Abstract
Mitochondrial oxidative metabolism plays a key role in meeting energetic demands of cells by oxidative phosphorylation (OxPhos). Here, we have briefly discussed (i) the dynamic relationship that exists among glycolysis, the tricarboxylic acid (TCA) cycle, and OxPhos; (ii) the evidence of impaired OxPhos (i.e. mitochondrial dysfunction) in breast cancer; (iii) the mechanisms by which mitochondrial dysfunction can predispose to cancer; and (iv) the effects of host and environmental factors that can negatively affect mitochondrial function. We propose that impaired OxPhos could increase susceptibility to breast cancer via suppression of the p53 pathway, which plays a critical role in preventing tumorigenesis. OxPhos is sensitive to a large number of factors intrinsic to the host (e.g. inflammation) as well as environmental exposures (e.g. pesticides, herbicides and other compounds). Polymorphisms in over 143 genes can also influence OxPhos system. Therefore, declining mitochondrial oxidative metabolism with age due to host and environmental exposures could be a common mechanism predisposing to cancer.
Keywords: Mitochondrial metabolism, Oxidative phosphorylation, OxPhos, inflammation, tumor suppressor p53, breast cancer
1. Introduction
Metabolism is central to cellular physiology. It provides energy in the form of adenosine 5′-triphosphate (ATP) and building blocks (other nucleotides, lipids, amino acids, etc.) for different cellular processes including cell growth, proliferation, and supporting functions of differentiated cells such as milk synthesis in lactating mammary glands [1,2]. The overall cellular metabolism relies on glycolysis, the tricarboxylic acid (TCA, also known as citric acid or Krebs) cycle, and oxidative phosphorylation (OxPhos). The reactions of glycolysis, the TCA cycle and OxPhos occur in the cytoplasm, mitochondrial matrix, and at mitochondrial inner membrane, respectively. Although occurring at distinct subcellular locations, these processes are interdependent (Fig. 1). Their relationship is tunable based on catabolic and anabolic demands of cells [3]. One of the key factors that regulate this relationship in real time is ATP-demand. In this review, we will briefly discuss their relationship and the role of impaired mitochondrial metabolism in breast cancer. Emphasis is given to OxPhos because it is susceptible to host and environmental factors that may contribute to a majority of breast cancers [4].
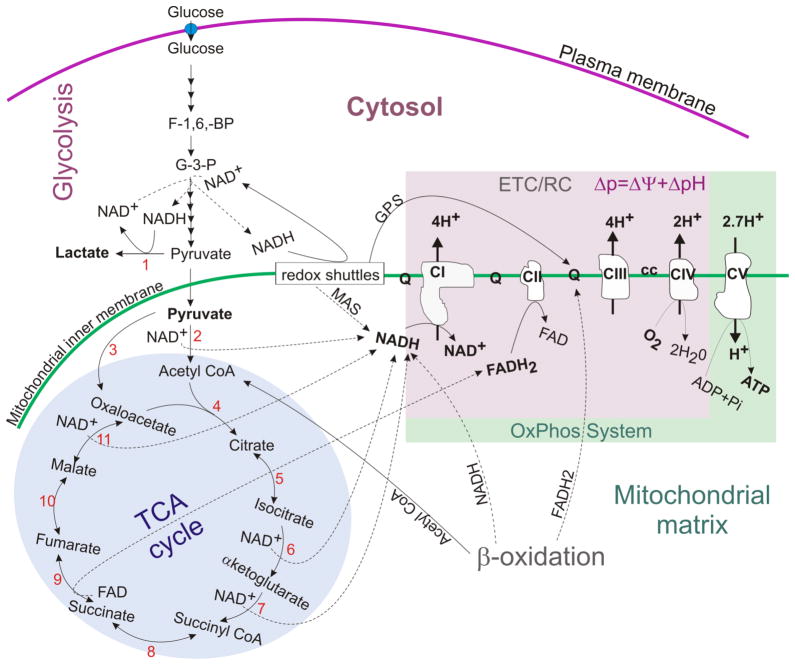
Figure 1. An overview of cellular metabolism showing the links among glycolysis, the TCA cycle and OxPhos.
Glycolysis is linked with the TCA cycle and OxPhos via pyruvate and redox shuttles (MAS, GPS). CI-V: Complexes I–V; ETC/RC: electron transport chain/respiratory chain made up by C-IV; OxPhos system: oxidative phosphorylation system made by CI-V (or RC + CV). Ubiquinone (Q) and cytochrome c (cc) are electron carriers; MAS: malate-aspartate redox shuttle; GPS: glycerol-3-phosphate redox shuttle. Numbers indicate selected enzymes- 1: lactate dehydrogenase (LDH); 2: pyruvate dehydrogenase (PDH); 3: pyruvate carboxylase (PC)- an anaplerotic enzyme; 4: citrate synthase (CS); 5: aconitase; 6: isocitrate dehydrogenase (IDH3: NAD+-dependent shown; IDH1, 2: NADP+-dependent not shown); 7: α-ketoglutarate dehydrogenase (KGDH); 8: succinyl-CoA synthase (SCS: ATP, GTP-dependent); 9: succinate dehydrogenase (SDH or Complex II); 10: fumarate hydratase or fumarase (FH); 11: malate dehydrogenase (MDH). Broken thin lines indicate reactions generating electron donors (NADH and FADH2) and the solid thin lines indicate reactions oxidizing NADH and FADH2.
2. An overview of cellular metabolism
In a typical mammalian cell glucose is metabolized via glycolysis to generate pyruvate. Glycolysis produces 2 net ATP molecules per glucose using the reactions catalyzed by phosphoglycerate kinase and pyruvate kinase, which are [1, 3-bisphosphoglycerate + ADP ⇔ 3-phosphoglycerate + ATP] and [phosphoenolpyruvate + ADP + H+ ⇔ pyruvate + ATP] respectively. This mode of ATP production is called substrate-level phosphorylation as it uses the chemical energy liberated from a substrate. In intact cells, this can be slowed down by a limitation in NAD+ supply [5], because the reaction upstream to phosphoglycerate kinase is catalyzed by an NAD+-dependent enzyme, glyceraldehyde-3-phosphate dehydrogenase [glyceraldehyde-3-phosphate + Pi + NAD+ ⇔ 1,3-bisphosphoglycerate + NADH + H+]. If the cytosolic NAD+ pool is completely reduced to NADH by glyceraldehyde-3-phosphate dehydrogenase, the NAD+ will become limiting. Therefore, NAD+ must be regenerated in cytosol to permit continued operation of glycolysis. Cytosolic NAD+ is regenerated using lactate dehydrogenase and NADH redox shuttles [6]. Lactate dehydrogenase reduces pyruvate into lactate by using an NADH [pyruvate + NADH + H+ ⇔ lactate + NAD+]. Conversion of glucose to lactate is favored when oxygen is limiting. However, even in the presence of excess oxygen nearly all mammalian cells convert a significant fraction of pyruvate into lactate. The fraction of pyruvate converted into lactate is higher in proliferative and tumor cells than in differentiated cells [7]. Apart from hypoxic conditions, genetic and drug-induced OxPhos deficiencies can also up-regulate pyruvate conversion into lactate. Therefore, release of lactate by cells provides a surrogate for real-time measurements of glycolysis [8].
OxPhos, the process of making ATP by consuming oxygen (i.e. respiration), is the major source of ATP in animal cells. It is carried out by five multimeric enzyme complexes (I–V) with the help of electron carriers (ubiquinone and cytochrome c) and electron donors (NADH and FADH2). Complexes I to IV constitute the respiratory chain, which establishes H+ gradient across the inner mitochondrial membrane. This gradient is established by Complexes I, III and IV, which pump out 4, 4, and 2 H+ respectively while transferring electrons liberated from NADH and FADH2 oxidations (Fig. 1). The H+ gradient is also known as proton motive force (Δp) and has two components- the mitochondrial membrane potential (Δψm) and pH gradient (ΔpH) [9]. The potential energy of Δp drives ATP synthesis by Complex V, a rotary motor that makes 3ATP/rotation while using 2.7H+/ATP [10]. After accounting for the expense of inorganic phosphate (Pi) and adenine nucleotides exchange, the net cost of making 1 ATP molecule is thought to be 3.7 H+ [10]. Thus, 10 NADH and 2 FADH2 generated from complete oxidation of 1 glucose molecule can produce about 30 ATP molecules via OxPhos. However, in reality the actual number will be lower than the calculated value due to H+ leak across the inner mitochondrial membrane.
Other fuels such as fatty acids, ketone bodies and amino acids also support OxPhos.β-oxidation of fatty acids generates acetyl-CoA, NADH and FADH2, which can support the TCA cycle and OxPhos (Fig. 1). β-oxidation derived FADH2 feeds electrons to Complex III via an electron transferring flavoprotein-ubiquinone oxidoreductase [9]. Both the TCA cycle and β-oxidation are feedback inhibited by NADH buildup. Therefore, β-oxidation is not sustainable without functional Complex I. Ketone bodies such as acetoacetate and β-hydroxybutyrate also support the TCA cycle and OxPhos. Amino acids oxidation can also support OxPhos. Before entering the TCA cycle, amino acids are converted to pyruvate (Ala, Ser, Gly, Thr, Cys, Trp), acetyl-CoA (Leu, Ile, Lys, Phe, Tyr, Trp, Thr), α-ketoglutarate (Glu, Gln, Pro, His, Arg), succinyl-CoA (Met, Ile, Val), fumarate (Phe, Tyr), or oxaloacetate (Asp, Asn) [3]. While all amino acids can enter TCA cycle, the oxidation of glutamine is a major contributor to OxPhos [11]. Clearly, the relative use of glucose, glutamine or other fuels will depend on catabolic and anabolic needs of cells [3]. For example, branched chain amino acid metabolism plays an important role in mammary epithelial cell physiology during lactation. The branched chain amino acids (Leu, Ile, Val) are metabolized to produce glutamate, glutamine, aspartate, alanine, and asparagine to be secreted in milk [12,13].
OxPhos is involved in a dynamic relationship with the TCA cycle and glycolysis. In real-time, inhibition of OxPhos with oligomycin up-regulates glycolysis and slows down the TCA cycle ([11,14,15]; unpublished data, C. Kim and N. Yadava). Oligomycin is a Complex V inhibitor. Respiratory chain defects result in auxotrophy for asparagine and CO2 due to blockage of the TCA cycle [16–18]. The CO2 auxotrophy is due to feedback inhibition of the NAD+-dependent pyruvate and α-ketoglutarate dehydrogenases. The asparagine auxotrophy is thought to be due to a limitation in oxaloacetate production. The requirement of asparagine clearly underscores the interplay between anabolism and OxPhos via the TCA cycle. Although OxPhos is the major source of ATP production, cells with <8% respiratory activity can meet their total ATP demand by glycolysis alone. As long as glucose supply is not limiting even complete OxPhos deficiency does not impair growth and proliferation of cells [11,19]. In respiration-competent lung fibroblasts ~40% of ATP is derived from OxPhos supported by glucose and glutamine oxidations [11]. The contribution of OxPhos to cellular ATP pool is expected to vary with tissue type. When OxPhos is suppressed due to hypoxia or the presence of inhibitors, glutamine can undergo reductive metabolism without any ATP production [20,21]. Tumor cells use this pathway of reductive glutamine metabolism (carboxylation) to produce citrate for lipid synthesis [21]. Citrate breakdown by ATP-citrate lyase following export from mitochondria into the cytoplasm provides acetyl-CoA for fatty acid synthesis [citrate + ATP + CoA + H2O ⇔ acetyl-CoA + ADP + Pi + oxaloacetate]. ATP-citrate lyase inhibition suppresses tumor cell growth [22]. Citrate production in mitochondria is limited by oxaloacetate and acetyl-CoA availability (Fig. 1). However, citrate exit from the TCA cycle will limit oxaloacetate production. Therefore, anaplerosis (filling-in) of the TCA cycle with oxaloacetate will be required. Glutamine oxidation is a major contributor to oxaloacetate pool in tumor cells. This is clear from the elimination of glutamine dependence of tumor cells in the presence of glucose by pyruvate carboxylase over expression [23]. Pyruvate carboxylase converts pyruvate into oxaloacetate [pyruvate + HCO3− + ATP ⇔ oxaloacetate + ADP + Pi]. Therefore, apart from glutamine dependence tumors may have pyruvate carboxylase up regulated [24]. The relative dependence on pyruvate carboxylase vs. glutamine metabolism for citrate synthesis may vary in different tumors.
Integrity of the OxPhos system is critical for optimal energy production, but functional assembly of OxPhos system is a very complicated process. Nuclear and mitochondrial genomes encode 45, 4, 11, 13, and 16 subunits of Complexes I, II, III, IV and V, respectively. The mitochondrial –(mt)DNA-encoded subunits are ND1-6 and ND-4L of Complex I; Cyt b of Complex III; COI-III of Complex IV; and ATP6 and ATP8 of Complex V. These 13 proteins are translated within mitochondria using a protein translation machinery similar to prokaryotes [25]. Thus, OxPhos system is vulnerable to antibiotics that inhibit bacterial protein synthesis. MtDNA also encodes 22 tRNAs and 2 rRNAs that constitute the mitochondrial protein synthesis system with nuclear-(n)DNA-encoded ribosomal proteins and other factors [25]. Between 2–10 assembly factors are required for the assembly of individual OxPhos complexes [26]. The final number of assembly factors for any given complex is not yet settled. The formation of super-complexes adds another layer of complexity to the OxPhos system. This also requires additional proteins such as HIG2A [27,28]. Over 143 genes (89 structural proteins, 30 assembly factors, 22 tRNAs and 2 rRNAs) are directly involved in OxPhos system biogenesis [26].
3. Evidence for impaired metabolism in breast cancer
Metabolic reprogramming is a common feature of the majority of cancers. Increased glycolysis in cancer was suggested by Otto Warburg over 90 year ago [29,30]. Warburg proposed that an “irreversible injury” to the OxPhos system could lead to metabolic shift towards glycolysis (the Warburg effect) thereby causing cancer development [29]. His observations offered the premise for using 2-fluoro (18F)-2′-deoxyglucose-positron emission tomography (FDG-PET) in cancer staging and monitoring [31,32]. The FDG signal is suggested to be proportional to the rate of glycolysis in viable cells and reports the relatively increased demand for glucose in tumors [33]. While the success of FDG-PET in diagnosis of primary breast cancers is variable, it has shown increased glucose uptake in metastatic breast cancers [34]. The redox scanning technique based on cryogenic NADH/flavoprotein fluorescence, which reports OxPhos activity in cells, has also revealed alterations in mitochondrial metabolism [35,36].
The Warburg hypothesis predicts that impaired OxPhos or mitochondrial dysfunction should be a common feature of cancer cells. To validate this hypothesis, several studies have examined mutations in mtDNA, which is relatively more vulnerable to damage than nuclear nDNA. A large number of mtDNA mutations in breast and other cancers have been identified [37–47]. Mutations in DNA polymerase-γ (POLG) that is involved in mtDNA replication result in reduced OxPhos activity due to mtDNA depletion/mutation in breast cancers [48]. The strongest support for the Warburg hypothesis has come from germ line mutations in Complex II genes [49–53]. Mutations in genes encoding Complex II subunits and assembly factor(s) predispose to hereditary paraganglioma/phaeochromocytoma syndrome [51–53]. Complex II germ line variants also modify breast and thyroid cancer risks in Cowden and Cowden-like syndrome [54]. A survey of COSMIC (catalogue of somatic mutations in cancers) database shows over 26 Complex I genes mutated in different cancers with ≤1% frequency (unpublished observations, N. Yadava). In ductal-breast carcinoma, somatic missense mutations have been found in several Complex I genes such as Ndufa2, Ndufa3 and Ndufa8.
Functional analyses of OxPhos in cancers are relatively rare. Almost complete loss of Complex I (mostly due to mtDNA mutations) is associated with renal and thyroid tumors [43,55]. While mtDNA mutations in the regulatory regions are expected to result in multiple deficiencies of OxPhos, the amino acid altering mtDNA mutations affect Complex I more frequently (>50%) [46]. Mutations in ND4 (C12084T) and ND5 (A13966G) subunits affect Complex I function and influence metastatic properties of MDA-MB-231 breast cancer cell line [56]. The ND3 polymorphism (G10398A) associated with aggressive breast cancer in African-American women also alters Complex I activity [57]. A few other studies also suggest impaired OxPhos in breast cancers [58,59]. Expression of the nuclear-encoded NDUFS3 subunit of Complex I is associated with breast cancer invasiveness [60]. Its increased expression may be a compensatory response to OxPhos deficiency. This is not unexpected because the NDUFS3 subunit enters into Complex I assembly very early to facilitate additions of other subunits [61]. Reduced expression of the β-subunit of Complex V (β-F1-ATPase) in cancer is often accompanied by an increase in glyceraldehyde-3-phosphate dehydrogenase. A bioenergetic signature derived from the relative expressions of β-F1-ATPase and glyceraldehyde-3-phosphate dehydrogenase has high prognostic value for breast and other cancers [62,63]. Reduced β-F1-ATPase and increased inhibitory factor 1 (IF1) and uncoupling proteins (e.g. UCP2) levels suggest that OxPhos efficiency is compromised in breast cancer cells [64,65]. A few studies also suggest metabolic interdependence between stromal fibroblasts and cancer cells. In this relationship fibroblasts provide nutrients (e.g. lactate) to tumor cells [66–68]. Whether metabolites shuttling is used for bioenergetic or other purposes remains to be clearly resolved. It is possible that limited NADH production in cancer cell mitochondria or a partial OxPhos deficiency force cells to switch their reliance on NADH redox shuttles. Lactate use via reversal of lactate dehydrogenase will support redox-shuttles by generating NADH. If respiration is primarily supported by the glycerol-3-phosphate shuttle (GPS) in cancer cells (Fig. 1), then substantial respiratory activity with less ATP yield is expected because only 6 H+ are pumped out per FADH2 oxidation vs. 10 per NADH oxidation.
4. Cancer development and altered metabolism
Cancer development is a multistep process involving over 4 rate-limiting stochastic events [69,70]. Somatic mutations, failures of negative regulatory feedback for proliferation and contact inhibition, senescence and the corruption of signaling pathways (an inhibitory pathway becoming promoter) are key events that occur at different stages of tumorigenesis. These events confer several hallmark capabilities in cancer cells. Six of these capabilities were described in the year 2000 by Hanahan and Weinberg [69]: (i) sustaining proliferative signaling, (ii) evading growth suppressors, (iii) resisting cell death, (iv) enabling replicative immortality, (v) inducing angiogenesis, and (vi) activating invasion and metastasis. Reprogramming of energy metabolism (vii) and evading immune destruction (viii) have been added to this list as emerging hallmark capabilities [70]. They are intricately linked and enabled by genomic instability, epigenetic modifications, and tumor-promoting inflammation [70]. Activation and suppression of signaling molecules such as B-Raf, Akt/PKB, mTOR, Ras, Myc, PTEN, pRB, p53, HIF1, NF-κB etc. are thought to result in acquisitions of the hallmark capabilities [70,71]. Whether modest changes in metabolism alone can alter multiple signaling pathways in otherwise normal cells and give them cancer hallmark capabilities remains to be elucidated.
Apart from a secondary role, impaired mitochondrial metabolism may also play a primary role in cancer development. Observations that mtDNA mutation/depletion enhance tumorigenicity and metastatic potential have confirmed the secondary role of mitochondrial dysfunction [56,72,73]. Assuming that metabolic alterations are a consequence of transformation, the majority of studies have focused on the effects of tumor suppressors and oncogenes on cellular metabolism to explain the Warburg effect [74–78]. This subject has been discussed in several reviews [7,79,80]. It is noted that while tumor suppressors promote oxidative metabolism, the oncogenes favor glycolysis, and they have opposing effects on glycolysis and OxPhos. Altered functions of tumor suppressors and oncogenes reprogram the metabolism of transformed cells to meet their anabolic demands [7,79,80]. The idea that altered metabolism may play a primary role in cancer development is gaining momentum [81]. Alterations in activities of enzymes such as pyruvate kinase (PKM2) and isocitrate dehydrogenase (IDH1, 2), and others are thought to enhance cellular transformation. The associations of heritable polymorphisms with the risks of breast and other cancers suggest that mitochondrial dysfunction could play a causative role in tumorigenesis [44,45,54,82]. Two recent studies suggest that mitochondrial dysfunction can predispose to tumorigenesis. One is based on maternal transmission of mtDNA mutations originated in mice due to haploinsufficiency of mitochondrial helicase mSuv3 [83]. The other is based on a Complex I deficient model due to a mutation in the mtDNA-encoded ND6 subunit [84]. However, it remains to be determined whether a primary OxPhos defect can predispose to mammary tumorigenesis. The observations of mtDNA and p53 mutations together in the majority of breast and other cancers support a causal link between mitochondrial dysfunction and cancer development [45,85,86].
Mitochondrial dysfunction may promote cancer development by conferring hallmark capabilities: (i) by suppressing p53 and PTEN function [87,88], (ii) activating AKT1 and HIF1α pathways [87,89–91], (iii) inducing extracellular matrix remodeling for invasion [73,92], (iv) providing resistance to cell death [88,93], (v) shifting metabolism towards glycolysis [94], and controlling inflammation [95]. Mitochondrial proteins such as SIRT3 (a protein deacetylase), CHCHD4 (a component of disulfide relay system regulating electron transport to cytochrome c), and NDUFA4L2 (a hypoxia inducible Complex I inhibitor) are clearly implicated in regulating both tumorigenesis and cellular respiration [96–98]. Impairments of the OxPhos system are linked with changes in cellular redox status, reactive oxygen species (ROS) levels, bioenergetics and ionic homeostasis. These changes in cellular physiology are linked with regulations of different pathways including p53 and AKT [54,87,99]. Both p53 and AKT are involved in early events of mammary epithelial cell transformation [100,101].
There is a possibility that declining OxPhos function could play a causative role in increased cancer incidence with age. The Yadava laboratory has shown that OxPhos deficiency can suppress p53 expression/function in several cell types [88]. The p53 suppression by OxPhos deficiency is a reversible phenomenon. Ablation of Complex I provides protection against γ-radiation induced apoptosis when glucose is not limiting in different cells including a mouse mammary epithelial cell line TM40A (unpublished, S. Compton and N. Yadava) [88]. The suppression of p53 is not limited to Complex I deficiency as inhibition of mitochondrial protein synthesis and Complex II-deficiency also result in reduced p53 expression/function (unpublished, C. Kim and N. Yadava, [88]). In mice the mtDNA mutation rate is linked to premature aging and p53 function declines with age [102,103]. Mitochondrial dysfunction is suggested to play a causative role in aging and other diseases including cancer [104]. As noted above, there is evidence for predisposition to tumorigenesis by mtDNA mutations in mice [83,84]. Therefore, there is a possibility that functional decline of p53 mediated by reduced mitochondrial function could predispose to cancer [88,103]. Even modest impairments of mitochondrial function can significantly suppress p53 function [88]. In humans, germ line mutations in the TP53 gene result in Li-Fraumeni syndrome, which is characterized by a strong predisposition to a spectrum of cancers with breast cancer being the most common in women [105]. Furthermore, studies with p53 knockout mice have clearly demonstrated the critical role p53 plays in tumor prevention [106–108].
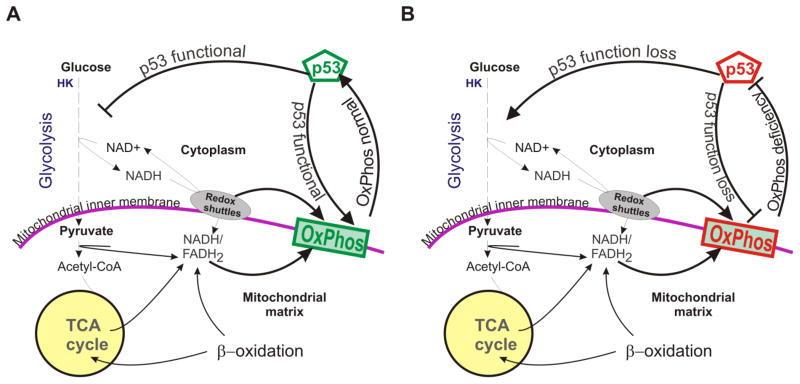
The relationship between p53 and metabolism is quite complicated. While it has been observed that basal levels of p53 promote mitochondrial metabolism, p53 activation following telomere dysfunction reduces mitochondrial biogenesis [76,109]. p53 is also reversibly suppressed by OxPhos deficiency [88]. The accepted view is that p53 suppresses glycolysis and promotes oxidative metabolism in proliferating cells (Fig. 2A) [110]. Positive regulation of TIGAR is primarily associated with negative regulation of glycolysis [77]. P53 promotes OxPhos by modulating Complex IV biogenesis [76], mtDNA copy number [111] and mtDNA maintenance [112]. Therefore p53 function loss is thought to cause a metabolic shift toward glycolysis (Fig. 2B). Suppressing p53 in the presence of OxPhos deficiency may not be in the best interest of cells because it may result in bioenergetic crises. From this point of view, a negative regulation of p53 by mitochondrial dysfunction in not unexpected [88]. It is possible that p53 and mitochondrial function act together to maintain cellular homeostasis and when their coordination is lost pathological consequences result. Alterations in p53 can also affect other signaling pathways such as AKT and NF-κB, which make mammary epithelial cells susceptible to oncogenic transformation [100,101,113,114]. Alterations in the AKT and NF-κB pathways can influence cellular metabolism and cell fate [115–117].
Figure 2. Relationship between the tumor suppressor p53 and metabolism.
A) p53’s relationship with glycolysis and OxPhos in normal cells. B) p53’s relationship with glycolysis and OxPhos in cells with OxPhos deficiency. Because p53 is suppressed by OxPhos deficiency, its negative and positive effects on glycolysis and OxPhos would be removed.
P53 primarily functions as a transcription factor that regulates genes implicated in a variety of cellular processes such as cell cycle arrest, DNA repair, apoptosis, senescence, autophagy, metabolism, oxidative/redox stress, angiogenesis and many more relevant to normal physiology and pathology [118]. P53 prevents cell proliferation when conditions are not favorable such as in the presence of acute DNA damage or oncogene activation [119]. In addition to acting as a transcription factor, p53 protein also acts directly to regulate cell fate [120–122]. This regulation is tissue- and stimulus-specific [123]. It remains to be explored whether variation in mitochondrial metabolism influences cell-specific properties of p53.
Mitochondrial dysfunction may result in genetic and epigenetic changes. Complexes I, II and III produce reactive oxygen species (ROS) when electron transfer is impaired [124–126]. Hypoxia, OxPhos deficiencies, high Δp, NADH/NAD+ and ubiquinone (reduced/oxidized) influence ROS homeostasis [127]. Increased ROS production due to OxPhos deficiency can result in genomic instability [128]. Additionally, alterations in the dTTP nucleotide pool due to OxPhos deficiency can also cause genomic instability [129]. Mitochondrial proteins such as Sirt3, CHCHD4 and NDUFA4L2 directly link OxPhos activity with hypoxic response, which plays an important role in cancer development [96–98,130]. ROS are thought to play major role in breast tumorigenesis. This is supported by a study showing that catalase expression in mitochondria can suppress invasive breast cancers in mice [131]. Catalase is an enzyme that degrades hydrogen peroxide (H2O2). Oxidative stress, particularly in stromal fibroblasts, is suggested to drive breast cancer development [132]. It is associated with the loss of caveolin-1 (Cav-1) in stromal fibroblasts [133,134]. Cav-1 deficiency is associated with cholesterol localization to mitochondria, which results in impairment of OxPhos and oxidative stress [135–137]. Hypoxia and OxPhos deficiency induce reductive glutamine metabolism that provides growth advantages [20,21,81]. The relationship between mitochondrial function and epigenetic changes have been recently described [47,81,138]. DNA methylation and histone acetylation that impart epigenetic influences are linked with mitochondrial metabolism [139,140].
5. Host and environmental factors affecting mitochondrial metabolism
Genetic alterations, polymorphisms and stochastic changes in expression of over 143 genes may exert influence on mitochondrial function [26,141]. nDNA and mtDNA polymorphisms are linked with breast cancer risk [44,45,54,142]. While polymorphic variations can directly affect the function of a protein, they are expected to differ in severity based on interactions between nDNA- and mtDNA-encoded subunits of OxPhos complexes. This possibility is clearly exemplified by mutational analysis of the MWFE (or NDUFA1) subunit of Complex I, which interacts with mtDNA-encoded proteins [19,143].
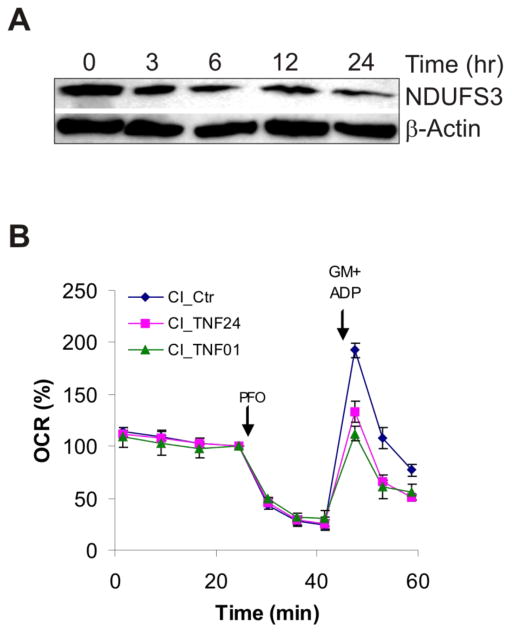
In addition to genetic factors, inflammation can play a significant role in suppressing mitochondrial metabolism. Thus, chronic inflammation very often associated with obesity/metabolic syndrome may facilitate cancer initiation and progression by suppressing OxPhos [144,145]. Inflammation can occur through localized secretion of factors such as TNFα, Wnt and leptin, which impair OxPhos [146–150]. Inflammation and oxidative stress are linked with TNFα-mediated damage to OxPhos system in ob/ob mice via protein nitration [151]. Often Complexes I, III and IV are affected by inflammatory cytokines. The Yadava Laboratory has found that low levels of TNFα (10 ng/ml) that do not induce cell death can reduce the levels of NDUFS3 subunit of Complex I in HEK293 cells (Fig. 3A). In primary human mammary epithelial cells (HMECs), TNFα exposure causes 30–40% reduction in Complex I-dependent respiration (Fig. 3B). The effect of TNFα could be seen even with 1 hr exposure suggesting that mitochondrial metabolism of HMECs is highly susceptible to inflammatory cytokines. Apart from TNFα, local macrophages can also secrete Wnts that promote epithelial-to-mesenchymal transition (EMT) and stem cell like behavior in breast cancer cells [152,153]. Interestingly, exposure to Wnts has also been shown to inhibit OxPhos and enhance glycolysis through the down-regulation of Complex IV subunits [150]. Thus, inflammation may influence cancer progression though alterations of mitochondrial metabolism.
Figure 3. Effect of inflammatory cytokine TNFα on Complex I (CI).
A) Effect of TNFα (10 ng/ml) on CI subunit NDUFS3 expression in human HEK293 cells. (B) Effect of TNFα (10 ng/ml) on CI function in human mammary epithelial cells (HMECs). Primary HMECs (passage 3, 50,000/well) were permeabilized with 1 nM PFO (perfringolysin-O) and then glutamate and malate (10 mM each) were added with 1 mM ADP (GM+ADP) to measure CI-dependent oxygen consumption rates (OCR). Relative OCR is shown after setting the basal rate before PFO addition to 100%. OCR was measured using a 24-well Extracellular Flux (XF24) Analyzer from Seahorse Bioscience (Billerica, MA). CI_Ctr, CI_TNF01, and CI_TNF24 show CI-dependent respiration in control, 1 hr and 24 hr TNFα-treated HMECs. Respiration rates were measured in a Ca2+-free buffer. The measurement of respiration in PFO-permeabilized cells is a novel method (unpublished, C. Kim, A. Heuck and N. Yadava).
A reciprocal relationship may exist between mitochondrial metabolism and inflammation. As noted above, while inflammatory cytokines can suppress mitochondrial function, there is a possibility that mitochondrial dysfunction can alter immune response. Immune cells carefully modulate their metabolism during development and activation, which often involves extensive proliferation. It is suggested that during proliferation metabolism of T-lymphocytes switches toward glycolysis [154]. Switching back from glycolysis to OxPhos supported by β-oxidation is linked with establishment of the memory T-cells [155]. During the contraction phase, the T-cells that cannot utilize OxPhos fail to form memory T-cells. Other immune cells, such as dendritic cells, also exhibit a similar switch to glycolysis upon toll-like receptor (TLR)-induced maturation [156]. Mitochondrial function also affects cytokine production from macrophages in response to lipopolysaccharides (LPS)-stimulation [157], and positively modulates MHC class I antigens expression on the cell surface [158]. Thus, diminished OxPhos activity in tumor cells may protect them by reducing MHC class I expression. MtDNA-variants are known to affect transplantation outcomes [159,160]. Additionally mitochondria-derived peptides may result in an inflammatory response [161,162]. P53 and NF-κB pathways that regulate inflammatory and autoimmune responses may also exert their influence on the immune response via altering cellular metabolism [76,77,117,163]. Taken together there is sufficient evidence to suggest that mitochondrial metabolism may play an important role in regulating immune response. Thus, it is possible that inflammation-induced mitochondrial dysfunction can sustain inflammation by altering properties of local immune cells and protect tumor cells by immune avoidance. Although, a transient exposure to inflammatory cytokine IL-6 is found sufficient to promote MCF10A cells transformation with 100% efficiency, the role of impaired oxidative metabolism in this process remains to be explored [113].
The effect of hormones on mitochondria provides a possible link by which endocrine disrupting chemicals can influence the outcome of breast cancer and other diseases[164]. Of particular interest are estrogens that regulate glycolysis, the TCA cycle and OxPhos in a sex-specific specific manner [165]. Estrogens regulate mitochondrial biogenesis and function both directly and indirectly [166]. Humans are exposed to a host of estrogenic compounds that occur naturally (e.g. isoflavones enriched in soy products). In addition, industrial contaminants (e.g. BPA) and agricultural chemicals (e.g. atrazine, permethrins) have been shown to have estrogenic activities. These compounds interact with a complex estrogen signaling pathway composed of 2 isoforms of estrogen receptors (ERα and ERβ) that act as transcription factors in the nucleus. These receptors form homo- and hetero-dimers, which bind promoters of target genes to either enhance or inhibit transcription depending on the presence of co-activators or co-repressors [167,168]. Estrogenic ligands show selectivity in binding to ERα and ERβ resulting in differences in transcriptional responses, effects on target genes and cellular behavior [169–171]. However, these estrogen receptors can also localize to membranes and activate distinct signaling cascades referred to as “non-genomic” signaling. Functional estrogen receptors are also found in mitochondria [165,172,173]. The effects of estrogenic compounds vary greatly in tissues depending on the complement of receptors.
Lipophilic compounds of natural and synthetic origins, to which humans are often exposed, can cause mitochondrial dysfunction [174]. Agricultural chemicals such as pesticides are known to inhibit OxPhos [175]. Use of rotenone, a common pesticide, is linked with Parkinson’s disease in rodents and humans [175,176]. Pyrethrins are commonly used to control insects. While these compounds break down quickly, residues can persist when used in poorly ventilated areas. Following treatment with insecticides, pyrethrins are detectable in cows’ milk [177]. Pyrethroids such as permethrin and cyhalothrin were shown to inhibit Complex I activity at picomolar levels [178]. Atrazine, a common herbicide, has estrogenic activity, and thus, it may also have significant consequences for mammary gland development. Although in utero exposure can alter mammary gland development in neonates by increasing the number of terminal end buds, this effect was transient [179]. Nonetheless, long-term exposure may have implications for breast cancer risk in women through its presumed estrogenic effects [180,181]. Atrazine’s herbicidal activity is due to its inhibitory action on photosystem Complex II in chloroplasts, which can also inhibit OxPhos [182]. As modest OxPhos-deficiency results in a near complete abrogation of the tumor suppressor p53 activity (unpublished data C. Kim and N. Yadava; [88]), the oncogenic effects of environmental contaminants may be related to their inhibitory effects on mitochondrial function.
6. Conclusion
Glycolysis, the TCA cycle and OxPhos are engaged in a dynamic relationship, which is primarily controlled by ATP demand (Fig. 1). Normal cells switch their metabolism from glycolysis toward OxPhos and vice versa based on their energetic needs and fuel availability. Glycolysis can alone supply sufficient ATP via substrate level phosphorylation for cellular growth and proliferation[11]. Thus, OxPhos is not essential as long as glucose entry into glycolysis is not limited and the ATP-demand does not exceed the capacity of glycolysis [11,183]. In proliferative and tumor cells, increased demands for nucleotides such as NADPH and lipids synthesis are thought to favor a metabolic switch toward glycolysis resulting in the Warburg effect [7,30]. The modification of breast cancer risk by genetic polymorphisms affecting the OxPhos system suggests that impaired mitochondrial metabolism could play a causative role in breast cancer development [44,45,54,142]. Several observations with cell lines showing promotion of cancer hallmark capabilities indicate that mitochondrial dysfunction could play a primary role in tumorigenesis. Two studies show that mice bearing mtDNA mutations are indeed predisposed to tumorigenesis [83,84]. The vulnerability of the OxPhos system to host factors (e.g. age, inflammatory cytokines) and environmental factors (e.g. pesticides, natural and synthesis compounds) may increase the susceptibility to breast cancer. We propose that the suppression of p53 by mitochondrial dysfunction could play a key role in enhanced breast cancer susceptibility [88].
Acknowledgments
This work was supported by institutional start-up and translational funds from CEAR at the PVLSI supported by an award (A00000000004448) from Massachusetts Technology Collaborative as administrator of the John Adams Innovation Institute to N. Y. We thank Maureen Lahti for critical reading of the manuscript.
Abbreviations
- NAD+ and NADH
oxidized and reduced nicotinamide adenine dinucleotides, respectively
- NADP+ and NADPH
phosphorylated forms of NAD+ and NADPH, respectively
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- Pi
inorganic phosphate
- FAD and FADH2
oxidized and reduced flavin adenine dinucleotides, respectively
- nDNA
nuclear-DNA
- mtDNA
mitochondrial-DNA
- OxPhos
oxidative phosphorylation
- TCA
tricarboxylic acid
- CoA
coenzyme A
Reference List
- 1.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 2.Lei J, Feng D, Zhang Y, Zhao FQ, Wu Z, San GA, et al. Nutritional and regulatory role of branched-chain amino acids in lactation. Front Biosci. 2012;17:2725–2739. doi: 10.2741/4082. [DOI] [PubMed] [Google Scholar]
- 3.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 4.Benson JR, Jatoi I. The global breast cancer burden. Future Oncol. 2012;8:697–702. doi: 10.2217/fon.12.61. [DOI] [PubMed] [Google Scholar]
- 5.Ying W, Alano CC, Garnier P, Swanson RA. NAD+ as a metabolic link between DNA damage and cell death. J Neurosci Res. 2005;79:216–223. doi: 10.1002/jnr.20289. [DOI] [PubMed] [Google Scholar]
- 6.Bender K, Newsholme P, Brennan L, Maechler P. The importance of redox shuttles to pancreatic beta-cell energy metabolism and function. Biochem Soc Trans. 2006;34:811–814. doi: 10.1042/BST0340811. [DOI] [PubMed] [Google Scholar]
- 7.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 9.Nicholls DG, Ferguson SJ. Bioenergetics. 3. Academic Press; London: 2002. [Google Scholar]
- 10.Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci U S A. 2010;107:16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnelly M, Scheffler IE. Energy metabolism in respiration-deficient and wild type Chinese hamster fibroblasts in culture. J Cell Physiol. 1976;89:39–51. doi: 10.1002/jcp.1040890105. [DOI] [PubMed] [Google Scholar]
- 12.Li P, Knabe DA, Kim SW, Lynch CJ, Hutson SM, Wu G. Lactating porcine mammary tissue catabolizes branched-chain amino acids for glutamine and aspartate synthesis. J Nutr. 2009;139:1502–1509. doi: 10.3945/jn.109.105957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lei J, Feng D, Zhang Y, Dahanayaka S, Li X, Yao K, et al. Regulation of leucine catabolism by metabolic fuels in mammary epithelial cells. Amino Acids. 2012 doi: 10.1007/s00726-012-1302-2. [DOI] [PubMed] [Google Scholar]
- 14.Nicholls DG, Shepherd D, Garland PB. A continuous recording technique for the measurement of carbon dioxide, and its application to mitochondrial oxidation and decarboxylation reactions. Biochem J. 1967;103:677–691. doi: 10.1042/bj1030677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholls DG, Darley-Usmar VM, Wu M, Jensen PB, Rogers GW, Ferrick DA. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 2010 doi: 10.3791/2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ditta G, Soderberg K, Landy F, Scheffler IE. The selection of Chinese hamster cells deficient in oxidative energy metabolism. Somatic Cell Genet. 1976;2:331–344. doi: 10.1007/BF01538838. [DOI] [PubMed] [Google Scholar]
- 17.DeFrancesco L, Werntz D, Scheffler IE. Conditionally lethal mutations in chinese hamster cells. Characterization of a cell line with a possible defect in the Krebs cycle. J Cell Physiol. 1975;85:293–305. doi: 10.1002/jcp.1040850216. [DOI] [PubMed] [Google Scholar]
- 18.DeFrancesco L, Scheffler IE, Bissell MJ. A respiration-deficient Chinese hamster cell line with a defect in NADH-coenzyme Q reductase. J Biol Chem. 1976;251:4588–4595. [PubMed] [Google Scholar]
- 19.Yadava N, Potluri P, Smith EN, Bisevac A, Scheffler IE. Species-specific and mutant MWFE proteins. Their effect on the assembly of a functional mammalian mitochondrial complex I. J Biol Chem. 2002;277:21221–21230. doi: 10.1074/jbc.M202016200. [DOI] [PubMed] [Google Scholar]
- 20.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci U S A. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, et al. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christian BE, Spremulli LL. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta. 2012;1819:1035–1054. doi: 10.1016/j.bbagrm.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghezzi D, Zeviani M. Assembly factors of human mitochondrial respiratory chain complexes: physiology and pathophysiology. Adv Exp Med Biol. 2012;748:65–106. doi: 10.1007/978-1-4614-3573-0_4. [DOI] [PubMed] [Google Scholar]
- 27.Schagger H. Respiratory chain supercomplexes. IUBMB Life. 2001;52:119–128. doi: 10.1080/15216540152845911. [DOI] [PubMed] [Google Scholar]
- 28.Chen YC, Taylor EB, Dephoure N, Heo JM, Tonhato A, Papandreou I, et al. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15:348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 30.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 31.Weber WA, Schwaiger M, Avril N. Quantitative assessment of tumor metabolism using FDG-PET imaging. Nucl Med Biol. 2000;27:683–687. doi: 10.1016/s0969-8051(00)00141-4. [DOI] [PubMed] [Google Scholar]
- 32.Kelloff GJ, Hoffman JM, Johnson B, Scher HI, Siegel BA, Cheng EY, et al. Progress and promise of FDG-PET imaging for cancer patient management and oncologic drug development. Clin Cancer Res. 2005;11:2785–2808. doi: 10.1158/1078-0432.CCR-04-2626. [DOI] [PubMed] [Google Scholar]
- 33.Kostakoglu L, Goldsmith SJ. 18F-FDG PET evaluation of the response to therapy for lymphoma and for breast, lung, and colorectal carcinoma. J Nucl Med. 2003;44:224–239. [PubMed] [Google Scholar]
- 34.Avril N, Rose CA, Schelling M, Dose J, Kuhn W, Bense S, et al. Breast imaging with positron emission tomography and fluorine-18 fluorodeoxyglucose: use and limitations. J Clin Oncol. 2000;18:3495–3502. doi: 10.1200/JCO.2000.18.20.3495. [DOI] [PubMed] [Google Scholar]
- 35.Xu HN, Tchou J, Chance B, Li LZ. Imaging the redox States of human breast cancer core biopsies. Adv Exp Med Biol. 2013;765:343–349. doi: 10.1007/978-1-4614-4989-8_48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu HN, Nioka S, Glickson JD, Chance B, Li LZ. Quantitative mitochondrial redox imaging of breast cancer metastatic potential. J Biomed Opt. 2010;15:036010. doi: 10.1117/1.3431714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandra D, Singh KK. Genetic insights into OXPHOS defect and its role in cancer. Biochim Biophys Acta. 2010;1807:620–625. doi: 10.1016/j.bbabio.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simonnet H, Demont J, Pfeiffer K, Guenaneche L, Bouvier R, Brandt U, et al. Mitochondrial complex I is deficient in renal oncocytomas. Carcinogenesis. 2003;24:1461–1466. doi: 10.1093/carcin/bgg109. [DOI] [PubMed] [Google Scholar]
- 39.bu-Amero KK, Alzahrani AS, Zou M, Shi Y. High frequency of somatic mitochondrial DNA mutations in human thyroid carcinomas and complex I respiratory defect in thyroid cancer cell lines. Oncogene. 2005;24:1455–1460. doi: 10.1038/sj.onc.1208292. [DOI] [PubMed] [Google Scholar]
- 40.Dasgupta S, Soudry E, Mukhopadhyay N, Shao C, Yee J, Lam S, et al. Mitochondrial DNA mutations in respiratory complex-I in never-smoker lung cancer patients contribute to lung cancer progression and associated with EGFR gene mutation. J Cell Physiol. 2011 doi: 10.1002/jcp.22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theodoratou E, Din FV, Farrington SM, Cetnarskyj R, Barnetson RA, Porteous ME, et al. Association between common mtDNA variants and all-cause or colorectal cancer mortality. Carcinogenesis. 2010;31:296–301. doi: 10.1093/carcin/bgp237. [DOI] [PubMed] [Google Scholar]
- 42.Gasparre G, Hervouet E, de LE, Demont J, Pennisi LF, Colombel M, et al. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum Mol Genet. 2008;17:986–995. doi: 10.1093/hmg/ddm371. [DOI] [PubMed] [Google Scholar]
- 43.Mayr JA, Meierhofer D, Zimmermann F, Feichtinger R, Kogler C, Ratschek M, et al. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin Cancer Res. 2008;14:2270–2275. doi: 10.1158/1078-0432.CCR-07-4131. [DOI] [PubMed] [Google Scholar]
- 44.Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- 45.Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007;249:249–255. doi: 10.1016/j.canlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 46.Plak K, Czarnecka AM, Krawczyk T, Golik P, Bartnik E. Breast cancer as a mitochondrial disorder (Review) Oncol Rep. 2009;21:845–851. doi: 10.3892/or_00000293. [DOI] [PubMed] [Google Scholar]
- 47.Minocherhomji S, Tollefsbol TO, Singh KK. Mitochondrial regulation of epigenetics and its role in human diseases. Epigenetics. 2012;7:326–334. doi: 10.4161/epi.19547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh KK, Ayyasamy V, Owens KM, Koul MS, Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J Hum Genet. 2009;54:516–524. doi: 10.1038/jhg.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baysal BE, Willett-Brozick JE, Lawrence EC, Drovdlic CM, Savul SA, McLeod DR, et al. Prevalence of SDHB, SDHC, and SDHD germline mutations in clinic patients with head and neck paragangliomas. J Med Genet. 2002;39:178–183. doi: 10.1136/jmg.39.3.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 51.Bardella C, Pollard PJ, Tomlinson I. SDH mutations in cancer. Biochim Biophys Acta. 2011;1807:1432–1443. doi: 10.1016/j.bbabio.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 52.Kunst HP, Rutten MH, de Monnink JP, Hoefsloot LH, Timmers HJ, Marres HA, et al. SDHAF2 (PGL2-SDH5) and hereditary head and neck paraganglioma. Clin Cancer Res. 2011;17:247–254. doi: 10.1158/1078-0432.CCR-10-0420. [DOI] [PubMed] [Google Scholar]
- 53.Bayley JP, Kunst HP, Cascon A, Sampietro ML, Gaal J, Korpershoek E, et al. SDHAF2 mutations in familial and sporadic paraganglioma and phaeochromocytoma. Lancet Oncol. 2010;11:366–372. doi: 10.1016/S1470-2045(10)70007-3. [DOI] [PubMed] [Google Scholar]
- 54.Ni Y, He X, Chen J, Moline J, Mester J, Orloff MS, et al. Germline SDHx variants modify breast and thyroid cancer risks in Cowden and Cowden-like syndrome via FAD/NAD-dependant destabilization of p53. Hum Mol Genet. 2012;21:300–310. doi: 10.1093/hmg/ddr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zimmermann FA, Mayr JA, Neureiter D, Feichtinger R, Alinger B, Jones ND, et al. Lack of complex I is associated with oncocytic thyroid tumours. Br J Cancer. 2009;100:1434–1437. doi: 10.1038/sj.bjc.6605028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imanishi H, Hattori K, Wada R, Ishikawa K, Fukuda S, Takenaga K, et al. Mitochondrial DNA mutations regulate metastasis of human breast cancer cells. PLoS ONE. 2011;6:e23401. doi: 10.1371/journal.pone.0023401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kulawiec M, Owens KM, Singh KK. mtDNA G10398A variant in African-American women with breast cancer provides resistance to apoptosis and promotes metastasis in mice. J Hum Genet. 2009;54:647–654. doi: 10.1038/jhg.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Owens KM, Kulawiec M, Desouki MM, Vanniarajan A, Singh KK. Impaired OXPHOS complex III in breast cancer. PLoS ONE. 2011;6:e23846. doi: 10.1371/journal.pone.0023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Putignani L, Raffa S, Pescosolido R, Aimati L, Signore F, Torrisi MR, et al. Alteration of expression levels of the oxidative phosphorylation system (OXPHOS) in breast cancer cell mitochondria. Breast Cancer Res Treat. 2008;110:439–452. doi: 10.1007/s10549-007-9738-x. [DOI] [PubMed] [Google Scholar]
- 60.Suhane S, Berel D, Ramanujan VK. Biomarker signatures of mitochondrial NDUFS3 in invasive breast carcinoma. Biochem Biophys Res Commun. 2011;412:590–595. doi: 10.1016/j.bbrc.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenzie M, Ryan MT. Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life. 2010;62:497–502. doi: 10.1002/iub.335. [DOI] [PubMed] [Google Scholar]
- 62.Isidoro A, Casado E, Redondo A, Acebo P, Espinosa E, Alonso AM, et al. Breast carcinomas fulfill the Warburg hypothesis and provide metabolic markers of cancer prognosis. Carcinogenesis. 2005;26:2095–2104. doi: 10.1093/carcin/bgi188. [DOI] [PubMed] [Google Scholar]
- 63.Cuezva JM, Krajewska M, de Heredia ML, Krajewski S, Santamaria G, Kim H, et al. The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- 64.Sanchez-Cenizo L, Formentini L, Aldea M, Ortega AD, Garcia-Huerta P, Sanchez-Arago M, et al. The up-regulation of the ATPase Inhibitory Factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J Biol Chem. 2010 doi: 10.1074/jbc.M110.146480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ayyasamy V, Owens KM, Desouki MM, Liang P, Bakin A, Thangaraj K, et al. Cellular model of Warburg effect identifies tumor promoting function of UCP2 in breast cancer and its suppression by genipin. PLoS ONE. 2011;6:e24792. doi: 10.1371/journal.pone.0024792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sotgia F, Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe RC, Witkiewicz AK, et al. Mitochondrial metabolism in cancer metastasis: visualizing tumor cell mitochondria and the “reverse Warburg effect” in positive lymph node tissue. Cell Cycle. 2012;11:1445–1454. doi: 10.4161/cc.19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salem AF, Whitaker-Menezes D, Lin Z, Tanowitz HB, Al-Zoubi MS, Howell A, et al. Two-compartment tumor metabolism: Autophagy in the tumor microenvironment and oxidative mitochondrial metabolism (OXPHOS) in cancer cells. Cell Cycle. 2012;11:2545–2556. doi: 10.4161/cc.20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balliet RM, Capparelli C, Guido C, Pestell TG, Martinez-Outschoorn UE, Lin Z, et al. Mitochondrial oxidative stress in cancer-associated fibroblasts drives lactate production, promoting breast cancer tumor growth: understanding the aging and cancer connection. Cell Cycle. 2011;10:4065–4073. doi: 10.4161/cc.10.23.18254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 70.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 71.Perkins ND. The diverse and complex roles of NF-kappaB subunits in cancer. Nat Rev Cancer. 2012;12:121–132. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- 72.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kulawiec M, Safina A, Desouki MM, Still I, Matsui S, Bakin A, et al. Tumorigenic transformation of human breast epithelial cells induced by mitochondrial DNA depletion. Cancer Biol Ther. 2008;7:1732–1743. doi: 10.4161/cbt.7.11.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007;21:1037–1049. doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 77.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 78.Yang D, Wang MT, Tang Y, Chen Y, Jiang H, Jones TT, et al. Impairment of mitochondrial respiration in mouse fibroblasts by oncogenic H-RAS(Q61L) Cancer Biol Ther. 2010:9. doi: 10.4161/cbt.9.2.10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 80.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 81.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shen L, Wei J, Chen T, He J, Qu J, He X, et al. Evaluating mitochondrial DNA in patients with breast cancer and benign breast disease. J Cancer Res Clin Oncol. 2011;137:669–675. doi: 10.1007/s00432-010-0912-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen PL, Chen CF, Chen Y, Guo XE, Huang CK, Shew JY, et al. Mitochondrial genome instability resulting from SUV3 haploinsufficiency leads to tumorigenesis and shortened lifespan. Oncogene. 2012 doi: 10.1038/onc.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hashizume O, Shimizu A, Yokota M, Sugiyama A, Nakada K, Miyoshi H, et al. Specific mitochondrial DNA mutation in mice regulates diabetes and lymphoma development. Proc Natl Acad Sci U S A. 2012;109:10528–10533. doi: 10.1073/pnas.1202367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lahiry L, Saha B, Chakraborty J, Adhikary A, Mohanty S, Hossain DM, et al. Theaflavins target Fas/caspase-8 and Akt/pBad pathways to induce apoptosis in p53-mutated human breast cancer cells. Carcinogenesis. 2009 doi: 10.1093/carcin/bgp240. [DOI] [PubMed] [Google Scholar]
- 86.Gochhait S, Bhatt A, Sharma S, Singh YP, Gupta P, Bamezai RN. Concomitant presence of mutations in mitochondrial genome and p53 in cancer development - a study in north Indian sporadic breast and esophageal cancer patients. Int J Cancer. 2008;123:2580–2586. doi: 10.1002/ijc.23817. [DOI] [PubMed] [Google Scholar]
- 87.Pelicano H, Xu RH, Du M, Feng L, Sasaki R, Carew JS, et al. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J Cell Biol. 2006;175:913–923. doi: 10.1083/jcb.200512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Compton S, Kim C, Griner NB, Potluri P, Scheffler IE, Sen S, et al. Mitochondrial Dysfunction Impairs Tumor Suppressor p53 Expression/Function. J Biol Chem. 2011;286:20297–20312. doi: 10.1074/jbc.M110.163063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Porcelli AM, Ghelli A, Ceccarelli C, Lang M, Cenacchi G, Capristo M, et al. The genetic and metabolic signature of oncocytic transformation implicates HIF1{alpha} destabilization. Hum Mol Genet. 2009;19:1019–1032. doi: 10.1093/hmg/ddp566. [DOI] [PubMed] [Google Scholar]
- 90.Sun W, Zhou S, Chang SS, McFate T, Verma A, Califano JA. Mitochondrial mutations contribute to HIF1alpha accumulation via increased reactive oxygen species and up-regulated pyruvate dehydrogenease kinase 2 in head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:476–484. doi: 10.1158/1078-0432.CCR-08-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Briere JJ, Favier J, Benit P, El GV, Lorenzato A, Rabier D, et al. Mitochondrial succinate is instrumental for HIF1alpha nuclear translocation in SDHA-mutant fibroblasts under normoxic conditions. Hum Mol Genet. 2005;14:3263–3269. doi: 10.1093/hmg/ddi359. [DOI] [PubMed] [Google Scholar]
- 92.van WC, Sun Y, Cheung HS, Moraes CT. Oxidative phosphorylation dysfunction modulates expression of extracellular matrix--remodeling genes and invasion. Carcinogenesis. 2006;27:409–418. doi: 10.1093/carcin/bgi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dey R, Moraes CT. Lack of oxidative phosphorylation and low mitochondrial membrane potential decrease susceptibility to apoptosis and do not modulate the protective effect of Bcl-x(L) in osteosarcoma cells. J Biol Chem. 2000;275:7087–7094. doi: 10.1074/jbc.275.10.7087. [DOI] [PubMed] [Google Scholar]
- 94.Acebo P, Giner D, Calvo P, Blanco-Rivero A, Ortega AD, Fernandez PL, et al. Cancer abolishes the tissue type-specific differences in the phenotype of energetic metabolism. Transl Oncol. 2009;2:138–145. doi: 10.1593/tlo.09106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kamp DW, Shacter E, Weitzman SA. Chronic inflammation and cancer: the role of the mitochondria. Oncology (Williston Park) 2011;25:400–10. 413. [PubMed] [Google Scholar]
- 96.Yang J, Staples O, Thomas LW, Briston T, Robson M, Poon E, et al. Human CHCHD4 mitochondrial proteins regulate cellular oxygen consumption rate and metabolism and provide a critical role in hypoxia signaling and tumor progression. J Clin Invest. 2012;122:600–611. doi: 10.1172/JCI58780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Haigis MC, Deng CX, Finley LW, Kim HS, Gius D. SIRT3 is a mitochondrial tumor suppressor: a scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res. 2012;72:2468–2472. doi: 10.1158/0008-5472.CAN-11-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tello D, Balsa E, Acosta-Iborra B, Fuertes-Yebra E, Elorza A, Ordonez A, et al. Induction of the mitochondrial NDUFA4L2 protein by HIF-1alpha decreases oxygen consumption by inhibiting Complex I activity. Cell Metab. 2011;14:768–779. doi: 10.1016/j.cmet.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 99.Asher G, Lotem J, Kama R, Sachs L, Shaul Y. NQO1 stabilizes p53 through a distinct pathway. Proc Natl Acad Sci U S A. 2002;99:3099–3104. doi: 10.1073/pnas.052706799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dimri G, Band H, Band V. Mammary epithelial cell transformation: insights from cell culture and mouse models. Breast Cancer Res. 2005;7:171–179. doi: 10.1186/bcr1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao JJ, Gjoerup OV, Subramanian RR, Cheng Y, Chen W, Roberts TM, et al. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell. 2003;3:483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 102.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 103.Feng Z, Hu W, Teresky AK, Hernando E, Cordon-Cardo C, Levine AJ. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci U S A. 2007;104:16633–16638. doi: 10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gonzalez KD, Noltner KA, Buzin CH, Gu D, Wen-Fong CY, Nguyen VQ, et al. Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol. 2009;27:1250–1256. doi: 10.1200/JCO.2008.16.6959. [DOI] [PubMed] [Google Scholar]
- 106.Hinkal G, Parikh N, Donehower LA. Timed somatic deletion of p53 in mice reveals age-associated differences in tumor progression. PLoS ONE. 2009;4:e6654. doi: 10.1371/journal.pone.0006654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 108.Kemp CJ, Wheldon T, Balmain A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat Genet. 1994;8:66–69. doi: 10.1038/ng0994-66. [DOI] [PubMed] [Google Scholar]
- 109.Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 111.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, et al. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shamanin VA, Androphy EJ. Immortalization of human mammary epithelial cells is associated with inactivation of the p14ARF-p53 pathway. Mol Cell Biol. 2004;24:2144–2152. doi: 10.1128/MCB.24.5.2144-2152.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Coloff JL, Macintyre AN, Nichols AG, Liu T, Gallo CA, Plas DR, et al. Akt-dependent glucose metabolism promotes Mcl-1 synthesis to maintain cell survival and resistance to Bcl-2 inhibition. Cancer Res. 2011;71:5204–5213. doi: 10.1158/0008-5472.CAN-10-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Plas DR, Thompson CB. Akt-dependent transformation: there is more to growth than just surviving. Oncogene. 2005;24:7435–7442. doi: 10.1038/sj.onc.1209097. [DOI] [PubMed] [Google Scholar]
- 117.Mauro C, Leow SC, Anso E, Rocha S, Thotakura AK, Tornatore L, et al. NF-kappaB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat Cell Biol. 2011;13:1272–1279. doi: 10.1038/ncb2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brady CA, Attardi LD. p53 at a glance. J Cell Sci. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 120.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 121.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 122.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 123.Jackson JG, Post SM, Lozano G. Regulation of tissue- and stimulus-specific cell fate decisions by p53 in vivo. J Pathol. 2011;223:127–136. doi: 10.1002/path.2783. [DOI] [PubMed] [Google Scholar]
- 124.Miwa S, Brand MD. The topology of superoxide production by complex III and glycerol 3-phosphate dehydrogenase in Drosophila mitochondria. Biochim Biophys Acta. 2005;1709:214–219. doi: 10.1016/j.bbabio.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 125.Lambert AJ, Brand MD. Superoxide production by NADH:ubiquinone oxidoreductase (complex I) depends on the pH gradient across the mitochondrial inner membrane. Biochem J. 2004;382:511–517. doi: 10.1042/BJ20040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Quinlan CL, Orr AL, Perevoshchikova IV, Treberg JR, Ackrell BA, Brand MD. Mitochondrial Complex II Can Generate Reactive Oxygen Species at High Rates in Both the Forward and Reverse Reactions. J Biol Chem. 2012;287:27255–27264. doi: 10.1074/jbc.M112.374629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sung HJ, Ma W, Wang PY, Hynes J, O’Riordan TC, Combs CA, et al. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat Commun. 2010;1:5. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Desler C, Munch-Petersen B, Stevnsner T, Matsui S, Kulawiec M, Singh KK, et al. Mitochondria as determinant of nucleotide pools and chromosomal stability. Mutat Res. 2007;625:112–124. doi: 10.1016/j.mrfmmm.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 130.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goh J, Enns L, Fatemie S, Hopkins H, Morton J, Pettan-Brewer C, et al. Mitochondrial targeted catalase suppresses invasive breast cancer in mice. BMC Cancer. 2011;11:191. doi: 10.1186/1471-2407-11-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Martinez-Outschoorn UE, Balliet RM, Rivadeneira DB, Chiavarina B, Pavlides S, Wang C, et al. Oxidative stress in cancer associated fibroblasts drives tumor-stroma co-evolution: A new paradigm for understanding tumor metabolism, the field effect and genomic instability in cancer cells. Cell Cycle. 2010;9:3256–3276. doi: 10.4161/cc.9.16.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sotgia F, Martinez-Outschoorn UE, Howell A, Pestell RG, Pavlides S, Lisanti MP. Caveolin-1 and cancer metabolism in the tumor microenvironment: markers, models, and mechanisms. Annu Rev Pathol. 2012;7:423–467. doi: 10.1146/annurev-pathol-011811-120856. [DOI] [PubMed] [Google Scholar]
- 134.Sotgia F, Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Lisanti MP. Understanding the Warburg effect and the prognostic value of stromal caveolin-1 as a marker of a lethal tumor microenvironment. Breast Cancer Res. 2011;13:213. doi: 10.1186/bcr2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bosch M, Mari M, Herms A, Fernandez A, Fajardo A, Kassan A, et al. Caveolin-1 deficiency causes cholesterol-dependent mitochondrial dysfunction and apoptotic susceptibility. Curr Biol. 2011;21:681–686. doi: 10.1016/j.cub.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bosch M, Mari M, Gross SP, Fernandez-Checa JC, Pol A. Mitochondrial cholesterol: a connection between caveolin, metabolism, and disease. Traffic. 2011;12:1483–1489. doi: 10.1111/j.1600-0854.2011.01259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Garcia-Ruiz C, Mari M, Colell A, Morales A, Caballero F, Montero J, et al. Mitochondrial cholesterol in health and disease. Histol Histopathol. 2009;24:117–132. doi: 10.14670/HH-24.117. [DOI] [PubMed] [Google Scholar]
- 138.Wallace DC. The epigenome and the mitochondrion: bioenergetics and the environment. Genes Dev. 2010;24:1571–1573. doi: 10.1101/gad.1960210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Smiraglia DJ, Kulawiec M, Bistulfi GL, Gupta SG, Singh KK. A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol Ther. 2008;7:1182–1190. doi: 10.4161/cbt.7.8.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 142.Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67:4687–4694. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- 143.Potluri P, Davila A, Ruiz-Pesini E, Mishmar D, O’Hearn S, Hancock S, et al. A novel NDUFA1 mutation leads to a progressive mitochondrial complex I-specific neurodegenerative disease. Mol Genet Metab. 2009;96:189–195. doi: 10.1016/j.ymgme.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sampey BP, Freemerman AJ, Zhang J, Kuan PF, Galanko JA, O’Connell TM, et al. Metabolomic profiling reveals mitochondrial-derived lipid biomarkers that drive obesity-associated inflammation. PLoS ONE. 2012;7:e38812. doi: 10.1371/journal.pone.0038812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Perfield JW, Lee Y, Shulman GI, Samuel VT, Jurczak MJ, Chang E, et al. Tumor progression locus 2 (TPL2) regulates obesity-associated inflammation and insulin resistance. Diabetes. 2011;60:1168–1176. doi: 10.2337/db10-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Park J, Kusminski CM, Chua SC, Scherer PE. Leptin receptor signaling supports cancer cell metabolism through suppression of mitochondrial respiration in vivo. Am J Pathol. 2010;177:3133–3144. doi: 10.2353/ajpath.2010.100595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Remels AH, Gosker HR, Schrauwen P, Hommelberg PP, Sliwinski P, Polkey M, et al. TNF-alpha impairs regulation of muscle oxidative phenotype: implications for cachexia? FASEB J. 2010;24:5052–5062. doi: 10.1096/fj.09-150714. [DOI] [PubMed] [Google Scholar]
- 148.Samavati L, Lee I, Mathes I, Lottspeich F, Huttemann M. Tumor necrosis factor alpha inhibits oxidative phosphorylation through tyrosine phosphorylation at subunit I of cytochrome c oxidase. J Biol Chem. 2008;283:21134–21144. doi: 10.1074/jbc.M801954200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Suematsu N, Tsutsui H, Wen J, Kang D, Ikeuchi M, Ide T, et al. Oxidative stress mediates tumor necrosis factor-alpha-induced mitochondrial DNA damage and dysfunction in cardiac myocytes. Circulation. 2003;107:1418–1423. doi: 10.1161/01.cir.0000055318.09997.1f. [DOI] [PubMed] [Google Scholar]
- 150.Lee SY, Jeon HM, Ju MK, Kim CH, Yoon G, Han SI, et al. Wnt/Snail signaling regulates cytochrome C oxidase and glucose metabolism. Cancer Res. 2012;72:3607–3617. doi: 10.1158/0008-5472.CAN-12-0006. [DOI] [PubMed] [Google Scholar]
- 151.Garcia-Ruiz I, Rodriguez-Juan C, az-Sanjuan T, del HP, Colina F, Munoz-Yague T, et al. Uric acid and anti-TNF antibody improve mitochondrial dysfunction in ob/ob mice. Hepatology. 2006;44:581–591. doi: 10.1002/hep.21313. [DOI] [PubMed] [Google Scholar]
- 152.Ojalvo LS, Whittaker CA, Condeelis JS, Pollard JW. Gene expression analysis of macrophages that facilitate tumor invasion supports a role for Wnt-signaling in mediating their activity in primary mammary tumors. J Immunol. 2010;184:702–712. doi: 10.4049/jimmunol.0902360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maciver NJ, Jacobs SR, Wieman HL, Wofford JA, Coloff JL, Rathmell JC. Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J Leukoc Biol. 2008;84:949–957. doi: 10.1189/jlb.0108024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Krawczyk CM, Holowka T, Sun J, Blagih J, Amiel E, DeBerardinis RJ, et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115:4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kasahara E, Sekiyama A, Hori M, Hara K, Takahashi N, Konishi M, et al. Mitochondrial density contributes to the immune response of macrophages to lipopolysaccharide via the MAPK pathway. FEBS Lett. 2011;585:2263–2268. doi: 10.1016/j.febslet.2011.05.049. [DOI] [PubMed] [Google Scholar]
- 158.Charni S, de BG, Rathore MG, Aguilo JI, van den Elsen PJ, Haouzi D, et al. Oxidative phosphorylation induces de novo expression of the MHC class I in tumor cells through the ERK5 pathway. J Immunol. 2010;185:3498–3503. doi: 10.4049/jimmunol.1001250. [DOI] [PubMed] [Google Scholar]
- 159.Hanekamp JS, Okumi M, Tena A, Arn S, Yamada K, Sachs DH. Cytoplasmic inheritance of transplantation antigens in animals produced by nuclear transfer. Transplantation. 2009;88:30–37. doi: 10.1097/TP.0b013e3181a9ed5b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ishikawa K, Toyama-Sorimachi N, Nakada K, Morimoto M, Imanishi H, Yoshizaki M, et al. The innate immune system in host mice targets cells with allogenic mitochondrial DNA. J Exp Med. 2010;207:2297–2305. doi: 10.1084/jem.20092296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Raoof M, Zhang Q, Itagaki K, Hauser CJ. Mitochondrial peptides are potent immune activators that activate human neutrophils via FPR-1. J Trauma. 2010;68:1328–1332. doi: 10.1097/TA.0b013e3181dcd28d. [DOI] [PubMed] [Google Scholar]
- 163.Johnson RF, Witzel II, Perkins ND. p53-dependent regulation of mitochondrial energy production by the RelA subunit of NF-kappaB. Cancer Res. 2011;71:5588–5597. doi: 10.1158/0008-5472.CAN-10-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.St-Hilaire S, Mandal R, Commendador A, Mannel S, Derryberry D. Estrogen receptor positive breast cancers and their association with environmental factors. Int J Health Geogr. 2011;10:32. doi: 10.1186/1476-072X-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen JQ, Brown TR, Russo J. Regulation of energy metabolism pathways by estrogens and estrogenic chemicals and potential implications in obesity associated with increased exposure to endocrine disruptors. Biochim Biophys Acta. 2009;1793:1128–1143. doi: 10.1016/j.bbamcr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rosca MG, Vazquez EJ, Chen Q, Kerner J, Kern TS, Hoppel CL. Oxidation of Fatty acids is the source of increased mitochondrial reactive oxygen species production in kidney cortical tubules in early diabetes. Diabetes. 2012;61:2074–2083. doi: 10.2337/db11-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chang EC, Charn TH, Park SH, Helferich WG, Komm B, Katzenellenbogen JA, et al. Estrogen Receptors alpha and beta as determinants of gene expression: influence of ligand, dose, and chromatin binding. Mol Endocrinol. 2008;22:1032–1043. doi: 10.1210/me.2007-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhao C, Dahlman-Wright K, Gustafsson JA. Estrogen receptor beta: an overview and update. Nucl Recept Signal. 2008;6:e003. doi: 10.1621/nrs.06003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hodges-Gallagher L, Valentine CD, El BS, Kushner PJ. Estrogen receptor beta increases the efficacy of antiestrogens by effects on apoptosis and cell cycling in breast cancer cells. Breast Cancer Res Treat. 2008;109:241–250. doi: 10.1007/s10549-007-9640-6. [DOI] [PubMed] [Google Scholar]
- 170.Kuiper GG, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, et al. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor beta. Endocrinology. 1998;139:4252–4263. doi: 10.1210/endo.139.10.6216. [DOI] [PubMed] [Google Scholar]
- 171.Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 172.Gavrilova-Jordan LP, Price TM. Actions of steroids in mitochondria. Semin Reprod Med. 2007;25:154–164. doi: 10.1055/s-2007-973428. [DOI] [PubMed] [Google Scholar]
- 173.Pedram A, Razandi M, Wallace DC, Levin ER. Functional estrogen receptors in the mitochondria of breast cancer cells. Mol Biol Cell. 2006;17:2125–2137. doi: 10.1091/mbc.E05-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hollerhage M, Matusch A, Champy P, Lombes A, Ruberg M, Oertel WH, et al. Natural lipophilic inhibitors of mitochondrial complex I are candidate toxins for sporadic neurodegenerative tau pathologies. Exp Neurol. 2009;220:133–142. doi: 10.1016/j.expneurol.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 175.Sherer TB, Richardson JR, Testa CM, Seo BB, Panov AV, Yagi T, et al. Mechanism of toxicity of pesticides acting at complex I: relevance to environmental etiologies of Parkinson’s disease. J Neurochem. 2007;100:1469–1479. doi: 10.1111/j.1471-4159.2006.04333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 177.Sereda B, Bouwman H, Kylin H. Comparing water, bovine milk, and indoor residual spraying as possible sources of DDT and pyrethroid residues in breast milk. J Toxicol Environ Health A. 2009;72:842–851. doi: 10.1080/15287390902800447. [DOI] [PubMed] [Google Scholar]
- 178.Gassner B, Wuthrich A, Scholtysik G, Solioz M. The pyrethroids permethrin and cyhalothrin are potent inhibitors of the mitochondrial complex I. J Pharmacol Exp Ther. 1997;281:855–860. [PubMed] [Google Scholar]
- 179.Hovey RC, Coder PS, Wolf JC, Sielken RL, Jr, Tisdel MO, Breckenridge CB. Quantitative assessment of mammary gland development in female Long Evans rats following in utero exposure to atrazine. Toxicol Sci. 2011;119:380–390. doi: 10.1093/toxsci/kfq337. [DOI] [PubMed] [Google Scholar]
- 180.Simpkins JW, Swenberg JA, Weiss N, Brusick D, Eldridge JC, Stevens JT, et al. Atrazine and breast cancer: a framework assessment of the toxicological and epidemiological evidence. Toxicol Sci. 2011;123:441–459. doi: 10.1093/toxsci/kfr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lasserre JP, Fack F, Revets D, Planchon S, Renaut J, Hoffmann L, et al. Effects of the endocrine disruptors atrazine and PCB 153 on the protein expression of MCF-7 human cells. J Proteome Res. 2009;8:5485–5496. doi: 10.1021/pr900480f. [DOI] [PubMed] [Google Scholar]
- 182.Lim S, Ahn SY, Song IC, Chung MH, Jang HC, Park KS, et al. Chronic exposure to the herbicide, atrazine, causes mitochondrial dysfunction and insulin resistance. PLoS ONE. 2009;4:e5186. doi: 10.1371/journal.pone.0005186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yadava N, Nicholls DG. Spare respiratory capacity rather than oxidative stress regulates glutamate excitotoxicity after partial respiratory inhibition of mitochondrial complex I with rotenone. J Neurosci. 2007;27:7310–7317. doi: 10.1523/JNEUROSCI.0212-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]