Abstract
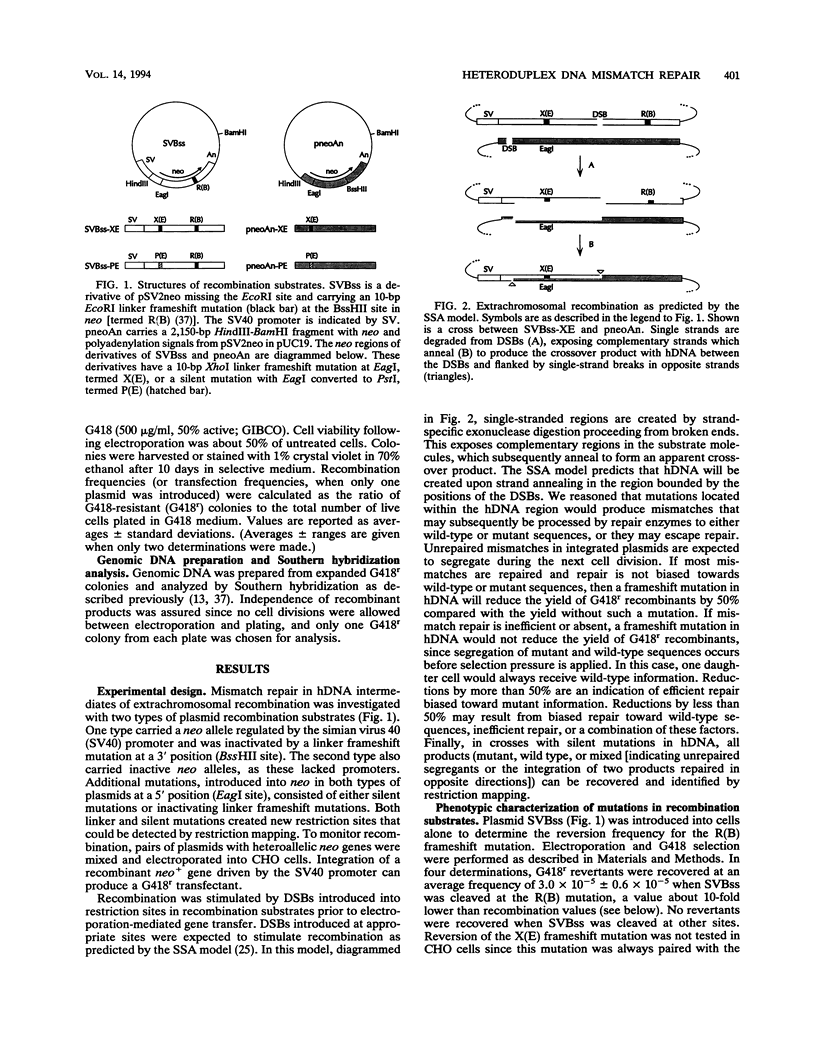
Previous work indicated that extrachromosomal recombination in mammalian cells could be explained by the single-strand annealing (SSA) model. This model predicts that extrachromosomal recombination leads to nonconservative crossover products and that heteroduplex DNA (hDNA) is formed by annealing of complementary single strands. Mismatched bases in hDNA may subsequently be repaired to wild-type or mutant sequences, or they may remain unrepaired and segregate following DNA replication. We describe a system to examine the formation and mismatch repair of hDNA in recombination intermediates. Our results are consistent with extrachromosomal recombination occurring via SSA and producing crossover recombinant products. As predicted by the SSA model, hDNA was present in double-strand break-induced recombination intermediates. By placing either silent or frameshift mutations in the predicted hDNA region, we have shown that mismatches are efficiently repaired prior to DNA replication.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. A., Eliason S. L. Recombination of homologous DNA fragments transfected into mammalian cells occurs predominantly by terminal pairing. Mol Cell Biol. 1986 Sep;6(9):3246–3252. doi: 10.1128/mcb.6.9.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayares D., Ganea D., Chekuri L., Campbell C. R., Kucherlapati R. Repair of single-stranded DNA nicks, gaps, and loops in mammalian cells. Mol Cell Biol. 1987 May;7(5):1656–1662. doi: 10.1128/mcb.7.5.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayares D., Spencer J., Schwartz F., Morse B., Kucherlapati R. Homologous recombination between autonomously replicating plasmids in mammalian cells. Genetics. 1985 Oct;111(2):375–388. doi: 10.1093/genetics/111.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya N. P., Maher V. M., McCormick J. J. Ability of structurally related polycyclic aromatic carcinogens to induce homologous recombination between duplicated chromosomal sequences in mouse L cells. Mutat Res. 1989 Apr;211(2):205–214. doi: 10.1016/0027-5107(89)90003-1. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya N. P., Maher V. M., McCormick J. J. Effect of nucleotide excision repair in human cells on intrachromosomal homologous recombination induced by UV and 1-nitrosopyrene. Mol Cell Biol. 1990 Aug;10(8):3945–3951. doi: 10.1128/mcb.10.8.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya N. P., Maher V. M., McCormick J. J. Intrachromosomal homologous recombination in human cells which differ in nucleotide excision-repair capacity. Mutat Res. 1990 Feb;234(1):31–41. doi: 10.1016/0165-1161(90)90028-m. [DOI] [PubMed] [Google Scholar]
- Bollag R. J., Elwood D. R., Tobin E. D., Godwin A. R., Liskay R. M. Formation of heteroduplex DNA during mammalian intrachromosomal gene conversion. Mol Cell Biol. 1992 Apr;12(4):1546–1552. doi: 10.1128/mcb.12.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag R. J., Waldman A. S., Liskay R. M. Homologous recombination in mammalian cells. Annu Rev Genet. 1989;23:199–225. doi: 10.1146/annurev.ge.23.120189.001215. [DOI] [PubMed] [Google Scholar]
- Brenner D. A., Smigocki A. C., Camerini-Otero R. D. Double-strand gap repair results in homologous recombination in mouse L cells. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1762–1766. doi: 10.1073/pnas.83.6.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D. A., Smigocki A. C., Camerini-Otero R. D. Effect of insertions, deletions, and double-strand breaks on homologous recombination in mouse L cells. Mol Cell Biol. 1985 Apr;5(4):684–691. doi: 10.1128/mcb.5.4.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Seidman M. M. Intramolecular recombination between transfected repeated sequences in mammalian cells is nonconservative. Mol Cell Biol. 1986 Jul;6(7):2520–2526. doi: 10.1128/mcb.6.7.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W. P., Nickoloff J. A. Preferential repair of UV damage in highly transcribed DNA diminishes UV-induced intrachromosomal recombination in mammalian cells. Mol Cell Biol. 1994 Jan;14(1):391–399. doi: 10.1128/mcb.14.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W. P., Nickoloff J. A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992 Jan;200(1):81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- Grilley M., Holmes J., Yashar B., Modrich P. Mechanisms of DNA-mismatch correction. Mutat Res. 1990 Sep-Nov;236(2-3):253–267. doi: 10.1016/0921-8777(90)90009-t. [DOI] [PubMed] [Google Scholar]
- Hare J. T., Taylor J. H. One role for DNA methylation in vertebrate cells is strand discrimination in mismatch repair. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7350–7354. doi: 10.1073/pnas.82.21.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellgren D., Luthman H., Lambert B. Induced recombination between duplicated neo genes stably integrated in the genome of CHO cells. Mutat Res. 1989 Jan;210(1):197–206. doi: 10.1016/0027-5107(89)90059-6. [DOI] [PubMed] [Google Scholar]
- Heywood L. A., Burke J. F. Mismatch repair in mammalian cells. Bioessays. 1990 Oct;12(10):473–477. doi: 10.1002/bies.950121004. [DOI] [PubMed] [Google Scholar]
- Heywood L. A., Burke J. F. Repair of single nucleotide DNA mismatches transfected into mammalian cells can occur by short-patch excision. Mutat Res. 1990 Jul;236(1):59–66. doi: 10.1016/0921-8777(90)90033-2. [DOI] [PubMed] [Google Scholar]
- Holmes J., Jr, Clark S., Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M., de Villiers J., Weber F., Schaffner W. High frequency of homologous recombination in mammalian cells between endogenous and introduced SV40 genomes. Cell. 1985 Dec;43(3 Pt 2):695–703. doi: 10.1016/0092-8674(85)90242-9. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N., Abraham J. A. Involvement of double-strand chromosomal breaks for mating-type switching in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1984;49:77–88. doi: 10.1101/sqb.1984.049.01.011. [DOI] [PubMed] [Google Scholar]
- Kolodkin A. L., Klar A. J., Stahl F. W. Double-strand breaks can initiate meiotic recombination in S. cerevisiae. Cell. 1986 Aug 29;46(5):733–740. doi: 10.1016/0092-8674(86)90349-1. [DOI] [PubMed] [Google Scholar]
- Kucherlapati R. S., Eves E. M., Song K. Y., Morse B. S., Smithies O. Homologous recombination between plasmids in mammalian cells can be enhanced by treatment of input DNA. Proc Natl Acad Sci U S A. 1984 May;81(10):3153–3157. doi: 10.1073/pnas.81.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K. M., Sternberg N. L. Extrachromosomal recombination in mammalian cells as studied with single- and double-stranded DNA substrates. Mol Cell Biol. 1987 Jan;7(1):129–140. doi: 10.1128/mcb.7.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Intermolecular recombination between DNAs introduced into mouse L cells is mediated by a nonconservative pathway that leads to crossover products. Mol Cell Biol. 1990 Jan;10(1):103–112. doi: 10.1128/mcb.10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Repair of double-stranded DNA breaks by homologous DNA fragments during transfer of DNA into mouse L cells. Mol Cell Biol. 1990 Jan;10(1):113–119. doi: 10.1128/mcb.10.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Mudgett J. S., Taylor W. D. Recombination between irradiated shuttle vector DNA and chromosomal DNA in African green monkey kidney cells. Mol Cell Biol. 1990 Jan;10(1):37–46. doi: 10.1128/mcb.10.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D. K., White M. A., Petes T. D. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989 Jul 27;340(6231):318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- Nairn R. S., Humphrey R. M., Adair G. M. Transformation depending on intermolecular homologous recombination is stimulated by UV damage in transfected DNA. Mutat Res. 1988 Jul;208(3-4):137–141. doi: 10.1016/0165-7992(88)90049-8. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Chen E. Y., Heffron F. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7831–7835. doi: 10.1073/pnas.83.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A., Reynolds R. J. Transcription stimulates homologous recombination in mammalian cells. Mol Cell Biol. 1990 Sep;10(9):4837–4845. doi: 10.1128/mcb.10.9.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A., Singer J. D., Hoekstra M. F., Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J Mol Biol. 1989 Jun 5;207(3):527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol Cell Biol. 1992 Dec;12(12):5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Siddiqi I., Kolodkin A. L., Stahl F. W. Intra-chromosomal gene conversion induced by a DNA double-strand break in Saccharomyces cerevisiae. J Mol Biol. 1988 May 20;201(2):247–260. doi: 10.1016/0022-2836(88)90136-2. [DOI] [PubMed] [Google Scholar]
- Seidman M. M. Intermolecular homologous recombination between transfected sequences in mammalian cells is primarily nonconservative. Mol Cell Biol. 1987 Oct;7(10):3561–3565. doi: 10.1128/mcb.7.10.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K. Y., Chekuri L., Rauth S., Ehrlich S., Kucherlapati R. Effect of double-strand breaks on homologous recombination in mammalian cells and extracts. Mol Cell Biol. 1985 Dec;5(12):3331–3336. doi: 10.1128/mcb.5.12.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Stahl F. W. Roles of double-strand breaks in generalized genetic recombination. Prog Nucleic Acid Res Mol Biol. 1986;33:169–194. doi: 10.1016/s0079-6603(08)60023-9. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Folger K. R., Capecchi M. R. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986 Feb 14;44(3):419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- Tsujimura T., Maher V. M., Godwin A. R., Liskay R. M., McCormick J. J. Frequency of intrachromosomal homologous recombination induced by UV radiation in normally repairing and excision repair-deficient human cells. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1566–1570. doi: 10.1073/pnas.87.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Y., Maher V. M., Liskay R. M., McCormick J. J. Carcinogens can induce homologous recombination between duplicated chromosomal sequences in mouse L cells. Mol Cell Biol. 1988 Jan;8(1):196–202. doi: 10.1128/mcb.8.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss U., Wilson J. H. Repair of single-stranded loops in heteroduplex DNA transfected into mammalian cells. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1619–1623. doi: 10.1073/pnas.84.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Waldman A. S. An examination of the effects of double-strand breaks on extrachromosomal recombination in mammalian cells. Genetics. 1992 Dec;132(4):1081–1093. doi: 10.1093/genetics/132.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]