SUMMARY
DNA methylation in mammals is highly dynamic during germ cell and pre-implantation development but is relatively static during development of somatic tissues. 5-hydroxymethylcytosine (5hmC), created by oxidation of 5-methylcytosine (5mC) by Tet proteins and most abundant in brain, is thought to be an intermediate towards 5mC demethylation. We investigated patterns of 5mC and 5hmC during neurogenesis in the embryonic mouse brain. 5hmC levels increase during neuronal differentiation. In neuronal cells, 5hmC is not enriched at enhancers but associates preferentially with gene bodies of activated neuronal function-related genes. Within these genes, gain of 5hmC is often accompanied by loss of H3K27me3. Enrichment of 5hmC is not associated with substantial DNA demethylation suggesting that 5hmC is a stable epigenetic mark. Functional perturbation of the H3K27 methyltransferase Ezh2 or of Tet2 and Tet3 leads to defects in neuronal differentiation suggesting that formation of 5hmC and loss of H3K27me3 cooperate to promote brain development.
INTRODUCTION
Methylation of cytosine at CpG sequences is an epigenetic modification linked to gene regulation and developmental processes (Suzuki and Bird, 2008). It has been assumed that patterns of 5mC display a certain level of plasticity during development (Bernstein et al., 2007; Mohn and Schubeler, 2009). This model proposes that 5mC needs to be rapidly removed by DNA demethylation processes at certain developmental stages. Mechanisms of active DNA demethylation have remained controversial, however (Ooi and Bestor, 2008; Wu and Zhang, 2010). Tet protein-mediated oxidation of 5mC resulting in the formation of 5hmC is a plausible intermediate step in a replication-independent DNA demethylation pathway (Guo et al., 2011; Tahiliani et al., 2009; Wu and Zhang, 2010). To address the potential biological significance of a putative C → 5mC → 5hmC → C methylation-demethylation pathway, analysis of 5mC and 5hmC at two temporal stages in development or in two linearly related cell types becomes essential. 5hmC is particularly abundant in mammalian brain tissue (Jin et al., 2011b; Kriaucionis and Heintz, 2009; Munzel et al., 2010; Szulwach et al., 2011; Szwagierczak et al., 2010). In this study, we investigated the in vivo patterns of 5mC and 5hmC at the developmental transition of neural progenitor cells (NPCs) from self-renewal to differentiation, comparing global changes of the two modified cytosines between NPCs and daughter neurons. We used a dual reporter strategy by generating transgenic mice in which NPCs are labeled with GFP expressed from the nestin promoter and differentiated neurons are labeled with RFP expressed from the doublecortin promoter. The use of a differentiation reporter in conjunction with a progenitor cell-specific promoter helps alleviate the problem of “carryover” of GFP from a primitive cell to progeny, thus allowing effective co-purification of NPCs and daughter neurons from the brain (Wang et al., 2011). We mapped the distribution of cytosine modifications and several key histone methylation marks during this important developmental step and studied the role of Tet proteins and the Polycomb complex in this in vivo system.
RESULTS
5hmC increases during neuronal differentiation and associates with genes important for neuron function
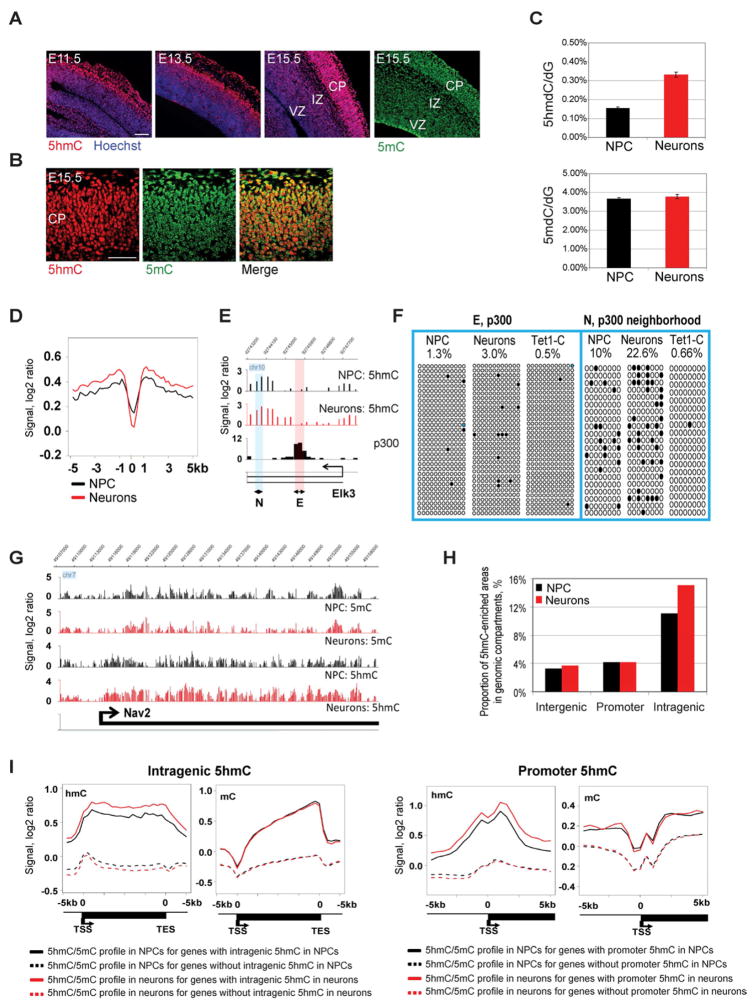
Our data revealed a dynamic change of 5hmC during neurogenesis. First, immunostaining showed a noticeable increase of 5hmC level in association with neuronal differentiation. NPCs in the ventricular zone (VZ) and young neurons in the intermediate zone (IZ) of the cortex contain lower levels of 5hmC, whereas maturing neurons in the cortical plate (CP) are enriched with 5hmC (Fig. 1A). The higher cell density in the forming cortical plate gives an apparent minor increase for staining of 5mC as well, but in the case of 5hmC, the difference is much stronger (and consistent in multiple sections). 5mC levels did not show an obvious reduction as might be anticipated if 5hmC acts as an intermediate leading to substantial DNA demethylation (Fig. 1A). In the CP, 5mC appeared to be present in smaller speckles, most likely indicating pericentromeric heterochromatin, whereas 5hmC was distributed throughout nuclei as a euchromatic mark (Fig. 1B). Second, LC-MS/MS quantification of cytosine modifications in NPCs and neurons isolated using the dual reporter approach indicated a doubling of 5hmC levels in neurons in comparison to NPCs (p<0.0001, t-test), whereas 5mC levels remain unchanged (p=0.22, t-test) (Fig. 1C). Next, we performed antibody-based 5hmC immunoprecipitation (hMeDIP) combined with mouse whole-genome tiling array analysis to examine the gene-specific distribution of 5hmC during neurogenesis. We did not see a bias of hMeDIP towards CA-containing repeat sequences as was previously suggested (Matarese et al., 2011) (Fig. S1A), and 5hmC profiling by hMeDIP or by T4 glycosyltransferase-based 5hmC enrichment (Song et al., 2011) gave similar data (Fig. S1B and S1C). For several DNA regions, hMeDIP patterns were also confirmed by Tet-assisted bisulfite (TAB) sequencing (Fig. 1F; see also Fig. 3A), which is able to distinguish between 5hmC and 5mC (Yu et al., 2012).
Figure 1. Global changes of 5hmC during neuronal differentiation.
A. Immunohistochemistry staining of mouse brain from the start of neurogenesis (E11.5) to a peak stage of neurogenesis (E15.5) with anti-5hmC or anti-5mC antibody. 5hmC level in the cortex increases with neuronal differentiation, whereas 5mC level shows little change. VZ, ventricular zone; IZ, intermediate zone; CP, cortical plate; Scale bars indicate 100 μm.
B. Co-staining of E15.5 mouse brain with anti-5hmC and anti-5mC antibodies. CP, cortical plate. Scale bars indicate 100 μm.
C. LC-MS/MS quantification of 5hmdC and 5mdC in neural progenitor cells and neurons. Levels of 5mdC and 5hmdC are represented relative to dG levels in each cell type.
D. 5hmC is depleted at enhancers (p300 sites) genome-wide. The composite profiles of 5hmC at p300 sites and their flanking areas from −5 kb to +5 kb are shown. The location of p300 sites was previously determined in mouse embryonic forebrain (Visel et al., 2009).
E. Representative snapshot of missing 5hmC at a p300 binding site in the Elk3 gene. The locations of regions E and N analyzed by TAB sequencing are indicated.
F. TAB sequencing of the intragenic Elk3 p300 binding site and flanking region in NPCs, neurons and non-glycosylated control with DNA from neurons (Tet1-C). The percentage of unconverted cytosines (5hmC) after TAB sequencing is indicated. Black circles represent hydroxymethylated CpGs; open circles represent unmethylated CpGs. CpGs with undefined methylation status were marked with blue color.
G. Snapshots of 5hmC and 5mC profiles in NPC and differentiating neurons in a representative genomic region encompassing the neuronal differentiation gene Nav2.
H. Proportion of 5hmC-enriched areas in genomic compartments during neuronal differentiation. The percentage of 5hmC-covered areas in genomic compartments was determined as the total length of 5hmC-enriched sequences in the compartment divided by the total length of the compartment.
I. Composite profiles of 5hmC and 5mC patterns in genes characterized by 5hmC enrichment or lack of 5hmC enrichment in gene body and in promoter regions. 5-hmC-enriched genes were identified by a sliding window approach (see Methods). The black solid lines indicate average levels of 5hmC or 5mC in NPCs for genes with 5hmC enrichment detected in NPCs. The black dotted lines show average levels of 5hmC or 5mC in NPCs for genes without 5hmC enrichment in NPCs. The red solid lines reflect average levels of 5hmC or 5mC in neurons for genes with 5hmC enrichment in neurons. The red dotted lines show average levels of 5hmC or 5mC in neurons for genes without 5hmC enrichment in neurons.
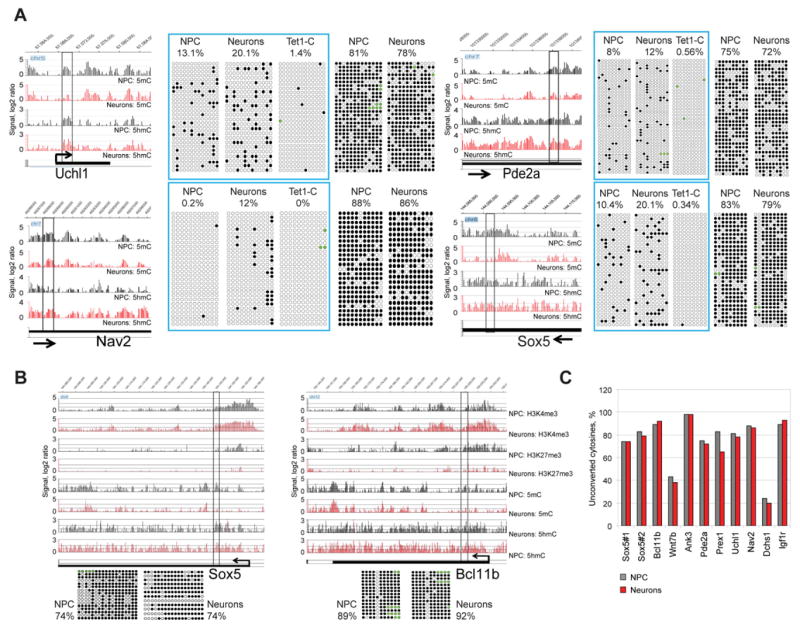
Figure 3. 5hmC gain is not linked to DNA demethylation.
A. Detection of 5hmC changes and changes of unmodified cytosines during neuronal differentiation by single-base resolution analysis. Four intragenic regions were analyzed by TAB sequencing and regular bisulfite sequencing in NPCs and neurons. Snapshots indicate patterns of 5hmC and 5mC in NPCs and neurons in the analyzed regions and surrounding areas. The analyzed regions and percentage of unconverted cytosines after TAB sequencing and bisulfite sequencing are shown. TAB data for specific 5hmC sites are framed and contain a control of 5mC/5hmC conversion by Tet1 (Tet1-C). Tet1-C is unglycosylated DNA from neurons after TAB sequencing. Black circles represent methylated or hydroxymethylated CpGs; open circles represent unmethylated CpGs. CpGs with undefined methylation status were marked with green color.
B. Bisulfite sequencing analysis of two DNA regions located in the Sox5 and Bcl11b genes, which become activated during neuronal differentiation. Snapshots indicate changes of epigenetic profiles associated with gene activation. The analyzed regions and percentage of unconverted cytosines after bisulfite sequencing are indicated. Black circles represent methylated or hydroxymethylated CpGs; open circles represent unmethylated CpGs. CpGs with undefined methylation status were marked with green color.
C. Summary of bisulfite sequencing for 11 DNA regions in NPCs and neurons. For each gene, the percentage of unconverted cytosines (5mC and 5hmC) is indicated.
In light of previous reports showing enrichment of 5hmC at enhancers in ES cells (Yu et al., 2012), we analyzed the 5hmC pattern at p300 binding sites previously mapped in mouse forebrain at embryonic day 11.5 (Visel et al., 2009). It was shown that p300 binding accurately predicts enhancers in vivo (Visel et al., 2009). In contrast to ES cells, cortical NPCs and neurons show an absence of 5hmC at p300 sites (Fig. 1D, 1E, 1F and Fig. S1D). However, sequences adjacent to p300 sites are characterized by 5hmC occupancy, which becomes enhanced during differentiation. The absence of 5hmC at p300 sites was confirmed by TAB sequencing for two p300 binding sites, an intragenic site in the Elk3 gene and a site located upstream of the Sox5 promoter (Fig. 1F and Fig. S1D). As expected, p300 sites are associated with an increased CpG density in comparison to surrounding areas (Fig. S1E).
The hMeDIP data revealed enrichment of 5hmC at promoters and gene bodies (Fig. S1F). We observed an increase of 5hmC signal, mostly in intragenic regions, during neuronal differentiation (Fig. 1G–I) but little change of 5mC (Fig. 1I). Levels of 5hmC are clearly dependent on 5mC because sequences that are lacking 5hmC also have very low levels of 5mC (dotted lines in Fig. 1I). The data also indicate that the profiles of 5mC and 5hmC are similar towards the 3′ends of genes but show distinctly opposite trends near promoters (Fig. 1I).
We found that 5hmC levels at promoters or intragenic regions are differentially correlated with CpG density and gene activity. Genes with highest intragenic CpG density are enriched with 5hmC in the gene body (Fig. S2A and S2B) whereas genes with moderate or low CpG density at the promoter and low transcriptional activity are characterized by frequent accumulation of 5hmC near the promoter (Fig. S2C and S2D).
We inspected the genes strongly enriched with intragenic 5hmC. The intragenic 5hmC-rich genes included 2782 genes and 3879 genes in NPC and in neurons, respectively; 1988 genes were common to both (Fig. S2E, Table S1). Intragenic 5hmC–enriched genes are associated with higher transcript levels than the rest of the genes, a trend that becomes more obvious in neurons (Fig. S2F). This group of genes shows strong enrichment in genes expressing in brain (Fig. S2G) and includes many genes critical for neuronal differentiation, migration or axon guidance, for example, Nav2 (Fig. 1G) and Robo1, Dab1, Dclk1, Tbr1, Sox5, Bcl11b, Satb2, Myt1l, Wnt7b, Cdk5r1, Rab3a and Efna3 (Table S1, Table S2 and Fig. S2H).
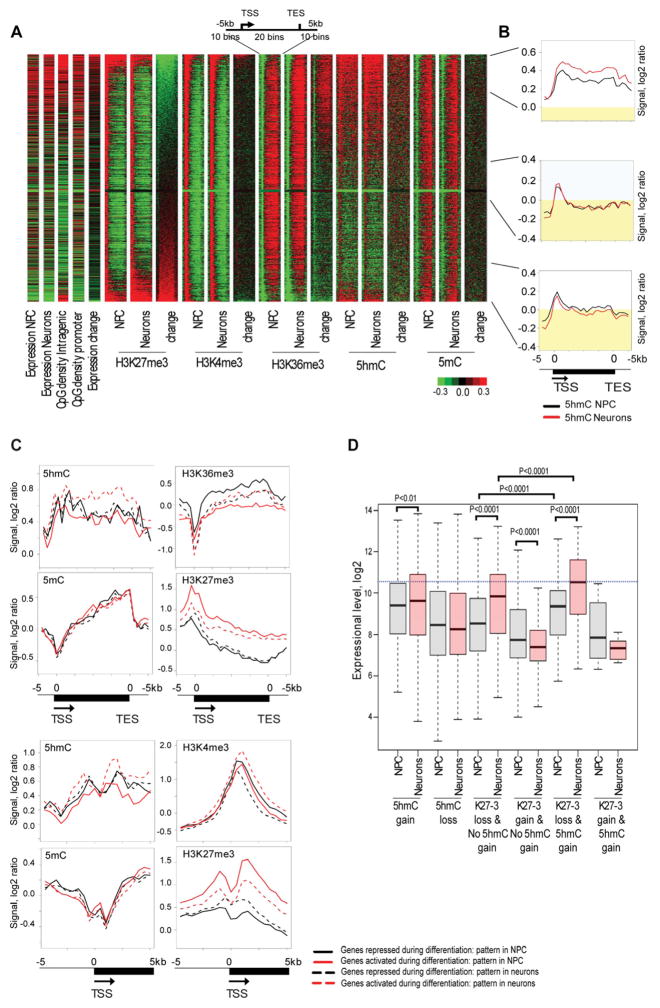
Changes of 5hmC are negatively correlated with changes of H3K27me3
To evaluate whether and how 5hmC dynamics may work in concert with chromatin modifications during neurogenesis, we profiled H3K4me3, H3K36me3 and H3K27me3 histone marks. Recently, a negative crosstalk between H3K27me3 and 5mC was described where loss of 5mC caused by Dnmt3a knockout resulted in accumulation of H3K27me3 (Wu et al., 2010). Our heat-maps and composite profiles show that the 5mC mark did not change significantly with gain and loss of intragenic H3K27me3 during neuronal differentiation (Fig. 2). We thus hypothesized that a negative association exists for H3K27me3 and 5hmC, and this was evident when sorting genes by changes of intragenic H3K27me3 during neuronal differentiation; loss of H3K27me3 is associated with gain of 5hmC in gene bodies (Fig. 2A). Composite profiles of the 15% of genes with the most intensive loss of intragenic H3K27me3 during differentiation detected a strong increase of intragenic 5hmC (Fig. 2B). Conversely, gain of intragenic H3K27me3 is linked to a loss of 5hmC in gene bodies.
Figure 2. Gain of intragenic 5hmC is associated with loss of H3K27me3 during neuronal differentiation.
A. Heatmap analysis of 5mC, 5hmC and histone methylation marks. The heat-map for chromatin modifications and their changes during neuronal differentiation was generated for genes larger than 2 kb and represents a region containing the gene body and the surrounding area (−5 kb to [gene body] to +5 kb). The heat-map contains information about expression in NPCs, expression in neurons, intragenic CpG density, promoter CpG density, expression changes during neuronal differentiation, histone modifications, 5hmC and 5mC patterns for the analyzed cell type and changes of these epigenetic marks during neuronal differentiation (neurons versus NPC). All analyzed genes were sorted by intragenic H3K27me3 changes. Green color indicates a low level or loss, black represents no change and red specifies an increase or high level of a mark. The top 15% of genes with the greatest loss of intragenic H3K27me3, 15% of genes with an intermediate state of H3K27me3 changes and top 15% of the genes with highest gain of intragenic H3K27me3 during neuronal differentiation are indicated on the right.
B. Composite profiles of the 5hmC mark in the top 15% of genes with greatest loss of intragenic H3K27me3, 15% of genes with intermediate state of H3K27me3 changes and top 15% of the genes with highest gain of intragenic H3K27me3 during neuronal differentiation (from top to bottom). The black line indicates 5hmC levels in undifferentiated cells and red color represents 5hmC in neurons. Yellow indicates 5hmC signal below zero.
C. Epigenetic changes associated with differential expression of genes marked by 5hmC. This analysis was done for genes, which have 5hmC enrichment in the gene body and/or promoters and belong to the top 25% activated or top 25% repressed genes during neuronal differentiation. Composite profiles were generated for promoter regions (−5 kb to TSS to +5 kb) and entire genes (−5 kb to TSS to gene body to TES plus 5 kb). Red solid lines indicate the status of epigenetic marks in NPCs for genes, which are activated during neuronal differentiation. Red dotted lines reflect the status of epigenetic marks in neurons for genes, which are activated during differentiation. Black solid lines indicate the status of epigenetic marks in NPCs for genes, which are repressed during differentiation. Black dotted lines reflect the status of epigentic marks in neurons for genes, which are repressed during differentiation.
D. Gain of intragenic 5hmC and loss of H3K27me3 characterizes genes activated during neuronal differentiation. Gene expression levels in NPCs and in neurons were plotted for six groups of genes: genes which gained intragenic 5hmC during neuronal differentiation, genes which lost intragenic 5hmC, genes which lost H3K27me3 at the TSS and its adjacent intragenic region (−0.5 kb<TSS<4.5 kb) and did not gain 5hmC, genes which gained H3K27me3 at the TSS and its adjacent intragenic region and did not gain intragenic 5hmC, genes which gained intragenic 5hmC and lost H3K27me3 at the TSS and its adjacent intragenic region and genes which gained intragenic 5hmC with simultaneous gain of H3K27me3 at promoters. P-values for significant differences between groups (t-test) are shown.
Differential expression of many genes during neural development is characterized by epigenetic switches associated with H3K27me3, H3K4me3, H3K36me3 and 5hmC
Loss of H3K27me3 and gain of intragenic 5hmC appear to be features of many genes that become activated and are important for the progression of neuronal differentiation. Composite profiles for the most activated and repressed genes during neuronal differentiation showed that gene activation is often linked to decrease of H3K27me3 in gene bodies and promoters, gain of H3K4me3 at promoters and gain of intragenic 5hmC (Fig. 2C). Many neuronal differentiation regulators including Sox5, Bcl11b, Wnt7b, Ank3, Mir124a and Myt1l undergo an epigenetic switch, which is characterized by a loss or reduction of promoter bivalent status due to decrease of H3K27me3 occupation at promoters (Table S1). On the other hand, gene repression is associated with increase of H3K27me3 at the TSS, decrease of H3K4me3 levels at promoters and loss of H3K36me3 in gene bodies (Fig. 2C). Accumulation of the H3K27me3 mark is mainly linked to silencing of genes responsible for maintaining an undifferentiated state such as Sox2, Pax6, Smad3, Rest, Jag1, Wwtr1, Prdm16, Nr2e1, Hes1, Notch1, Ccnd1, Rhoc and Bmp7 (Table S3). Analysis of genes gaining intragenic 5hmC indicated a link to neuronal differentiation and axonogenesis (Table S3). In addition, transcriptional analysis revealed that accumulation of intragenic 5hmC together with loss of H3K27me3 is associated with the most significant gene activation during neuronal differentiation (Fig. 2D).
Gene activation is associated with an increase of 5hmC in gene bodies but no evidence for substantial DNA demethylation
To evaluate if 5hmC accumulation is associated with demethylation of cytosines, we performed bisulfite sequencing analysis on 11 regions that were abundant with 5hmC and apparently lost some 5mC during neuronal differentiation (Fig. 3 and Fig. S3). Bisulfite sequencing cannot distinguish between 5mC and 5hmC but distinguishes both modified cytosines from cytosine (Huang et al., 2010; Jin et al., 2010). We did not see a substantial conversion of 5mC or 5hmC into C in 10 out 11 examined regions, where nine regions are located in genes (Nav2, Sox5, Bcl11b, Wnt7b, Ank3, Pde2a, Prex1 and Uchl1), which become activated during neuronal differentiation. Most (8/11) of the analyzed DNA regions indicated no change of unconverted cytosine or minimal loss, below 5% of the total analyzed CpGs, and 2 fragments were associated with increase of modified cytosine between 3–4% (Fig. 3C). In order to determine the exact frequency of 5hmC at selected loci, we performed TAB sequencing of 5 intragenic regions located in the Sox5, Nav2, Uchl1, Pde2a and Elk3 genes (Fig. 1E, N region) and Fig. 3A). This data indicated that 5hmC frequency is indeed doubling at some intragenic regions and reaches up to 20% of all CpGs in the analyzed regions and 25% of modified cytosines (5hmC level according to TAB sequencing divided by number of all modified cytosines detected by regular bisulfite sequencing). This data indicated that the rise of 5hmC can have a significant impact on sequence-specific 5mC levels during neuronal differentiation and also suggests substantial stability of this modification in the genome.
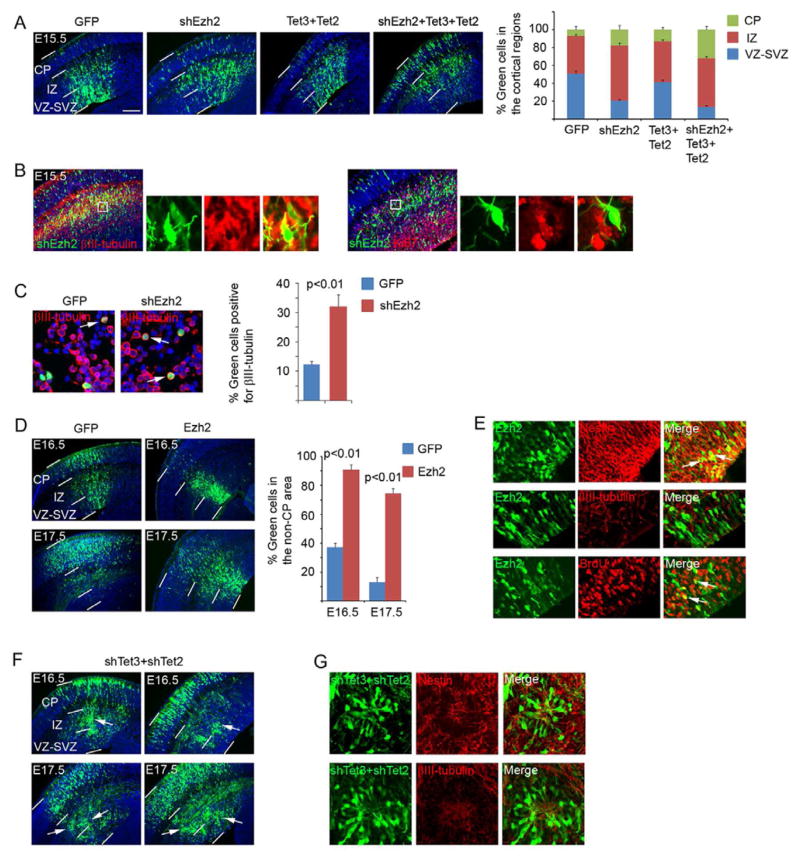
Polycomb and Tet proteins regulate the normal progression of neuronal differentiation in the cortex
We next explored the functional significance of the dynamics of 5hmC and histone marks in relation to the developmental switch of neuronal differentiation. We targeted Tet proteins and the Ezh2 subunit of the Polycomb complex. In situ hybridization showed that Tet3 followed by Tet2 are the Tet genes most highly expressed in the cortex (Fig. S4A). Their expression levels were upregulated during neuronal differentiation (Fig. S4A and S4B) in parallel with the increase of 5hmC levels (Fig. 1A). Expression of Ezh2 is reduced (RNA in situ based on genepaint.org and Fig. S4B) during neurogenesis. We thus asked whether overexpression of Tet (Fig. S4C) or inhibition of Ezh2 might be necessary for neuronal differentiation to proceed. RNA interference (RNAi) of Ezh2 in the cortex via in utero electroporation-mediated expression of an shRNA (Fig. S4E and S4F) caused more cortical cells to translocate from the VZ into the IZ and CP (Fig. 4A), an indication of differentiation of the affected NPCs (Murai et al., 2010; Qiu et al., 2008). This was supported by the observation that cells having moved into the CP were positive for neuronal marker β-III-tubulin but lacked dividing cell marker Ki67 (Fig. 4B). Consistent with the observed cell distribution, quantification of acutely dissociated cells derived from electroporated cortices showed an increase of β-III-tubulin positive cells in the shEzh2-expressing population (Fig. 4C). Over-expression of Tet3 and Tet2 caused a similar but less pronounced trend of early neuronal differentiation and induced a stronger effect when combined with knockdown of Ezh2 (Fig. 4A and Fig. S4C). We next asked whether prolonged expression of Ezh2 (Fig. S4D) or inhibition of Tet2 and Tet3 by shRNA (Fig. S4E and S4F) might prevent differentiation. We found that, in contrast to the progression of neuronal differentiation in GFP control cells, for which differentiated progeny have progressively moved into the CP (Qiu et al., 2008), over-expression of Ezh2 caused many cells to remain in the VZ and IZ (Fig. 4D). Ezh2-expressing cells were positive for nestin but negative for β-III-tubulin and some cells could incorporate BrdU (Fig. 4E), suggesting that expression of Ezh2 could block differentiation. Reduction of Tet gene expression via co-electroporation of shRNAs directed against Tet3 and Tet2 (Ito et al., 2010) did not cause an overall shift of cell distribution as in the case of Ezh2 over-expression, but revealed a distinct effect (Fig. 4F). Knockdown of Tet3 and Tet2 often (8/16 brains) led to abnormal accumulation of cell clusters along the radial axis in the IZ and VZ, whereas GFP control cells (Fig. 4D) rarely (1/14 brains) produced such a phenomenon (p=0.017; Fisher’s exact test, two-tailed). Clustered cells did not express neuronal marker β-III-tubulin and some of the cells showed expression of nestin in their processes (Fig. 4G), suggesting a defect in the progression of differentiation.
Figure 4. Functional analyses of Ezh2, Tet2 and Tet3 in the embryonic cortex.
A. DNA plasmids (shEzh2, Tet2 and Tet3 cDNAs; all carry an ubiquitin promoter-GFP expression cassette for visualization of the transfected cells) were introduced into the cortex at E13.5 via in utero electroporation. The brains were collected at E15.5 for analysis. Distributions (percentage) of transfected cells in different radial regions of the cortex were scored. Scale bar indicates 100 μm. CP, cortical plate; IZ, intermediate zone; VZ-SVZ, ventricular-subventricular zone.
B. Immunostaining of the brains electroporated with shEzh2 for βIII-tubulin or Ki67 expression.
C. Cortical cells derived from E15.5 brains (electroporated at E13.5) were plated on poly-D-lysine-coated coverslips for 2 hours, fixed, and stained for βIII-tubulin. Arrows indicate examples of positive cells. Percentage of βIII-tubulin positive green cells was scored.
D. Expression plasmids carrying Ezh2 cDNA were electroporated at E13.5 and the transfected brains were analyzed at E16.5 and E17.5. Percentage of cells remaining in the non-CP area (including the IZ and VZ) was scored.
E. Immunostaining of the brains electroporated with Ezh2 for nestin, β 3III-tubulin and incorporated BrdU with a 30 min pulse labeling.
F. shRNAs of Tet3 and Tet2 were co-electroporated at E13.5 and the transfected brains were analyzed at E16.5 and E17.5. White arrows indicate the clusters of cells in the cortex.
G. Immunostaining of the brains electroporated with shRNAs of Tet3 and Tet2 for nestin or βIII-tubulin.
DISCUSSION
Taken together, our data suggest that formation of 5hmC along gene bodies is associated with gene activation during neuronal differentiation. The purpose of intragenic 5mC oxidation remains unclear. One possibility is that 5hmC is more strongly antagonizing Polycomb-mediated repression and H3K27me3 formation than 5mC, thus allowing efficient gene activation to occur and to be maintained. We find that many activated genes functioning in neuronal differentiation lose the Polycomb mark near promoters and in gene bodies concomitant with a gain of 5hmC. However, it is unlikely that the two processes are always directly linked, since loss of H3K27me3 is commonly observed at promoters and just downstream of the TSS whereas gain of 5hmC occurs along the entire gene body length. In agreement with this model, our functional data suggest that Polycomb and Tet proteins may act in sequence in regulation of neurogenesis. Loss-of-function of Ezh2 promoted neuronal differentiation, whereas gain-of-function of Ezh2 did the opposite, indicating a role of Ezh2 in guiding NPCs’ decision to either self-renew or differentiate. Loss-of-function or gain-of-function of Tets did not swing the fate choice of NPCs, but appeared to affect the ability of NPCs to complete the differentiation process. Furthermore, in agreement with the functional data, we observed that prominent changes of H3K27me3 and 5hmC marks in relation to neuronal differentiation occurred in cell fate determinants or neuronal function-related genes, respectively. Thus collectively, our data suggest that Polycomb functions to regulate the switch of NPCs from expansion to differentiation while Tet proteins are involved in maintaining the proper progression of the differentiation process after its initiation.
Our data indicates that 5hmC patterns in neuronal cells are dissimilar to those in ES cells which are associated with high levels of Tet1 and Tet2 and low levels of Tet3 (Koh et al., 2011). According to TAB sequencing data for ES cells, 5hmC is abundant at enhancers (p300 sites) and underrepresented in gene bodies (Yu et al., 2012). In contrast, cortical NPCs and neurons at E15.5 are characterized by low levels of 5hmC at p300 sites and by enrichment of 5hmC in gene bodies. These facts may suggest that Tet1 may play a prominent role in 5hmC formation at enhancers in ES cells whereas Tet3 activity may be associated with intragenic 5hmC deposition during neuronal development. This information also suggests that cortical NPCs and neurons are able to maintain their enhancers unmethylated by mechanisms other than 5mC oxidation or that 5hmC turnover is more rapid at neuronal enhancers.
5hmC is thought to be an intermediate in enzyme-catalyzed active DNA demethylation. However, we could not substantiate a primary role of 5hmC in demethylation during neurogenesis. It is possible that 5hmC may slowly be converted to cytosine over time during the maturation of neurons; nevertheless, it appears that 5hmC is a rather stable epigenetic mark, as previously suggested from studies of early embryo development and tumorigenesis, in which passive, replication-dependent loss of 5hmC was observed (Gu et al., 2011; Inoue and Zhang, 2011; Iqbal et al., 2011; Jin et al., 2011a). 5hmC may be recognized by specific proteins, or, alternatively, it may represent a negative signal that interferes with protein complexes that bind to either 5mC, such as methyl-CpG binding proteins (Jin et al., 2010), or to CpG-rich DNA regions in general, such as the Polycomb complex. However, one publication reported that MBD3, one MBD family member, binds to 5hmC (Yildirim et al., 2011). Moreover, a very recent publication has suggested that the methyl-CpG binding protein MeCP2, an abundant protein in the brain, can in fact bind to 5hmC and accumulates in gene bodies (Mellen et al., 2012) where it may organize a nuclease-sensitive chromatin structure at active genes. Whatever the exact mechanism will be, formation of 5hmC along gene bodies appears to be an important signal that is linked to increased expression of genes critical for the neuronal differentiation process. Defects in this pathway or even subtle aberrations could manifest themselves in neurodevelopmental or neurological disorders.
EXPERIMENTAL PROCEDURES
Purification of neural progenitors and differentiating neurons
Purification of E15.5 cortical NPCs and neurons using a double reporter strategy and characterization of the purified cells were reported previously (Wang et al., 2011). Briefly, we bred heterozygous Nestin-GFP mice with homozygous DCX-RFP mice to yield both GFP/RFP double positive and RFP single positive littermate embryos, which were used for isolating GFP+RFP- cells (NPCs) and RFP+ cells (neurons), respectively.
Immunohistochemistry
The immunohistochemistry staining was done as described previously (Jin et al., 2011a; Qiu et al., 2008). Each antibody staining was performed on multiple brain sections and the experiment was repeated one or more times. We used the following primary antibodies: anti-5hmC (Active Motif, #39769, 1:1,000); anti-5mC (Diagenode, #BI-MECY-0100; 1:200); anti-nestin rat-401 (Developmental Studies Hybridoma Bank, 1:100); anti–βIII tubulin (Sigma-Aldrich, #T 8660, 1:400); anti-bromodeoxyuridine (BrdU) (Sigma-Aldrich, #B2531, 1:500); anti-Ki67 (BD Pharmingen; #550609; 1:20).
RNA in situ hybridization and Western blot
For RNA in situ hybridization, digoxigenin-labeled cDNAs were synthesized from linearized pBluescript constructs containing C-terminal fragments of Tet1, Tet2 and Tet3 cDNAs and hybridized to embryonic forebrain coronal sections with further detection by an anti-digoxigenin AP (alkaline phosphatase) approach. For Western blot, transfected HEK cells or purified cortical cells were lysed in 2x SDS sample buffer, sonicated and boiled. Proteins were separated by SDS gel electrophoresis, transferred to PVDF membranes and incubated with Ezh2 (#39934, Active Motif), α-tubulin (T6199, Sigma-Aldrich) and rabbit anti-Tet3 antibody raised against an N-terminal 102 amino acid peptide of mouse CXXC-Tet3 fused to GST.
Electroporation and phenotype analysis
In utero electroporation-mediated functional assays and acutely dissociated cell assays were performed as previously described (Qiu et al., 2008). For overexpression, cDNAs encoding full-length Ezh2, Tet2, and CXXC-Tet3 were cloned into a CAG promoter plasmid.
shRNA efficiency screening
shRNAs were expressed under control of a mouse U6 promoter in pNUTS vector additionally containing EGFP expressed from the ubiquitin promoter. Candidate shRNAs were tested with targets cloned in psi-CHECK vector in transfected HEK293 cells, using a dual luciferase reporter assay (Promega, E1910). The sequences of shRNA for Tet transcripts were previously described by Ito and colleagues (Ito et al., 2010). The shRNA for Ezh2 (Open Biosystems) had the following sequence:
5′TTGAGTACTGTGGGCAATTTATTCAAGAGATAAATTGCCCACAGTACTCAA.
LC-MS/MS quantification of 5hmdC and 5mdC in genomic DNA
Global 5hmdC and 5mdC levels were measured by liquid chromatography tandem mass spectrometry (LC-MS/MS) using stable isotope-labeled internal standards as described previously (Jin et al., 2011a).
Profiling of methylated and hydroxymethylated cytosines
For analysis of 5mC, the methylated CpG island recovery assay (MIRA) was used as described previously (Rauch et al., 2006). Immunopreciptated and input DNA were 5′-end- phosphorylated and blunt-ended with END-It DNA end-repair kit (Epicentre Biotechnologies) with further generation of overhanging A ends by Klenow Exo- (New England Biolabs) and DNA ligation with an overhanging “T” linker (5′-GCGGTGACCCGGGAGATCTGAATTCT, 5′-GAATTCAGATC) with T4 ligase (New England Biolabs). The genome amplification procedure was done as previously described (Hahn et al., 2008). For hMeDIP, linker ligated DNA was immunoprecipitated with anti-5hmC antibody (#39769, Active Motif; Carlsbad, CA).
After genome amplification, DNA obtained after MIRA or hMeDIP were hybridized versus input DNA on Mouse ChIP-chip 2.1M whole-genome tiling arrays and for additional biological replicates on tiling arrays of mouse chr7 (NimbleGen). For 5hmC profiling by a glycosylation-based method (Song et al., 2011), we used the Hydroxymethyl Collector™ Kit (Active Motif) according to the manufacturer’s protocol. DNA was amplified and hybridized onto tiling arrays of mouse chr7 (NimbleGen).
Sequencing of modified cytosines
TAB conversion was performed by using the 5hmC TAB-Seq Kit (Wisegene) with two rounds of Tet1 oxidation. Bisulfite conversion of TAB-treated and untreated DNA was performed by using the EZ DNA methylation-Gold kit (Zymo Research) according to the manufacturer’s instructions. PCR products were cloned by using CloneJet PCR cloning kit (ThermoScientific). The efficiency of Tet1 conversion was validated by TAB sequencing of DNA from neurons without glycosylation (Tet1-C in Figure 1F and Fig. 3A).
Chromatin immunoprecipitation (ChIP)
The ChIP protocol was described previously (Hahn et al., 2008). The following antibodies were used: anti-H3K4me3 (39159, Active Motif), anti-H3K36me3 (ab9050, Abcam) and anti-H3K27me3 (07–449, Milipore). For obtaining the H3K4me3 profile, the anti-H3K4me3 antibodies were pre-blocked with an H3K9me3 peptide (Abcam). After genome amplification, immunopreciptated DNA was hybridized versus input DNA on Mouse ChIP-chip 2.1M whole genome tiling arrays (NimbleGen) and for additional biological replicates on chr7 tiling arrays.
Bioinformatics analyses of profiling data
All analyses were performed using R statistical language, except gene ontology analysis, which was performed using DAVID annotation tools, and the heatmaps were generated with Java Treeview v2.0. Refseq genes were downloaded from the UCSC mm9 annotation database. A detailed description of individual bioinformatics analyses can be found in the Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
5hmC levels increase in gene bodies of neuronal function related genes in neurogenesis;
5hmC gain is accompanied by H3K27me3 loss at promoters and gene bodies;
Gain of 5hmC is not associated with substantial DNA demethylation;
Polycomb and Tet protein together promote proper progression of neurogenesis
Acknowledgments
This work was supported by NIH grants MH094599 from NIMH (Q.L. and G.P.P) and NS075393 from NINDS (Q.L.).
Footnotes
ACCESSION NUMBERS
The accession number for the 5mC, 5hmC, H3K27me3, H3K4me3 and H3K36me3 profiling data reported in this paper is GSE38118 (Gene Expression Omnibus).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–10289. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011a;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011b;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarese F, Carrillo-de Santa Pau E, Stunnenberg HG. 5-Hydroxymethylcytosine: a new kid on the epigenetic block? Mol Syst Biol. 2011;7:562. doi: 10.1038/msb.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 Binds to 5hmC Enriched within Active Genes and Accessible Chromatin in the Nervous System. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Schubeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 2009;25:129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Murai K, Qiu R, Zhang H, Wang J, Wu C, Neubig RR, Lu Q. Galpha subunit coordinates with ephrin-B to balance self-renewal and differentiation in neural progenitor cells. Stem Cells. 2010;28:1581–1589. doi: 10.1002/stem.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Qiu R, Wang X, Davy A, Wu C, Murai K, Zhang H, Flanagan JG, Soriano P, Lu Q. Regulation of neural progenitor cell state by ephrin-B. J Cell Biol. 2008;181:973–983. doi: 10.1083/jcb.200708091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature neuroscience. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang H, Young AG, Qiu R, Argalian S, Li X, Wu X, Lemke G, Lu Q. Transcriptome analysis of neural progenitor cells by a genetic dual reporter strategy. Stem Cells. 2011;29:1589–1600. doi: 10.1002/stem.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.