Abstract
The synucleins are a family of proteins involved in numerous neurodegenerative pathologies [α-synuclein and β-synuclein (βS)], as well as in various types of cancers [γ-synuclein (γS)]. While the connection between α-synuclein and Parkinson's disease is well established, recent evidence links point mutants of βS to dementia with Lewy bodies. Overexpression of γS has been associated with enhanced metastasis and cancer drug resistance. Despite their prevalence in such a variety of diseases, the native functions of the synucleins remain unclear. They have a lipid-binding motif in their N-terminal region, which suggests interactions with biological membranes in vivo. In this study, we used fluorescence correlation spectroscopy to monitor the binding properties of βS and γS to model membranes and to determine the free energy of the interactions. Our results show that the interactions are most strongly affected by the presence of both anionic lipids and bilayer curvature, while membrane fluidity plays a very minor role. Quantifying the lipid-binding properties of βS and γS provides additional insights into the underlying factors governing the protein–membrane interactions. Such insights not only are relevant to the native functions of these proteins but also highlight their contributions to pathological conditions that are either mediated or characterized by perturbations of these interactions.
Keywords: intrinsically disordered proteins, amphipathic, dementia with Lewy bodies, Parkinson's disease, aggregation
Introduction
The synuclein family of proteins is composed of α-synuclein (αS), β-synuclein (βS), and γ-synuclein (γS). They are expressed predominantly in neurons, although their localization within the cell varies. Both αS and βS are found ubiquitously in presynaptic terminals throughout the brain, while γS tends to be localized in more differentiated nerve cells, and away from the forebrain.1 The synucleins have also been found in other various cell types including liver (γS), skeletal muscle (αS, βS, and γS), and lung (αS) cells.2 αS is the most well studied of the synuclein family primarily due to its link to Parkinson's disease.3,4 Despite their similarities to αS, significantly less is known about βS and γS. However, recent studies found that mutations of βS were linked directly to learning deficits and motor dysfunction as well as being capable of enhancing the pathogenic state of αS, thereby promoting further neurodegeneration as seen in α-synucleopathies.5 In fact, two variants of βS, carrying either a V70M or a P123H mutation, have been independently identified in cases of dementia with Lewy bodies (DLB).6 Alternatively, wild-type (WT) βS is known to have an inhibitory effect on αS aggregation,7 a property that can be exploited in the development of novel therapeutics against various neurological pathologies. The synucleins are not only associated with neurodegenerative disorders. Several studies implicate γS in various types of cancers. Most notably, the role of γS has been investigated in breast carcinomas where it was shown that it was capable of conferring drug resistance to the cancerous cells and of promoting metastasis.8–10 Consequently, the interest in all the synucleins for their potential roles in a broad array of physiological functions and in pathogenic mechanisms is growing rapidly.
The synucleins have generally been described as intrinsically disordered proteins or IDPs, although recent studies suggest that, at least, αS might exist in a partially structured conformation under physiological conditions.11–13 Their primary sequence can be divided into two segments: (1) an N-terminal region rich in basic amino acid residues, which mediates interactions with membranes, and (2) a net negatively charged C-terminal region that has no well-established function, though there have been reports of its involvement in protein–protein interactions and of it possessing anti-aggregation properties14,15 (Fig. 1). Further sequence analyses of the synucleins indicate the presence of several repeat motifs in their N-terminal region that share significant similarities with lipid-binding motifs found in apolipoproteins.16,17 Those motifs, imperfect repeats of KTKEGV, are unevenly distributed among the synucleins: αS and γS each have six, while βS has only five. Other sequence differences include the central region of βS, which lacks a stretch of 11 residues, corresponding to residues 73–83 of the non-amyloid β component region of αS2,18 (Fig. 1). Furthermore, γS has a shorter C-terminal region, resulting in a slightly less negative tail. These sequence variations may be responsible for differing native functions of the synucleins.
Fig. 1.
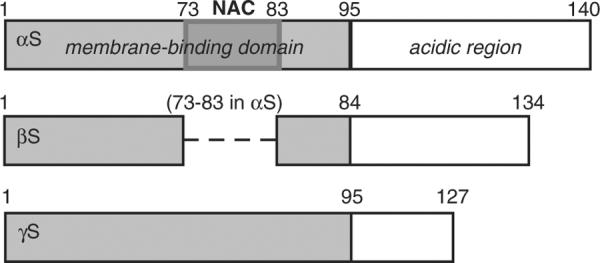
Schematic of the synuclein proteins. The N-terminal lipid-binding region of each protein is represented by the gray box. The broken line in βS depicts the missing 11 residues within the region corresponding to the non-amyloid β component portion (NAC, darker-gray box in αS).
While monomer proteins are unstructured in solution, the N-terminal region of all of the synucleins undergoes a conformational change to α-helical structure upon binding to lipid bilayers19 (Fig. S1). Thus, with their conformational conservation, their high level of sequence similarity, and the overlap in their expression pattern, the synucleins may carry out redundant physiological roles. Interestingly, relative to single and double αS, βS, and γS knockout models, triple-knockout mice exhibited more severe phenotypes.20,21 Particularly, loss of the three synuclein isoforms had a significant effect on synapse morphology and the size of the synaptic terminal.20 Yet, despite this, existing studies of the interactions of βS and γS with model membranes are relatively sparse and they have thus far only explored the qualitative nature of these interactions.19 In this study, we use fluorescence correlation spectroscopy (FCS) to monitor the interactions of βS and γS with liposomes. As with αS, both βS and γS displayed a significant preference for highly curved lipid bilayers. Moreover, both proteins show increased affinity for liposomes with higher anionic content, emphasizing the importance of electrostatics in mediating the protein–membrane interactions. However, weak interactions are also observed with liposomes formed only with neutral lipids, supporting that other factors contribute to the stabilization of these interactions. Modifying the bilayer fluidity by introducing nonlamellar-prone lipids has only moderate effects on the binding affinity of the proteins for the liposomes. Interestingly, we find a 5-fold increase in the partitioning free energies of βS disease mutants relative to WT βS, suggesting that the stimulated neurodegeneration attributed to those mutants may be linked to an increased association with biological membranes.
Results and Discussion
Variation of liposome surface charge
Both proteins were site-specifically labeled at residue 9 with Alexa 488 fluorophore. The liposomes were unlabeled so that they are only visible when protein is bound to them. Free βS and γS in solution have fast diffusion times (~530±3.2 μs and ~511.4±4.6μs, respectively) relative to the large liposomes (~5.5 ms for ~90-nm-diameter liposomes) (Fig. S3). This difference was exploited to detect protein–liposome interactions using FCS; binding of protein to the liposomes resulted in a shift to the right of the autocorrelation curve relative to that of the free protein (Fig. 2a). Binding curves were generated by fitting the autocorrelation curves to determine the fraction of free protein for each lipid concentration and then plotting the fraction bound as a function of lipid concentration (Fig. 2b and Supplemental Data).
Fig. 2.
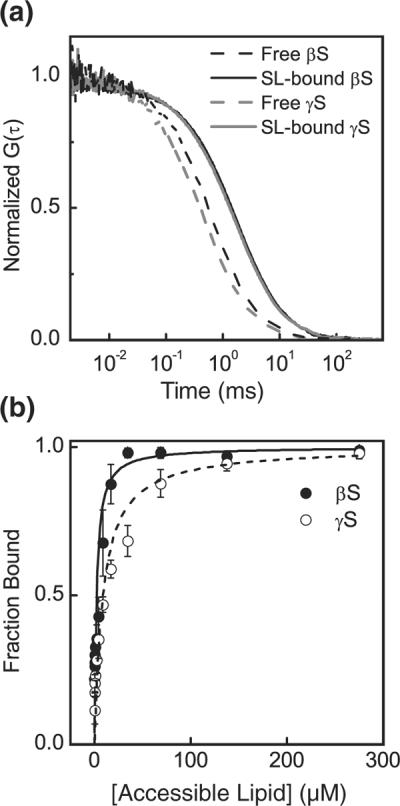
(a) Normalized FCS curves indicating binding of βS and γS to small liposomes (SL) made of 50:50 POPC: POPS. Free βS, black broken line; SL-bound βS, black continuous line; free γS, gray broken line; SL-bound γS, gray continuous line. (b) By fitting the autocorrelation curves with Eq. (1), we can calculate the fraction of bound protein and construct the binding isotherm. βS (filled circles); γS (open circles). Lines are fits to Eq. (3).
We investigated the role of electrostatics in membrane binding of βS and γS by measuring their affinity for liposomes made of varying ratios of zwitterionic 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and anionic 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) lipids. For both proteins, the affinity of the binding interaction decreased with decreasing amounts of anionic lipids (Fig. 3a and Table 1). This is not unexpected as modifying the membrane surface potential, which scales linearly with membrane anionic content, can affect the interactions of peripheral proteins with the lipid bilayer.22,23 Here, we found that the partitioning of both proteins into liposomes increased significantly when the anionic content was increased from 25% to 75% (Fig. 3). A similar dependence on membrane anionic content has been noted for αS,24 and it is generally observed for binding of peripheral proteins mediated by electrostatics.25,26 The free energy of partitioning scales linearly with the surface potential (Ψo) [Eq. (S3)] as the anionic content of the liposomes is increased from 25% to 75%, but it deviates from linearity at Ψo=0 (Fig. 3c). Strikingly, the free energy of binding for 100% POPC liposomes indicates a significantly stronger interaction than predicted by a linear extrapolation of the higher-anionic-content curve to 0 (~2 kcal/mol higher; Fig. 3c). The magnitudes of the binding energies for βS and γS for 100% POPC liposomes are consistent with our previous report for αS27 and suggest that, in addition to electrostatic and hydrophobic interactions, other factors contribute to the binding energy for the synuclein proteins (see Supplemental Data for more details).
Fig. 3.
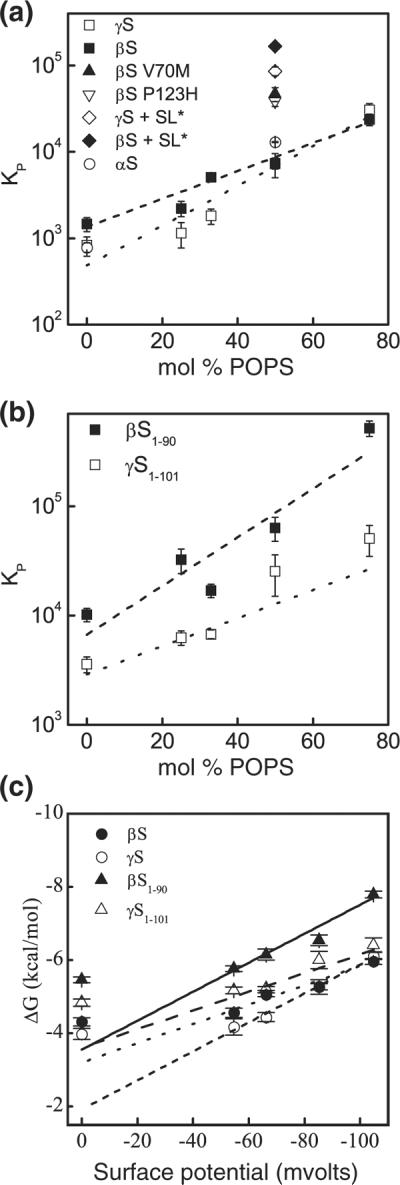
Molar partition coefficients (KP) of the interactions of βS and γS with respect to the liposome surface charge and size. (a) Full-length constructs: WT βS (filled squares), WT γS (open squares), V70M βS (filled triangle), P123H βS (inverted open triangle), WT αS (open circles),27 βS+SL* (SL: ~46nm liposomes) (filled diamond), and γS+SL* (open diamond). (b) Truncated constructs: βS1–90 (filled squares) and γS1–101 (open squares). Continuous and broken lines, linear fits of the corresponding data. (c) Dependence of the free energy of partitioning on surface potential of the liposomes [Eq. (S3)]. WT βS (filled circles); WT γS(open circles); βS1–90 (filled triangles); γS1–101 (open triangles). Lines are fits to Eq. (S2).
Table 1.
Effect of liposome surface charge and size on binding
|
Kd,app (μM) |
ΔG (kcal/mol) |
|||
|---|---|---|---|---|
| mol% POPS | βS | γS | βS | γS |
| 0 | 938±175 | 1765.2±666.3 | −4.31±0.11 | −3.98±0.15 |
| 25 | 502.8±86.4 | 781.4±271.8 | −4.56±0.12 | −4.17±0.22 |
| 33 | 262.2±22.7 | 842±218 | −5.05±0.05 | −4.44±0.12 |
| 50 | 212±58 | 199.8±69.1 | −5.27±0.08 | −5.26±0.20 |
| 36.8±13.2 (V70M) | −6.36±0.10 (V70M) | |||
| 43.9 ±12.6 (P123H) | −6.25 ±0.07 (P123H) | |||
| 7.9±0.2a | 15.5±1.9a | −7.11±0.02a | −6.73±0.07a | |
| 75 | 60.5±19.4 | 51.9±18.3 | −5.96±0.08 | −6.11±0.10 |
Apparent dissociation constants (Kd,app) were calculated from Eq. (3), and free energies of partitioning (ΔG) were calculated from ΔG=−RTlnKP.
Calculated for ~50-nm-diameter liposomes.
To further investigate the role of electrostatics, we determined the influence of the C-terminal region on binding by creating truncated constructs of both proteins that spanned their putative membrane-binding domains (βS1–90 and γS1–101) and measured their partitioning into POPC:POPS liposomes (0–75 mol% POPS) (Fig. 3b). As seen with the full-length proteins, the affinity of truncated βS and γS decreases with decreasing anionic lipid content. Notably, the free-energy difference between full-length protein and truncated protein is positive, indicating that the C-terminus has a destabilizing effect on the protein–liposome interactions (Table 2 and Materials and Methods), despite the fact that the C-terminal domain of the proteins is not directly involved in protein–membrane interactions.19,28–30 We observed a nearly 2-fold difference between the energetic contribution of the C-terminal domain of βS relative to that of γS. The C-terminus of βS is longer and more negatively charged than that of γS (net charge of the removed C-terminal segment: −16 and −5 for βS and γS, respectively), and these results suggest that it may exert a greater electrostatic repulsion near the liposome surface, thereby influencing the protein partitioning in the membrane. As with the full-length proteins, the relationship between free energy of binding and Ψo is linear for truncated constructs and liposomes consisting of 25–75% POPS (Fig. 3c), deviating from this relationship as Ψo→0 (100% POPC liposomes) (Fig. 3c). If we consider the linear parts of the curves to represent the primarily electrostatic contribution to the binding energy, then we would expect the slopes of these lines to scale roughly with the charge of the protein multiplied by the Faraday constant31 (Supplemental Data for details). However, while the charges of the full-length and truncated proteins are significantly different (actual charges: βS=−14.8, βS1–90=1.2, γS=−6, and γS1–101 =−0.1), the slopes of these lines are very similar (~0.03). Moreover, with the exception of βS1–90, the effective charge of each of the constructs derived from the linear fit (effective charges: βS=1.4, βS1–90=1.2, γS=1.1, and γS1–101 = 1.7) is very similar in magnitude and does not correspond to the actual charge. While an effective charge that differs from the actual charge is not unexpected,31–33 it suggests that, while the anionic C-terminus alters the actual magnitude of the binding interaction, the N-terminus of the protein is largely responsible for the response of βS and γS to changes in the anionic content of the liposomes.
Table 2.
Energetic contribution of the C-terminus on binding
| βS |
γS |
|||
|---|---|---|---|---|
| mol% POPS | ΔGtrunc (kcal/mol) | ΔΔGC-term (kcal/mol) | ΔGC-trunc (kcal/mol) | ΔΔGC-term (kcal/mol) |
| 0 | −5.46±0.08 | 1.15±0.14 | −4.84±0.10 | 0.86±0.18 |
| 25 | −6.15±0.15 | 1.59±0.19 | −5.17±0.09 | 1.00±0.24 |
| 33 | −5.76±0.08 | 0.71 ±0.09 | −5.22±0.05 | 0.78±0.13 |
| 50 | −6.54±0.15 | 1.27±0.12 | −6.00±0.24 | 0.74±0.31 |
| 75 | −7.79±0.09 | 1.83±0.12 | −6.41±0.19 | 0.30±0.22 |
The energetic contribution was calculated as ΔΔGC-term=ΔGFL−ΔGtrunc with ΔΔGC-term>0 indicating that the C-terminus destabilizes protein–liposome interactions. ΔΔGC-term, energetic contribution of the C-terminal segment; ΔGtrunc, free energy of partitioning of the truncated constructs; ΔGFL, free energy of partitioning of the full-length proteins.
In order to gain additional insight into the electrostatics-mediated interactions of βS and γS with the lipid bilayer, we constructed an 11/3 helical wheel projection model for the lipid-binding regions of both proteins (Fig. 4a and b), based on a similar model constructed for αS.26,34 When projected onto the 11/3 model, the residue distribution results in polar and nonpolar residues confined to opposite sides of the helix, the two sides separated by a string of positively charged residues, as noted for the lipid-binding region of certain apolipoproteins.26 In both models, there is a stretch of threonine residues located on the hydrophobic side of the amphipathic helix, a motif identified in αS as important both for curvature-sensing ability and for anionic lipid binding.35 The N-terminus/liposome contact in both βS and γS is similarly mediated by positively charged residues lining the polar/nonpolar interface of the amphipathic helix.
Fig. 4.
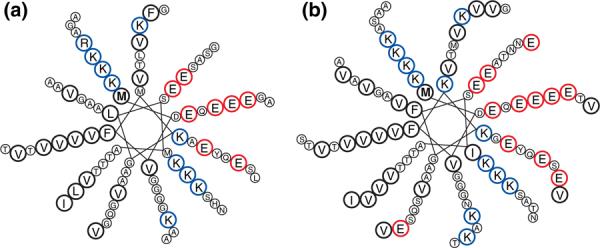
Model for topologies of βS and γS upon liposome binding. Having a high extent of sequence homology with αS and with studies confirming their interactions with model membranes, we expect the N-terminal segments of βS and γS to form a helix with a topology similar to that of αS. 11/3 helical wheel projection of the N-terminal region of βS, residues 1–84 (a), and γS, residues 1–95 (b). The boldfaced “M” residue indicates the first amino acid of each protein; blue circles designate the lysine residues delineating the interface between the solvent-facing side of the helix rich in acidic residues (red circles) and its membrane-facing side rich in highly hydrophobic residues (bold black circles).
The initial step in membrane binding of an amphipathic helix is thought to be via long-range electrostatic interactions,36 whereby the positively charged segment of the protein is attracted to negatively charged regions on the target membrane. Therefore, it is a mechanistic advantage to possess a quantitative description of this mandatory step in the protein–membrane interactive process. There is a strong dependence of the binding affinity on electrostatics, as the affinity of the proteins is highest for liposomes containing the highest anionic lipid content. However, weak interactions are observed with electrically neutral liposomes (~1 mM), suggesting the contributions of additional factors to the protein–membrane interactions. Thus, in the absence of surface charge, other major factors modulating these interactions can be the physicochemical characteristics of the interacting species,37 such as hydrophobicity,25,38 steric hindrance, presence of defects in the membranes,38–41 or folding of the protein upon binding to the liposome.42,43 In that context, it is interesting that the fact that βS lacks an 11-residue stretch, essentially hydrophobic, in its lipid-binding region does not impair its binding to POPC-only vesicles relative to that of αS and γS.
Recently, two variants of βS have been identified in two independent cases of DLB.6 Those two mutants, βS V70M and βS P123H, have been linked directly to lysosomal pathologies. Particularly, the P123H mutant transfected into murine brain caused neurodegeneration and also enhanced αS aggregation.5 To determine whether the pathogenic quality of these mutants was associated with membrane-binding affinity, we compared WT βS with the disease mutants by also monitoring the partitioning of these mutants into 50:50 POPC:POPS liposomes (Fig. 3a). There was an approximately 5-fold increase in the binding affinities of both mutants compared to WT βS (36.8±13.2μM for V70M, 43.9±12.6 μM for P123H, and 212±58μM for WT). Notably, a similar increase in binding affinity was observed for the DLB-linked αS mutant E46K relative to WT.27 While it was not unexpected that the V70M mutation located within the membrane-binding domain of βS altered the binding affinity, the increase cannot simply be explained by differences in membrane partitioning, as the energetics (ΔG)ofvaline relative to methionine are comparable.44 However, the linear side chain of methionine, as opposed to the branched valine, allows stronger van der Waals interactions with the bilayer core. Moreover, valine has a higher conformational entropy cost relative to methionine (0.39 versus 0.13kcal/mol, respectively) for localization in an α-helix.45 The combination of these factors could explain the observed increase in affinity for the V70M construct. Experimental studies based on spin-labeling analysis, as well as computational studies, have identified the V70 residue in αS as penetrating the hydrocarbon core of the bilayer,29,46 supporting that a methionine substitution could result in an enthalpy-driven increase in affinity, in agreement with the nonclassical membrane binding of an amphipathic moiety.47,48 The change in affinity due to the C-terminal P123H mutation is less intuitive, as it is not located in the region of the protein that interacts directly with the lipid bilayer. However, we have recently observed that C-terminal modifications to αS were capable of altering its affinity for lipid bilayers via a non-electrostatic mechanism49; thus, it is possible that this is a general mechanism for modulating membrane interactions for the synuclein proteins. The noted increase in lipid-binding affinity for both disease mutants may be relevant to their observed ability to enhance synucleopathies and to their reported toxic gain of function characterized by a higher propensity to self-aggregate both in vivo and in vitro.5,50,51 Indeed, numerous studies have looked into the role of lipid bilayers, or lipids in general, which are readily accessible in the intracellular environment, in fostering protein aggregation.52–55
Variation of liposome size
Membrane curvature is an important parameter of the lipid membrane that often influences interactions between amphipathic proteins and lipid bilayers.26,40 The ability of the lipid membrane to form curved surfaces is exploited in various biological processes, such as protein transport, which requires high-curvature transport vesicles, or vesicle fusion and fission.37 We determined the sensitivity of βS and γS to liposome curvature by monitoring the effect of liposome size on the protein–membrane interactions by comparing binding to small (~46 nm diameter) relative to large (~100 nm diameter) liposomes made of 50:50 POPC:POPS. Both βS and γS showed similar affinities to the 100-nm liposomes (212±58 μM and 199.8±69.1μM, respectively) and to the 46-nm liposomes (7.9±0.2μM and 15.5±1.9μM, respectively), with a significantly larger affinity for the small liposomes (Fig. 3a) as has been observed previously for αS.27 We do note that small sonicated liposomes are not directly comparable to the larger liposomes formed by extrusion because they are nonequilibrium structures that may be problematic for thermodynamic measurements.56,57 We found no evidence of large-scale instability such as liposome fusion or aggregation in our assays (Supplemental Data and Fig. S2); however, these approaches are not sensitive to variable lipid packing found in these liposomes. Thus, the higher affinity may be attributed not only to an intrinsic preference for more highly curved lipid surfaces but also due to the nonequilibrium nature of the sonicated liposome bilayer. The synucleins are primarily presynaptic proteins, where their function may potentially involve cellular events requiring membrane remodeling, which often results in areas of high curvature. In fact, a study recently reported on the ability of αS and βS to generate membrane curvature and yield tubules and vesicles that have a similar morphology to structures produced by amphiphysin, which is a known curvature-inducing protein with an active role in endocytosis.58
Effect of membrane defects and fluidity
Previous studies have proposed that packing defects in the membrane promote protein–membrane interactions by providing additional binding sites to the protein moieties.38,59 We monitored the effect of membrane defects by introducing the neutral nonlamellar-prone 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) into POPC: POPS mixtures. Because of their unique geometry, phosphatidylethanolamine lipids tend to cause defects in the bilayer. This results in changes in monolayer curvature or surface tension, which is required in regions of the membrane undergoing remodeling,60–62 like during vesicle fusion or fission. The anionic content of the liposomes was held constant at 25%, while the POPC:POPE molar ratio was varied, with their total ratio maintained at 75%. Although the resulting higher propensity to defects is expected to favor the amphipathic helix insertion within the bilayer,61,63,64 increasing POPE concentration only resulted in a minor increase in binding affinity of βS and γS (Fig. 5a). This lack of or minor dependence on phosphatidylethanolamine has also been observed for other peripheral proteins, including Gβ or GRK2.61
Fig. 5.
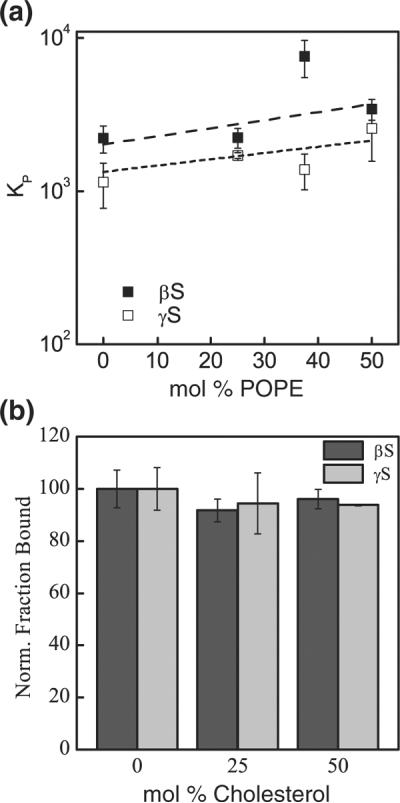
Variation of membrane fluidity. (a) Binding to POPE-containing liposomes. Broken lines, linear fits of the corresponding data. βS, filled squares; γS, open squares. (b) Binding to cholesterol-containing liposomes. The interactions of βS and γS with 50:50 POPC:POPS is monitored at different molar ratios of cholesterol. Data are presented as the ratio of the amount of protein bound in the presence of cholesterol to that bound in the absence of cholesterol. Dark-gray bars, βS; light-gray bars, γS.
Like membrane defects, membrane fluidity has also been suggested to influence protein–membrane interactions. Particularly, a preferential binding of αS for gel-phase lipids has been observed in previous studies.26,27,38,39 Here, cholesterol, a ubiquitous component of biological membranes,65,66 was used to modulate the fluidity of the bilayer and to monitor the resulting effect on protein–membrane interactions. Starting with 50:50 POPC:POPS liposomes, cholesterol was directly substituted for POPC over a range of 25–50% cholesterol molar ratio. Both βS and γS were relatively insensitive to the presence of cholesterol in the lipid bilayer (Fig. 5b). Similarly, we did not observe any change in affinity in the presence of cholesterol when the cholesterol was added to a constant background of 100% POPS or 50:50 POPC:POPS liposomes (Fig. S7). Lastly, we evaluated the effect of increasing 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG), which forms a gel phase at room temperature in fluid-phase 1,2-dioleoyl-sn-glycero-3-phosphoglycerol bilayers. Neither βS nor γS exhibited significant changes in binding affinity with increasing DPPG content in the membrane (Fig. S8).
Cholesterol, when introduced in a fluid-phase bilayer, reduces its permeability by intercalating between lipid molecules thereby decreasing the flexibility of the surrounding acyl chains.61 The insensitivity of both proteins to increasing cholesterol content interactions would suggest a superficial insertion of these proteins within the bilayer upon contact. Cholesterol can inhibit the formation of nonlamellar structures favoring defects in the bilayer38,67,68 and, as such, would be expected to reduce the partitioning of the proteins into the liposomes.
Conclusions and Biological Significance
Here, we have investigated the effects of several factors on the interactions of βS and γS with model membranes (summarized in Table 3). Similar to αS, an important factor in initiating contact between βS and γS with the model membranes was surface charge, although we also observed low affinity binding to neutral membranes. Those observations are in line with the cytosolic localization of the synucleins and the fact that the cytosol-facing leaflet of biological membranes is particularly rich in negatively charged phospholipids. We established and quantified the modulation of the synuclein–membrane interactions by lipid composition and membrane curvature, both properties commonly identified as important in mediating such interactions, as well as in facilitating the reversible association of proteins with biological membranes. Though a common link among amphipathic helices is their ability to bind lipid membranes, how they respond to variations in parameters such as surface charge density and liposome size differs widely.
Table 3.
Summary of the effects of membrane and protein properties on protein–liposome interactions
| Electrostatic/physicochemical changes | βS | γS |
|---|---|---|
| Decreasing anionic lipid content | Decreases binding | Decreases binding |
| Increasing membrane curvature | Increases binding | Increases binding |
| Increasing POPE content | Minor effect | Minor effect |
| Increasing cholesterol content | No effect | No effect |
| Increasing gel-phase content | No effect | No effect |
| Truncating C-terminus of protein | Increases binding | Increases binding |
The interactions of βS and γS with biological membranes are important to understand the physiological functions of these proteins, as well as their potential contribution to the pathogenesis of various neurological disorders. When compared to the MARCKS peptide, which has a +13 charge and whose function involves direct interactions with negatively charged PIP2 lipids, interactions between the synuclein proteins and anionic liposomes are considerably weaker (for 25% POPS liposomes, Kd,app~600μM for synuclein compared to Kd,app~0.1μM for MARCKS).25 Larger membrane-binding proteins actively involved in a wide variety of cellular functions, including cell signaling processes, have also been observed to bind with much higher affinities, for example, in the order of 0.01μM for annexin V or 0.6μM for Src.69–72 However, a more relevant comparison may be with other amphipathic peripheral membrane-binding proteins. For example, an amphipathic helix from an N-BAR domain binds with similar affinities to anionic lipids as we have observed for βS and γS.73 These relatively low affinities emphasize the transient nature of the protein–membrane interactions for the synuclein proteins, which is consistent with the primarily cytosolic localization of these proteins. Yet, the fact that the synucleins are soluble proteins that possess lipid-binding abilities could suggest a role in signaling processes, where transitory associations with biological membranes are often required. There are a number of peripheral membrane-binding proteins known to be involved in signal transduction. However, in contrast to the synucleins, they bind readily to liposomes made of only neutral phospholipids and they showed no evidence of binding to negatively charged phospholipids.61 Still, both βS and γS bind preferentially to smaller liposomes (~46nm), which mimic the size of transport vesicles inside the cells. While it is somewhat speculative, there is growing evidence to support a role for βS and γS in signaling events such as neurotransmitter packaging and release, as suggested by their potential role in modulating monoamine transporters74,75 and their involvement in various signal transduction pathways.8,76–78 Further studies need to be performed to establish the role of the synucleins in such processes. Lastly, our data indicate that the βS disease mutants, V70M and P123H, similar to DLB-associated E46K αS, have an increased affinity for lipid membranes relative to WT, which could be a contributing factor to the stimulated neurodegeneration observed in model systems carrying those mutations.
Materials and Methods
Expression, purification, and labeling of recombinant (β/γ)-synuclein
βS and γS were expressed as His-tagged constructs with a tobacco etch virus cleavage site separating the purification tag from the protein sequence from Escherichia coli as described previously.28 The cell lysates were incubated with Ni-NTA agarose resin (Qiagen, Valencia, CA) and then washed with increasing concentrations of imidazole to elute the proteins. The His-tag was removed by incubation with tobacco etch virus protease at a 2:1 protein-to-protease ratio at room temperature overnight. Cleaved proteins were separated from uncleaved proteins by a second incubation with the Ni-NTA resin. The cleaved protein was collected in the flow-through of the column and was separated from remaining contaminants on a size-exclusion column (Superdex 75, GE Healthcare). Both labeling and disease mutants were produced using a protocol based on the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). For site-specific labeling with the thiol-reactive Alexa 488 maleimide fluorophore, we introduced a cysteine residue at residue 9 for both proteins and all variants studied here. Although a common polymorphism of γS (V110E) has been reported previously,79 the recombinant γS used in this study retained a valine residue at position 110.
Unilamellar liposomes preparation
The lipids used were POPS, POPC, POPE, 1,2-dioleoyl-sn-glycero-3-phosphoglycerol, DPPG, and cholesterol (Avanti Polar Lipids, Alabaster, AL). Lipids were dissolved to 10–20mg/mL in chloroform and stored at −20°C in glass vials. Aliquots were removed and mixed at the desired ratio in a glass vial, dried to a thin film under a nitrogen stream, and any residual chloroform was removed by placing the samples in a vacuum-evacuated desiccator for at least 2 h. The resulting lipid film was rehydrated in Mops buffer [20mM Mops, 147mM NaCl, and 2.7mM KCl (pH7.4)] for 1h at room temperature. Liposomes were prepared by extruding the rehydrated samples 21 times through two stacked 50-nm polycarbonate membranes (Whatman Nuclepore Track Etch, Maidstone, UK) in a Liposofast extruder (Avestin, Ottawa, Canada), resulting in average liposome diameters of ~100–130nm. Solutions containing DPPG were heated 5–10°C above its melting temperature (Tm=41°C) before extrusion.
To prepare the small liposomes, we first rehydrated the lipid film in 500μL of working buffer for 1h at room temperature. The total volume of the lipid solution was brought to 5 mL for a total lipid concentration of 500μM. The mixture was sonicated on ice, with a flat tip probe, for 30 min, pulsing and pausing for 5 s. After sonication, an 8000g spin for 15min at 4°C was used to pellet large aggregates, the small liposomes remaining in the supernatant. The stability of the small liposomes during the course of our experiments was monitored using FCS (Supplemental Data and Fig. S2).
For all liposome preparations, the total lipid concentrations were determined by assaying the lipid solutions for inorganic phosphate.80,81 All lipid compositions are given as relative percentages of the different components, based on molar concentrations. The size of the liposomes was determined by dynamic light scattering (Supplemental Data and Table S1).
Fluorescence correlation spectroscopy
FCS measurements were taken using a laboratory-built instrument based around an inverted microscope using an Olympus IX71 microscope as described previously,27 using a 488-nm-wavelength laser as the excitation source. All measurements were made in eight-well chambered cover glasses (Nunc, Rochester, NY) passivated with polylysine-conjugated polyethylene glycol to prevent protein adsorption to chamber surfaces. Before their addition to the liposomes, the protein samples were incubated for 5 min with tris(2-carboxyethyl)phosphine to avoid cysteine-mediated dimer formation that could result from incomplete labeling of the proteins. The protein–liposome samples were allowed to equilibrate for 5 min before each measurement. All measurements were taken under equilibrium conditions (Figs. S5 and S6) at room temperature (20.5±0.1 °C) in Mops buffer (pH 7.0), 140 mM NaCl, and 2.7 mM KCl, in the presence of 2 mM tris(2-carboxyethyl)phosphine. The reported data are from at least two separate binding measurements collected under similar conditions. Twenty-five25 autocorrelation curves, at 10 s each, were measured for each sample and later averaged. Each average curve (Fig. 2a) was fitted to Eq. (1), which describes the diffusion of two distinct species in a three-dimensional Gaussian volume82,83:
| (1) |
where N is the number of proteins, f is the fraction of protein free in solution, and α is the average brightness of the liposomes relative to a single protein. As has been noted,25,27,84 in general, the values of α derived from fitting do not appear to accurately reflect the actual brightness of the liposomes. Thus, all binding curves were derived from the free protein contribution to the autocorrelation curve, which is not directly dependent upon α (additional discussion is in Supplemental Data).
The structure factor, s, which is the ratio of the radial to axial dimensions of the focal volume, was determined as a floating parameter from measurements of Alexa 488 hydrazide (s=0.2) and fixed for all subsequent fitting. The diffusion times (τD), τfree and τbound, for free protein and liposome-bound protein, respectively, were measured independently and obtained by fitting the autocorrelation curves to Eq. (2), which describes the diffusion of one species in the focal volume. Briefly, ten 10-s-long autocorrelation curves were measured for a solution containing 100nM protein, averaged, and fit with Eq. (2) to yield the translational diffusion time of the free protein (τfree). The diffusion time of the slow-diffusing liposomes (τbound) was determined with 100nM protein under saturating lipid concentrations, collecting twenty-five 10-s-long autocorrelation curves.
| (2) |
The averaged autocorrelation curve for each liposome concentration was fitted locally using Eq. (1), with fixed values of s and τfree and all other parameters (N, α, and τbound) floating within set limits based on the experimental conditions.
Binding analysis
The fraction of free protein f, independent of the brightness term α, generated from Eq. (1) was used to calculate the fraction of protein bound (1–f) (Supplemental Data). We used (1–f), represented as y in Eq. (3), to construct binding isotherms (Fig. 2b). The binding curves were fitted to Eq. (3), yielding an apparent dissociation constant, Kd,app (Supplemental Data and Fig. S4).
| (3) |
x is the concentration of accessible lipids, which is equal to slightly more than half of the total lipid concentration, considering that the liposomes were extruded through 50-nm pores, as well as taking into account the dimensions of the bilayer and the lipid molecule. Details of the calculations and fit equations were published previously.27 The Kd,app obtained was used to calculate the molar partition coefficient, KP, KP=[Kd,app×νlip×NA]−1 and where νlip represents the volume of 1 mol lipid85 and NA is Avogadro's number. The molar partition coefficient, a dimensionless parameter, then allowed the calculation of the Gibbs free energy of partitioning, ΔG, with ΔG=−RTlnKP (Supplemental Data). Repeating measurements at lipid concentrations corresponding to the region of the binding curve where 0.2<f<0.8 allowed direct calculation of KP relative to the fraction of free protein (f) and the total lipid (LTot) concentration, according to Eq. (4).
| (4) |
Another commonly used approach to calculate the free energy of partitioning is to use the mole-fraction-based partition coefficient,56 which is discussed in Supplemental Data (Table S2). The surface potential of the liposomes, Ψo, was calculated as described in Supplemental Data.
Statistical analysis
All results are reported as mean±standard error from at least two independent measurements. The standard error (δ) in Kd,app as determined from fittinginOrig in (OriginLab, Northampton, MA) was propagated in determining the error in KP (δKP), defined as δKP=δKd,app×[(Kd,app)2×νlip×NA]−1. The standard error in each Gibbs free energy was calculated as (RT/KP)×δKP. In the caseswhere the KP was determined from individual data points, the standard error was calculated as σ, where σ is the standard deviation.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health predoctoral training at the interface of Chemistry and Biology grant to V.C.D. (NIH 5T32 GM067543), the National Institutes of Health predoctoral program in Biophysics grant to V.C.D. (NIH 5T32 GM008283), and the National Science Foundation project grant to E.R. (NSF 0919853). We thank David Eliezer for providing the template plasmids for βS and γS, Andrew Miranker for helpful discussions, and Tobias Baumgart for comments on the manuscript.
Abbreviations used
- αS
α-synuclein
- βS
β-synuclein
- γS
γ-synuclein
- DLB
dementia with Lewy bodies
- FCS
fluorescence correlation spectroscopy
- DPPG
1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPS
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine
- WT
wild type
Footnotes
Supplementary Data Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2012.08.008
References
- 1.Chandra S, Fornai F, Kwon H, Yazdani U, Atasoy D, Liu X, et al. Double-knockout mice for α- and β-synucleins: effect on synaptic functions. Proc. Natl Acad. Sci. USA. 2004;101:14966–14971. doi: 10.1073/pnas.0406283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lavedan C. The synuclein family. Genome Res. 1998;8:871–880. doi: 10.1101/gr.8.9.871. [DOI] [PubMed] [Google Scholar]
- 3.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 4.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 5.Fujita M, Sugama S, Sekiyama K, Sekigawa A, Tsukui T, Nakai M, et al. A β-synuclein mutation linked to dementia produces neurodegeneration when expressed in mouse brain. Nat. Commun. 2010;1:110. doi: 10.1038/ncomms1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohtake H, Limprasert P, Fan Y, Onodera O, Kakita A, Takahashi H, et al. β-Synuclein gene alterations in dementia with Lewy bodies. Neurology. 2004;63:805–811. doi: 10.1212/01.wnl.0000139870.14385.3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto M, Rockenstein E, Mante M, Mallory M, Masliah E. β-Synuclein inhibits α-synuclein aggregation: a possible role as an anti-Parkinsonian factor. Neuron. 2001;32:213–223. doi: 10.1016/s0896-6273(01)00462-7. [DOI] [PubMed] [Google Scholar]
- 8.Pan Z-Z, Bruening W, Giasson BI, Lee VMY, Godwin AK. γ-Synuclein promotes cancer cell survival and inhibits stress- and chemotherapy drug-induced apoptosis by modulating MAPK pathways. J. Biol. Chem. 2002;277:35050–35060. doi: 10.1074/jbc.M201650200. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad M, Attoub S, Singh MN, Martin FL, El-Agnaf OMA. γ-Synuclein and the progression of cancer. FASEB J. 2007;21:3419–3430. doi: 10.1096/fj.07-8379rev. [DOI] [PubMed] [Google Scholar]
- 10.Singh VK, Zhou Y, Marsh JA, Uversky VN, Forman-Kay JD, Liu J, Jia Z. Synuclein-γ targeting peptide inhibitor that enhances sensitivity of breast cancer cells to antimicrotubule drugs. Cancer Res. 2007;67:626–633. doi: 10.1158/0008-5472.CAN-06-1820. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Perovic I, Chittuluru J, Kaganovich A, Nguyen LT, Liao J, et al. A soluble α-synuclein construct forms a dynamic tetramer. Proc. Natl Acad. Sci. USA. 2011;108:17797–17802. doi: 10.1073/pnas.1113260108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartels T, Choi JG, Selkoe DJ. α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477:107–110. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trexler AJ, Rhoades E. N-terminal acetylation is critical for forming α-helical oligomer of α-synuclein. Protein Sci. 2012;21:601–605. doi: 10.1002/pro.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertoncini CW, Rasia RM, Lamberto GR, Binolfi A, Zweckstetter M, Griesinger C, Fernandez CO. Structural characterization of the intrinsically unfolded protein β-synuclein, a natural negative regulator of α-synuclein aggregation. J. Mol. Biol. 2007;372:708–722. doi: 10.1016/j.jmb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Rivers RC, Kumita JR, Tartaglia GG, Dedmon MM, Pawar A, Vendruscolo M, et al. Molecular determinants of the aggregation behavior of α- and β-synuclein. Protein Sci. 2008;17:887–898. doi: 10.1110/ps.073181508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 17.Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- 18.Uversky VN, Li J, Souillac P, Millett IS, Doniach S, Jakes R, et al. Biophysical properties of the synucleins and their propensities to fibrillate. J. Biol. Chem. 2002;277:11970–11978. doi: 10.1074/jbc.M109541200. [DOI] [PubMed] [Google Scholar]
- 19.Sung Y-H, Eliezer D. Secondary structure and dynamics of micelle bound β- and γ-synuclein. Protein Sci. 2006;15:1162–1174. doi: 10.1110/ps.051803606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greten-Harrison B, Polydoro M, Morimoto-Tomita M, Diao L, Williams AM, Nie EH, et al. αβγ-Synuclein triple knockout mice reveal age-dependent neuronal dysfunction. Proc. Natl Acad. Sci. USA. 2010;107:19573–19578. doi: 10.1073/pnas.1005005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burré J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Südhof TC. α-Synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329:1663–1667. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivotto M, Arcangeli A, Carla M, Wanke E. Electric fields at the plasma membrane level: a neglected element in the mechanisms of cell signalling. BioEssays. 1996;18:495–504. doi: 10.1002/bies.950180612. [DOI] [PubMed] [Google Scholar]
- 23.Yeung T, Grinstein S. Lipid signaling and the modulation of surface charge during phagocytosis. Immunol. Rev. 2007;219:17–36. doi: 10.1111/j.1600-065X.2007.00546.x. [DOI] [PubMed] [Google Scholar]
- 24.Rhoades E, Ramlall TF, Webb WW, Eliezer D. Quantification of α-synuclein binding to lipid vesicles using fluorescence correlation spectroscopy. Biophys. J. 2006;90:4692–4700. doi: 10.1529/biophysj.105.079251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rusu L, Gambhir A, McLaughlin S, Rädler J. Fluorescence correlation spectroscopy studies of peptide and protein binding to phospholipid vesicles. Biophys. J. 2004;87:1044–1053. doi: 10.1529/biophysj.104.039958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of α-synuclein secondary structure upon binding to synthetic membranes. J. Biol. Chem. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 27.Middleton ER, Rhoades E. Effects of curvature and composition on α-synuclein binding to lipid vesicles. Biophys. J. 2010;99:2279–2288. doi: 10.1016/j.bpj.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eliezer D, Kutluay E, Bussell R, Jr., Browne G. Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 29.Jao CC, Der-Sarkissian A, Chen J, Langen R. Structure of membrane-bound α-synuclein studied by site-directed spin labeling. Proc. Natl Acad. Sci. USA. 2004;101:8331–8336. doi: 10.1073/pnas.0400553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human α-synuclein. J. Biol. Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 31.Ladokhin AS, White SH. Protein chemistry at membrane interfaces: non-additivity of electrostatic and hydrophobic interactions. J. Mol. Biol. 2001;309:543–552. doi: 10.1006/jmbi.2001.4684. [DOI] [PubMed] [Google Scholar]
- 32.Beschiaschvili G, Seelig J. Melittin binding to mixed phosphatidylglycerol/phosphatidylcholine membranes. Biochemistry. 1990;29:52–58. doi: 10.1021/bi00453a007. [DOI] [PubMed] [Google Scholar]
- 33.Beschiaschvili G, Baeuerle H-D. Effective charge of melittin upon interaction with POPC vesicles. Biochim. Biophys. Acta, Biomembr. 1991;1068:195–200. doi: 10.1016/0005-2736(91)90210-y. [DOI] [PubMed] [Google Scholar]
- 34.Bussell R, Jr., Eliezer D. A structural and functional role for 11-mer repeats in α-synuclein and other exchangeable lipid binding proteins. J. Mol. Biol. 2003;329:763–778. doi: 10.1016/s0022-2836(03)00520-5. [DOI] [PubMed] [Google Scholar]
- 35.Pranke IM, Morello V, Bigay J, Gibson K, Verbavatz JM, Antonny B, Jackson CL. α-Synuclein and ALPS motifs are membrane curvature sensors whose contrasting chemistry mediates selective vesicle binding. J. Cell Biol. 2011;194:89–103. doi: 10.1083/jcb.201011118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 37.Dowhan W, Bogdanov M, Mileykovskaya E. Functional roles of lipids in membranes. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. 5th Elsevier; Amsterdam, The Netherlands: 2008. pp. 1–37. [Google Scholar]
- 38.Shvadchak VV, Falomir-Lockhart LJ, Yushchenko DA, Jovin TM. Specificity and kinetics of α-synuclein binding to model membranes determined with fluorescent excited state intramolecular proton transfer (ESIPT) probe. J. Biol. Chem. 2011;286:13023–13032. doi: 10.1074/jbc.M110.204776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamp F, Beyer K. Binding of α-synuclein affects the lipid packing in bilayers of small vesicles. J. Biol. Chem. 2006;281:9251–9259. doi: 10.1074/jbc.M512292200. [DOI] [PubMed] [Google Scholar]
- 40.Nuscher B, Kamp F, Mehnert T, Odoy S, Haass C, Kahle PJ, Beyer K. α-Synuclein has a high affinity for packing defects in a bilayer membrane. J. Biol. Chem. 2004;279:21966–21975. doi: 10.1074/jbc.M401076200. [DOI] [PubMed] [Google Scholar]
- 41.Cui H, Lyman E, Voth GA. Mechanism of membrane curvature sensing by amphipathic helix containing proteins. Biophys. J. 2011;100:1271–1279. doi: 10.1016/j.bpj.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ladokhin AS, White SH. Folding of amphipathic α-helices on membranes: energetics of helix formation by melittin. J. Mol. Biol. 1999;285:1363–1369. doi: 10.1006/jmbi.1998.2346. [DOI] [PubMed] [Google Scholar]
- 43.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 44.Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc. Natl Acad. Sci. USA. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lomize A, Pogozheva I, Lomize M, Mosberg H. The role of hydrophobic interactions in positioning of peripheral proteins in membranes. BMC Struct. Biol. 2007;7:44. doi: 10.1186/1472-6807-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seelig J, Ganz P. Nonclassical hydrophobic effect in membrane binding equilibria. Biochemistry. 1991;30:9354–9359. doi: 10.1021/bi00102a031. [DOI] [PubMed] [Google Scholar]
- 48.Seelig J. Titration calorimetry of lipid-peptide interactions. Biochim. Biophys. Acta, Rev. Biomembr. 1997;1331:103–116. doi: 10.1016/s0304-4157(97)00002-6. [DOI] [PubMed] [Google Scholar]
- 49.Sevcsik E, Trexler AJ, Dunn JM, Rhoades E. Allostery in a disordered protein: oxidative modifications to α-synuclein act distally to regulate membrane binding. J. Am. Chem. Soc. 2011;133:7152–7158. doi: 10.1021/ja2009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei J, Fujita M, Nakai M, Waragai M, Watabe K, Akatsu H, et al. Enhanced lysosomal pathology caused by β-synuclein mutants linked to dementia with Lewy bodies. J. Biol. Chem. 2007;282:28904–28914. doi: 10.1074/jbc.M703711200. [DOI] [PubMed] [Google Scholar]
- 51.Fujita M, Sekigawa A, Sekiyama K, Sugama S, Hashimoto M. Neurotoxic conversion of β-synuclein: a novel approach to generate a transgenic mouse model of synucleinopathies? J. Neurol. 2009;256:286–292. doi: 10.1007/s00415-009-5246-8. [DOI] [PubMed] [Google Scholar]
- 52.Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond SJ, Smart EJ, et al. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc. Natl Acad. Sci. USA. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer β-amyloid precursor protein depends on lipid rafts. J. Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandra S, Gallardo G, Fernández-Chacón R, Schlüter OM, Südhof TC. α-Synuclein cooperates with CSPα in preventing neurodegeneration. Cell. 2005;123:383–396. doi: 10.1016/j.cell.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 55.Knight JD, Hebda JA, Miranker AD. Conserved and cooperative assembly of membrane-bound α-helical states of islet amyloid polypeptide. Biochemistry. 2006;45:9496–9508. doi: 10.1021/bi060579z. [DOI] [PubMed] [Google Scholar]
- 56.White SH, Wimley WC, Ladokhin AS, Hristova K. Protein folding in membranes: determining energetics of peptide–bilayer interactions. Methods Enzymol. 1998;295:62–87. doi: 10.1016/s0076-6879(98)95035-2. [DOI] [PubMed] [Google Scholar]
- 57.Ladokhin AS, Jayasinghe S, White SH. How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Anal. Biochem. 2000;285:235–245. doi: 10.1006/abio.2000.4773. [DOI] [PubMed] [Google Scholar]
- 58.Varkey J, Isas JM, Mizuno N, Jensen MB, Bhatia VK, Jao CC, et al. Membrane curvature induction and tubulation are common features of synucleins and apolipoproteins. J. Biol. Chem. 2010;285:32486–32493. doi: 10.1074/jbc.M110.139576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatzakis NS, Bhatia VK, Larsen J, Madsen KL, Bolinger PY, Kunding AH, et al. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat. Chem. Biol. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 60.Epand RM. Lipid polymorphism and protein–lipid interactions. Biochim. Biophys. Acta, Rev. Biomembr. 1998;1376:353–368. doi: 10.1016/s0304-4157(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 61.Escribá PV, Ozaita A, Ribas C, Miralles A, Fodor E, Farkas T, García-Sevilla JA. Role of lipid polymorphism in G protein–membrane interactions: nonlamellar-prone phospholipids and peripheral protein binding to membranes. Proc. Natl Acad. Sci. USA. 1997;94:11375–11380. doi: 10.1073/pnas.94.21.11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rinia HA, de Kruijff B. Imaging domains in model membranes with atomic force microscopy. FEBS Lett. 2001;504:194–199. doi: 10.1016/s0014-5793(01)02704-1. [DOI] [PubMed] [Google Scholar]
- 63.Soulages JL, Salamon Z, Wells MA, Tollin G. Low concentrations of diacylglycerol promote the binding of apolipophorin III to a phospholipid bilayer: a surface plasmon resonance spectroscopy study. Proc. Natl Acad. Sci. USA. 1995;92:5650–5654. doi: 10.1073/pnas.92.12.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho C, Slater SJ, Stubbs CD. Hydration and order in lipid bilayers. Biochemistry. 1995;34:6188–6195. doi: 10.1021/bi00018a023. [DOI] [PubMed] [Google Scholar]
- 65.Liu J-P, Tang Y, Zhou S, Toh BH, McLean C, Li H. Cholesterol involvement in the pathogenesis of neurodegenerative diseases. Mol. Cell. Neurosci. 2010;43:33–42. doi: 10.1016/j.mcn.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 66.Miller WL, Bose HS. Early steps in steroidogenesis: intracellular cholesterol trafficking. J. Lipid Res. 2011;52:2111–2135. doi: 10.1194/jlr.R016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cullis PR, de Kruijff B. Polymorphic phase behaviour of lipid mixtures as detected by 31P NMR. Evidence that cholesterol may destabilize bilayer structure in membrane systems containing phosphatidylethanolamine. Biochim. Biophys. Acta, Biomembr. 1978;507:207–218. doi: 10.1016/0005-2736(78)90417-0. [DOI] [PubMed] [Google Scholar]
- 68.Cheetham JJ, Wachtel E, Bach D, Epand RM. Role of the stereochemistry of the hydroxyl group of cholesterol and the formation of nonbilayer structures in phosphatidylethanolamines. Biochemistry. 1989;28:8928–8934. doi: 10.1021/bi00448a036. [DOI] [PubMed] [Google Scholar]
- 69.Tait JF, Gibson DF, Smith C. Measurement of the affinity and cooperativity of annexin V-membrane binding under conditions of low membrane occupancy. Anal. Biochem. 2004;329:112–119. doi: 10.1016/j.ab.2004.02.043. [DOI] [PubMed] [Google Scholar]
- 70.Posokhov YO, Rodnin MV, Lu L, Ladokhin AS. Membrane insertion pathway of annexin B12: thermodynamic and kinetic characterization by fluorescence correlation spectroscopy and fluorescence quenching. Biochemistry. 2008;47:5078–5087. doi: 10.1021/bi702223c. [DOI] [PubMed] [Google Scholar]
- 71.Davidson WS, Ghering AB, Beish L, Tubb MR, Hui DY, Pearson K. The biotin-capture lipid affinity assay: a rapid method for determining lipid binding parameters for apolipoproteins. J. Lipid Res. 2006;47:440–449. doi: 10.1194/jlr.D500034-JLR200. [DOI] [PubMed] [Google Scholar]
- 72.Sigal CT, Zhou W, Buser CA, McLaughlin S, Resh MD. Amino-terminal basic residues of Src mediate membrane binding through electrostatic interaction with acidic phospholipids. Proc. Natl Acad. Sci. USA. 1994;91:12253–12257. doi: 10.1073/pnas.91.25.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fernandes F, Loura LM, Chichón FJ, Carrascosa JL, Fedorov A, Prieto M. Role of helix 0 of the N-BAR domain in membrane curvature generation. Biophys. J. 2008;94:3065–3073. doi: 10.1529/biophysj.107.113118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Surguchov A. Molecular and cellular biology of synucleins. Int. Rev. Cell Mol. Biol. 2008;270:225–317. doi: 10.1016/S1937-6448(08)01406-8. [DOI] [PubMed] [Google Scholar]
- 75.Oaks AW, Sidhu A. Synuclein modulation of monoamine transporters. FEBS Lett. 2011;585:1001–1006. doi: 10.1016/j.febslet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jenco JM, Rawlingson A, Daniels B, Morris AJ. Regulation of phospholipase D2: selective inhibition of mammalian phospholipase D isoenzymes by α- and β-synucleins. Biochemistry. 1998;37:4901–4909. doi: 10.1021/bi972776r. [DOI] [PubMed] [Google Scholar]
- 77.Pan Z-Z, Godwin AK. Involvement of RHO GTPases and ERK in synuclein-γ enhanced cancer cell motility. Int. J. Oncol. 2006;29:1201–1205. [PubMed] [Google Scholar]
- 78.Gorbatyuk OS, Li S, Nha Nguyen F, Manfredsson FP, Kondrikova G, Sullivan LF, et al. α-Synuclein expression in rat substantia nigra suppresses phospholipase D2 toxicity and nigral neuro-degeneration. Mol. Ther. 2010;18:1758–1768. doi: 10.1038/mt.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lavedan C, Leroy E, Dehejia A, Buchholtz S, Dutra A, Nussbaum RL, Polymeropoulos MH. Identification, localization and characterization of the human γ-synuclein gene. Hum. Genet. 1998;103:106–112. doi: 10.1007/s004390050792. [DOI] [PubMed] [Google Scholar]
- 80.Fiske CH, Subbarow Y. The colorimetric determination of phosphorus. J. Biol. Chem. 1925;66:375–400. [Google Scholar]
- 81.Chen PS, Toribara TY, Warner H. Microdetermination of phosphorus. Anal. Chem. 1956;28:1756–1758. [Google Scholar]
- 82.Rigler R, Mets U, Widegren J, Kask P. Fluorescence correlation spectroscopy with high count rate and low-background-analysis of translational diffusion. Eur. Biophys. J. Biophys. Lett. 1993;22:169–175. [Google Scholar]
- 83.Thompson NL. Fluorescence correlation spectroscopy. In: Lakowicz J, editor. Topics in Fluorescence Microscopy. Plenum Press; New York,NY: 1991. pp. 337–378. [Google Scholar]
- 84.Melo AM, Prieto M, Coutinho A. The effect of variable liposome brightness on quantifying lipid–protein interactions using fluorescence correlation spectroscopy. Biochim. Biophys. Acta, Biomembr. 2011;1808:2559–2568. doi: 10.1016/j.bbamem.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 85.Kučerka N, Tristram-Nagle S, Nagle J. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J. Membr. Biol. 2006;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


