups (Digo, Duruma, Giriama, Jibana and Ribe). DNA was extracted from the swabs using MasterAmp™ DNA Extraction Solution (Epicentre Technologies, Madison, WI). Then, mtDNA hypervariable region I (HVRI) was amplified using a touchdown PCR protocol and either of the primer sets (L15985 5′-GCACCCAAAGCTAAGATTCTAA-3′ and H404 5′-AAAGTGCATACCGCCAAAAG-3′or L15926 5′-TCAAAGCTTACACCAGTCTTGTAAACC-3′ and H16498 5′-CCTGAAGTAGGAACCAGATG-3′). The amplified product was sequenced in both directions using the BigDye Terminator Cycle Sequencing Kit, version 3.1 (Applied Biosystems) and analyzed on an ABI 3730 DNA sequencer. Sequences were edited and aligned in Sequencher 4.1.4 (GeneCodes). The nucleotide positions (nps) between 16024 and 16383 relative to the Cambridge Reference Sequence (Anderson et al. 1981) were edited. When T→C transition at np 16189 was present, nps 16182 and 16183 were excluded from analysis because of heteroplasmy (Bendall and Sykes, 1995; Pfeiffer et al., 1999). The Taita and Mijikenda mtDNA sequences were compared to sequences from 38 published population samples from sub-Saharan Africa, grouped by region and language spoken. The samples included 24 Bantu, 4 Nilo-Saharan, 8 Afro-Asiatic, and 2 Khoisan groups (Fig. S1 and Table S1). The 24 Bantu populations included 4 east African, 9 central African, and 11 southeastern African populations. We sequenced one Kikuyu sample and included it with the Kikuyu samples analyzed by Watson et al. (1997). Each mtDNA sequence was assigned to a haplogroup and given a place of origin based on Salas et al. (2002) and Kivisild et al. (2004).
Statistical methods
Estimates of within-population genetic diversity, haplotype diversity (h) and parameter θ=2Nfeμ (θk, θS and θπ), two tests of molecular neutrality (Tajima’s D and Fu’s Fs), and mismatch distribution, were performed in Arlequin 3.5 (Excoffier et al., 2005). The value of θk is based on the relationship between the number of sequences (k) and the sample size (Ewens 1972), while θS is based on the relationship between the number of segregating sites (S) and the sample size (Watterson 1975). The θπ is a measure of mean pairwise differences (π) (Tajima, 1983). The θk and θS values are sensitive to recent demographic events, while θπ values reflect more ancient events (Tajima, 1989; Rogers, 1995; Helgason et al., 2000; Helgason et al., 2003). Large negative Tajima’s D and Fu’s Fs values indicate recent population expansion, but Fu’s Fs is considered to be more a sensitive than Tajima’s D (Excoffier and Schneider, 1999). Mutation heterogeneity in mtDNA HVRI reduces number of S and has an opposite effect on Tajima’s D, while it has smaller effect on number of k (Aris-Brosou and Excoffier, 1996). To understand the pattern of population subdivision, we estimated pairwise genetic distances between populations (ΦST) and performed Exact Tests of population differentiation and Analysis of Molecular Variance (AMOVA). Mantel tests were undertaken in Arlequin to test whether an isolation-by-distance model explained the observed mtDNA variation.
SPSS statistical software was used to visualize population pairwise genetic distances through Multidimensional scaling (MDS) analysis and to test correlations between genetic diversity (h, θk, θS, and θπ) and the geographical distances from the center of Bantu expansion in order to identify spatial patterns under two models of expansion. In the first model, we measured the distance from the expansion core to the center of each Bantu population to approximate a one-step expansion process. We used Douala, a coastal city in northwestern Cameroon, to represent the center of the expansion. We also modeled a two-step expansion process, using the southern tip of the Lake Victoria as the center of an eastern Bantu expansion, calculating the distance from Douala to the southern tip of the Lake Victoria, and then from there to the centers of each east Bantu population.
Migration rates were estimated by two methods. M=2Nfem under a spatial expansion model was estimated using Arlequin. Assuming that population size was stable, the migration rates necessary to produce the observed mismatch distribution were estimated using an infinite-island model, which is equivalent to the continent-island model (Excoffier, 2004). Migration rates, 2Nfem, between each pair of populations were also estimated using maximum-likelihood method implemented with MIGRATE (Beerli and Felsenstein, 2001). The migration rate estimates were obtained using the averages of three independent runs. Each run had 10 short chains (10,000 genealogies per chain) and three long chains (100,000 genealogies per chain) with increments of 20 and 200 steps respectively. The first 100,000 trees in each chain were discarded. Instead of sampling more genealogies, Metropolis coupled Markov Chain Monte Carlo, or ‘heating’ was used to explore a wider genealogical space by setting four temperatures (1, 1.5, 3, 6) and long chains were replicated.
RESULTS
Haplogroups and haplogroup frequencies
A total of 126 different haplotypes was found among the Taita and Mijikenda samples (Table S2), and most were assigned to mtDNA haplogroups as defined in previous phylogeographic analyses (Watson et al., 1997; Salas et al., 2002; Kivisild et al., 2004; van Oven and Kayser, 2009). Assignment to smaller subclade was difficult because of the limited number of diagnostic mutations within HVRI. For example, three L3 haplotypes (n=3, 0.7%) lacked the diagnostic mutations that define L3 sub-haplogroups.
Table 1 shows the distribution of these haplogroups in Bantu-speaking groups in central, east and southeastern Africa, and also Nilo-Saharan and Afro-Asiatic groups in East Africa. Generally, frequencies of the haplogroups of proposed west/central African origin in east African populations are lower than in central African Bantu population. Haplogroup L1c is common in the central African Bantu group (32.5%), but less common in the east African Bantu group (<6.7%). L1c is rare in non-Bantu east African populations, and only one single L1c haplotype has been reported in the Nilo-Saharan Dinka (Krings et al., 1999). Haplogroup L3e is also common in central African Bantu populations, but uncommon in east Africa. The Mijikenda are the exception with an L3e frequency of 23.6%. This haplogroup is also extremely rare in non-Bantu east Africa groups (0.0–2.0%).
Table 1.
Mitochondrial DNA HPG frequency (%) of Bantu and non-Bantu east African populations
| HPGs | Proposed Origina | Taita | Mijikenda | E.A. Bantub,c | C.A. Bantu | S.A. Bantuc | S-E. N-Sc | N-E. N-Sc | S-E. A-Ac | N-E. A-Ac |
|---|---|---|---|---|---|---|---|---|---|---|
| L0a | East Africa | 26.1 | 16.9 | 14.8 | 9.9 | 24.8 | 37.2 | 11.7 | 26.0 | 7.1 |
| L0d | Khoisan (SA) | 0.6 | 2.6 | 0.0 | 0.0 | 5.0 | 1.1 | 0.0 | 2.0 | 0.0 |
| L0f | East Africa | 13.4 | 2.1 | 9.4 | 0.0 | 0.0 | 4.3 | 0.8 | 30.0 | 0.3 |
| L1b | West Africa | 1.9 | 3.1 | 0.0 | 6.2 | 1.2 | 0.0 | 4.7 | 0.0 | 2.0 |
| L1c | Central Africa | 4.5 | 6.7 | 3.9 | 32.5 | 5.3 | 0.0 | 0.8 | 0.0 | 0.0 |
| L2a | Wide Spread | 5.7 | 14.4 | 7.0 | 14.5 | 32.9 | 8.5 | 17.2 | 4.0 | 13.8 |
| L2 other | West/Central Africa | 0.0 | 0.5 | 2.3 | 5.1 | 2.9 | 0.0 | 0.0 | 0.0 | 1.7 |
| L3b and L3d | West/Central Africa | 7.0 | 12.8 | 14.8 | 7.6 | 8.4 | 2.1 | 2.3 | 0.0 | 3.0 |
| L3e | Central Africa | 5.1 | 23.6 | 7.8 | 15.4 | 15.4 | 0.0 | 0.8 | 2.0 | 0.0 |
| L3f | East Africa | 7.6 | 7.7 | 0.8 | 6.0 | 2.4 | 5.3 | 8.6 | 0.0 | 5.4 |
| L3i | East Africa | 4.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.6 | 0.0 | 1.3 |
| L3x | East Africa | 0.6 | 1.0 | 0.8 | 0.0 | 0.0 | 0.0 | 0.8 | 2.0 | 4.0 |
| L4 | East Africa | 12.1 | 6.2 | 15.6 | 1.3 | 0.0 | 13.8 | 5.5 | 10.0 | 6.4 |
| Other | 10.8 | 2.6 | 22.8 | 1.5 | 1.7 | 27.7 | 45.2 | 24.0 | 55.0 | |
|
| ||||||||||
| n | 157 | 195 | 128 | 1180 | 416 | 94 | 128 | 50 | 297 | |
The places of origin for mtDNA haplogroups were previously proposed (Kivisild et al. 2004; Salas et al. 2002; Watson et al. 1997).
East African Bantu populations include the Hutu, Kikuyu, Sukuma, and Turu.
The grouping of populations as shown on the Table S1 (east African Bantu, central African Bantu, southeastern African Bantu, south east African Nilo-Saharan, north east African Nilo-Saharan, south east African Afro-Asiatic, and north east African Afro-Asiatic populations).
Haplogroups of east African origin have been found in higher frequencies in east African Bantu-speaking populations than in central African Bantu populations, and the Taita have higher frequencies of east African haplogroups (L0a, L0f, and L4) than are found in the Mijikenda sample. Haplogroup L0a is most common in southeastern Nilo-Saharan populations (37.2%) and is relatively common in east African Bantu populations (14.8–26.1%). Haplogroup L0f is common in both southeastern Afro-Asiatic groups (30.0%) and east African Bantu populations (13.4% in the Taita, 9.4% in previously sampled east African Bantu populations). Haplogroup L4 is another common haplogroup in east African Bantu and non-Bantu groups, but is found only in low frequencies in central African Bantu populations. Haplogroups L3i and L3x are rare in East African populations and absent in central African Bantu populations.
Four M1a haplotypes (n=7) were found in the Taita and Mijikenda samples. M1 is a non-L haplogroup of east African or Middle Eastern origin (Quintana-Murci et al., 1999; Kivisild et al., 2004) and it is rare among previously published east African Bantu samples (Watson et al., 1997; Knight et al., 2003; Castrì et al., 2009; Tishkoff et al., 2007). The Sukuma are the only other east African Bantu population where this haplogroup has been reported (Knight et al., 2003), but the haplogroup is relatively common in east African Afro-Asiatic and Nilo-Saharan populations (e.g., 8.5% in the Nubians, 8.9% in the Tigrais, and 15.8% in the Amhara) (Krings et al., 1999; Kivisild et al., 2004).
The overall proportion of mtDNA haplogroups of east African origin in east African Bantu-speaking populations is substantial. Over 50% of east African Bantu populations haplogroups (~ 66% for Taita and ~ 55% for other east African Bantu populations) are of east African origin. The Mijikenda are less reflective of this trend having retained more a larger proportion of central/western African mtDNA haplogroups (~ 47%) and fewer east African haplogroups (~ 34%). In contrast, the contribution of east African mtDNA to southeastern African Bantu populations is much smaller (~28%). While the frequency of east African haplogroups in east African Bantu populations is generally large, the frequency of central African mtDNA haplogroups in non-Bantu-speaking east African populations is minimal (<10%).
Within-population genetic diversity and population expansion
The Taita, Mijikenda, and other east African Bantu-speaking populations are genetically diverse in comparison to the central African and Bantu-speaking populations (Table 2). The east African populations tend to have larger θS and θπ values than populations from other areas of Africa, and the Taita are one of the more genetically diverse Bantu-speaking populations. The Taita have the largest θS value and the third largest θπ value among the Bantu-speaking populations. The Mijikenda are not genetically very diverse. They have small θk and θπ, but relatively large θS. Although gene diversity is negatively correlated with distance across all measures, none are statistically significant (Table S3). Figure S2 illustrates that this lack of this correlation is largely due to the greater genetic diversity observed among east African Bantu-speaking populations. No east African Bantu population has a significantly negative Tajima’s D value, but all of the east African Bantu-speaking populations analyzed, except for the Turu, show evidence of population expansion with significant Fu’s Fs values.
Table 2.
African population summary statistics
| Populations | h | θk (95% CI) | θS (SD) | θπ (SD) | Tajima’s D | Fs | Ma |
|---|---|---|---|---|---|---|---|
| Taita | 0.981 | 61.419 (44.209–84.846) | 14.936 (3.676) | 11.946 (6.013) | −1.091 | −24.330** | 38.975 |
| Mijikenda | 0.952 | 36.651 (26.798–49.814) | 12.605 (3.065) | 9.930 (5.048) | −1.068 | −24.436** | 16.781 |
|
| |||||||
| East Africa - South | |||||||
|
| |||||||
| Bantu | |||||||
| Kikuyu | 0.993 | 133.893 (44.790–450.024) | 12.447 (4.316) | 9.650 (5.100) | −1.325 | −14.172** | NA |
| Sukuma | 0.988 | 78.779 (34.916–190.912) | 13.160 (4.317) | 10.843 (5.634) | −1.145 | 14.350** | 66.575 |
| Hutu | 0.979 | 45.529 (24.384–87.209) | 11.852 (3.720) | 12.139 (6.216) | −0.595 | −11.737* | 94.190 |
| Turu | 0.951 | 19.273 (9.487–39.686) | 10.185 (3.486) | 9.770 (5.128) | −0.712 | −3.545 | 15.792 |
| Nilo-Saharan | |||||||
| Turkana | 0.994 | 198.007 (77.407–565.238) | 14.612 (4.619) | 12.469 (6.398) | −1.098 | −23.478** | |
| Datoga | 0.984 | 58.278 (33.955–101.877) | 14.529 (4.235) | 12.453 (6.327) | −1.020 | −19.557** | |
| Afro-Asiatic | |||||||
| Burunge | 0.937 | 20.951 (11.214–39.355) | 10.472 (3.390) | 11.353 (5.851) | −0.368 | −4.036 | |
| Iraqw | 0.924 | 9.317 (3.286–27.264) | 9.603 (4.021) | 13.022 (7.108) | −0.370 | 0.838 | |
| Khoisan | |||||||
| Sandawe | 0.831 | 16.604 (10.394–26.206) | 9.634 (2.751) | 9.103 (4.689) | −0.679 | −6.058 | |
| Hadza | 0.796 | 16.401 (10.499–25.292) | 8.956 (2.514) | 6.292 (3.336) | −1.283 | −11.600* | |
|
| |||||||
| East Africa - North | |||||||
|
| |||||||
| Nilo-Saharan | |||||||
| Dinka | 0.995 | 228.871 (98.623–584.887) | 14.107 (4.288) | 10.332 (5.330) | −1.379 | −24.807** | |
| Nubia | 0.975 | 63.755 (40.859–100.331) | 14.665 (4.007) | 9.760 (5.013) | −1.481* | −24.785** | |
| Afro-Asiatic | |||||||
| Gurage | 1.000 | NA | 11.952 (4.313) | 9.989 (5.311) | −1.196 | −15.273** | |
| Tigrais | 0.994 | 162.663 (81.383–346.177) | 14.764 (4.355) | 9.868 (5.090) | −1.536* | −24.843** | |
| Oromo | 0.992 | 154.674 (60.007–443.620) | 14.045 (4.556) | 10.466 (5.444) | −1.420* | −21.188** | |
| Amhara | 0.994 | 147.293 (99.453–220.952) | 17.722 (4.478) | 10.558 (5.364) | −1.629* | −24.558** | |
| Somali | 0.992 | 99.676 (38.058–288.718) | 11.156 (3.842) | 8.242 (4.391) | −1.387 | −15.986** | |
| Afar | 0.975 | 30.015 (10.753–90.518) | 11.753 (4.521) | 11.183 (6.006) | −0.899 | −2.896 | |
|
| |||||||
| Central Africa | |||||||
|
| |||||||
| Bantu | |||||||
| Bassa | 0.991 | 99.974 (50.799–207.105) | 13.652 (4.161) | 11.165 (5.733) | −1.119 | −24.283** | 105.188 |
| Ngoumba | 0.991 | 90.184 (45.619–187.442) | 11.954 (3.717) | 10.701 (5.514) | −0.900 | −23.242** | 107.463 |
| Mbundu | 0.989 | 73.060 (37.653–147.902) | 13.174 (4.079) | 11.062 (5.692) | −1.060 | −19.271** | 88.444 |
| Bamileke | 0.988 | 63.764 (34.821–120.307) | 11.942 (3.654) | 9.379 (4.864) | −1.166 | −22.539** | 95.218 |
| Ewondo | 0.983 | 58.673 (33.389–105.392) | 11.679 (3.517) | 12.331 (6.276) | −0.479 | −18.723** | 56.891 |
| Bakaka | 0.983 | 56.184 (31.460–102.721) | 12.725 (3.841) | 11.807 (6.031) | −0.823 | −17.540** | 65.416 |
| Sanga | 0.970 | 29.755 (14.512–62.785) | 9.340 (3.203) | 10.783 (5.617) | −0.161 | −5.913* | 27.757 |
| Bateke | 0.945 | 15.913 (9.074–27.687) | 9.377 (2.919) | 7.736 (4.067) | −1.003 | −5.416* | 18.569 |
| Bubi | 0.908 | 7.469 (3.946–13.796) | 7.089 (2.329) | 7.629 (4.025) | 0.148 | 0.208 | 9.978 |
P < 0.001,
P < 0.05
Migration rates, M=2Nfem under a spatial expansion model, were estimated only for Bantu populations
Sequence variation in two east African haplogroups (L0a and L4) and two central African haplogroups (L1c and L3e) in east African populations was examined (Table 3). Both of the east African haplogroups are very diverse and show evidence of expansion with significant Tajima’s D and Fu’s Fs P-values. On the other hand, the central African haplogroups found in east Africa are much less diverse and do not show evidence of population expansions.
Table 3.
Sequence variation of two east African haplogroups (L0 and L4) and two central African haplogroups (L1c and L3e) in east African populations
| n | h | θk (95% CI) | θS(SD) | θπ (SD) | Tajima’s D | Fs | |
|---|---|---|---|---|---|---|---|
| L0a | 180 | 0.917 | 33.036 (23.832–45.481) | 9.190 (2.342) | 3.660 (2.060) | −1.905* | −25.976** |
| L4 | 178 | 0.890 | 31.393 (22.560–43.366) | 10.250 (2.579) | 4.666 (2.543) | −1.804* | −25.601** |
|
| |||||||
| L1c | 26 | 0.908 | 13.929 (6.640–29.404) | 8.386 (2.994) | 8.869 (4.707) | −0.334 | −2.051 |
| L3e | 70 | 0.697 | 3.426 (1.732–6.465) | 2.905 (1.057) | 1.978(1.255) | −1.028 | −2.131 |
P < 0.001,
P < 0.05
We compared Taita and Mijikenda mismatch distributions with those of the Turkana and Shona (Fig. S3). The Turkana, a Nilo-Saharan group from Kenya, have a large θπ value, similar to those of the Taita. The well-sampled (n=59) Shona are a genetically diverse Bantu-speaking population from southeastern Africa. They have larger θπ value than the Mijikenda, but not the Taita. The spatial expansion model predicts that two populations from the same spatial expansion wave will have similar mismatch distributions (Excoffier, 2004), so the Bantu populations at the expansion periphery should have similar mismatch distributions. The Taita’s unimodal mismatch distribution is similar to that of the Turkana, not the Shona, which suggests that as a result of gene flow, the Taita’s mismatch distribution mimics a signature of a very ancient expansion observed in non-Bantu east African populations. Conversely, the Mijikenda and Shona have similar mismatch distributions despite their geographical distance and different cultural history.
Population differentiation
The MDS analysis was used to illustrate the pairwise genetic distances (ΦST). The stress value (0.279) was within an acceptable range (Sturrock and Rocha, 2000). The central and southeastern African Bantu population samples are distributed in the lower half of the MDS plot with the non-Bantu east African found above. The east African Bantu populations (except for the Turu) cluster in the center of the plot and are located between non-Bantu east African populations and central African and southeastern African Bantu populations (Fig. 2). The Taita are plotted with the Hutu (Bantu) and Turkana (Nilo-Saharan) between the Tanzanian Nilo-Saharan and Afro-Asiatic populations on one side and the central African Bantu populations on the other side. The Mijikenda are plotted between the central African Bantu and the northern east African populations, but are closer to the central African Bantu populations.
Figure 2.
Multidimensional scaling plot of Bantu and east African populations.
Exact tests based on haplogroup frequencies show that the east African Bantu populations are genetically differentiated from each other (Table S4). The Taita and Mijikenda are genetically different from each other and from other east African Bantu populations. The genetic heterogeneity among east African Bantu groups is shown in the AMOVA results as well. The east African Bantu group has greater among-population variance than the southeastern African Bantu and east African Afro-Asiatic groups with a significant ΦST P-value (Table S5).
Female gene flow
We tested an isolation-by-distance model using the Mantel test to examine gene flow among neighboring populations. A significant correlation between pairwise population genetic distances (ΦST) and geographical distances (P-value = 0.001) was observed.
Migration rates, M=2Nfem, were estimated to quantify the intensity of gene flow. The migration rates estimated using the spatial expansion model of mismatch distribution are shown in Table 2. The genetically diverse populations, especially those with large θk values, have large M values and the populations with low genetic diversity have small M values. The Taita, Mijikenda, and other east African Bantu populations have small to moderate θk and M values, when compared to central African Bantu populations. A migration rate could not be obtained from the Kikuyu, possibly because of poor fit of the data to the spatial expansion model.
The migration rates (2Nfem) obtained from MIGRATE analyses show that female gene flow among different ethnic and language groups was common (Table 4). As the geographic proximity and close social ties would predict, the migration rate estimated between the Taita and Mijikenda (2Nfem = 28.071) is high. The estimated migration rates of the Taita and Mijikenda with other Bantu group are higher than with non-Bantu groups. The Taita, Mijikenda, and other east African Bantu group have from moderate to high migration rates with neighboring Nilo-Saharan and Afro-Asiatic groups.
Table 4.
Migration rates (2Nfem) estimated for Bantu and east African populations using MIGRATE
| Taita | Mijikenda | East Bantu | Other Bantu | Nilo-Saharan | N.E. Afro-Asiatic | |
|---|---|---|---|---|---|---|
| Taita | ||||||
| Mijikenda | 28.071 | |||||
| East African Bantu | 25.925 | 18.696 | ||||
| Other Bantu1 | 25.453 | 36.270 | 37.884 | |||
| Nilo-Saharans | 18.750 | 17.319 | 48.221 | 23.737 | ||
| N.E. Afro-Asiatic | 6.123 | 9.704 | 18.074 | 11.588 | 42.136 | |
| S.E. Afro-Asiatic | 15.367 | 6.600 | 37.249 | 11.422 | 63.710 | 13.556 |
The population groupings are based on their language and geographical location and see the listing of populations on Table S1.
Other Bantu populations include central African and southeastern African Bantu populations
DISCUSSION
In order to better understand the Bantu expansion processes in east Africa, we evaluated the interactions that took place between the Bantu- and non-Bantu-speaking populations in east Africa. Contrary to many previous genetic studies of Bantu-speaking populations (Salas et al., 2002; Berniell-Lee et al., 2009; Alves et al., 2011), our mtDNA study of two Bantu-speaking ethnic groups in the northeastern periphery of the Bantu expansion found mtDNA haplogroup frequencies similar to non-Bantu-speaking populations in east Africa, increased genetic diversity, and great genetic differentiation. Large migration rate estimates suggest that observed mtDNA genetic pattern resulted from intensive and/or constant interactions with the non-Bantu speaking neighboring ethnic groups.
Genetic studies have used linguistic and cultural homogeneity in Bantu-speaking populations to explain observed reduced genetic diversity in southeastern Bantu periphery and genetic homogeneity in the Bantu-speaking populations, and proposed that the Bantu expansion was demic expansion (Cavalli-Sforza et al., 1994; de Filippo et al., 2012). Y chromosome data shows reduced Y chromosome STR diversity and male effective population size in southeastern Africa (Thomas et al., 2000; Pereira et al., 2002; Coelho et al., 2009). de Filippo et al. (2012) also found statistically significant negative correlation between mtDNA genetic diversity and distance. In this study, mtDNA genetic diversity estimates does show a negative correlation with distance from the center of the Bantu expansion, but the correlation was statistically not significant.
Many genetic studies have also shown genetic homogeneity in Bantu-speaking populations (Excoffier et al., 1987; Cavalli-Sforza et al., 1994; Salas et al., 2002; Berniell-Lee et al., 2009; Alves et al., 2011; Schlebusch et al., 2012). Contrary to these studies, Montano et al. (2011) recently reported that Y chromosome variation in the Bantu populations in central Africa shows great genetic heterogeneity. Using 1000 single nucleotide polymorphisms genotyped across the genome, Sikora et al. (2011) similarly found that the Bantu group from southeastern African showed unexpectedly high genetic differentiation. Although mtDNA studies have often noted genetic homogeneity, Castrì and colleagues (2009) found that east African Bantu populations were genetically different from the central and southeastern African Bantu groups. Our study supports these recent studies, finding that 1) east African Bantu-speaking populations are genetically different from central African Bantu groups, and 2) east African Bantu populations are genetically differentiated from each other.
Migration rates estimated in this study support the observed haplogroup frequencies and suggest intensive or constant gene flow between the Bantu- and non-Bantu-speaking populations. Unequal effective population size and/or asymmetric gene flow between two groups may explain the observed genetic variation in east Africa. First, female effective population size may have been smaller in east African Bantu-speaking populations than in non-Bantu east African populations. The previously published Afro-Asiatic populations from northern east Africa and Nilo-Saharan from east Africa included in our study have larger genetic diversity estimates than the sampled east African Bantu populations, possibly reflecting larger female effective population sizes in non-Bantu-speaking east African populations. Furthermore, the small genetic diversity of two central African haplogroups analyzed suggests that founder effects occurred during the initial Bantu settlement of east Africa. The large non-Bantu pastoralist and foraging populations were likely already present in east Africa when the Bantu migrants arrived. The unequal effective population size would also explain the differences in genetic contribution from the Bantu migrants between east Africa and southeastern Africa where the landscape was more sparsely populated. Second, the gene flow among Bantu and non-Bantu East African populations may have been asymmetrical or unidirectional. Scozzari and colleagues (1999) and Underhill and colleagues (2001) argue that the Y chromosome E3a (E-M2) haplogroup was brought to east and southeastern Africa by the Bantu migrants. The frequency of this Y chromosome haplogroup is high in many east African Bantu populations (ranging from 42–83% in sampled populations), but east African Y chromosome haplogroups are relatively common as well (Luis et al., 2004; Tishkoff et al., 2007). If gene flow was simply asymmetric, we would expect the male Bantu genetic contribution to non-Bantu east Africans groups to be larger than the female Bantu contribution. Genetic researchers (Knight et al., 2003; Wood et al., 2005; Tishkoff et al., 2007) have not found this expectation to hold true, and the contribution of Bantu Y chromosome to non-Bantu east African populations is small. Two Nilo-Saharan populations, the Kenyan Masai and Tanzanian Datoga, have low frequencies of the central African Y chromosome haplogroup E3a (~11–16%) (Knight et al., 2003; Wood et al., 2005; Tishkoff et al., 2007), but this haplogroup is rare or absent in other east African populations (Semino et al., 2002; Hassan et al., 2008). Both mtDNA and Y chromosome data suggest that gene flow was unidirectional from non-Bantu to Bantu populations (Wood et al., 2005).
Our analysis of mtDNA variation in northeastern periphery of Bantu expansion provides a new insight on the processes of Bantu expansion and interactions that took place between the Bantu- and non-Bantu-speaking populations in east Africa. The central African Bantu populations initially experienced demographic expansion (Batini et al., 2007), but the population size of the ancestral Bantu speaking groups that migrated into east Africa may have been small and they experienced founder effects that reduced the west and central African haplogroup variation. After their arrival, individual east African Bantu groups experienced varying evolutionary histories, so today east African Bantu-speaking populations are genetically diverse and heterogeneous for different reasons. Some Bantu populations (e.g., the Kikuyu and Sukuma) have large θk values because their numbers quickly grew again once they acquired new technologies such as metallurgy and/or new domesticates such as grain crops, cattle and sheep (Schoenbrun, 1993; Phillipson, 2005). In some cases, Bantu populations like the Mijikenda maintained close relationships with other Bantu populations that reduced the effects of genetic drift and maintained diversity. In other cases, Bantu groups like the Taita and Hutu interacted with their non-Bantu neighbors and new haplogroups increased their genetic diversity through gene flow that occurred both during and after the initial Bantu spatial expansion into east Africa (Vansina, 1995). The Taita oral history indicates that Bantu speaking migrant groups absorbed the foragers who were living in the Taita Hills when they arrived (Bravman, 1998; Kusimba, 2009). Incomplete sampling may help explain the smaller migration rates observed in the Taita because some of the non-Bantu populations with whom they are known to have interacted, for example, the Masai, have not yet been sampled.
CONCLUSION
The genetic studies generally strengthen the argument for some form of migration by west/central Bantu-speakers into sub-Saharan Africa, but the mtDNA studies also indicate that individual Bantu-speaking populations experienced varying evolutionary histories in east Africa. Although expanding Bantu populations may have experienced founder effects as they first entered east Africa, there were extensive female gene flow and exchange both among Bantu groups and between Bantu speaking groups and their non-Bantu neighbors. These interactions with neighboring groups resulted in high levels of genetic diversity observed in some Bantu speaking-populations in east Africa. The Taita and Mijikenda remind us that even groups with similar backgrounds who live near each other today experienced unique demographic histories.
Supplementary Material
Acknowledgments
Grant Sponsorship: University of Illinois at Chicago Provost Research Award (K.B. and K.B.B.), NCI training grants: Cancer Education and Career Development Program (K.B.), J. William Fulbright fellowship, Explorer’s Club and Sigma Xi Grants In Aid of Research (K.B.B.)
We would like to thank the Mijikenda and Taita volunteers who provided the samples. We would like to thank Betsy Abrams, Mary Ashley, Geoffrey Hayes, and Crystal Patil for reviewing early versions of the manuscript and for helpful discussions. We also would like to thank to the Pritzker Lab at the Field Museum, Chicago for providing sequencing facilities for this research. This research project was supported by the University of Illinois at Chicago Provost Research Award, J. William Fulbright fellowship, Explorer’s Club, Sigma Xi Grants In Aid of Research, and NCI Training Program: Cancer Education and Career Development Program (5R25 CA057699).
Contributor Information
Ken Batai, Cancer Education and Career Development Program, Institute for Health Research and Policy, University of Illinois at Chicago, Chicago, IL 60607. Institute of Human Genetics, College of Medicine, University of Illinois at Chicago, Chicago, IL 60607.
Kara B. Babrowski, U.S. Department of State, Washington, D.C. 20520
Juan Pablo Arroyo, Department of Anthropology, University of South Florida, Tampa, FL 33620.
Chapurukha M. Kusimba, Department of Anthropology, The Field Museum, Chicago, IL 60607
Sloan R. Williams, Department of Anthropology, University of Illinois at Chicago, Chicago, IL, 60607
Literature Cited
- Alves I, Coelho M, Gignoux C, Damasceno A, Prista A, Rocha J. Genetic homogeneity across Bantu-speaking groups from Mozambique and Angola challenges early split scenarios between East and West Bantu populations. Hum Biol. 2011;83:13–38. doi: 10.3378/027.083.0102. [DOI] [PubMed] [Google Scholar]
- Anderson A, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–470. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Aris-Brosou S, Excoffier L. The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism. Mol Biol Evol. 1996;13:494–504. doi: 10.1093/oxfordjournals.molbev.a025610. [DOI] [PubMed] [Google Scholar]
- Bastin Y, Coupez A, Mann M. Continuity and divergence in the Bantu languages: perspectives from a lexicostatistic study. Tervuren: MRAC; 1999. [Google Scholar]
- Batini C, Coia V, Battaggia C, Rocha J, Pilkington MM, Spedini G, Comas D, Destro-Bisol G, Calafell F. Phylogeography of the human mitochondrial L1c haplogroup: genetic signatures of the prehistory of Central Africa. Mol Phylogenet Evol. 2007;43:635–644. doi: 10.1016/j.ympev.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall KE, Sykes BC. Length heteroplasmy in the first hypervariable segment of the human mtDNA control region. Am J Hum Genet. 1995;57:248–256. [PMC free article] [PubMed] [Google Scholar]
- Berniell-Lee G, Calafell F, Bosch E, Heyer E, Sica L, Mouguiama-Daouda P, van der Veen L, Hombert J-M, Quintana-Murci L, Comas D. Genetic and demographic implications of the Bantu expansion: insights from human paternal lineages. Mol Biol Evol. 2009;26:1581–1589. doi: 10.1093/molbev/msp069. [DOI] [PubMed] [Google Scholar]
- Blench R. Archaeology, language, and the African Past. Lanham, MD: AltaMira Press; 2006. [Google Scholar]
- Bostoen K. Pots, words and the Bantu problem: on lexical reconstruction and early African history. J Afr Hist. 2007;48:173–199. [Google Scholar]
- Bravman B. Making ethnic ways: communities and their transformations in Taita, Kenya, 1800–1950. Portsmouth, NH: Heinemann; 1998. [Google Scholar]
- Castrì L, Tofanelli S, Garagnani P, Bini C, Fosella X, Pelotti S, Paoli G, Pettener D, Luiselli D. mtDNA variability in two Bantu-speaking populations (Shona and Hutu) from Eastern Africa: implications for peopling and migration patterns in sub-Saharan Africa. Am J Phys Anthropol. 2009;140:302–311. doi: 10.1002/ajpa.21070. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi R, Piazza A. The history and geography of human genes. Princeton, NJ: Princeton University Press; 1994. [Google Scholar]
- Coelho M, Sequeira F, Luiselli D, Beleza S, Rocha J. On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola. BMC Evol Biol. 2009;9:80. doi: 10.1186/1471-2148-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, La Fratta R, Santolamazza P, Sellitto D, Pascone R, Moral P, Watson E, Guida V, Colomb EB, Zaharova B, et al. Phylogeographic analysis of haplogroup E3b (E-M215) Y chromosomes reveals multiple migratory events within and out of Africa. Am J Hum Genet. 2004;74:1014–1022. doi: 10.1086/386294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Filippo C, Barbieri C, Whitten M, Mpoloka SW, Gunnarsdóttir ED, Bostoen K, Nyambe T, Beyer K, Schreiber H, de Knijff P, et al. Y-chromosomal variation in sub-Saharan Africa: insights into the history of Niger-Congo groups. Mol Biol Evol. 2011;28:1255–1269. doi: 10.1093/molbev/msq312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Filippo C, Bostoen K, Stoneking M, Pakendorf B. Bringing together linguistic and genetic evidence to test the Bantu expansion. Proc Biol Sci. 2012;279:3256–3263. doi: 10.1098/rspb.2012.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert MK. The Bantu problem and African archaeology. In: Stahl AB, editor. African archaeology. Malden, MA: Blackwell Publishing; 2005. pp. 301–326. [Google Scholar]
- Ehret C. Language and history. In: Heine B, Nurse D, editors. African languages: an introduction. Cambridge: Cambridge University Press; 2000. pp. 272–297. [Google Scholar]
- Ehret C. Bantu expansions: re-envisioning a central problem of early African history. Int J Afr Hist Stud. 2001;34:5–41. [Google Scholar]
- Ehret C. Language family expansion: broadening our understandings of cause from an African perspective. In: Bellwood P, Renfrew C, editors. Examining the farming/language dispersal hypothesis. Cambridge: University of Cambridge Press; 2002. pp. 163–176. [Google Scholar]
- Ehret C, Nurse D. The Taita Cushites. SUGIA. 1981;3:125–168. [Google Scholar]
- Ewens WJ. The sampling theory of selectively neutral alleles. Theor Popul Biol. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- Excoffier L. Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol. 2004;13:853–864. doi: 10.1046/j.1365-294x.2003.02004.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Pellegrini B, Sanchez-Mazas A, Simon C, Langaney A. Genetics and history of Sub-Saharan Africa. Yearb Phys Anthropol. 1987;30:151–194. [Google Scholar]
- Excoffier L, Schneider S. Why hunter-gatherer populations do not show signs of Pleistocene demographic expansions. Proc Natl Acad Sci USA. 1999;96:10597–10602. doi: 10.1073/pnas.96.19.10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JH. The language of Africa. Bloomington: Indiana University Press; 1963. [Google Scholar]
- Greenberg JH. Linguistic evidence for Bantu origins. J Afr Hist. 1972;13:189–216. [Google Scholar]
- Guthrie M. Some developments in the prehistory of the Bantu languages. J Afr Hist. 1962;3:273–282. [Google Scholar]
- Hassan HY, Underhill PA, Cavalli-Sforza LL, Ibrahim ME. Y-chromosome variation among Sudanese: restricted gene flow, concordance with language, geography, and history. Am J Phys Anthropol. 2008;137:316–323. doi: 10.1002/ajpa.20876. [DOI] [PubMed] [Google Scholar]
- Helgason A, Nicholson GJ, Stefánsson K, Donnelly P. A reassessment of genetic diversity in Icelanders: strong evidence from multiple loci for relative homogeneity caused by genetic drift. Ann Hum Genet. 2003;67:281–297. doi: 10.1046/j.1469-1809.2003.00046.x. [DOI] [PubMed] [Google Scholar]
- Helgason A, Siguroardottir S, Gulcher JR, Ward R, Stefansson K. mtDNA and the origin of the Icelanders: deciphering signals of recent population history. Am J Hum Genet. 2000;66:999–1016. doi: 10.1086/302816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnesbusch T, Nurse D, Mould MJ. Studies in the classification of eastern Bantu languages. Hamburg: Helmut Buske Verlag; 1981. [Google Scholar]
- Holden CJ. Bantu language trees reflect the spread of farming across sub-Saharan Africa: a maximum-persimony analysis. Proc Biol Sci. 2002;269:793–799. doi: 10.1098/rspb.2002.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden CJ, Gray RD. Rapid radiation, borrowing and dialect continua in the Bantu languages. In: Forster P, Renfrew C, editors. Phylogenetic methods and the prehistory of languages. Cambridge: McDonald Institute for Archaeological Research; 2006. [Google Scholar]
- Kivisild T, Reidla M, Metspalu E, Rosa A, Brehm A, Pennarun E, Parik J, Geberhiwot T, Usanga E, Villems R. Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the Gate of Tears. Am J Hum Genet. 2004;75:752–770. doi: 10.1086/425161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight A, Underhill PA, Mortensen HM, Zhivogtovsky LA, Lin AA, Henn BM, Louis D, Ruhlen M, Mountain JL. African Y chromosome and mtDNA divergence provides insight into the history of Click languages. Curr Biol. 2003;13:464–473. doi: 10.1016/s0960-9822(03)00130-1. [DOI] [PubMed] [Google Scholar]
- Krings M, Halim Salem A-e, Bauer K, Geisert H, Malek AK, Chaix L, Simon C, Welsby D, Di Rienzo A, Utermann G, et al. mtDNA analysis of Nile River valley populations: a genetic corridor of a barrier to migration. Am J Hum Genet. 1999;64:1166–1176. doi: 10.1086/302314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusimba CM. Practicing postcolonial archaeology in East Africa from the United States. In: Schmidt PR, editor. Postcolonial archaeology in Africa. Santa Fe, NM: School of Advanced Research Press; 2009. pp. 57–76. [Google Scholar]
- Luis JR, Rowold DJ, Regueiro M, Caeiro B, Cinnioglu C, Roseman C, Underhill PA, Cavalli-Sforza LL, Herrera RJ. The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet. 2004;74:532–544. doi: 10.1086/382286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maho J. A classification of the Bantu languages: an update of Guthrie’s referential system. In: Nurse D, Philippson G, editors. The Bantu languages. London: Routledge; 2003. pp. 639–651. [Google Scholar]
- Montano V, Ferri G, Marcari V, Batini C, Anyaele O, Destro-Bisol G, Comas D. The Bantu expansion revisited: a new analysis of Y chromosome variation in Central Western Africa. Mol Ecol. 2011;20:2693–2708. doi: 10.1111/j.1365-294X.2011.05130.x. [DOI] [PubMed] [Google Scholar]
- Nurse D, Philippson G, editors. The Bantu languages. London: Routledge; 2003. [Google Scholar]
- Pereira L, Gusmao L, Alves C, Amorim A, Prata MJ. Bantu and European Y-lineages in sub-Saharan Africa. Ann Hum Genet. 2002;66:369–378. doi: 10.1017/S0003480002001306. [DOI] [PubMed] [Google Scholar]
- Pereira L, Macaulay V, Torroni A, Scozzari R, Prata MJ, Amorin A. Prehistoric and historic traces in the mtDNA of Mozambique: insights into the Bantu expansions and the slave trade. Ann Hum Genet. 2001;65:439–458. doi: 10.1017/S0003480001008855. [DOI] [PubMed] [Google Scholar]
- Pfeiffer H, Brinkmann B, Hühne J, Rolf B, Morris AA, Steighner R, Holland MM, Forster P. Expanding the forensic German mitochondrial DNA control region database: genetic diversity as a function of sample size and microgeography. Int J Legal Med. 1999;112:291–298. doi: 10.1007/s004140050252. [DOI] [PubMed] [Google Scholar]
- Philippson G, Montlahuc M-L. Kilimanjaro Bantu. In: Nurse D, Philippson G, editors. The Bantu languages. London: Routledge; 2003. pp. 470–500. [Google Scholar]
- Phillipson DW. Iron Age history and archaeology in Zambia. J Afr Hist. 1974;15:1–25. [Google Scholar]
- Phillipson DW. The chronology of the Iron Age in Bantu Africa. J Afr Hist. 1975;16:321–342. [Google Scholar]
- Phillipson DW. Archaeology and Bantu linguistics. World Archaeol. 1976;8:65–82. [Google Scholar]
- Phillipson DW. Later prehistory of eastern and southern Africa. London: Heinemann; 1977. [Google Scholar]
- Phillipson DW. African archaeology. Cambridge: Cambridge University Press; 2005. [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt H-J, Passarino G, McElreavey K. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet. 1999;23:437–441. doi: 10.1038/70550. [DOI] [PubMed] [Google Scholar]
- Robertson JH, Bradley R. A new paradigm: the African Early Iron Age without Bantu migrations. Hist Afr. 2000;27:287–323. [Google Scholar]
- Rogers AR. Genetic evidence for a Pleistocene population expansion. Evolution. 1995;49:608–615. doi: 10.1111/j.1558-5646.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- Salas A, Richards M, De le Fe T, Lareu M-V, Sobrino B, Sánchez-Diz P, Macaulay V, Carracedo A. The making of the African mtDNA landscape. Am J Hum Genet. 2002;71:1082–1111. doi: 10.1086/344348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadeberg TC. Historical linguistics. In: Nurse D, Philippson G, editors. The Bantu languagues. London: Routledge; 2003. pp. 143–163. [Google Scholar]
- Schlebusch CM, Skoglund P, Sjödin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MGB, et al. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science. 2012;338:374–379. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenbrun DL. We are what we eat: ancient agriculture between the Great Lakes. J Afr Hist. 1993;34:1–31. [Google Scholar]
- Semino O, Sanchiara Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA. Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet. 2002;70:265–268. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora M, Laayouni H, Calafell F, Comas D, Bertranpetit J. A genomic analysis identifies a novel component in the genetic structure of sub-Saharan African populations. Eur J Hum Genet. 2011;19:84–88. doi: 10.1038/ejhg.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear TT. Traditional myths and historian’s myths: variations on the Singwaya theme of Mijikenda origins. Hist Afr. 1974;1:67–84. [Google Scholar]
- Spear TT. Traditional myths and linguistic analysis: Singwaya revisited. Hist Afr. 1977;4:229–264. [Google Scholar]
- Spear TT. Traditions of origin and their interpretation: the Mijikenda of Kenya. Athens, OH: Ohio University Center for International Studies; 1981. [Google Scholar]
- Sturrock K, Rocha J. A multidimensional scaling stress evaluation table. Field Methods. 2000;12:49–60. [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Parfitt T, Weiss DA, Skorecki K, Wilson JF, le Roux M, Bradman N, Goldstein DB. Y Chromosomes traveling south: the Cohen modal haplotype and the origins of the Lemba—the Black Jews of southern Africa. Am J Hum Genet. 2000;66:674–686. doi: 10.1086/302749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Gonder MK, Henn BM, Mortensen H, Knight A, Gignoux C, Fernandopulle N, Lema G, Nyambo TB, Ramakrishnan U, et al. History of Click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol Biol Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo J-M, Doumbo O, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- Vansina J. New linguistic evidence and ‘the Bantu Expansion’. J Afr Hist. 1995;36:173–195. [Google Scholar]
- Watson E, Bauer K, Aman R, Weiss G, von Haeseler A, Pääbo S. mtDNA sequence diversity in Africa. Am J Hum Genet. 1996;59:437–444. [PMC free article] [PubMed] [Google Scholar]
- Watson E, Forster P, Richards M, Bandelt H-J. Mitochondrial footprints of human expansions in Africa. Am J Hum Genet. 1997;61:691–704. doi: 10.1086/515503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G-A. On the number of segregation sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Willis J. Mombasa, the Swahili, and the making of the Mijikenda. Oxford: Clarendon Press; 1993. [Google Scholar]
- Wood ET, Stover DA, Ehret C, Destro-Bisol G, Spedini G, McLeod H, Louie L, Bamshad M, Strassmann BI, Soodyal H, et al. Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur J Hum Genet. 2005;13:867–876. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.