Abstract
Aggregation of misfolded proteins is characteristic of a number of neurodegenerative diseases, including Huntington disease (HD). The CCT/TRiC (chaperonin containing TCP-1/TCP-1 ring) chaperonin complex can inhibit aggregation and cellular toxicity induced by expanded repeat Huntingtin (mHtt) fragments. The substrate-binding apical domain of CCT/TRiC subunit CCT1, ApiCCT1, is sufficient to inhibit aggregation of expanded repeat mHtt fragments in vitro, providing therapeutic promise for HD. However, a key hurdle in considering ApiCCT1 as a potential treatment is in delivery. Because ApiCCT1 has a region of similarity to the HIV Tat protein cell-transduction domain, we tested whether recombinant ApiCCT1 (ApiCCT1r) protein could enter cells following exogenous delivery and modulate an established panel of mHtt-mediated cell-based phenotypes. Cell fractionation studies demonstrate that exogenous ApiCCT1r can penetrate cell membranes and can localize to the nucleus, consistent with a strategy that can target both cytosolic and nuclear pathogenic events in HD. ApiCCT1r application does indeed modulate HD cellular phenotypes by decreasing formation of visible inclusions, fibrillar oligomers, and insoluble mHtt derived from expression of a truncated mHtt exon 1 fragment. ApiCCT1r also delays the onset of inclusion body formation as visualized via live imaging. ApiCCT1r reduces mHtt-mediated toxicity in immortalized striatal cells derived from full-length knock-in HD mice, suggesting that therapeutic benefit may extend beyond effects on aggregation. These studies provide the basis for a potentially robust and unique therapeutic strategy to target mHtt-mediated protein pathogenesis.
Huntington disease (HD) is a devastating neurodegenerative disorder that strikes in midlife and is characterized by movement abnormalities, psychiatric symptoms, and cognitive deficits, as well as by the accumulation of pathogenic proteins and peptides (1–3). Currently no disease-modifying therapy is available. The disease is caused by an abnormal CAG repeat expansion in the HD gene, leading to the production of an expanded polyglutamine repeat in the amino terminal domain of the Huntingtin protein (Htt) (2, 3). A hallmark of HD is the propensity for the mutant protein (mHtt) to misfold and aggregate (1). While the connection between large fibrillar deposits and neurodegeneration is not clear, accumulation and aggregation of mHtt is likely to result from a deficit in cellular quality control machinery and can be used as a surrogate outcome measure for disease progression.
Molecular chaperones, which are colocalized within mHtt inclusion bodies, suppress neurodegeneration in several animal models of protein misfolding diseases, including HD (4–7). For instance, overexpression of Hsp70 and Hsp40 suppresses neurodegeneration in animal models of polyglutamine diseases (8, 9). In parallel, deletion of Hsp70 markedly worsens pathogenesis in a mouse model of HD (10). Chaperones are thought to suppress the toxicity of disease-associated proteins through direct effects on their misfolding and clearance; however, the mechanisms are not well understood.
The 1MDa CCT/TRiC (TCP1-ring complex) chaperonin is an ATP-dependent, ring-shaped hetero-oligomeric chaperone that binds and folds newly translated polypeptides (11). A genome-wide RNAi screen in Caenorhabditis elegans identified six of the eight subunits of CCT (1, 2, 4–7) as suppressors of polyglutamine aggregation (12). CCT/TRiC expression also prevents mHtt aggregation in cell and yeast models of HD (13–16). Further, overexpression of one subunit of the TRiC/CCT complex, CCT1, is sufficient to inhibit aggregation and reduce mHtt-mediated toxicity in mouse N2a neuronal cells (15). CCT/TRiC can also suppress aggregation in vitro, using purified components (15). Reduction to just the 20 kDa substrate-binding apical domain of yeast CCT1 (ApiCCT1) inhibits aggregation of recombinant mHtt in vitro, potentially through its ability to bind to the N-terminal 17 amino acid domain of Htt. This domain is involved in multiple Htt functions and in aggregation kinetics of mHtt (15). However, the efficacy of this apical domain has not yet been tested in HD cell models.
Given the potential of ApiCCT1 as a therapeutic for the treatment of HD, we tested if recombinant yeast ApiCCT1 (ApiCCT1r) protein could alter the aggregation and neurotoxicity of mHtt in selected proof-of-concept assays when purified protein was directly added to the culture media of cellular models of HD. We hypothesized that ApiCCT1r could be directly taken up by cells given the potential similarity of a positively charged region in Api1 to the protein transduction domain of the HIV protein Tat. We establish here that exogenously delivered ApiCCT1r can penetrate cell membranes, supporting the idea that ApiCCT1r has potential therapeutic application without necessitating intracellular expression. Of note, ApiCCT1r can cofractionate with nuclear fractions, which is significant in the context of nuclear accumulation and toxicity mediated by mHtt (1, 12, 17, 18). Application of just the apical domain of recombinant yeast CCT1 to rat pheochromocytoma-derived PC12 cells stably expressing truncated mutant Htt exon 1 protein (mHttex1p) (14A2.6 cells) decreased the formation of oligomeric and insoluble mHtt and slowed the kinetics of inclusion body formation in 14A2.6 cells. Finally, ApiCCT1r rescued mHtt-mediated toxicity in mouse striatal STHdhQ109/HdhQ109 cells derived from a knock-in mouse model of HD. Together these results provide a rationale for future testing of extracellular infusion of ApiCCT1r or cell-based delivery of secreted forms of ApiCCT1 to alleviate HD phenotypes in vivo.
Results
Purified ApiCCT1r Is Taken Up by Cells Following Exogenous Delivery.
Chaperones target protein homeostasis cellular processes and have significant potential to modulate the protein misfolding and accumulation that appears key to many neurodegenerative diseases (19). Previous in vitro results suggest that expression of ApiCCT1 in neurons might reduce the misfolding and accumulation of mHtt. However, extensive molecular manipulations would be required for intracellular expression of ApiCCT1 in cells in the brain; therefore, strategies to deliver ApiCCT1 extracellularly and be taken up by cells could be highly beneficial (Fig. 1A). It is a well-established phenomenon that some exogenously delivered proteins, specifically cell-penetrating peptides (CPPs), can cross membranes to enter cells and even nuclei (20, 21). There are no specific primary sequences common to all CPPs; however, many have a high abundance of positively charged amino acids or an alternating pattern of charged and hydrophobic residues (22, 23). This includes one of the first and most highly studied CPPs, the transactivator of transcription (TAT) protein of HIV (20, 21). To evaluate the possibility of directly introducing ApiCCT1 into cells expressing mHtt, we compared the sequence of yeast ApiCCT1r and TAT. Fortuitously, we find that ApiCCT1r does contain a region with a similar abundance of positively charged amino acids (Fig. 1B), providing a rationale to test exogenous application of ApiCCT1.
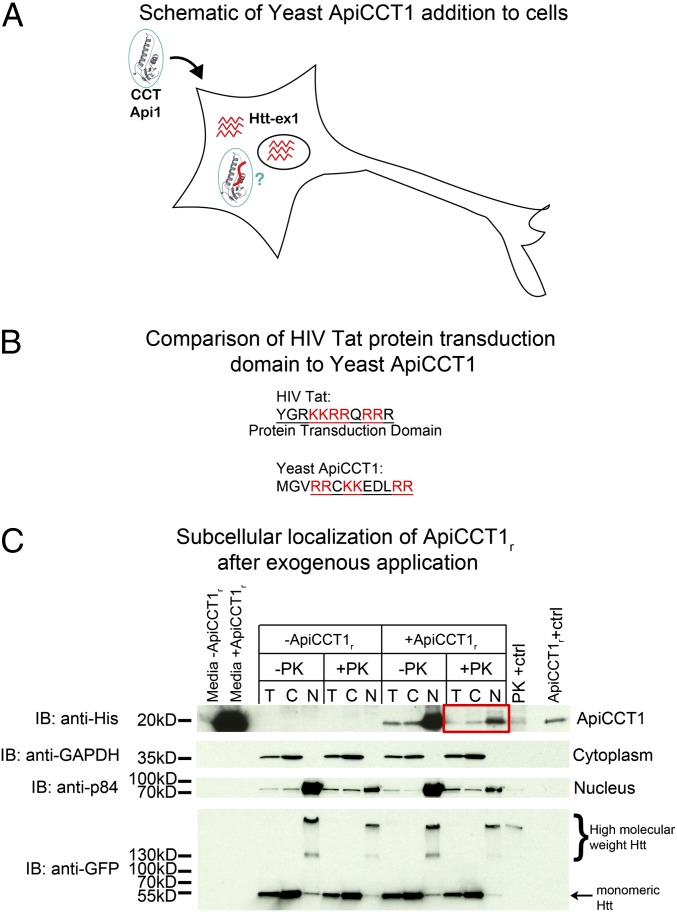
Fig. 1.
Exogenous ApiCCT1r enters cells and localizes to the cytosol and nucleus of PC12 cells. (A) Schematic of possible protein transduction by yeast ApiCCT1r. (B) Comparison of yeast ApiCCT1 to HIV Tat. (C) Fractionation of 14A2.6 treated with ApiCCT1r. Induced 14A2.6 cells treated with ApiCCT1r were treated with PK before lysis, then lysed (T) and fractionated into cytoplasmic (C) and nuclear (N) fractions, and analyzed by SDS/PAGE with Western blotting. Nuclei were also treated with PK before lysis. Media from the cells was also analyzed for the presence of ApiCCT1r. Lanes are labeled as follows: –PK, no PK treatment; Api1, no ApiCCT1r as a control; PK+ctrl, a positive control for PK treatment. In the latter case, the nuclear extract was treated with PK for 30 min on ice. The final lane is ApiCCT1r loaded as a standard. The blot was probed with anti-His antibody (ApiCCT1r), anti-GAPDH (cytoplasmic marker), anti-p84 (nuclear marker), and anti-EGFP (Htt). We loaded 30µg of total protein in each lane except for the media which only had 10 µg of protein loaded to avoid overexposure.
To test the hypothesis that ApiCCT1 can enter cells, a biochemical approach was used to determine whether ApiCCT1 cofractionated with cytosolic or nuclear cellular fractions. ApiCCT1r was added to the media of PC12 cells, Htt14A2.6, that inducibly express a truncated form of expanded repeat Htt exon 1 protein fused at the C terminus to enhanced green fluorescent protein (EGFP) (24), and cell fractionations performed. 14A2.6 cells induced with Ponasterone A were simultaneously treated with a single application of 1 µM ApiCCT1r for 48 h. Before lysis, cells were treated with 90 μg/mL proteinase K (PK) to digest any extracellular protein associated with the cell surface. Following lysis, the total extract (T) was fractionated into cytoplasmic (C) and nuclear (N) fractions. Intact nuclei were again treated with PK before lysis to remove any proteins bound to the exterior of the nuclear membrane. All fractions were analyzed by SDS/PAGE followed by Western analysis (Fig. 1C). Blots were probed with anti-His antibody (ApiCCT1r), anti-GAPDH (cytoplasmic marker), anti-p84 (nuclear marker), and anti-EGFP (mHtt). As expected, the majority of the ApiCCT1r remains in the media and a significant fraction appears trapped in the cell membrane as ApiCCT1r levels are lower upon PK treatment. This finding precluded analysis of cellular localization by immunofluorescence using available antibodies. However, ApiCCT1r is clearly visible in the PK-treated samples, indicating that the ApiCCT1r does enter cells (red box). Moreover, ApiCCT1r appears to localize both within the cytosolic and nuclear fractions, suggesting the availability of ApiCCT1 to act in both compartments. Although levels of ApiCCT1r appear to be higher in nuclear versus cytoplasmic fractions, this may be due to the fact that ApiCCT1 makes up a smaller percentage of the total protein in the cytoplasm. To ensure that the ApiCCT1r was not bound to aggregated mHtt or trapped within aggregates, the experiment was repeated using uninduced cells, where mHtt aggregates are absent. ApiCCT1r is still found within the cytoplasm and nuclei of these cells (Fig. S1), indicating that the entry of ApiCCT1r into cells is independent of the presence of mHtt aggregates.
Exogenous Delivery of ApiCCT1r Reduces Multiple Aggregation Species in PC12 Cells Stably Expressing Truncated Httex1p.
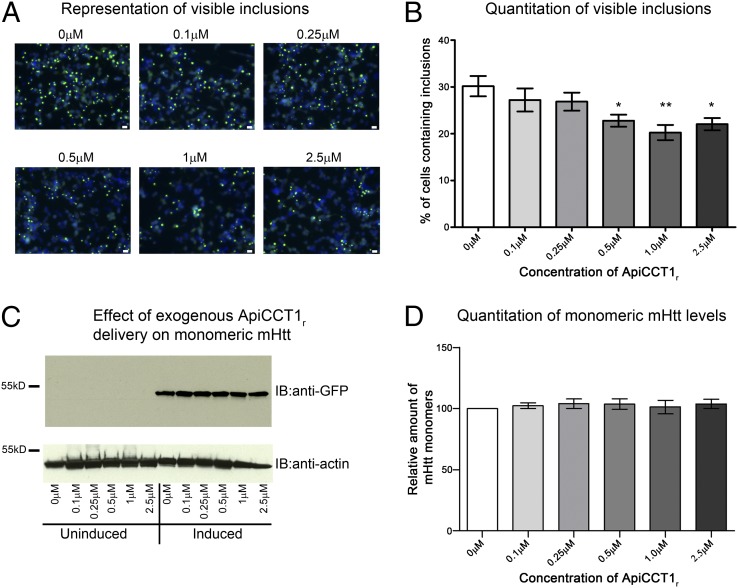
To evaluate the therapeutic potential of exogenous delivery of ApiCCT1r, a panel of established cell-based assays was used to monitor effects on specific aggregation species and cellular toxicity. We first tested whether ApiCCT1r could modulate visible inclusion formation, which is a highly utilized first assessment of altered aggregation. Exogenous delivery to 14A2.6 cells significantly decreased the number of visible inclusions following induction and ApiCCT1r application (one-way ANOVA: F = 3.933, P = 0.0037; Dunnett’s multiple comparison test: 0 µM vs. 0.5 µM, P < 0.05; 0 µM vs. 1 µM, P < 0.01; and 0 µM vs. 2.5 µM, P < 0.05) (Fig. 2 A and B). To determine whether this reduction is accompanied by a concomitant increase in monomeric, soluble Htt, Htt monomer levels from the same experiments were examined by Western analysis. ApiCCT1r does not appear to alter monomer levels (one-way ANOVA: F = 0.1908, P = 0.9604) or expression levels (Fig. 2 C and D and qPCR), suggesting that ApiCCT1r does not interfere with Htt expression and does not alter the steady-state levels of monomeric mHtt and no obvious differences in cell numbers following ApiCCT1r delivery were observed.
Fig. 2.
ApiCCT1r reduces the number of cells containing visible inclusions but does not increase levels of monomeric mHtt in 14A2.6 cells. (A and B) The 14A2.6 cells were treated with ApiCCT1r at the concentrations listed for 48 h. Cells were then fixed, imaged at 20× magnification, and the number of cells containing inclusion bodies was counted by fluorescence microscopy. (A) Representative images used for quantitation. Scale bar, 20 μm. (B) ApiCCT1r significantly decreases the number of visible inclusions in the 14A2.6 cells (Dunnett’s multiple comparison test, *P < 0.05, **P < 0.01). Error bars represent group means ± SEM. (C and D) The 14A2.6 cells were treated with ApiCCT1r at the concentrations listed for 48 h. SDS/PAGE analysis of lysates shows no significant differences in monomeric mHtt levels with ApiCCT1r treatment. Blots were probed with anti-EGFP (mHtt) and anti-actin (loading control). Error bars represent group means ± SEM.
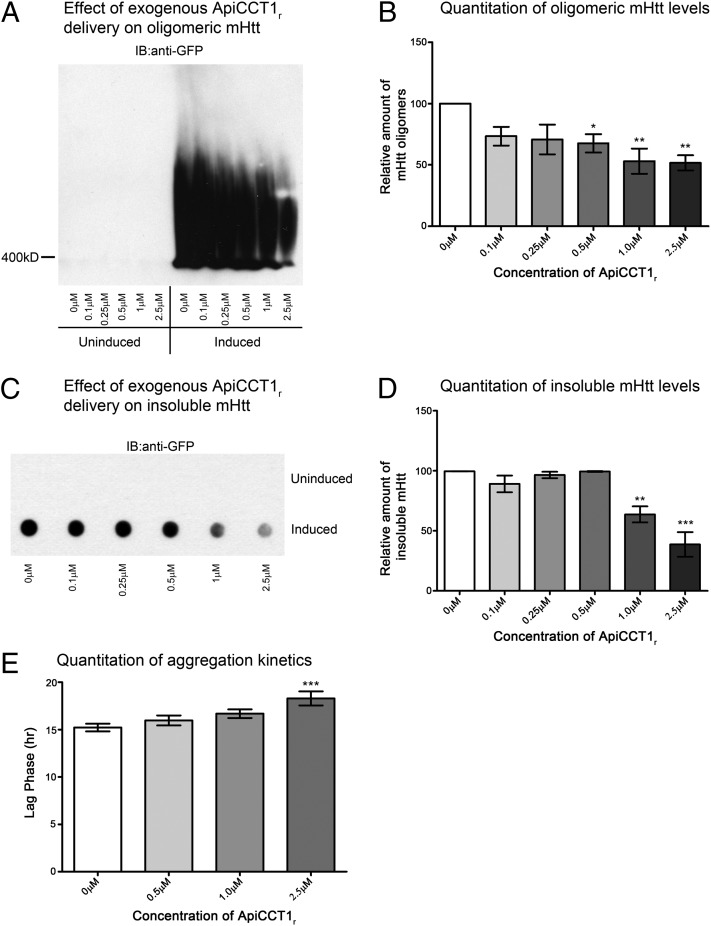
Insoluble Htt aggregates and mHtt oligomers are hallmarks of HD (1, 17, 25). In particular, oligomeric mHtt species are thought to be a major source of toxicity in affected neurons (25). Accordingly, mHtt oligomers and insoluble Htt species were next evaluated to determine whether exogenous delivery of ApiCCT1r influences accumulation of these forms. We used SDS agarose gel electrophoresis (AGE) to resolve oligomeric species, as this approach seems to preferentially resolve fibrillar oligomers of mHtt (26–29). Equivalent amounts of protein from cell lysates were loaded on SDS–AGE gels. We observed that ApiCCT1 application caused a dose-dependent decrease in both the level of mHtt oligomers (one-way ANOVA: F = 4.548, P = 0.0047; Dunnett’s multiple comparison test: 0 µM vs. 0.5 µM, P < 0.05; 0 µM vs. 1 µM, P < 0.01; 0 µM vs. 2.5 µM, P < 0.01) (Fig. 3 A and B) and the amount of SDS-insoluble mHtt (one-way ANOVA: F = 17.91, P < 0.0001; Dunnett’s multiple comparison test: 0 µM vs. 1 µM, P < 0.01; 0 µM vs. 10 µM, P < 0.001) (Fig. 3 C and D). These data indicate that exogenous ApiCCT1r application is able to reduce the formation of oligomeric and insoluble mHtt as well as large visible inclusions, suggesting that ApiCCT1r may have effects on both protein misfolding and/or protein accumulation and that exogenous application is sufficient to modulate these readouts.
Fig. 3.
ApiCCT1r decreases fibrillar oligomers and insoluble mHtt and delays inclusion body formation in 14A2.6 cells. (A–F) The 14A2.6 cells were treated with ApiCCT1r at the concentrations listed for 48 h. Cells were lysed and analyzed for different conformations of mHtt. (A and B) Lysates were analyzed by SDS-AGE with Western blotting and membranes were probed with anti-EGFP (mHtt). There is a dose-dependent decrease in the level of mHtt oligomers (Dunnett’s multiple comparison test, *P < 0.05, **P < 0.01). Error bars represent group means ± SEM. (C and D) ApiCCT1r significantly decreases the amount of SDS-insoluble mHtt (Dunnett’s multiple comparison test, **P < 0.01, ***P < 0.001) by filter-retardation assay. Membranes were probed with anti-EGFP antibody (mHtt). Error bars represent group means ± SEM. (E) ApiCCT1r delays the formation of visible inclusions in the 14A2.6 cells (Dunnett’s multiple comparison test, ***P < 0.001) by live cell imaging visualizing EGFP-tagged mHtt inclusion bodies. Error bars represent group means ± SEM.
ApiCCT1r Delays Inclusion Body Formation in 14A2.6 Cells.
To investigate whether kinetics of aggregation could be affected, a live-cell imaging assay system was used to determine whether altered kinetics is observed, specifically targeting the lag phase in inclusion formation (28). Strikingly, ApiCCT1r delays the formation of visible inclusions in the 14A2.6 cells (one-way ANOVA: F = 5.769, P = 0.0011; Dunnett’s multiple comparison test: 0 µM vs. 2.5 µM, P < 0.001) from 15.2 h to 18.3 h (Fig. 3E and Movie S1), suggesting that ApiCCT1r might be delaying the formation of the “seed” that initiates the formation of visible inclusions. These data further support altered protein homeostasis as causative of inclusion formation and suggest that, from a therapeutic standpoint, delivery of ApiCCT1 could delay the onset of HD phenotypes.
Exogenous ApiCCT1r Reduces mHtt-Mediated Toxicity in a Knock-in Cell Model of HD.
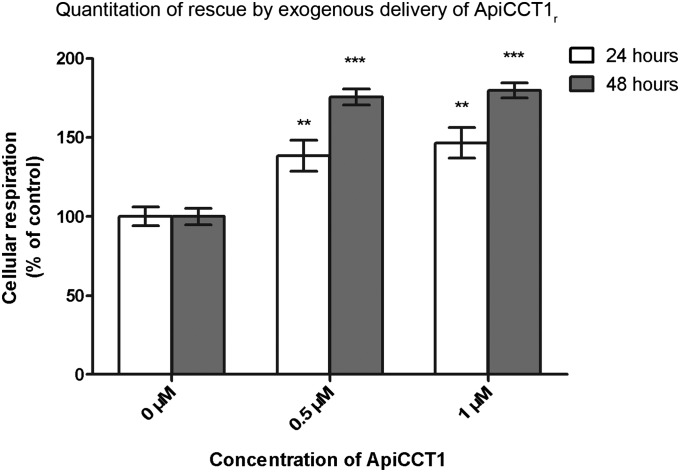
Overexpression of the CCT1 subunit can reduce mHtt-mediated toxicity in mouse neuronal N2a cells (15). We next determined if the apical domain of CCT1 alone could also ameliorate mHtt-mediated toxicity in cells and whether exogenous application of this would be sufficient. To test neurotoxicity, striatal STHdhQ109/HdhQ109 cells were used. These are striatal progenitor cells immortalized by a temperature-sensitive large T antigen, derived from a knock-in mouse model of expanded polyglutamines (30) and expressing full-length mHtt within the endogenous mouse locus. Upon temperature shift and serum withdrawal, mHtt lines show decreased viability and increased cell death compared with the same cells at time 0 immediately before this shift (31). This control is used instead of comparing to a line expressing wild-type HTT due to clonal variability between lines. Cell viability was measured using an XTT (sodium 2,3,-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)-carbonyl]-2H-tetrazolium inner salt) colorimetric cell proliferation assay to detect altered cellular metabolic activities. Results are presented as percent relative to cells not treated with ApiCCT1r following shift to nonpermissive conditions. Exogenous delivery of ApiCCT1r reduced mHtt-mediated toxicity in STHdhQ109/HdhQ109 cells, depicted graphically as an increase in cellular respiration (XTT), at both 24 and 48 h after temperature shift and addition of ApiCCT1r (24 h one-way ANOVA: F = 8.154, P = 0.0013; 48 h one-way ANOVA: F = 78.91, P < 0.0001) (Fig. 4). This corresponds to a percent rescue of 38.4% (0.5 μM) and 46.5% (1 μM) at 24 h and 75.5% (0.5 μM) and 79.7% (1 μM) at 48 h. Post hoc analysis with Dunnett’s multiple comparison test revealed significant differences between the control (0 µM) and treatment (0.5 and 1 µM) groups at 24 h (P < 0.01 and P < 0.01, respectively) and 48 h (P < 0.001 and P < 0.001, respectively). This effect is not due simply to changes in proliferation, as these cells have significantly reduced cell division following inactivation of the large T antigen following shift to nonpermissive conditions. Interestingly, this effect persists after 48 h, suggesting that the exogenous ApiCCT1r may have a sustained effect on cell survival.
Fig. 4.
ApiCCT1r reduces mHtt-mediated toxicity in a knock-in cell model of HD. ApiCCT1r increases survival of STHdhQ109/HdhQ109 cells at both 24 and 48 h after treatment and temperature shift (Dunnett’s multiple comparison test, *P < 0.05, ***P < 0.001) by XTT assay. Samples were normalized to the vehicle control. Values over 100% indicate higher o.d. and therefore increased respiration likely due to increased cell survival. Error bars represent group means ± SEM.
Discussion
Protein homeostasis mechanisms and protein accumulation is impacted in HD and other neurodegenerative diseases and targeted modulation of the chaperone network has been considered an attractive therapeutic target (19). Previous studies demonstrated that overexpression of CCT1 can alter HD phenotypes in cell models of HD and that the apical subunit of yeast CCT1 (ApiCCT1r) alone interacts with recombinant mHtt in vitro and inhibits aggregation (15, 16). Therefore, if exogenous delivery of ApiCCT1r to cells could modulate intracellular aggregation and toxicity, the need for overexpression of the protein inside the cell would be circumvented and provide a rational option for delivery to the brain. We investigated this possibility in a series of targeted proof-of-concept assays and find that exogenous delivery of ApiCCT1r can enter cells and modulate cellular phenotypes.
There is established precedence for exogenous proteins crossing cell membranes and entering cells by several different mechanisms direct penetration, pore formation, endocytosis, and transporter-mediated cell entry (20, 21). Experiments are ongoing to determine the mechanism of ApiCCT1r cell entry with the goal of optimizing the amount of protein that can enter cells in culture and ultimately in brain. These experiments will include mutating the positive charged amino acid domain of yeast ApiCCT1 to determine if this domain is required. Additionally, testing whether exogenous application of human ApiCCT1r also results in cell entry and phenotypic modulation is in progress. As ApiCCT1r does not contain a known nuclear localization signal (32), perhaps the ability to penetrate membranes similar to CPPs might explain the ability of ApiCCT1r to cofractionate with nuclear proteins. Alternatively, the small size of this chaperone domain would allow it to passively diffuse through the nuclear pore (33). The finding that exogenous ApiCCT1r delivery can be taken up by cells and localizes to both the cytosol and nucleus are highly significant given that both compartments appear to contribute to pathogenic effects mediated by chronic mHTT expression (34), in particular nuclear accumulation of mHtt exerts toxic effects (1, 12, 17, 18).
ApiCCT1r may be directly interacting with mHtt to alter aggregation and toxicity, consistent with the previous finding that ApiCCT1r can by itself suppress formation of insoluble mHtt aggregates and interacts with the first 17 amino acids of Htt, which regulates aggregation kinetics and other Htt functions (16). Given that toxicity is ameliorated in a cell model expressing full-length mHtt in which inclusions are not observed, it is likely that ApiCCT1r exerts direct effects on mHtt-mediated phenotypes beyond interference with aggregation. Experiments to evaluate whether there is a direct association between mHTT and exogenously delivered ApiCCT1r as anticipated and whether ApiCCT1r selectively binds to specific aggregation intermediates of mHTT are in progress. It is also possible that ApiCCT1r also exerts extracellular effects. Further study is required to elucidate the mechanism of action of this chaperone protein upon cellular mHTT-mediated phenotypes.
When a cell-based assay designed to screen for altered inclusion body formation was performed (24), exogenous ApiCCT1r protein reduced the formation of visible aggregates, consistent with previous in vitro results. Using this assay, ApiCCT1r was also found to significantly delay the formation of inclusion bodies. While it is not yet known if this delay is protective, it is encouraging that ApiCCT1r might be able to slow the onset of HD phenotypes.
ApiCCT1r decreased oligomeric and insoluble mHtt in an inducible cell model of HD. It is of interest to compare these findings with previous work demonstrating that the entire TRiC/CCT complex increased the formation of nontoxic, SDS-soluble mHtt oligomers (13). Additionally, Hsp70 and Hsp40 were required for the assembly of the nontoxic oligomers, suggesting that for this system, increased expression of both of these chaperones might be necessary for the formation of the nontoxic oligomeric species. Of note, the SDS-AGE technique used here specifically resolves fibrillar SDS-resistant oligomers, raising the possibility that SDS-resistant and SDS-soluble oligomers, respectively, represent toxic and benign oligomeric species.
The decrease in both oligomeric and insoluble mHtt is important as it is not currently known whether insoluble or oligomeric mHtt induce toxicity in HD. Given that oligomeric mHtt levels are decreased, ApiCCT1r appears to be intervening at the initiating events in the aggregation pathway. Further, ApiCCT1r does not alter the steady-state levels of monomeric mHtt or the expression of the transgene. Because there is no accumulation of monomeric mHtt, which might be expected given the decreased inclusion body formation, it is possible that clearance of the protein might be increased. Alternatively, ApiCCT1r might be remodeling mHtt conformation to either refold misfolded mHtt or produce nontoxic species that are off-pathway to the formation of fibrillar and amyloid species. Further studies to determine the effect of ApiCCT1r on protein folding, conformation, and clearance, such as misfolding of sensor proteins such as dihydrofolate reductase (35), proteasomal activity, or autophagic flux (36) assays will be necessary to understand the mechanisms by which ApiCCT1r alters aggregation and toxicity.
While mechanisms underlying ApiCCT1r cell uptake and modulation of mHtt aggregation have yet to be determined, the data described here provide proof-of-concept evidence that exogenous delivery of just the 20 kDa apical domain of CCT1 is sufficient to alter a panel of cellular mHTT phenotypes and could have profound therapeutic potential without the need to create approaches for intracellular expression of the protein. Indeed, there is support for the feasibility of direct protein delivery in vivo with administration of DJ-1 protein to the brains of 6-Hydroxydopamine (6-OHDA) lesioned rat models of Parkinson’s disease resulting in neuroprotective outcomes (33, 37). Further optimization to increase the efficiency of ApiCCT1r transduction into cells and nuclei might also increase the potency of ApiCCT1r as a therapeutic reagent and is under current investigation. Given the data presented here, the efficacy of in vivo delivery into striatum of HD mouse models can now be investigated and optimized, including cell transplantation-based secretion of ApiCCT1, viral injections of cDNAs encoding secreted forms of ApiCCT1, and direct protein delivery of recombinant protein.
Materials and Methods
Inducible PC12 Cells.
The Htt14A2.6 PC12 line was generated and propagated as described in ref. 24.
Protein.
The 6×-his tagged ApiCCT1r (MVPGYALNCTVASQAMPKRIAGGNVKIACLDLNLQKARMAMGVQINIDDPEQLEQIRKREAGIVLERVKKIIDAGAQVVLTTKGIDDLCLKEFVEAKIMGVRRCKKEDLRRIARATGATLVSSMSNLEGEETFESSYLGLCDEVVQAKFSDDECILIKGTSKAAAAALEHHHHHH) was used as a stock of 87 µM in apical buffer [50 mM Hepes, pH 7.5, 300 mM NaCl, 10% (vol/vol) glycerol, 1 mM DTT]. Apical buffer was used as a vehicle control. Purification was conducted as previously described (15, 16).
Cellular Fractionation.
Cell fractionation was adapted from ref. 38, but the protocol was terminated after the lysing of the nuclei as purification of nucleoli was not required. PK treatment was performed as described in ref. 39. Briefly, 14A2.6 cells were grown as described above. At 24 h after plating, cells were induced with Ponasterone A (PA) and treated with 6×-His-tagged ApiCCT1r. Cells were then allowed to grow for 48 h without additional treatment with ApiCCT1r. Cells were harvested and spun down at 1,000× g for 5 min at 4 C. A small volume of media was retained to determine the amount of ApiCCT1r present in the media. A reference volume (RV) of cells was determined and cells were resuspended in 15 RV of cold buffer (10 mM Tris, pH 7.4, 10 mM NaCl, and 1 mM MgCl2). The samples were divided in half, and 90 µg/mL PK was added to one set, while the other set was treated with a buffer control (10 mM Tris⋅HCl, pH 7.5). After 30 min of incubation on ice, the PK was inactivated by addition of 500 µg/mL PMSF. Cells were lysed by adding Nonidet P-40 to a final concentration of 0.3% and homogenized in a glass Dounce homogenizer. Crude total cell lysate was then removed for future analysis (T). Remaining lysates were centrifuged at 1,200× g for 5 min at 4 C, and the supernatant (cytoplasmic fraction) was saved for future analysis (C). The nuclear pellet was resuspended in 10 RV of 250 mM sucrose containing 10 mM MgCl2. A total of 10 RV of 880 mM sucrose solution containing 5 mM MgCl2 was added under the nuclear fraction, and nuclei were purified by centrifuging at 1,200× g for 10 min at 4 C through this sucrose cushion. The purified nuclear pellet was resuspended in 10 RV of 340 mM sucrose solution containing 5 mM MgCl2. PK treatment was performed as before on the samples previously treated with PK. After stopping the PK reaction, protease inhibitor (Roche) was added to each sample, and the nuclei were lysed by sonicating 3× for 30 s at 40 V with 5 min on ice in between pulses. SDS/PAGE analysis was performed on all samples as above except samples were run on a 15% Criterion gel (BioRad), and because the ApiCCT1r was so abundant in the media, only 10 µg of protein was loaded from the media to avoid extreme overexposure. All other samples were 30 µg of total protein. Samples were then analyzed by SDS/PAGE as described below.
Immunocytochemistry and Quantitation of Inclusion Bodies.
14A2.6 cells were grown as described above on UV-treated coverslips for 24 h and then induced with 2.5 µM PA for 48 h. Cells were then fixed in 2% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS, and nuclei were stained with 4′,6-diamidino-2-phenylindole. Fluorescent microscopy was performed using Axiovision software and a Zeiss AxioObserver.Z1 microscope. A minimum of 500 cells were counted from ∼5–6 fields in three independent experiments for each data point at 20× magnification. Aggregation is expressed as the percentage of cells with visible inclusions versus total number of cells.
SDS/PAGE of PC12 Cell Homogenates.
mHtt expression was induced with 2.5 µM PA and simultaneously treated with listed concentrations of ApiCCT1r for 48 h with no subsequent addition of ApiCCT1r. Three independent experiments were performed. At 48 h postinduction, cells were lysed in radioimmune precipitation assay buffer [10 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA (pH 8.0), 1% Nonidet P-40, 0.5% SDS] containing Complete Protease Inhibitor (Roche Diagnostics). A DC protein assay (Bio-Rad) was performed to determine protein concentration. A total of 30 μg lysate was added in a 1:1 ratio to loading buffer (125 mM Tris, pH 6.8, 20% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.004% bromophenol blue) and loaded onto a 10% Criterion gel (Bio-Rad). The gel was then run at 125 V until the dye front reached the bottom of the gel. It was then wet-transferred to a nitrocellulose membrane, blocked for 1 h in StartingBlock T20 (TBS) Blocking Buffer (Pierce) at room temperature and probed with anti-EGFP (Clontech, 1:1,000), anti-Actin (Sigma Monoclonal Anti-Actin Clone AC-40 Cat No. A-4700, 1:5,000), anti-His antibody (Sigma, 1:1,000), anti-GAPDH (Imgenex, 1:1,000), or anti-p84 (AbCam, 1:1,000). Peroxidase-conjugated AffiniPure goat anti-rabbit or anti-mouse secondary antibodies (Jackson ImmunoResearch Laboratories) were used at 1:20,000 for 1 h at room temperature. Blots were detected using PICO detection reagent (Pierce).
SDS-AGE of PC12 Cell Homogenates.
The analysis of mHtt by SDS-AGE with Western analysis was performed as previously described (26–29).
Filter Retardation Analysis of PC12 Cell Homogenates.
The same lysates used in the SDS/PAGE and SDS-AGE assays were also analyzed for SDS-insoluble mHtt by filter-retardation assays as described in refs. 27, 28, 40.
Quantification of Western Blots.
Densitometry of the autoradiographs was performed using ImageJ software (National Institutes of Health). Measurements were normalized to control samples treated with vehicle in all of the assays as there is no loading control for SDS-AGE or filter-retardation assays.
Live Cell Imaging.
Time-lapse microscopy was performed as described in ref. 28. The 14A2.6 cells were grown as above, and 5 × 105 cells were plated onto collagen-coated glass-bottom microwell dishes (MatTek, P35G-1.5–14-C). Cells were induced with 2.5 µM PA and simultaneously treated with listed concentration of ApiCCT1r at 24 h postplating and allowed to equilibrate in the imaging system. The amount of equilibration time was variable between experiments, and this difference in time was accounted for in the time elapsed for analysis. Cells were then allowed to grow for 48 h in a VivaView live cell imaging system (Olympus). Cells were imaged every 10 min for 48 h at 20× magnification every in nine different locations per well to allow for sufficient cell numbers in analysis. Three independent experiments were performed. The “lag phase” of inclusion formation is shown as the number of hours before the appearance of the first inclusion body.
Knock-In Cell Line.
Homozygous mutant STHdhQ109/HdhQ109 cell lines (gift from E. Cattaneo, University of Milano, Milan) were plated in 24-well plates (0.75 × 105 cells/well) in complete medium (DMEM / 5% glucose, 10% FBS) at 33 C as described previously (31). Experiments were plated to have five technical replicates per concentration per experiment, with each independent experiment repeated three times. Cells were treated with listed concentrations of ApiCCT1r concurrently with the addition of low serum medium and temperature shift with no subsequent treatments throughout the duration of the study.
Cell Survival Assay.
Cells were treated as described above. XTT (sodium 2,3,-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)-carbonyl]-2H-tetrazolium inner salt) assays were performed at 24 and 48 h after the temperature shift. At those time points, cells were incubated for 4 h with XTT and phenazine methosulfate (0.2 mg/mL and 0.1 µg/mL, respectively; Sigma-Aldrich), and absorbance was read at 450 nm. Higher values in the treated groups indicate increased cellular respiration suggestive of increased cell survival.
Statistical Analysis.
All statistical analyses were performed using GraphPad Prism 5.04 software. All data are expressed as mean ± SE of measure. P < 0.05 was considered to be statistically significant in all cases. Statistical comparisons of results were performed by performing one-way ANOVA analysis followed by Dunnett’s multiple comparison tests.
Supplementary Material
Acknowledgments
We thank Dr. Jack Reidling for helpful discussions of the manuscript and Dr. Christopher Sontag and Dr. Brian Cummings for advice and technical assistance with live cell imaging. Funding was provided by National Institutes of Health Grants PN2EY016525 and NS52789 (to L.M.T.) and AG00538 (to C.G.G.) and the Hereditary Disease Foundation and Huntington’s Disease Society of America (to L.M.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222663110/-/DCSupplemental.
References
- 1.Davies SW, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90(3):537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 2.The Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Gusella JF, MacDonald ME. Huntington’s disease. Semin Cell Biol. 1995;6(1):21–28. doi: 10.1016/1043-4682(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 4.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6(1):11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 5.Muchowski PJ, et al. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97(14):7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol. 2011;3(8):a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voisine C, Pedersen JS, Morimoto RI. Chaperone networks: Tipping the balance in protein folding diseases. Neurobiol Dis. 2010;40(1):12–20. doi: 10.1016/j.nbd.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warrick JM, et al. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23(4):425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 9.Cummings CJ, et al. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum Mol Genet. 2001;10(14):1511–1518. doi: 10.1093/hmg/10.14.1511. [DOI] [PubMed] [Google Scholar]
- 10.Wacker JL, et al. Loss of Hsp70 exacerbates pathogenesis but not levels of fibrillar aggregates in a mouse model of Huntington’s disease. J Neurosci. 2009;29(28):9104–9114. doi: 10.1523/JNEUROSCI.2250-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: Protein folding in the chamber of secrets. Trends Cell Biol. 2004;14(11):598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, Dunlap JR, Andrews RB, Wetzel R. Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum Mol Genet. 2002;11(23):2905–2917. doi: 10.1093/hmg/11.23.2905. [DOI] [PubMed] [Google Scholar]
- 13.Behrends C, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23(6):887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Kitamura A, et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8(10):1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 15.Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8(10):1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tam S, et al. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol. 2009;16(12):1279–1285. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 18.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95(1):55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 19.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 20.Koren E, Torchilin VP. Cell-penetrating peptides: Breaking through to the other side. Trends Mol Med. 2012;18(7):385–393. doi: 10.1016/j.molmed.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Lindgren M, Hällbrink M, Prochiantz A, Langel U. Cell-penetrating peptides. Trends Pharmacol Sci. 2000;21(3):99–103. doi: 10.1016/s0165-6147(00)01447-4. [DOI] [PubMed] [Google Scholar]
- 22.Derossi D, et al. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J Biol Chem. 1996;271(30):18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 23.Vivès E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272(25):16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 24.Apostol BL, et al. A cell-based assay for aggregation inhibitors as therapeutics of polyglutamine-repeat disease and validation in Drosophila. Proc Natl Acad Sci USA. 2003;100(10):5950–5955. doi: 10.1073/pnas.2628045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 26.Legleiter J, et al. Mutant huntingtin fragments form oligomers in a polyglutamine length-dependent manner in vitro and in vivo. J Biol Chem. 2010;285(19):14777–14790. doi: 10.1074/jbc.M109.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sontag EM, et al. Methylene blue modulates huntingtin aggregation intermediates and is protective in Huntington’s disease models. J Neurosci. 2012;32(32):11109–11119. doi: 10.1523/JNEUROSCI.0895-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sontag EM, et al. Detection of mutant huntingtin aggregation conformers and modulation of SDS-soluble fibrillar oligomers by small molecules. Journal of Huntington's Disease. 2012;1(1):127–140. doi: 10.3233/JHD-2012-129004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss A, et al. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington’s disease. J Neurochem. 2008;104(3):846–858. doi: 10.1111/j.1471-4159.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- 30.Trettel F, et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9(19):2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 31.Apostol BL, et al. CEP-1347 reduces mutant huntingtin-associated neurotoxicity and restores BDNF levels in R6/2 mice. Mol Cell Neurosci. 2008;39(1):8–20. doi: 10.1016/j.mcn.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32(Web Server issue):W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inden M, et al. PARK7 DJ-1 protects against degeneration of nigral dopaminergic neurons in Parkinson’s disease rat model. Neurobiol Dis. 2006;24(1):144–158. doi: 10.1016/j.nbd.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Benn CL, et al. Contribution of nuclear and extranuclear polyQ to neurological phenotypes in mouse models of Huntington’s disease. Hum Mol Genet. 2005;14(20):3065–3078. doi: 10.1093/hmg/ddi340. [DOI] [PubMed] [Google Scholar]
- 35.Mayhew M, et al. Protein folding in the central cavity of the GroEL-GroES chaperonin complex. Nature. 1996;379(6564):420–426. doi: 10.1038/379420a0. [DOI] [PubMed] [Google Scholar]
- 36.Liggett A, Crawford LJ, Walker B, Morris TC, Irvine AE. Methods for measuring proteasome activity: Current limitations and future developments. Leuk Res. 2010;34(11):1403–1409. doi: 10.1016/j.leukres.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Sun SY, An CN, Pu XP. DJ-1 protein protects dopaminergic neurons against 6-OHDA/MG-132-induced neurotoxicity in rats. Brain Res Bull. 2012;88(6):609–616. doi: 10.1016/j.brainresbull.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 38.Hacot S, et al. Isolation of nucleoli. Curr Protoc Cell Biol. 2001;Chapter 3:Unit3.36. doi: 10.1002/0471143030.cb0336s47. [DOI] [PubMed] [Google Scholar]
- 39.Miyata N, Fujiki Y. Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol Cell Biol. 2005;25(24):10822–10832. doi: 10.1128/MCB.25.24.10822-10832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherzinger E, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90(3):549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.