Abstract
Background
Btau_4.0 and UMD3.1 are two distinct cattle reference genome assemblies. In our previous study using the low density BovineSNP50 array, we reported a copy number variation (CNV) analysis on Btau_4.0 with 521 animals of 21 cattle breeds, yielding 682 CNV regions with a total length of 139.8 megabases.
Results
In this study using the high density BovineHD SNP array, we performed high resolution CNV analyses on both Btau_4.0 and UMD3.1 with 674 animals of 27 cattle breeds. We first compared CNV results derived from these two different SNP array platforms on Btau_4.0. With two thirds of the animals shared between studies, on Btau_4.0 we identified 3,346 candidate CNV regions representing 142.7 megabases (~4.70%) of the genome. With a similar total length but 5 times more event counts, the average CNVR length of current Btau_4.0 dataset is significantly shorter than the previous one (42.7 kb vs. 205 kb). Although subsets of these two results overlapped, 64% (91.6 megabases) of current dataset was not present in the previous study. We also performed similar analyses on UMD3.1 using these BovineHD SNP array results. Approximately 50% more and 20% longer CNVs were called on UMD3.1 as compared to those on Btau_4.0. However, a comparable result of CNVRs (3,438 regions with a total length 146.9 megabases) was obtained. We suspect that these results are due to the UMD3.1 assembly's efforts of placing unplaced contigs and removing unmerged alleles. Selected CNVs were further experimentally validated, achieving a 73% PCR validation rate, which is considerably higher than the previous validation rate. About 20-45% of CNV regions overlapped with cattle RefSeq genes and Ensembl genes. Panther and IPA analyses indicated that these genes provide a wide spectrum of biological processes involving immune system, lipid metabolism, cell, organism and system development.
Conclusion
We present a comprehensive result of cattle CNVs at a higher resolution and sensitivity. We identified over 3,000 candidate CNV regions on both Btau_4.0 and UMD3.1, further compared current datasets with previous results, and examined the impacts of genome assemblies on CNV calling.
Keywords: Cattle genome, Breed, Copy number variation (CNV), Single nucleotide polymorphism (SNP)
Background
Genomic structural variation including copy number variation (CNV) has been intensively studied in human [1-4] and rodents [5-8]. Dozens of human and mouse CNV studies have demonstrated that some CNVs are associated with phenotypic traits and diseases [9-12]. Initial CNV reports also appeared in domesticated animals, including dog [13-15], cattle [16], chicken [17,18], pig [19,20], sheep, and goat [21,22]. Recent bovine CNV studies have generated several cattle CNV maps using various approaches [23-26]. In our previous study, we performed an analysis of CNV using the Bovine HapMap SNP genotyping data, including 539 animals of 21 modern cattle breeds and 6 outgroups [27]. Efforts to explore the association between cattle CNV and economical traits have been published [28,29], even though the actual functional mechanisms are not yet well defined.
CNV can be identified using various approaches, including comparative genomic hybridization (CGH) array, SNP array, and next-generation sequencing. Compared to other approaches, the advantages of SNP array include its low cost, dense coverage, and high throughput. Substantial genotyping data have been produced from genome-wide association studies, which can be directly exploited for the CNV analysis. A wide range of algorithms of CNV discovery based on SNP array has been developed, including CNVPartition, QuantiSNP [30], PennCNV [31], Birdsuite [32], Cokgen [33], and others. Reviews of the strengths and weaknesses of these algorithms have been published [34,35]. As one of the leading methods, PennCNV incorporates multiple sources of information, including total signal intensity and allelic intensity ratio at each SNP marker, the distance between neighboring SNPs, and the allele frequency of SNPs. PennCNV also integrates a computational approach by fitting regression models with GC content to overcome "genomic waves" [36,37]. Furthermore, PennCNV is capable of considering pedigree information (parent-offspring trios) to improve call rates and accuracy of breakpoint prediction as well as to infer chromosome-specific SNP genotypes in CNVs.
The availability of two alternative cattle reference genomes (Btau_4.0 and UMD3, [38,39]) has opened new avenues of cattle genome research. With the advent of next-generation sequencing, more high density SNP arrays were made commercially available, including Illumina BovineHD BeadChip with more than 750,000 SNPs (Van Tassell et al., unpublished; [40]), which is 15-fold denser as compared to the previous BovineSNP50 array. Furthermore, then-published CNV results were incorporated during the BovineHD design phase to increase its coverage in variable genomic regions.
Based on this high density BovineHD SNP array, our goals in this study were to perform high resolution CNV analyses on both Btau_4.0 and UMD3.1, to compare them with the previous results, and to examine the impacts of genome assemblies on CNV calling.
Results and Discussion
Cattle CNV identification
As described previously [27], we performed CNV calling on both Btau_4.0 and UMD3.1 assemblies. Due to mapping uncertainty, we excluded chrX and chrUn from our analysis.
On the Btau_4.0 assembly, 34,311 CNVs were detected with an average of 51 events for each animal (Table 1, Additional file 1: Table S2, Figure 1A and Additional file 2: Figure S1). The average CNV length was 39,953 bp. For subspecies/groups such as the Taurine, Indicine, Composite (Taurine × Indicine) and African breeds, the average CNV events per sample were 45, 65, 53 and 66 respectively (i.e. T:I:C:A = 45:65:53:66). Indicine and African breeds had the most CNVs identified. Within each subspecies/group, the numbers of unique CNVs ranged from 2.3 to 5.1 per sample, indicating that the majority of CNVs were shared at least by two individuals within the same subspecies/groups.
Table 1.
CNVs or CNVRs on Btau_4.0 and UMD3.1
| Subspecies/groups | Sample | Count | Unique | Gain | Loss | Total Length |
|---|---|---|---|---|---|---|
|
Btau_4.0 |
|
|
|
|
|
|
|
Taurine |
447 |
20,302 (45.4) |
1,044 (2.3) |
7,916 (17.7) |
12,386 (27.7) |
814,447,018 (40,117) |
|
Indicine |
113 |
7,352 (65.1) |
309 (2.7) |
2,595 (23.0) |
4,757 (42.1) |
266,853,291 (36,297) |
|
Taurine × Indicine |
67 |
3,569 (53.3) |
198 (3.0) |
1,508 (22.5) |
2,061 (30.8) |
159,735,770 (44,756) |
|
African Breeds |
47 |
3,088 (65.7) |
240 (5.1) |
1,248 (26.6) |
1,840 (39.1) |
129,777,675 (42,026) |
|
Total |
674 |
34,311 (50.9) |
1,791 (2.7) |
13,267 (19.7) |
21,044 (31.2) |
1,370,813,754 (39,953) |
|
CNVR |
674 |
3,346 a |
1,316 b |
986 c |
2,051 c |
142,718,107 (42,653) |
|
UMD3.1 |
|
|
|
|
|
|
|
Taurine |
434 |
32,445 (74.8) |
1,052 (2.4) |
12,419 (28.6) |
20,026 (46.1) |
1,551,624,380 (47,823) |
|
Indicine |
97 |
8,715 (89.8) |
327 (3.4) |
3,102 (32.0) |
5,613 (57.9) |
394,966,292 (45,320) |
|
Taurine × Indicine |
63 |
5,332 (84.6) |
237 (3.8) |
2,268 (36.0) |
3,064 (48.6) |
256,749,182 (48,153) |
|
African Breeds |
36 |
3,212 (89.2) |
230 (6.4) |
1,197 (33.3) |
2,015 (56.0) |
151,559,764 (47,185) |
|
Total |
630 |
49,704 (78.9) |
1,846 (2.9) |
18,986 (30.1) |
30,718 (48.8) |
2,354,899,618 (47,378) |
| CNVR | 630 | 3,438 a | 1,360 d | 1,054 e | 2,042 e | 146,905,950 (42,730) |
Numbers in parentheses are values normalized by sample counts, except in the case of the parentheses values in the "total length" column, which are average lengths normalized by CNV counts. a These numbers represent nonredundent CNVR counts. In Btau_4.0, b 1,316 out of 3,346 CNVRs were unique to one sample, 2,030 CNVRs were shared by at least 2 individuals and 179 of 2,030 multiple events had a frequency > =5%. c Besides the 986 gain and 2,051 loss CNVRs, there were 309 CNVRs containing both loss and gain events. In UMD3.1, d 1,360 out of 3,438 CNVRs were unique to one sample, 2,078 CNVRs were shared by at least 2 individuals and 230 of 2,078 multiple events had a frequency > =5%. e Besides the 1,054 gain and 2,042 loss CNVRs, there were 342 CNVRs containing both loss and gain events.
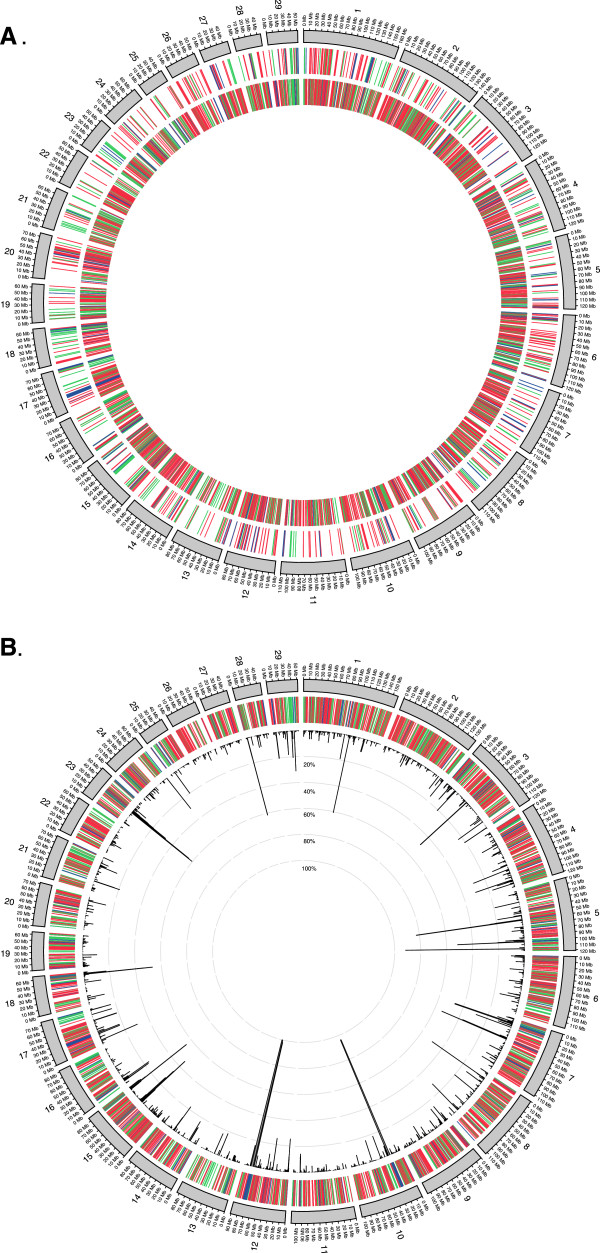
Figure 1.
Cattle copy number variations derived from SNP arrays on Btau_4.0 and UMD3.1. A. On Btau_4.0, CNV regions (682 events, 139.8 Mb) derived from BovineSNP50 assay are shown in the outer circle in green (gain), red (loss) and dark blue (both), while the inner circle shows the CNV regions (3,346 events, 142.7 Mb) derived from BovineHD assay. B. On UMD3.1, CNV regions (3,438 events, 146.9 Mb) derived from BovineHD assay are shown in the outer circle in green (gain), red (loss) and dark blue (both), while the inner circle shows their frequencies.
When we merged CNVs into nonredundent CNV regions (CNVRs), a total of 3,346 events were identified covering 142.7 Mb of polymorphic sequence, corresponding to 5.61% of the autosomal genome sequence (142.7/2,545.9 Mb) and 4.89% of the whole cattle genome (142.7/2,918.0 Mb, Table 1, Figure 1A and Additional file 3: Table S3). These CNVRs were comprised of 2,051 loss events, 986 gain and 309 both (loss and gain within the same region), ranging from 1,018 to 5,552,622 bp (Additional file 3: Table S3). Loss events are approximately 2.1-fold more common than gain events, but have smaller sizes than gain events on average (28.5 kb vs. 37.6 kb).
Furthermore, 1,316 CNVRs were found in only one sample (Unique), 2,030 CNVRs were shared at least by two animals (Multiple or more), and 179 events had a frequency >5% (Additional file 3: Table S3). These results suggest that segregating CNVs exist among these subspecies, breeds and groups, which is consistent with our earlier results [26,27].
Strikingly, the mean and median CNVR lengths were significantly shorter, 42,653 and 15,794 bp respectively, when compared to values derived from our previous low density SNP50 array study (mean: 204,965 and median: 131,179 bp). With a similar total length of ~140 Mb, our new dataset contains almost five times the number of detected CNVs (3,346) than our previous study (682), suggesting that the BovineHD array provides higher resolution and sensitivity for CNV discovery. Also the current CNV results seem to be more uniformly distributed, with more events detected in the centromere and telomere regions compared to the previous results (For example, chr10 and chr20 in Figure 1A).
We also made CNV calls using the UMD3.1 assembly (Figure 1B and Table 1). For UMD3.1, 630 animals passed the PennCNV quality filtering, 44 fewer as compared to Btau_4.0. When comparing results between Btau_4.0 and UMD3.1, more CNVs of longer length were found on UMD3.1 than those on Btau_4.0 (Additional file 1: Table S4 and Additional file 4: Table S5). However, the relative differences in CNV counts across distinct species/breeds were preserved (i.e. T:I:C:A ratio: 45:65:53:66 vs. 75:90:85:89) and the merged CNV region results (count and length) were consistent.
Compared to the Btau_4.0 results, 44.86% more CNVs (49,704) were detected within the placed autosomes and an average of 54.90% more events (79) were obtained for each sample on UMD3.1 (Additional file 1: Table S4). The average length of CNV was also 18.58% larger (47,378 bp) when using the UMD3.1 assembly coordinates; however, we identified a similar number of CNVRs (3,438 events) compared to Btau_4.0 CNVRs. The total lengths of CNVRs were similar with comparable statistics (146,905,950 bp; only 2.93% larger, Additional file 4: Table S5). When we assessed the difference in CNVR type calls (gain, loss and both), counts for each category varied less than 70 between the Btau_4.0 and UMD3.1 assemblies.
These results were not unexpected in light of the attributes of these two assemblies. As both assemblies are based on the same raw whole-genome shotgun reads, the most obvious difference between the two is that the Btau_4.0 unplaced contigs from chrUn are now placed on UMD3.1. Another difference between them is that local duplication artifacts, or duplicated regions that were artificially created on the Btau_4.0 assembly, were removed on UMD3.1. As our previous segmental duplication analyses detected, 267 Mb of duplicated sequence on Btau_4.0 were considered to be artifacts from a failure to merge high identity alleles on the reference assembly [41]. Our results were supported by independent FISH experiments [42] and similar observations reported by Zimin et al [39]. In summary, UMD3.1 seems to be more favorable than Btau_4.0 in terms of placing unplaced contigs and removing the unmerged alleles. As a result, most of CNVs calls derived from chrUn of Btau_4.0 could be recovered in UMD3.1 results. It is interesting to note even though UMD3.1’s CNV calls are more abundant and larger, the merged CNVR results were almost equivalent in terms of count and length. However, the exact effects of local assembly differences (besides UMD3.1’s placing unplaced contigs and removing the unmerged alleles) on CNV calling (count and length) warrant more detailed investigations in the future.
Quality assessment of selected CNV Events
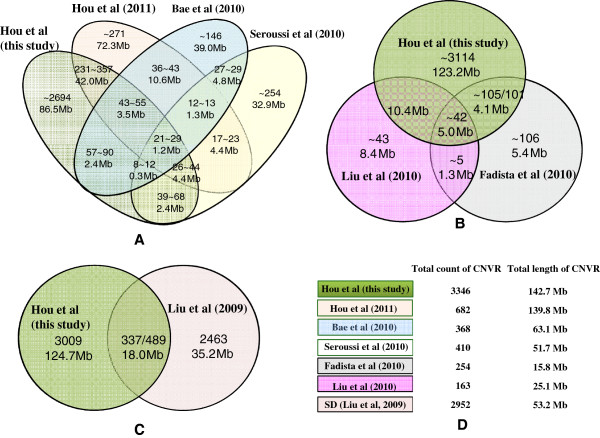
Since most existing cattle CNV studies were based on Btau_4.0, we first compared the identified CNV regions with previous published CNVs dataset [23-26] and the segment duplication (SD) dataset [41] based on Btau_4.0 (Figure 2). Since calls on chrX were not always included in these studies, we only compared the CNVRs detected within the autosomes. We found that 789 of our 3346 CNVRs (65.1 Mb) overlapped with all combined nonredundant published data. Detailed information of each comparison was displayed in Figure 2. Approximately 14% of our new CNV calls (482 out of 3346 CNVRs) overlapped with 51% (346 out of total 682 CNVR) of the CNV regions identified in 521 animals of 21 cattle breeds using BovineSNP50 arrays [27]. When considering the CNVR lengths, 36% of variable sequence space identified in this study intersected with 37% of the previous study, representing an overlap of 51.1 Mb. The discrepancy between these two studies could be due to a difference in samples as 674 animals of 27 cattle breeds were used in this study out of which, approximately 442 animals of 19 breeds (~65.6%) were shared by the later study. Additionally, SNP arrays that probed 763,572 SNPs were used in this study, while arrays with only 56,947 SNPs were used in the previous study. Only 46,475 SNPs were shared by both platforms.
Figure 2.
Comparisons between 3,346 autosomal CNVRs identified in this study and the other cattle CNVR datasets and the segmental duplication (SD) dataset on Btau_4.0. A, three CNVR datasets derived from SNP array (Hou et al., 2011, [24,25,27]); B, two CNVR datasets derived from array CGH studies [23,26]; C, the SD dataset [41]; D, the summaries and legends of existing cattle CNVR and SD datasets.
After comparison with other existing datasets, we found that 54.38% (77.6 Mb) of our CNVR calls were not reported in the literature. In order to confirm these novel CNVRs, we performed 56 quantitative PCR (qPCR) assays for 18 CNV loci in 12 animals (Additional file 5: Table S6). All primer coordinates on both assemblies were given in Additional file 5: Table S6, with the exception that primers for CNVR No. 1856 could only be placed on the Btau_4.0 assembly. Most of the CNV regions had two target amplicons placed near the probes in the CNVR regions. Out of 28 CNV loci and animal combinations, 23 (82.14%) had positive qPCR confirmations in at least one amplicon and 18 (64.30%) had positive results at both amplicons. If each PCR assay was counted separately, 73.21% (41/56) were in an agreement with the CNV status estimated by PennCNV (Additional file 5: Table S6), which is considerably higher than the previous validation rates between 48 to 60% [27].
CNVs overlap with segmental duplications and other genomic features
We also compared the identified CNV regions with the 2,952 SD (excluding chrX and chrUn) identified by WGAC and WSSD [41]. Agreeing with previous predictions regarding cattle SDs, a local tandem distribution pattern is predominant in our cattle CNVR dataset (Figure 1A). About 10.07% (337/3346) of CNV regions directly overlap with cattle SDs with an overlapping span of 18 Mb (12.61% of the total 142.7 Mb). Approximately 16.57% (489/2952) of the SDs (excluding chrX and chrUn) identified by WGAC and WSSD exhibit CNVs (Figure 2C). In comparison, 25.66% of the CNVRs (175/682) detected by using BovineSNP50 array overlap with cattle SDs, corresponding to 16.28 Mb (11.65% of the total 139.8 Mb) [27]. The fractions of CNVR calls that overlap with SDs were similar (12.61% vs. 11.65%) in both SNP array studies and they were significantly lower than the fraction of SD-overlapping CNVRs (58.90%, 96/163 or 60.56%, 15.2/25.1 Mb) as detected by using array CGH [26]. This lower overlap fraction probably reflects the fact that both SNP arrays have poor representation within cattle SD regions. SNP density on the BovineSNP50 array drops by one-third (from 21 probes/Mb in unique regions down to 14 probes/Mb) in SD regions while SNP density on the BovineHD array drops by over 61% (from 277 probes/Mb in unique regions down to 107 probes/Mb) in SD regions. We also overlapped our CNVRs with cattle Online Mendelian Inheritance in Animals (OMIA), Online Mendelian Inheritance in Man (OMIM) and cattle QTL datasets (Additional file 3: Table S3).
Gene content of CNV regions
Since UMD3.1 placed unplaced contigs and removed unmerged alleles, we focused on further characterization of the 3,438 high-confidence CNV regions from UMD3.1 autosomes. Additionally, we used both Btau_4.0 and UMD3.1 assemblies and obtained similar Panther and IPA results, as presented in Additional file 6: Table S7 and Additional file 7: Table S8.
We investigated the gene content of CNV regions derived from both Btau_4.0 and UMD3.1 (Table 2). Within the 3,438 CNVRs on UMD3.1, there were 939 unique cattle RefSeq genes and 2,153 Ensembl peptides, corresponding to 1,855 unique Ensembl genes (Table 2 and Additional file 4: Table S5). Approximately, 24% (817/3,438) of our CNVRs spanned cattle Refseq genes and 34% (1,165/3,438) of them overlapped with Ensembl genes. A detailed list of CNVR-gene overlapping is displayed in Additional file 4: Table S5. Beside those reported before such as ABCC4, ATP8A1, IGLL1, LYZ, PGR, SGCD, SCP2, WC1, ULBP, OR, ZNF280A, HLA-DQA and BLA-DQ, novel CNVR related genes or gene families include GBP6, SCMH1, GIMAP7, UGT2B10, ORM1 and others.
Table 2.
Gene contents of CNVRs on Btau_4.0 and UMD3.1
| Cattle RefSeq | in silico mapped human RefSeq | Ensembl genes | Glean genes | |
|---|---|---|---|---|
|
Btau_4.0 |
|
|
|
|
| #genes |
935 (836) |
5,549 (3,146) |
1,890 (1,788) |
2,536 (2,408) |
| #overlapped CNVR |
703 (21%) |
1,507 (45%) |
1,116 (33%) |
1,330 (40%) |
|
UMD3.1 |
|
|
|
|
| #genes |
1,062 (939) |
6,497 (3,828) |
1,964 (1,855) |
ND |
| #overlapped CNVR | 817 (24%) | 1,533 (45%) | 1,165 (34%) | ND |
Numbers in parentheses for #genes are the number of unique nonredundent gene counts, as one gene could show up multiple times in different CNVRs. Numbers in parentheses for #overlapped CNVR are the percentages of overlapped CNVR in the total CNVRs. ND: Glean genes on UMD3.1 were not determined.
We assigned PANTHER accessions to the overlapped peptides for CNVRs identified on both Btau_4.0 and UMD3.1 in this study. Similar statistically significant overrepresentations were observed for multiple categories (Additional file 6: Table S7). This set of copy number variable genes encompasses a wide spectrum of molecular functions, biological processes, pathways, cellular components and Panther protein classes. For example, immune system process, cellular defense response, response to stimulus, antigen processing and presentation, natural killer cell activation were among several enriched biological processes. G-protein coupled receptor activity was enriched while transcription factor and regulation activities were under represented in the Molecular Function terms.
Our UMD3.1 CNVRs overlapped with 3,828 unique in silico mapped human RefSeq genes (Table 2 and Additional file 4: Table S5), of which 3,669 were mapped as candidate genes for IPA analysis. A total of eleven networks with an IPA score greater than 10 were identified (Table 3). A score of 10 indicates that there was less than a 10-10 probability that the genes in the network were associated together by chance. The identified regulatory networks covered a broad range of physiological functions and processes, including inflammatory response, cell-to-cell signaling and interaction, cell development and cycle, lipid metabolism, normal development and function (embryo, organism, reproductive system, hematological system, skeletal and muscular system), and genetic disorder (Additional file 7: Table S8). Immunological and defense pathways were particularly enriched in IPA Pathway and Function analyses, further confirming the Panther results. The results of our functional analyses are in agreement with the findings of previous CNV studies [23-27]. Using the Btau_4.0 assembly, the IPA results are generally similar, with slight differences likely attributable to differences in assemblies and their annotations.
Table 3.
Network analyses using IPA based on UMD3.1
| ID | Score | Focus Molecules | Top Functions |
|---|---|---|---|
| 1 |
28 |
31 |
Reproductive System Development and Function, Cellular Development, Cellular Growth and Proliferation |
| 2 |
27 |
30 |
Inflammatory Response, Inflammatory Disease, Cell-To-Cell Signaling and Interaction |
| 3 |
27 |
30 |
Lipid Metabolism, Molecular Transport, Small Molecule Biochemistry |
| 4 |
27 |
30 |
Cellular Development, Cellular Growth and Proliferation, Decreased Levels of Albumin |
| 5 |
25 |
29 |
Cellular Assembly and Organization, Cellular Function and Maintenance, Cancer |
| 6 |
17 |
24 |
Gene Expression, DNA Replication, Recombination, and Repair, Cancer |
| 7 |
11 |
17 |
DNA Replication, Recombination, and Repair, Cancer, Gastrointestinal Disease |
| 8 |
11 |
19 |
Cell Cycle, Cancer, Genetic Disorder |
| 9 |
11 |
17 |
Cellular Development, Hematological System Development and Function, Hematopoiesis |
| 10 |
11 |
15 |
Cellular Growth and Proliferation, Tumor Morphology, Inflammatory Response |
| 11 | 10 | 14 | Embryonic Development, Organismal Development, Skeletal and Muscular System Development and Function |
Conclusion
In addition to single nucleotide polymorphisms (SNPs), CNVs have been revealed to be a substantial source of genetic variation in cattle. In this study, we performed two comprehensive CNV analyses based on Btau_4.0 and UMD3.1. When we compared our current and previous results across different SNP platforms on Btau_4.0, we detected higher resolution, sensitivity and PCR validation rate in these current CNV datasets. Our findings suggest that the use of high-density oligonucleotide arrays may allow more precise boundary information to be extracted for CNV detection. Therefore, the use of high-density SNP arrays combined with improved CNV calling algorithms seems to significantly improve the accuracy of CNV calling. As a consequence, more CNVs were identified and the CNVs identified from those SNP arrays were more accurate and with better defined boundaries.
In this study, we provide additional support to the observation that the distribution of CNVs varies among the four subspecies groups. In case of the complete absence of a particular allele of a SNP in a certain breed, the copy number of that SNP in the breed would be biased towards loss or deletion. While some of these breed differences could be related to the fact that the SNP markers were designed on the reference genome sequence (which was derived from the sequence of a Hereford cow of European origin; L1 Dominette 01449), our observation of differential CNV counts in different breeds and subspecies was largely consistent with their histories and divergences.
We also systematically evaluated current results across two existing cattle reference assemblies. Although we obtained comparable CNVR results, we found approximately 50% more and 20% longer CNVs on UMD3.1 as opposed to those on Btau_4.0. It is worth to note significant differences exist between different assemblies. Therefore it is critical to select appropriate assemblies and further validate the predicted variants experimentally.
Methods
Selection of cattle breeds and animals
Cattle selected in this study were composed of 686 individuals from 27 breeds originally, out of which, 674 distinct high quality genotyping results with call rate larger than 99.70% remained after PennCNV quality filtering. This panel included 447 animals from 18 taurine breeds, 113 animals from three breeds of predominantly indicine background, 67 animals from three breeds that are Taurine × Indicine Composites, and 47 animals from three African breeds (Additional file 1: Table S1). It is worth to note that for each subspecies, breeds or individuals within breeds were sampled from more than one continent to represent the global cattle population. This panel contained 41 trios where both parents and an offspring were genotyped.
Identification of cattle CNVs
For research use, the Infinium BovineHD BeadChip array features 786,799 evenly spaced SNPs probes based on assembly UMD3.1 and out of which, 783,617 were placed on autosomes and the X chromosome. This multi-sample genotyping panel delivers >99% call rate and >99.9% reproducibility, along with the ability to detect CNV ( http://www.illumina.com/Documents//products/datasheets/datasheet_bovineHD.pdf). Since we also surveyed on Btau_4.0, SNP probe coordinates were migrated from UMD3.1.0 to Btau_4.0 by using both UCSC liftOver ( http://hgdownload.cse.ucsc.edu/admin/exe/) and Blat tools. Approximately 97% (763,572/783,617) of the probes on autosomes and the X chromosome were converted successfully. CNV was detected using the PennCNV algorithm as described previously [27]. PennCNV incorporates multiple sources of information together, including Log R Ratio (LRR) and B Allele Frequency (BAF) at each SNP marker for each individual, more realistic models for state transition between different copy number states based on the distance between neighboring SNPs, population frequency of B allele (PFB), the allele frequency of SNPs, and the pedigree information where available, into a hidden Markov model (HMM). Both LRR and BAF of each SNP for each individual were exported from Illumina GenomeStudio Genotyping Module v1.9 software using the default clustering file. The PFB file was calculated as the average BAF for each marker in this population. Genomic waves, calculated as the GC content of the 1 Mb genomic region surrounding each marker (500 kb each side), were corrected by performing the -gcmodel option. Pedigree information including 41 trios were used to improve the accuracy of CNV identification. As described previously [27], PennCNV algorithm (with options: -test or -joint) was applied to autosomes (with option: -lastchr 29) to detect cattle CNV. After detection, 674 out of 686 animals passed the standard filtering of low-quality samples with the default cutoffs (standard deviation of LRR as 0.30, BAF drift as 0.01, and waviness factor as 0.05). The final CNVs set was the nonredundant combination of CNVs from the -joint results for family trio members and the -test results for unrelated individuals. CNVRs were determined by aggregating overlapping CNVs identified across samples.
qPCR validation
Primers were designed using Primer3 ( http://frodo.wi.mit.edu/primer3/) with a limitation of amplicon length to 150 bp to 250 bp, as well as CG clamp as 2. All other Primer3 settings were left at the default values. Primer information is shown in Additional file 5: Table S6. qPCR experiments were conducted using SYBR green chemistry in triplicate reactions, each with a reaction volume of 25 μl. All reactions were amplified on a BioRad MyIQ thermocycler. An intron-exon junction of the BTF3 gene was chosen as a reference location for all qPCR experiments. Analysis of resultant crossing thresholds (CT) was performed using the relative comparative CT method. Calibrations of CT values were derived from amplification of reference and test primers on a genomic DNA template derived from sequenced cow, L1 Dominette 01449, an European-origin Hereford. Since all reference and test primers did not overlap with any of Dominette’s CNV regions, two-copy states were assumed for both amplicons in Dominette. The copy number for each test region was calculated as 2(1+ddCT).
Gene content
Gene content of cattle CNV regions was assessed using cattle RefSeq and in silico mapped human RefSeq, the Glean consensus gene set (the UCSC Genome Browser website at http://genome.ucsc.edu/), Ensembl genes ( ftp://ftp.ensembl.org/pub/current_fasta/bos_taurus/pep/). We obtained a total of 26,977 and 22,118 bovine peptides from Ensembl on Btau_4.0 and UMD3.1 respectively. In addition, using the PANTHER classification system, we tested the hypothesis that the PANTHER biological process, molecular function, pathway, cellular component, and Panther protein class terms were under- or overrepresented in CNV regions after Bonferroni corrections [26]. It is worth noting that a portion of the genes in the bovine genome has not been annotated or has been annotated with unknown function, which may influence the outcome of this analysis. Overlapping between CNVRs and additional genomic features such as cattle OMIA, OMIM and cattle QTL datasets were performed as described [26].
In silico mapped human RefSeq genes in CNVRs were analyzed using Ingenuity Pathways Analysis (IPA) v9.0 (Ingenuity Systems, Redwood City, CA) as previously described [27]. The accessions of unique genes were imported into the software and subsequently mapped to their corresponding annotations in the Ingenuity Pathways Knowledge Base. The ''Core Analysis” function included in IPA ( http://www.ingenuity.com/) was used to analyze these genes in the context of networks, biological functions and Pathways. The networks accommodated these unique genes (also called focus molecules) were identified in comparison with the comprehensive global networks developed by IPA. The molecule network was illustrated with an assigned relevance score, the number of focus molecules, as well as the top function of the networks. In the process of analysis, each network was set to have a maximum of 35 molecules by default. We used only human genes and all confidence levels, including evidences of experimentally observed, predicted high or moderate confidence. The top significant biological functions and Pathway were listed.
Abbreviations
aCGH: array comparative genomic hybridization; CNV: copy number variation; CNVR: CNV region; CT: threshold cycle; DT: Dominette; NGS: next-generation sequencing; qPCR: quantitative PCR; QTL: Quantitative trait locus; OMIA: Online Mendelian Inheritance in Animals; OMIM: Online Mendelian Inheritance in Man; SD: segmental duplication; SNP: single nucleotide polymorphism.
Competing interests
M.L.H. and S.D. are employees of Pfizer Animal Genetics. A.E. is an employee of Illumina, Inc. The other authors declare that they have no competing interests.
Author contributions
GEL and YH conceived and designed the experiments. DMB, YH, CL, and JS performed in silico prediction, qPCR, and computational analyses. MLH, DAB, SF, AE, SD, GRW, TSS and CPVT collected samples and generated SNP array genotyping data. YH, GEL, and DMB wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Table S1. Numbers of subspecies, breeds, animals and trios used to call CNVs genotyped by BovineHD assay. Table S2. The summary of CNVs or CNVRs for each specie/breed based on Btau_4.0. Table S4. The summary of CNVs or CNVRs for each specie/breed based on UMD3.1.
Figure S1. Comparison of cattle copy number variations derived from BovineHD and BovineSNP50 assays on Batu_4.0.
Table S3. Btau_4.0 CNV regions, their frequencies, corresponding gene contents, QTL, OMIM, and OMIA overlapping information.
Table S5. UMD3.1 CNV regions, their frequencies, corresponding gene contents.
Table S6. The summary of PCR results.
Table S7. Over/Underrepresentation of PANTHER terms (molecular function, biological process, pathway, cellular component and PANTHER protein class) on Batu_4.0 and UMD3.1.
Table S8: Network, Biological function and Pathway analyses using IPA on Batu_4.0 and UMD3.1.
Contributor Information
Yali Hou, Email: houyali1210@gmail.com.
Derek M Bickhart, Email: Derek.Bickhart@ars.usda.gov.
Miranda L Hvinden, Email: miranda.hvinden@pfizer.com.
Congjun Li, Email: Congjun.Li@ars.usda.gov.
Jiuzhou Song, Email: songj88@umd.edu.
Didier A Boichard, Email: didier.boichard@jouy.inra.fr.
Sébastien Fritz, Email: sebastien.fritz@jouy.inra.fr.
André Eggen, Email: aeggen@illumina.com.
Sue DeNise, Email: Sue.Denise@pfizer.com.
George R Wiggans, Email: George.Wiggans@ars.usda.gov.
Tad S Sonstegard, Email: Tad.Sonstegard@ars.usda.gov.
Curtis P Van Tassell, Email: Curt.VanTassell@ars.usda.gov.
George E Liu, Email: George.Liu@ars.usda.gov.
Acknowledgements
We thank LABOGENA and other members of the Illumina Bovine HD SNP Consortium for sharing their samples. We would also like to thank R. Anderson, and A. Dimtchev for technical assistance. C.P.V.T. was supported by Agriculture and Food Research Initiative competitive grant no. 2009-65205-05635 from the USDA National Institute of Food and Agriculture Animal Genome Program. G.E.L. was supported by NRI/AFRI grants no. 2007-35205-17869 and 2011-67015-30183 from the USDA CSREES (now NIFA) and Project 1265-31000-098-00 from USDA-ARS. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. The USDA is an equal opportunity provider and employer.
References
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F. et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, MacDonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2009;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu FL, Bonnen PE, de Bakker PIW, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia XM, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis LR, Ren YR, Wheeler D, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarroll SA. et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, Chinwalla A, Conrad DF, Fu Y, Grubert F, Hajirasouliha I, Hormozdiari F, Iakoucheva LM, Iqbal Z, Kang S, Kidd JM, Konkel MK, Korn J, Khurana E, Kural D, Lam HYK, Leng J, Li R, Li Y, Lin CY, Luo R. et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, Eis PS, Shannon WD, Li X, McLeod HL, Cheverud JM, Ley TJ. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Cheng Z, Zollner S, Church DM, Eichler EE. Mouse segmental duplication and copy number variation. Nat Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev V, Saar K, Adamovic T, Verheul M, van Heesch SA, Cook S, Pravenec M, Aitman T, Jacob H, Shull JD, Hubner N, Cuppen E. Distribution and functional impact of DNA copy number variation in the rat. Nat Genet. 2008;40:538–545. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan XC, Nellaker C, Goodstadt L, Nicod J, Bhomra A, Hernandez-Pliego P, Whitley H, Cleak J, Dutton R, Janowitz D, Mott R, Adams DJ, Flint J. Sequence-based characterization of structural variation in the mouse genome. Nature. 2011;477:326–329. doi: 10.1038/nature10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, Hodges MD, Bhangal G, Patel SG, Sheehan-Rooney K, Duda M, Cook PR, Evans DJ, Domin J, Flint J, Boyle JJ, Pusey CD, Cook HT. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC. et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BHY, Cochrane L, Corsello C. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WK, Swartz JD, Rush LJ, Alvarez CE. Mapping DNA structural variation in dogs. Genome Res. 2009;19:500–509. doi: 10.1101/gr.083741.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas TJ, Cheng Z, Ventura M, Mealey K, Eichler EE, Akey JM. The genomic architecture of segmental duplications and associated copy number variants in dogs. Genome Res. 2009;19:491–499. doi: 10.1101/gr.084715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas TJ, Baker C, Eichler EE, Akey JM. A high-resolution integrated map of copy number polymorphisms within and between breeds of the modern domesticated dog. BMC Genomics. 2011;12:414. doi: 10.1186/1471-2164-12-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GE, Van Tassell CP, Sonstegard TS, Li RW, Alexander LJ, Keele JW, Matukumalli LK, Smith TP, Gasbarre LC. Detection of germline and somatic copy number variations in cattle. Dev Biol. 2008;132:231–237. doi: 10.1159/000317165. [DOI] [PubMed] [Google Scholar]
- Wang XF, Nahashon S, Feaster TK, Bohannon-Stewart A, Adefope N. An initial map of chromosomal segmental copy number variations in the chicken. BMC Genomics. 2010;11:351. doi: 10.1186/1471-2164-11-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M, Backstrom N, Skinner BM, Langley EJ, Bunzey SK, Ellegren H, Griffin DK. Copy number variation, chromosome rearrangement, and their association with recombination during avian evolution. Genome Res. 2010;20:503–511. doi: 10.1101/gr.103663.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista J, Nygaard M, Holm LE, Thomsen B, Bendixen C. A Snapshot of CNVs in the Pig Genome. PLoS One. 2008;3:e3916. doi: 10.1371/journal.pone.0003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramayo-Caldas Y, Castelló A, Pena NR, Alves E, Mercadé A, Souza AC, Fernández IA, Perez-Enciso M, Folch MJ. Copy number variation in the porcine genome inferred from a 60 k SNP BeadChip. BMC Genomics. 2010;11:593. doi: 10.1186/1471-2164-11-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi L, Martelli PL, Beretti F, Riggio V, Dall'olio S, Colombo M, Casadio R, Russo V, Portolano B. An initial comparative map of copy number variations in the goat (Capra hircus) genome. BMC Genomics. 2010;11:639. doi: 10.1186/1471-2164-11-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi L, Beretti F, Martelli PL, Colombo M, Dall'olio S, Occidente M, Portolano B, Casadio R, Matassino D, Russo V. A first comparative map of copy number variations in the sheep genome. Genomics. 2011;97:158–165. doi: 10.1016/j.ygeno.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Fadista J, Thomsen B, Holm LE, Bendixen C. Copy number variation in the bovine genome. BMC Genomics. 2010;11:284. doi: 10.1186/1471-2164-11-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JS, Cheong HS, Kim LH, NamGung S, Park TJ, Chun JY, Kim JY, Pasaje CF, Lee JS, Shin HD. Identification of copy number variations and common deletion polymorphisms in cattle. BMC Genomics. 2010;11:232. doi: 10.1186/1471-2164-11-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seroussi E, Glick G, Shirak A, Yakobson E, Weller JI, Ezra E, Zeron Y. Analysis of copy loss and gain variations in Holstein cattle autosomes using BeadChip SNPs. BMC Genomics. 2010;11:673. doi: 10.1186/1471-2164-11-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GE, Hou Y, Zhu B, Cardone MF, Jiang L, Cellamare A, Mitra A, Alexander LJ, Coutinho LL, Dell'Aquila ME, Gasbarre LC, Lacalandra G, Li RW, Matukumalli LK, Nonneman D, Regitano LCD, Smith TPL, Song J, Sonstegard TS, Van Tassell CP, Ventura M, Eichler EE, McDaneld TG, Keele JW. Analysis of copy number variations among diverse cattle breeds. Genome Res. 2010;20:693–703. doi: 10.1101/gr.105403.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Liu GE, Bickhart DM, Cardone MF, Wang K, Kim ES, Matukumalli LK, Ventura M, Song J, VanRaden PM, Sonstegard TS, Van Tassell CP. Genomic characteristics of cattle copy number variations. BMC Genomics. 2011;12:127. doi: 10.1186/1471-2164-12-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GE, Brown T, Hebert DA, Cardone MF, Hou YL, Choudhary RK, Shaffer J, Amazu C, Connor EE, Ventura M, Gasbarre LC. Initial analysis of copy number variations in cattle selected for resistance or susceptibility to intestinal nematodes. Mammalian Genome. 2011;22:111–121. doi: 10.1007/s00335-010-9308-0. [DOI] [PubMed] [Google Scholar]
- Hou Y, Liu GE, Bickhart DM, Matukumalli LK, Li C, Song J, Gasberre LC, Van Tassell CP, Sonstegard TS. Genomic regions showing copy number variations associate with resistance or susceptibility to gastrointestinal nematodes in Angus cattle. Funct Integr Genomics. 2011;12:81–92. doi: 10.1007/s10142-011-0252-1. [DOI] [PubMed] [Google Scholar]
- Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, Lee C, Nizzari MM, Gabriel SB, Purcell S, Daly MJ, Altshuler D. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavas G, Koyuturk M, Ozsoyoglu M, Gould MP, LaFramboise T. An optimization framework for unsupervised identification of rare copy number variation from SNP array data. Genome Biol. 2009;10:R119. doi: 10.1186/gb-2009-10-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchester L, Yau C, Ragoussis J. Comparing CNV detection methods for SNP arrays. Brief. Funct. Genomic Proteomic. 2009;8:353–366. doi: 10.1093/bfgp/elp017. [DOI] [PubMed] [Google Scholar]
- Pinto D, Darvishi K, Shi XH, Rajan D, Rigler D, Fitzgerald T, Lionel AC, Thiruvahindrapuram B, MacDonald JR, Mills R, Prasad A, Noonan K, Gribble S, Prigmore E, Donahoe PK, Smith RS, Park JH, Hurles ME, Carter NP, Lee C, Scherer SW, Feuk L. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29:512–U76. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Thorne NP, Valsesia A, Fitzgerald T, Redon R, Fiegler H, Andrews TD, Stranger BE, Lynch AG, Dermitzakis ET, Carter NP, Tavare S, Hurles ME. Breaking the waves: improved detection of copy number variation from microarray-based comparative genomic hybridization. Genome Biol. 2007;8:R228. doi: 10.1186/gb-2007-8-10-r228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin SJ, Li M, Hou C, Yang S, Glessner J, Hakonarson H, Bucan M, Maris JM, Wang K. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36:e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Bovine Genome Sequencing and Analysis Consortium. The Genome Sequence of Taurine Cattle: a window to ruminant biology and evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS, Marcais G, Roberts M, Subramanian P, Yorke JA, Salzberg SL. A whole-genome assembly of the domestic cow. Bos taurus. 2009;10:R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon G, Weber KL, Van Eenennaam AL, Golden BL, Medrano JF. Hot topic: Performance of bovine high-density genotyping platforms in Holsteins and Jerseys. Journal of Dairy Science. 2011;94:6116–6121. doi: 10.3168/jds.2011-4764. [DOI] [PubMed] [Google Scholar]
- Liu GE, Ventura M, Cellamare A, Chen L, Cheng Z, Zhu B, Li C, Song J, Eichler EE. Analysis of recent segmental duplications in the bovine genome. BMC Genomics. 2009;10:571. doi: 10.1186/1471-2164-10-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partipilo G, D’Addabbo P, Lacalandra GM, Liu GE, Rocchi M. Refinement of Bos Taurus sequence assembly based on BAC-FISH experiments. BMC Genomics. 2011;12:639. doi: 10.1186/1471-2164-12-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Numbers of subspecies, breeds, animals and trios used to call CNVs genotyped by BovineHD assay. Table S2. The summary of CNVs or CNVRs for each specie/breed based on Btau_4.0. Table S4. The summary of CNVs or CNVRs for each specie/breed based on UMD3.1.
Figure S1. Comparison of cattle copy number variations derived from BovineHD and BovineSNP50 assays on Batu_4.0.
Table S3. Btau_4.0 CNV regions, their frequencies, corresponding gene contents, QTL, OMIM, and OMIA overlapping information.
Table S5. UMD3.1 CNV regions, their frequencies, corresponding gene contents.
Table S6. The summary of PCR results.
Table S7. Over/Underrepresentation of PANTHER terms (molecular function, biological process, pathway, cellular component and PANTHER protein class) on Batu_4.0 and UMD3.1.
Table S8: Network, Biological function and Pathway analyses using IPA on Batu_4.0 and UMD3.1.