Abstract
Decades of studies of candidate genes show their complex role in aging-related traits. We focus on apolipoprotein E e2/3/4 polymorphism and ages at onset of cardiovascular diseases (CVD) and cancer in the parental and offspring generations of the Framingham Heart Study participants to gain insights on the role of age and gender across generations in genetic trade-offs. The analyses show that the apolipoprotein E e4 allele carriers live longer lives without cancer than the non-e4 allele carriers in each generation. The role of the e4 allele in onset of CVD is age- and generation-specific, constituting two modes of sexually dimorphic genetic trade-offs. In offspring, the e4 allele confers risk of CVD primarily in women and can protect against cancer primarily in men of the same age. In the parental generation, genetic trade-off is seen in different age groups, with a protective role of the e4 allele against cancer in older men and its detrimental role in CVD in younger women. The puzzling complexity of genetic mechanisms working in different genders, ages, and environments calls for more detail and systemic analyses beyond those adapted in current large-scale genetic association studies.
Introduction
Decades of studies of candidate genes show that they are not linked to aging-related traits in a straightforward fashion.1,2 Recent genome-wide association studies (GWAS) provide fundamentally the same conclusion by showing that the traits in late life are likely controlled by a relatively large number of common genetic variants (e.g., Teslovich et al.3), many of which have a tiny effect.4
The tiny effect of genes can be because they indeed confer small risks or/and they confer large risks, but in a complex fashion. For example, recent studies demonstrated that genes could show antagonistic pleiotropy (postulated by Williams5), when the same gene can be advantageous for fitness phenotypes in early life but become detrimental by conferring risks of diseases in old ages.2,6–10 In general, the same allele can work differently at different ages.2,11–15 The complex role of genes can readily result in underestimation of the effect.7 Furthermore, the same genes can confer risk for some traits but protect against the others, exhibiting so-called genetic trade-off.16–28 Again, trade-off among endophenotypes can result in a tiny or no overall effect on an upstream phenotype. For example, Kulminski et al. 17 show that the same allele of the apolipoprotein E (APOE) gene can confer risk for cardiovascular disease (CVD) but can protect against cancer that significantly alters estimates for life span. Sexual dimorphism is another important factor complicating the effect of genes on complex aging-related diseases and traits in humans.29,30
In this work, we extend the analyses in Kulminski et al.17 by focusing on data from the Framingham Heart Study original (FHS) and Offspring (FHSO) cohorts to elucidate the potential role of age and gender in trade-off of the APOE e4 allele on risks of premature onset of CVD and cancer (all sites but skin) across two human generations followed for up to 60 years.
Materials and Methods
Study design and population
The study design of the original (FHS) cohort (launched in 1948) and the FHSO cohort (launched 22 years later) has been described previously.31–35 In short, the FHS includes N=5209 respondents aged 28–62 years at baseline who have been followed biannually during about 60 years. The FHSO respondents (N=5124) aged 5–70 years at baseline were biological descendants (n=3514), their spouses (n=1576), and adopted offspring (n=34) of the FHS participants who have been followed for about 36 years. The FHS/FHSO participants were examined for the onset of CVD and cancer at regular examinations at the FHS clinic and surveillance of hospital admissions,33,34 currently through 2008. Biospecimens were mostly collected in the late 1980s and through 1990s from surviving participants.36,37 Accordingly, genotyped FHS and FHSO participants represent demographically unbiased samples of aged populations (File S1; Supplementary Data are available at www.liebertonline.com/rej/). The procedure used for the APOE genotyping is described by Lahoz et al.36 The limited-access data available for this study include information on the APOE e2/3/4 polymorphism for the 1258 FHS and 3924 FHSO participants.
Analysis
Following the results by Kulminski et al.,17 the analyses were focused on the effect of the APOE e4 (risk; e2/4, e3/4, and e4/4) allele contrasted by the non-e4 allele (e2/2, e2/3, and e3/3) genotypes on ages at onset of CVD (diseases of heart and stroke regardless of condition) and cancer (all sites regardless of condition; skin cancer was excluded).
The analyses were conducted using Kaplan–Meier empirical estimator and the Cox proportional hazards regression model. To better address the role of age in onset of diseases, we used age at event (cases) or the end of follow-up (censored individuals) in 2008 (or death) as a time variable. Given the lack of the APOE-related selection bias (see File S1), the analyses were focused on male and female participants of the baseline examinations in each cohort to maximize the sample size. Five cases of CVD and 3 cases of cancer that occurred prior baseline examination of the FHS genotyped participants, as well as 35 cases of CVD and 22 cases of cancer that occurred prior baseline examination of the FHSO genotyped participants, were excluded from the analyses. The Cox regression model was adjusted for age at baseline and sex, when applicable. To better understand potential modulating role of CVD or cancer on risks of premature (i.e., at younger ages) onset of cancer or CVD, we also conducted the analyses with the Cox model adjusted by alternative diseases (e.g., the model for onset of cancer was adjusted by the prevalence of CVD). We used a robust sandwich estimator in the Cox model to account for potential clustering (e.g., familial).38 Statistical analyses were conducted using SAS (release 9.3, Cary, NC).
Results
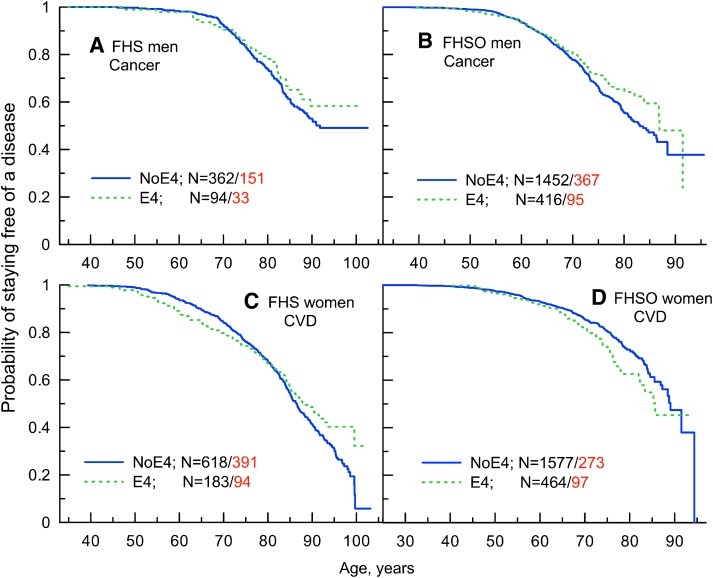
Mean ages at baseline and Kaplan–Meier age patterns of onset of cancer or CVD for male and female carriers of the e4 allele and non-carriers of this allele in the FHS and FHSO cohorts are presented in Figs. S1 and S2. Empirical screening of the age patterns of onset of cancer for men and women combined shows that the e4 carriers tend to live longer lives without cancer (see Fig. S1) in each cohort. Sex-specific analyses show that this protective effect is more characteristic for men (Fig. S1 and Fig. 1A,B).
FIG. 1.
Kaplan–Meier age patterns of probability original cohort of staying free of (A and B) cancer and (C and D) cardiovascular disease (CVD) for (A and C) the Framingham Heart Study (FHS) participants and (B and D) the Framingham Heart Study Offspring cohort (FHSO) participants carrying and not carrying the APOE e4 (E4) allele. N=m/k denotes the total number of carriers (m) and the number of events among them (k). (Color image available online at www.liebertpub.com/rej).
Empirical age patterns of onset of CVD for men and women combined show antagonistic effects with the changing role of the e4 allele in the onset of CVD over age in the FHS from detrimental in younger ages to protective in older ages. In the FHSO, we observed the detrimental effect of the e4 allele (Fig. S2). Sex-specific analyses suggested a leading role of the e4 allele in women (Fig. S2 and Fig. 1C,D).
Cox regression analyses show (Table 1) that both men and women carrying the e4 allele can live longer lives without cancer, although the relative risk (RR) is only marginally significant in men in the FHSO (RR=0.81, p=0.079). However, given the same type of empirical age patterns in each cohort (Fig. S1 and Fig. 1A,B), the data can be safely pooled together to increase sample size. In the pooled sample of the FHS and FHSO cohorts (Table 1), the estimates of the RRs of onset of cancer become significant for men and women combined (RR=0.87, p=0.046) and marginally significant for men (RR=0.82, p=0.055).
Table 1.
Relative Risks of Premature Onset of Cancer for the e4 Allele Carriers Compared to the Non-e4 Carriers
| Cohort | Age group | Gender | N totala | N cancer | RRb | 95% CI | P value |
|---|---|---|---|---|---|---|---|
| FHS | All ages | M&W | 1250 | 421 | 0.86 | 0.67–1.10 | 0.232 |
| Men | 452 | 182 | 0.85 | 0.58–1.25 | 0.415 | ||
| Women | 798 | 239 | 0.87 | 0.63–1.19 | 0.376 | ||
| FHSO | All ages | M&W | 3867 | 830 | 0.87 | 0.73–1.03 | 0.105 |
| Men | 1842 | 449 | 0.81 | 0.64–1.02 | 0.079 | ||
| Women | 2025 | 381 | 0.91 | 0.71–1.17 | 0.474 | ||
| FHS and FHSO | All ages | M&W | 5117 | 1251 | 0.87 | 0.76–1.00 | 0.046 |
| Men | 2294 | 631 | 0.82 | 0.67–1.00 | 0.055 | ||
| Women | 2823 | 620 | 0.90 | 0.74–1.09 | 0.278 | ||
| FHS and FHSO | >65 years | M&W | 3535 | 761 | 0.78 | 0.64–0.93 | 6.7E-03 |
| Men | 1567 | 419 | 0.74 | 0.58–0.95 | 0.017 | ||
| Women | 1968 | 342 | 0.82 | 0.63–1.08 | 0.165 |
Bold font highlights significant and marginally significant results.
All subjects with missing information on onset of either CVD or cancer were excluded.
Cox regression model was adjusted for age as well as for sex and cohort when applicable.
RR, Relative risk; CI, confidence interval; FHS, Framingham Heart Study original cohort; FHSO, Framingham Heart Study Offspring cohort; M&W, men and women; age group, “>65 years” denotes subjects who have being older than 65 years at cancer onset for cases or the end of follow-up in 2008 (or death) for censored individuals.
Empirical analyses (Fig. S1 and Fig. 1A,B) suggest, however, that the protective effect of the e4 allele is disproportionally shifted to onsets in older ages. Accordingly, the Cox regression model with proportional hazards likely provides underpowered estimates in this case. Note that conventional analyses of interactions with age at baseline do not address this problem. When we focus on onsets in older ages (representatively, being older than 65 years at cancer onset for cases or the end of follow up in 2008 for censored individuals; the estimates are robust to the choice of cutoff), we have considerably more compelling estimates (Table 1) in terms of the effect size and significance, e.g., RR=0.78, p=6.7×10−3 for men and women combined.
Analysis of the RR for onset of CVD without controlling for age specific in the FHS generation shows no significant effect in the sample of men and women combined and for each sex (Table 2, all ages). The situations for men and women are, however, qualitatively different. Empirical analysis indeed suggests that the e4 allele confers at most a tiny effect in men (Fig. S2C). This is clearly not the case in women for whom the lack of significant effect is because of complex modalities of gene action (Fig. 1C). To address the role of the observed disproportionality, we defined more homogeneous groups in the FHS as: “Younger,” being 75 years and younger at onset of CVD for cases or the end of follow-up in 2008 (or death) for censored individuals, and “older,” being older than 75 years at onset of CVD for cases or the end of follow-up in 2008 (or death) for censored individuals.
Table 2.
Relative risks of Premature Onset of CVD for the e4 Allele Carriers Compared to the Non-e4 Carriers
| Cohort | Age group | Gender | N totala | N CVD | RRb | 95% CI | P value |
|---|---|---|---|---|---|---|---|
| FHS | All ages | M&W | 1250 | 804 | 0.93 | 0.78–1.11 | 0.405 |
| Men | 452 | 322 | 1.08 | 0.83–1.41 | 0.547 | ||
| Women | 798 | 482 | 0.84 | 0.66–1.07 | 0.153 | ||
| FHS | ≤75 years | M&W | 397 | 379 | 1.49 | 1.18–1.88 | 7.5E-04 |
| Men | 190 | 178 | 1.25 | 0.90–1.72 | 0.178 | ||
| Women | 207 | 201 | 1.73 | 1.24–2.43 | 1.4E-03 | ||
| FHS | 76+ years | M&W | 853 | 425 | 0.78 | 0.60–1.00 | 0.051 |
| Men | 262 | 144 | 0.99 | 0.68–1.46 | 0.971 | ||
| Women | 591 | 281 | 0.69 | 0.50–0.95 | 0.025 | ||
| FHSO | All ages | M&W | 3867 | 931 | 1.22 | 1.05–1.42 | 9.0E-03 |
| Men | 1842 | 569 | 1.16 | 0.96–1.41 | 0.121 | ||
| Women | 2025 | 362 | 1.33 | 1.05–1.69 | 0.017 | ||
| FHSO | >65 years | M&W | 2206 | 388 | 1.22 | 0.96–1.54 | 0.105 |
| Men | 997 | 210 | 1.03 | 0.74–1.41 | 0.882 | ||
| Women | 1209 | 178 | 1.49 | 1.06–2.10 | 0.022 | ||
| FHS&FHSO | All ages | M&W | 5117 | 1735 | 1.08 | 0.96–1.21 | 0.201 |
| Men | 2294 | 891 | 1.13 | 0.97–1.32 | 0.130 | ||
| Women | 2823 | 844 | 1.03 | 0.87–1.22 | 0.761 |
Age groups “≤75 years” and “76+ years” denote subjects who have been 75 years or younger and 76 years or older at CVD onset for cases or the end of follow up in 2008 (or death) for censored individuals in the FHS cohort.
Age group “>65 years” denotes subjects who have being older than 65 years at CVD onset for cases or the end of follow up in 2008 (or death) for censored individuals in the FHSO cohort.
Bold font highlights significant and marginally significant results.
All subjects with missing information on onset of either CVD or cancer were excluded.
Cox regression model was adjusted for age as well as for sex and cohort when applicable.
CVD, Cardiovascular disease; RR, relative risk; CI, confidence interval; FHS, Framingham Heart Study original cohort; FHSO, Framingham Heart Study Offspring cohort; M&W, men and women.
The analyses of the “younger” group (Table 2) show a highly significant detrimental effect of the e4 allele in the sample of men and women combined (RR=1.49, p=7.5×10−4) and in women only (RR=1.73, p=1.4×10−3). Younger men can also be at higher risk of premature onset of CVD, although the effect is much smaller than in women and it is not significant. The analyses of the “older” group show significant protective effect of the e4 allele in women (RR=0.69, p=0.025) and marginally significant in men and women combined (Table 2, FHS). No effect is, however, seen in men.
In the FHSO cohort, the e4 allele confer risks of premature onset of CVD in men and women combined (RR=1.22, p=9.0×10−3) with a leading role in women (Table 2, FHSO). The effect becomes stronger in older women, defined as being older than 65 years at onset of CVD for cases or the end of follow-up in 2008 (or death) for censored individuals (Table 2). Importantly, given the complex role of genes in the etiology of traits with well-pronounced post-reproductive manifestation, increasing the sample size (by combining the FHS and the FHSO in this case) can readily result in no effect, just because extending sample can scale the heterogeneity in parallel (e.g., RR=1.08, p=0.201 for men and women combined; see Table 2).
Connection of the same allele with CVD and cancer raises the question of whether CVD and cancer can be developed in the same e4 carriers or not. To address this problem, we evaluated the confounding role of alternative diseases. The analyses show at most a minor role of CVD as a confounding factor in the models for onset of cancer (see Table S1) and of cancer in the models for onset of CVD (see Table S2).
Discussion
Extension of the analyses of genetic trade-off in the effect of the e4 allele on risks of premature onset of cancer and CVD presented in Kulminski et al.17 for the FHS original cohort helps to better characterize the puzzling sensitivity of this phenomenon to gender, ages, and generations. The analyses suggest that the e4 allele can be protective against cancer, with a more pronounced effect in men. This protective effect is more characteristic for cancers at older ages and it holds in the parental and offspring generations of the FHS participants.
Unlike cancer, the effect of the e4 allele on ages at onset of CVD is more pronounced in women. The analyses suggest that the role of this allele in the etiology of CVD can be sensitive to age and generations. In the parental generation of the FHS participants, we observed an antagonistic action of the e4 allele on onset of CVD in women across ages: The e4 allele can confer risks of developing CVD in younger women but protect against CVD in older women. In the offspring generation, the e4 allele can confer risks of developing CVD primarily in older women. Antagonistic action of the same allele at different ages is an important phenomenon that deserves more detailed analyses.39 Genetic trade-off is at most barely confounded by alternative diseases, primarily because of strong sexual dimorphism.
Thus, our analyses suggest two modes of sexually dimorphic genetic trade-offs. One mode is observed in the FHSO generation when the e4 allele confers risk of premature onset of CVD primarily in women and this allele can protect against cancer primarily in men of the same age. The other mode is pronounced in the FHS generation when genetic trade-off is seen in different age groups with a protective role of the e4 allele against cancer in older men and its detrimental role in CVD in younger women.
In relation to CVD, sexual dimorphism of the APOE gene is widely discussed in literature. For example, studies demonstrate significant heterogeneity in the effect of the e4 allele on coronary heart disease (CHD). Some studies document a more pronounced role of this allele in women,40 particularly in high-risk women defined by conventional risk factors.41 Other studies document a more substantial effect of this allele on CHD in men.42,43 Sexual dimorphism of the APOE gene is typically attributed to differential hormonal and insulin regulation in men and women.40
In relation to cancer, given that the mechanism of the protective effect of the e4 allele is unclear and that this effect is largely attributed to non-prostate and non-breast cancer sites,17,44 the mechanism of sexual dimorphism appears to be unclear as well. However, given that the protective effect of the e4 allele on cancer is more pronounced at older ages, this effect may, in part, be linked to the effect of the e4 allele on Alzheimer disease (AD). Indeed, the APOE e4 allele is well known to increase the risk of AD, whereas the risk of cancer is significantly smaller among AD patients, as has been recently shown.22,45,46 One potential biological mechanism linking smaller cancer risk in AD patients carrying the e4 allele could be related to inflammation. For example, carriers of the e4 allele have been shown to have lower C-reactive protein (CRP) levels,47 whereas the lower CRP levels have been shown to be associated with reduced overall cancer risk and better prognosis in a number of studies.48,49
Differential effects of genes on various phenotypes with well-pronounced post-reproductive manifestation at different ages have been extensively documented in candidate gene studies.11–15,40 Qualitative similarity of the effect of the e4 allele on onset of cancer across generations and dissimilarity across generations in the case of CVD support the hypothesis on the qualitative difference in the mechanisms underlying connection of the e4 allele with CVD and cancer.17 Given that the APOE gene is directly associated with diet and lifestyle50,51 and that these exposures experience dramatic changes across generations,52–55 we can hypothesize that in the case of CVD the mechanism is sensitive to modern environment, whereas in the case of cancer this is unlikely to be the case.
Our study also provides explicit evidence that the aging-related processes in different generations (which are proxies for changing environment) can crucially impact strength and significance of genetic effects on traits in late life. These results contribute to ongoing discussion about a major problem in current GWAS that have failed to explain the proportion of genetic variance contributing to complex phenotypes.56 This study strongly supports the view that the so-called problem of missing heritability “is overblown, and the focus on hits that are significant genome-wide is distracting attention from more general concerns over the ability of GWAS to fully describe the architecture of phenotypic variation” shaping genetic effects.56,57 Our results also support the view that “increasing the size of human disease cohorts is likely only to scale the heterogeneity in parallel”58 with non-obvious chances to gain more insights.
Overall, gender-, age-, and generation-specific genetic trade-off of the same e4 allele on seemingly unrelated diseases explicitly demonstrates the puzzling complexity of genetic mechanisms working in different ages, genders, and environments. Current GWAS strategies that are largely adapted to find universal genes working in different settings have to be revisited to reflect the real complexity of gene action on traits with well-pronounced post-reproductive manifestation.
Supplementary Material
Acknowledgments
The research reported in this paper was supported by award number R01AG030612 from the National Institute on Aging. The Framingham Heart Study (FHS) is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with the FHS Investigators. This manuscript was prepared using a limited access dataset obtained from the NHLBI. This manuscript was not prepared in collaboration with investigators of the FHS and does not necessarily reflect the opinions or views of the FHS, Boston University, or the NHLBI.
Author Disclosure Statement
No competing financial interests exist.
Author contributions: A.M.K. contributed to the study conception, design, statistical analysis, interpretation of the results, and writing the manuscript. I.C. contributed to the study design and interpretation of the results. K.G.A. contributed to statistical analyses. L.A. contributed to preparation of the data. S.V.U. contributed to final version and discussion of mechanisms of genetic trade-offs, and A.I.Y. contributed to assessing the logic of the final version and discussion of genetic trade-offs.
References
- 1.Finch CE. Tanzi RE. Genetics of aging. Science. 1997;278:407–411. doi: 10.1126/science.278.5337.407. [DOI] [PubMed] [Google Scholar]
- 2.Martin GM. Modalities of gene action predicted by the classical evolutionary biological theory of aging. Ann NY Acad Sci. 2007;1100:14–20. doi: 10.1196/annals.1395.002. [DOI] [PubMed] [Google Scholar]
- 3.Teslovich TM. Musunuru K. Smith AV. Edmondson AC. Stylianou IM. Koseki M. Pirruccello JP. Ripatti S. Chasman DI. Willer CJ. Johansen CT. Fouchier SW. Isaacs A. Peloso GM. Barbalic M. Ricketts SL. Bis JC. Aulchenko YS. Thorleifsson G. Feitosa MF. Chambers J. Orho-Melander M. Melander O. Johnson T. Li X. Guo X. Li M. Shin Cho Y. Jin Go M. Jin Kim Y. Lee JY. Park T. Kim K. Sim X. Twee-Hee Ong R. Croteau-Chonka DC. Lange LA. Smith JD. Song K. Hua Zhao J. Yuan X. Luan J. Lamina C. Ziegler A. Zhang W. Zee RY. Wright AF. Witteman JC. Wilson JF. Willemsen G. Wichmann HE. Whitfield JB. Waterworth DM. Wareham NJ. Waeber G. Vollenweider P. Voight BF. Vitart V. Uitterlinden AG. Uda M. Tuomilehto J. Thompson JR. Tanaka T. Surakka I. Stringham HM. Spector TD. Soranzo N. Smit JH. Sinisalo J. Silander K. Sijbrands EJ. Scuteri A. Scott J. Schlessinger D. Sanna S. Salomaa V. Saharinen J. Sabatti C. Ruokonen A. Rudan I. Rose LM. Roberts R. Rieder M. Psaty BM. Pramstaller PP. Pichler I. Perola M. Penninx BW. Pedersen NL. Pattaro C. Parker AN. Pare G. Oostra BA. O'Donnell CJ. Nieminen MS. Nickerson DA. Montgomery GW. Meitinger T. McPherson R. McCarthy MI. McArdle W. Masson D. Martin NG. Marroni F. Mangino M. Magnusson PK. Lucas G. Luben R. Loos RJ. Lokki ML. Lettre G. Langenberg C. Launer LJ. Lakatta EG. Laaksonen R. Kyvik KO. Kronenberg F. Konig IR. Khaw KT. Kaprio J. Kaplan LM. Johansson A. Jarvelin MR. Janssens AC. Ingelsson E. Igl W. Kees Hovingh G. Hottenga JJ. Hofman A. Hicks AA. Hengstenberg C. Heid IM. Hayward C. Havulinna AS. Hastie ND. Harris TB. Haritunians T. Hall AS. Gyllensten U. Guiducci C. Groop LC. Gonzalez E. Gieger C. Freimer NB. Ferrucci L. Erdmann J. Elliott P. Ejebe KG. Doring A. Dominiczak AF. Demissie S. Deloukas P. de Geus EJ. de Faire U. Crawford G. Collins FS. Chen YD. Caulfield MJ. Campbell H. Burtt NP. Bonnycastle LL. Boomsma DI. Boekholdt SM. Bergman RN. Barroso I. Bandinelli S. Ballantyne CM. Assimes TL. Quertermous T. Altshuler D. Seielstad M. Wong TY. Tai ES. Feranil AB. Kuzawa CW. Adair LS. Taylor HA., Jr. Borecki IB. Gabriel SB. Wilson JG. Holm H. Thorsteinsdottir U. Gudnason V. Krauss RM. Mohlke KL. Ordovas JM. Munroe PB. Kooner JS. Tall AR. Hegele RA. Kastelein JJ. Schadt EE. Rotter JI. Boerwinkle E. Strachan DP. Mooser V. Stefansson K. Reilly MP. Samani NJ. Schunkert H. Cupples LA. Sandhu MS. Ridker PM. Rader DJ. van Duijn CM. Peltonen L. Abecasis GR. Boehnke M. Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stranger BE. Stahl EA. Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 6.Williams PD. Day T. Antagonistic pleiotropy, mortality source interactions, and the evolutionary theory of senescence. Evolution. 2003;57:1478–1488. doi: 10.1111/j.0014-3820.2003.tb00356.x. [DOI] [PubMed] [Google Scholar]
- 7.Kulminski AM. Culminskaya I. Ukraintseva SV. Arbeev KG. Land KC. Yashin AI. Beta2-adrenergic receptor gene polymorphisms as systemic determinants of healthy aging in an evolutionary context. Mech Ageing Dev. 2010;131:338–345. doi: 10.1016/j.mad.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Summers K. Crespi BJ. Xmrks the spot: Life history tradeoffs, sexual selection and the evolutionary ecology of oncogenesis. Mol Ecol. 2010;19:3022–3024. doi: 10.1111/j.1365-294x.2010.04739.x. [DOI] [PubMed] [Google Scholar]
- 9.Alexander DM. Williams LM. Gatt JM. Dobson-Stone C. Kuan SA. Todd EG. Schofield PR. Cooper NJ. Gordon E. The contribution of apolipoprotein E alleles on cognitive performance and dynamic neural activity over six decades. Biol Psychol. 2007;75:229–238. doi: 10.1016/j.biopsycho.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Schnebel EM. Grossfield J. Antagonistic pleiotropy—an interspecific Drosophila comparison. Evolution. 1988;42:306–311. doi: 10.1111/j.1558-5646.1988.tb04134.x. [DOI] [PubMed] [Google Scholar]
- 11.De Benedictis G. Carotenuto L. Carrieri G. De Luca M. Falcone E. Rose G. Yashin AI. Bonafe M. Franceschi C. Age-related changes of the 3′ APOB-VNTR genotype pool in ageing cohorts. Ann Hum Genet. 1998;62(Pt 2):115–122. doi: 10.1046/j.1469-1809.1998.6220115.x. [DOI] [PubMed] [Google Scholar]
- 12.Ilveskoski E. Perola M. Lehtimaki T. Laippala P. Savolainen V. Pajarinen J. Penttila A. Lalu KH. Mannikko A. Liesto KK. Koivula T. Karhunen PJ. Age-dependent association of apolipoprotein E genotype with coronary and aortic atherosclerosis in middle-aged men: An autopsy study. Circulation. 1999;100:608–613. doi: 10.1161/01.cir.100.6.608. [DOI] [PubMed] [Google Scholar]
- 13.Yashin AI. Ukraintseva SV. De Benedictis G. Anisimov VN. Butov AA. Arbeev K. Jdanov DA. Boiko SI. Begun AS. Bonafe M. Franceschi C. Have the oldest old adults ever been frail in the past? A hypothesis that explains modern trends in survival. J Gerontol A Biol Sci Med Sci. 2001;56:B432–B442. doi: 10.1093/gerona/56.10.b432. [DOI] [PubMed] [Google Scholar]
- 14.Jarvik GP. Goode EL. Austin MA. Auwerx J. Deeb S. Schellenberg GD. Reed T. Evidence that the apolipoprotein E-genotype effects on lipid levels can change with age in males: A longitudinal analysis. Am J Hum Genet. 1997;61:171–181. doi: 10.1086/513902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergman A. Atzmon G. Ye K. MacCarthy T. Barzilai N. Buffering mechanisms in aging: A systems approach toward uncovering the genetic component of aging. PLoS Comput Biol. 2007;3:e170. doi: 10.1371/journal.pcbi.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulminski AM. Ukraintseva SV. Arbeev KG. Manton KG. Oshima J. Martin GM. Il'yasova D. Yashin AI. Health-protective and adverse effects of the apolipoprotein E epsilon2 allele in older men. J Am Geriatr Soc. 2008;56:478–483. doi: 10.1111/j.1532-5415.2007.01574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulminski AM. Culminskaya I. Ukraintseva SV. Arbeev KG. Arbeeva L. Wu D. Akushevich I. Land KC. Yashin AI. Trade-off in the effects of the apolipoprotein E polymorphism on the ages at onset of CVD and cancer influences human lifespan. Aging Cell. 2011;10:533–541. doi: 10.1111/j.1474-9726.2011.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crespi BJ. The origins and evolution of genetic disease risk in modern humans. Ann NY Acad Sci. 2010;1206:80–109. doi: 10.1111/j.1749-6632.2010.05707.x. [DOI] [PubMed] [Google Scholar]
- 19.Kulminski AM. Culminskaya IV. Ukraintseva SV. Arbeev KG. Akushevich I. Land KC. Yashin AI. Polymorphisms in the ACE and ADRB2 genes and risks of aging-associated phenotypes: The case of myocardial infarction. Rejuvenation Res. 2010;13:13–21. doi: 10.1089/rej.2009.0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K. Baldassano R. Zhang H. Qu HQ. Imielinski M. Kugathasan S. Annese V. Dubinsky M. Rotter JI. Russell RK. Bradfield JP. Sleiman PM. Glessner JT. Walters T. Hou C. Kim C. Frackelton EC. Garris M. Doran J. Romano C. Catassi C. Van Limbergen J. Guthery SL. Denson L. Piccoli D. Silverberg MS. Stanley CA. Monos D. Wilson DC. Griffiths A. Grant SF. Satsangi J. Polychronakos C. Hakonarson H. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum Mol Genet. 2010;19:2059–2067. doi: 10.1093/hmg/ddq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnes JA. Dix DJ. Collins BW. Luft C. Allen JW. Expression of inducible Hsp70 enhances the proliferation of MCF-7 breast cancer cells and protects against the cytotoxic effects of hyperthermia. Cell Stress Chaperones. 2001;6:316–325. doi: 10.1379/1466-1268(2001)006<0316:eoihet>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ukraintseva SV. Arbeev KG. Akushevich I. Kulminski A. Arbeeva L. Culminskaya I. Akushevich L. Yashin AI. Trade-offs between cancer and other diseases: Do they exist and influence longevity? Rejuvenation Res. 2010;13:387–396. doi: 10.1089/rej.2009.0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weilbach FX. Toyka KV. Does Down's syndrome protect against multiple sclerosis? Eur Neurol. 2002;47:52–55. doi: 10.1159/000047947. [DOI] [PubMed] [Google Scholar]
- 24.Finch CE. Evolution in health and medicine Sackler colloquium: Evolution of the human lifespan and diseases of aging: Roles of infection, inflammation, and nutrition. Proc Natl Acad Sci USA. 2010;107(Suppl 1):1718–1724. doi: 10.1073/pnas.0909606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charlesworth B. Evolution of senescence: Alzheimer's disease and evolution. Curr Biol. 1996;6:20–22. doi: 10.1016/s0960-9822(02)00411-6. [DOI] [PubMed] [Google Scholar]
- 26.Martin GM. APOE alleles and lipophylic pathogens. Neurobiol Aging. 1999;20:441–443. doi: 10.1016/s0197-4580(99)00078-0. [DOI] [PubMed] [Google Scholar]
- 27.Frazer KA. Murray SS. Schork NJ. Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 28.van Heemst D. Mooijaart SP. Beekman M. Schreuder J. de Craen AJ. Brandt BW. Slagboom PE. Westendorp RG. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp Gerontol. 2005;40:11–15. doi: 10.1016/j.exger.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Denton K. Baylis C. Physiological and molecular mechanisms governing sexual dimorphism of kidney, cardiac, and vascular function. Am J Physiol Regul Integr Comp Physiol. 2007;292:R697–R699. doi: 10.1152/ajpregu.00766.2006. [DOI] [PubMed] [Google Scholar]
- 30.Nedungadi TP. Clegg DJ. Sexual dimorphism in body fat distribution and risk for cardiovascular diseases. J Cardiovasc Transl Res. 2009;2:321–327. doi: 10.1007/s12265-009-9101-1. [DOI] [PubMed] [Google Scholar]
- 31.Dawber TR. The Framingham study: The Epidemiology of Atherosclerotic Disease. Harvard University Press; Cambridge, MA: 1980. [Google Scholar]
- 32.Gail MH. Johnson NL. Proceedings of the American Statistical Association: sesquicentennial invited papers session. American Statistical Association; Alexandria, VA. 1989. [Google Scholar]
- 33.Govindaraju DR. Cupples LA. Kannel WB. O'Donnell CJ. Atwood LD. D'Agostino RB., Sr. Fox CS. Larson M. Levy D. Murabito J. Vasan RS. Splansky GL. Wolf PA. Benjamin EJ. Genetics of the Framingham Heart Study population. Adv Genet. 2008;62:33–65. doi: 10.1016/S0065-2660(08)00602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Splansky GL. Corey D. Yang Q. Atwood LD. Cupples LA. Benjamin EJ. D'Agostino RB., Sr. Fox CS. Larson MG. Murabito JM. O'Donnell CJ. Vasan RS. Wolf PA. Levy D. The Third Generation Cohort of the National Heart, Lung, and Blood Institute's Framingham Heart Study: Design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 35.Cupples LA. Heard-Costa N. Lee M. Atwood LD. Genetics Analysis Workshop 16 Problem 2: The Framingham Heart Study data. BMC Proc. 2009;3(Suppl 7):S3. doi: 10.1186/1753-6561-3-s7-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lahoz C. Schaefer EJ. Cupples LA. Wilson PW. Levy D. Osgood D. Parpos S. Pedro-Botet J. Daly JA. Ordovas JM. Apolipoprotein E genotype and cardiovascular disease in the Framingham Heart Study. Atherosclerosis. 2001;154:529–537. doi: 10.1016/s0021-9150(00)00570-0. [DOI] [PubMed] [Google Scholar]
- 37.Myers RH. Schaefer EJ. Wilson PW. D'Agostino R. Ordovas JM. Espino A. Au R. White RF. Knoefel JE. Cobb JL. McNulty KA. Beiser A. Wolf PA. Apolipoprotein E epsilon4 association with dementia in a population-based study: The Framingham study. Neurology. 1996;46:673–677. doi: 10.1212/wnl.46.3.673. [DOI] [PubMed] [Google Scholar]
- 38.Lee EW. Wei LJ. Amato DA. Leurgans S. Cox-type regression-analysis for large numbers of small-groups of correlated failure time observations. Nato Adv Sci I E-App. 1992;211:237–247. [Google Scholar]
- 39.Kulminski AM. Culminskaya I. Arbeev KG. Ukraintseva SV. Stallard E. Arbeeva L. Yashin AI. The role of lipid-related genes, aging-related processes, and environment in healthspan. 2012. [DOI] [PMC free article] [PubMed]
- 40.Kolovou G. Damaskos D. Anagnostopoulou K. Cokkinos DV. Apolipoprotein E gene polymorphism and gender. Ann Clin Lab Sci. 2009;39:120–133. [PubMed] [Google Scholar]
- 41.Hirashiki A. Yamada Y. Murase Y. Suzuki Y. Kataoka H. Morimoto Y. Tajika T. Murohara T. Yokota M. Association of gene polymorphisms with coronary artery disease in low- or high-risk subjects defined by conventional risk factors. J Am Coll Cardiol. 2003;42:1429–1437. doi: 10.1016/s0735-1097(03)01062-3. [DOI] [PubMed] [Google Scholar]
- 42.Scuteri A. Bos AJ. Zonderman AB. Brant LJ. Lakatta EG. Fleg JL. Is the apoE4 allele an independent predictor of coronary events? Am J Med. 2001;110:28–32. doi: 10.1016/s0002-9343(00)00639-2. [DOI] [PubMed] [Google Scholar]
- 43.Frikke-Schmidt R. Tybjaerg-Hansen A. Steffensen R. Jensen G. Nordestgaard BG. Apolipoprotein E genotype: Epsilon32 women are protected while epsilon43 and epsilon44 men are susceptible to ischemic heart disease: The Copenhagen City Heart Study. J Am Coll Cardiol. 2000;35:1192–1199. doi: 10.1016/s0735-1097(00)00520-9. [DOI] [PubMed] [Google Scholar]
- 44.de Jong PT. Age-related macular degeneration. N Engl J Med. 2006;355:1474–1485. doi: 10.1056/NEJMra062326. [DOI] [PubMed] [Google Scholar]
- 45.Roe CM. Behrens MI. Xiong C. Miller JP. Morris JC. Alzheimer disease and cancer. Neurology. 2005;64:895–898. doi: 10.1212/01.WNL.0000152889.94785.51. [DOI] [PubMed] [Google Scholar]
- 46.Tabares-Seisdedos R. Dumont N. Baudot A. Valderas JM. Climent J. Valencia A. Crespo-Facorro B. Vieta E. Gomez-Beneyto M. Martinez S. Rubenstein JL. No paradox, no progress: Inverse cancer comorbidity in people with other complex diseases. Lancet Oncol. 2011;12:604–608. doi: 10.1016/S1470-2045(11)70041-9. [DOI] [PubMed] [Google Scholar]
- 47.Chasman DI. Kozlowski P. Zee RY. Kwiatkowski DJ. Ridker PM. Qualitative and quantitative effects of APOE genetic variation on plasma C-reactive protein, LDL-cholesterol, and apoE protein. Genes Immun. 2006;7:211–219. doi: 10.1038/sj.gene.6364289. [DOI] [PubMed] [Google Scholar]
- 48.Nikiteas NI. Tzanakis N. Gazouli M. Rallis G. Daniilidis K. Theodoropoulos G. Kostakis A. Peros G. Serum IL-6, TNFalpha and CRP levels in Greek colorectal cancer patients: Prognostic implications. World J Gastroenterol. 2005;11:1639–1643. doi: 10.3748/wjg.v11.i11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koukourakis MI. Kambouromiti G. Pitsiava D. Tsousou P. Tsiarkatsi M. Kartalis G. Serum C-reactive protein (CRP) levels in cancer patients are linked with tumor burden and are reduced by anti-hypertensive medication. Inflammation. 2009;32:169–175. doi: 10.1007/s10753-009-9116-4. [DOI] [PubMed] [Google Scholar]
- 50.Ordovas JM. Gender, a significant factor in the cross talk between genes, environment, and health. Gend Med. 2007;4(Suppl B):S111–S122. doi: 10.1016/s1550-8579(07)80052-0. [DOI] [PubMed] [Google Scholar]
- 51.Eichner JE. Dunn ST. Perveen G. Thompson DM. Stewart KE. Stroehla BC. Apolipoprotein E polymorphism and cardiovascular disease: A HuGE review. Am J Epidemiol. 2002;155:487–495. doi: 10.1093/aje/155.6.487. [DOI] [PubMed] [Google Scholar]
- 52.Neel JV. Weder AB. Julius S. Type II diabetes, essential hypertension, and obesity as “syndromes of impaired genetic homeostasis”: The “thrifty genotype” hypothesis enters the 21st century. Perspect Biol Med. 1998;42:44–74. doi: 10.1353/pbm.1998.0060. [DOI] [PubMed] [Google Scholar]
- 53.Kuningas M. Mooijaart SP. van Heemst D. Zwaan BJ. Slagboom PE. Westendorp RG. Genes encoding longevity: From model organisms to humans. Aging Cell. 2008;7:270–280. doi: 10.1111/j.1474-9726.2008.00366.x. [DOI] [PubMed] [Google Scholar]
- 54.Vijg J. Suh Y. Genetics of longevity and aging. Annu Rev Med. 2005;56:193–212. doi: 10.1146/annurev.med.56.082103.104617. [DOI] [PubMed] [Google Scholar]
- 55.Boomsma DI. Kempen HJ. Gevers Leuven JA. Havekes L. de Knijff P. Frants RR. Genetic analysis of sex and generation differences in plasma lipid, lipoprotein, and apolipoprotein levels in adolescent twins and their parents. Genet Epidemiol. 1996;13:49–60. doi: 10.1002/(SICI)1098-2272(1996)13:1<49::AID-GEPI5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 56.Eichler EE. Flint J. Gibson G. Kong A. Leal SM. Moore JH. Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibson G. Decanalization and the origin of complex disease. Nat Rev Genet. 2009;10:134–140. doi: 10.1038/nrg2502. [DOI] [PubMed] [Google Scholar]
- 58.MacRae CA. Vasan RS. Next-generation genome-wide association studies: Time to focus on phenotype? Circ Cardiovasc Genet. 2011;4:334–336. doi: 10.1161/CIRCGENETICS.111.960765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.