Abstract
Heterotrimeric G-proteins (α, β and γ subunits) are primarily involved in diverse signaling processes by transducing signals from an activated transmembrane G-protein coupled receptor (GPCR) to appropriate downstream effectors within cells. The role of α and β G-protein subunits in salinity and heat stress has been reported but the regulation of γ subunit of plant G-proteins in response to abiotic stress has not heretofore been described. In the present study we report the isolation of full-length cDNAs of two isoforms of Gγ [RGG1(I), 282 bp and RGG2(I), 453 bp] from rice (Oryza sativa cv Indica group Swarna) and described their transcript regulation in response to abiotic stresses. Protein sequence alignment and pairwise comparison of γ subunits of Indica rice [RGG(I)] with other known plant G-protein γ subunits demonstrated high homology to barley (HvGs) while soybean (GmG2) and Arabidopsis (AGG1) were least related. The numbers of the exons and introns were found to be similar between RGG1(I) and RGG2(I), but their sizes were different. Analyses of promoter sequences of RGG(I) confirmed the presence of stress-related cis-regulatory signature motifs suggesting their active and possible independent roles in abiotic stress signaling. The transcript levels of RGG1(I) and RGG2(I) were upregulated following NaCl, cold, heat and ABA treatments. However, in drought stress only RGG1(I) was upregulated. Strong support by transcript profiling suggests that γ subunits play a critical role via cross talk in signaling pathways. These findings provide first direct evidence for roles of Gγ subunits of rice G-proteins in regulation of abiotic stresses. These findings suggest the possible exploitation of γ subunits of G-protein machinery for promoting stress tolerance in plants.
Keywords: Rice G-protein γ subunits, abiotic stress, heterotrimeric G-proteins, real-time PCR, signal transduction
Introduction
Heterotrimeric G-protein is composed of α, β and γ subunits and constitute one of the most important components of cell signaling cascade. In eukaryotes, it participates in relaying a wide range of extracellular signals perceived through their G-protein coupled receptor (GPCR).1-4 The transmission of stimulus perceived by signaling machinery into the cell via membrane receptor occurs through signal transduction triad (receptor/transducer/effector).5 According to the typical paradigm, G-protein exists in inactive state. G-protein signaling initiates with binding of extracellular ligand that results in a conformational change in a G-protein coupled receptor (GPCR). Once activated by the GPCR, the Gα protein, which possesses a GDP/GTP-nucleotide binding site and GTP-hydrolase activity, changes its form to a structure that allows exchange of GDP for GTP. The GTP-bound Gα separates from the associated Gβγ dimer and thus freed Gα and Gβγ proteins can then interact with downstream effector molecules, alone or in combination, to transduce the signal. Subsequent to signal propagation, the intrinsic GTPase activity of Gα eventually results in hydrolysis of bound GTP to GDP, which inactivates Gα and allows its re-association with the Gβγ dimer to reform the inactive G-protein complex.6-8 Recently, the crystal structure of a self-activating G protein α subunit from Arabidopsis revealed its distinct mechanism of signal initiation from the well-established mechanism found in animals.9 Trusov et al.10 suggested that G protein gamma subunits provide functional selectivity in G β-gamma dimer signaling in Arabidopsis and suggested that some new elements also exist in the heterotrimeric G protein-signaling complex.
The genes for putative α, β and γ subunits of heterotrimeric G-protein have been isolated from various plant species in higher plants.11-13 Unlike in mammalians, plants have very small number of G-protein subunit genes reported.14 For example the model species Arabidopsis contain only a single canonical Gα gene, GPA1,11 one Gβ gene, AGB115 and three Gγ genes, AGG1, AGG2 and AGG3.16-18 Two Gα subunit genes in pea (PGA1 and PGA2)19,13 and four Gα subunit genes (GmGα1–4) in soybean20 were reported. The fully sequenced genome of rice contains only one conventional Gα, Gβ, three Gγ subunits (RGG1, RGG2 and RGG3); the RGG3 is a homolog of AGG3, also known as DEP118 and one GPCR.3 These limited number of components, however, regulate diverse signaling pathways, including hormone signaling, environmental sensing, ion channel regulation, disease response and cell death.13,21,22 Recently, Pandey23 has identified an elaborate network of G-proteins in soybean. Although additional, splice variants or non-conventional genes for Gγ subunit may also exist.20 The Gγ protein of the G-proteins is essential for its proper targeting at the plasma membrane and correct functioning.24 G-protein γ-subunit is also reported to be involved in guard cell K⁺-channel regulation and morphological development in Arabidopsis thaliana.18 Recently, a detail study of Arabidopsis G-protein interactome revealed a novel role for G-proteins in regulating cell wall modification.25 However, little is known about the role of Gγ subunits as an individual in stress conditions. In the present study, we have described the phylogenetic relationship, genomic organization, promoter analysis and transcript profile of RGG1 and RGG2 subunits of indica rice G-proteins under different abiotic stress treatments including high salt, cold, heat, drought and ABA.
Results
Cloning of RGG1(I) and RGG2(I) genes
The complete coding sequence of RGG1(I) and RGG2(I), were amplified by PCR using first-strand cDNA templates prepared from total RNA. Sequence analysis showed that the amplified DNA of 282 bp and 453 bp encoded full-length gene of RGG1(I) and RGG2(I), respectively. The deduced amino acid sequence revealed a protein consisting of 93 amino acid residues with a predicted molecular mass of about 10.49 kDa and pI 4.86 for RGG1(I), and a protein consisting of 150 amino acid residues with a predicted molecular mass of 16.83 kDa and pI 5.34 for RGG2(I).
In silico analysis of RGG1(I) and RGG2(I) proteins
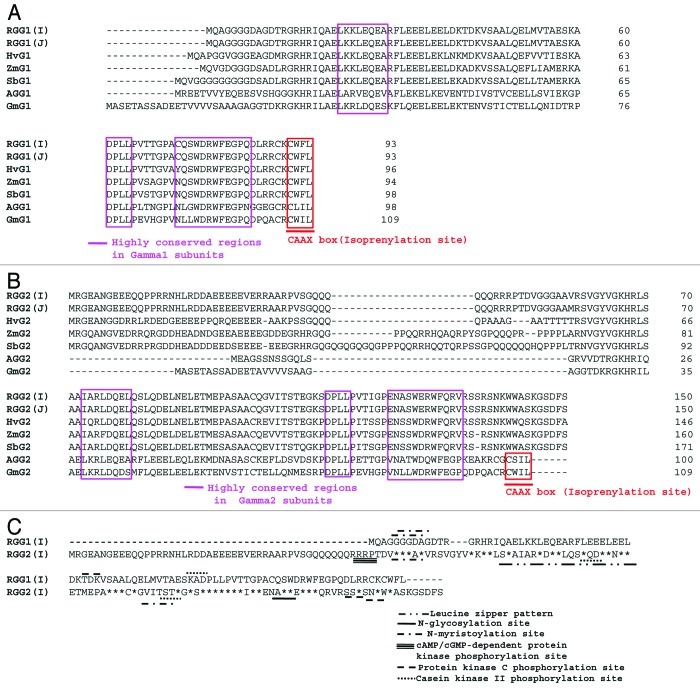
Amino acid sequence alignments of RGG1(I) and RGG2(I) subunits with their corresponding subunits from Japonica rice, maize, Sorghum, barley, Arabidopsis and soybean, is shown in Figure 1A and B. The RGG1(I) and RGG2(I) share 41% identity with each other. RGG1(I) is identical to RGG1(J) followed by 84% identity with barley (HvG1), 81% with sorghum (SbG1), 78% with ZmG1, 53% with soybean (GmG1), 45% with Arabidopsis (AGG1) and showed least homology of 30% with soybean (GmG1) (Table 1). The sequence of RGG1(I) contained all the reported conserved domains of the Gγ1 subunit (Fig. 1A). On the other hand, RGG2(I) shared 99% identity with RGG2(J), followed by 70% identity with sorghum (SbG2), 68% with both HvG2 and ZmG2, 37% with Arabidopsis (AGG2) while showing least homology of 30% with soybean (GmG2) (Table 1). RGG2 also has all the reported conserved domains of Gγ2 subunit except isoprenylation site (CAAX box) at C-terminus (Fig. 1B).
Figure 1. Amino acid sequence alignment of rice G-protein gamma subunits using ClustalW program (www.ebi.ac.uk/clustalw). Gaps were inserted to optimize the alignment are indicated by dashes. (A) RGG1(I) protein aligned with Japonica rice [RGG1(J); AK241226.1], maize (ZmG1; NP_001152725), barley (HvG1, AK359503), sorghum (SbG1, XP_002464204), Arabidopsis (AGG1; NP_567147.1) and soybean (GmG1; Glyma10 g03610). (B) RGG2(I) protein with Japonica rice (RGG2; NM_001052368.1), maize (ZmG2; NP_001151842), barley (HvG2, AK367089), sorghum (SbG2, XP_002451511), Arabidopsis (AGG2; AT3G22942.1) and soybean (GmG2; Glyma02 g16190). (C) RGG1(I) and RGG2(I) aligned together. Identical residues denoted by asterisk. Different motifs and patterns identified using Expasy PROSITE database.
Table 1. Amino acid sequence identity (%) of Indica rice [RGG(I)] gamma subunits (1 and 2) with corresponding proteins of Japonica rice [RGG(J)], maize (ZmG), Sorghum (SbG), barley (HvG), Arabidopsis (AGG) and soybean (GmG) using ClustalW 2.0 program.
| RGG1(I) | RGG1(J) | ZmG1 | SbG1 | HvG1 | AGG1 | GmG1 | |
|---|---|---|---|---|---|---|---|
| RGG1(I) |
*** |
100 |
78 |
81 |
84 |
45 |
53 |
| RGG1(J) |
|
*** |
78 |
81 |
84 |
45 |
53 |
| ZmG1 |
|
|
*** |
90 |
75 |
44 |
53 |
| SbG1 |
|
|
|
*** |
76 |
43 |
53 |
| HvG1 |
|
|
|
|
*** |
40 |
47 |
| AGG1 |
|
|
|
|
|
*** |
51 |
| GmG1 | *** |
| RGG2(I) | RGG2(J) | ZmG2 | SbG2 | HvG2 | AGG2 | GmG2 | |
|---|---|---|---|---|---|---|---|
| RGG2(I) |
*** |
99 |
68 |
70 |
68 |
37 |
30 |
| RGG2(J) |
|
*** |
68 |
70 |
68 |
37 |
30 |
| ZmG2 |
|
|
*** |
81 |
67 |
37 |
31 |
| SbG2 |
|
|
|
*** |
65 |
39 |
32 |
| HvG2 |
|
|
|
|
*** |
37 |
33 |
| AGG2 |
|
|
|
|
|
*** |
51 |
| GmG2 | *** |
The Expasy PROSITE database of protein families and domains revealed different motifs, patterns and biologically significant sites in RGG1(I) and RGG2(I) (Fig. 1C). RGG1(I) had two predicted potent N-myristoylation sites, viz 5–10: GGgdAG; 6–11: GGdaGD, one protein kinase C phosphorylation site, 42–44: TdK and one casein kinase II phosphorylation site, viz 58–61: SkaD. Whereas, different biologically significant sites predicted in RGG2(I) included two potent N-myristoylation sites, viz 53–58: GGgaAV; 102–107: GVitST, one cAMP- and cGMP-dependent protein kinase phosphorylation site, viz 47–50: RRpT, two casein kinase II phosphorylation sites, viz 82–85: SlqD; 105–108: TstE, one potential N-glycosylation site, viz 123–126: NASW, two protein kinase C phosphorylation sites, viz 135–137: SsR; 138–140: SnK and one Leucine zipper pattern, viz 69–90: LsaaiarLdqelqsLqdelneL.
Phylogenetic trees of RGG1(I) and RGG2(I)
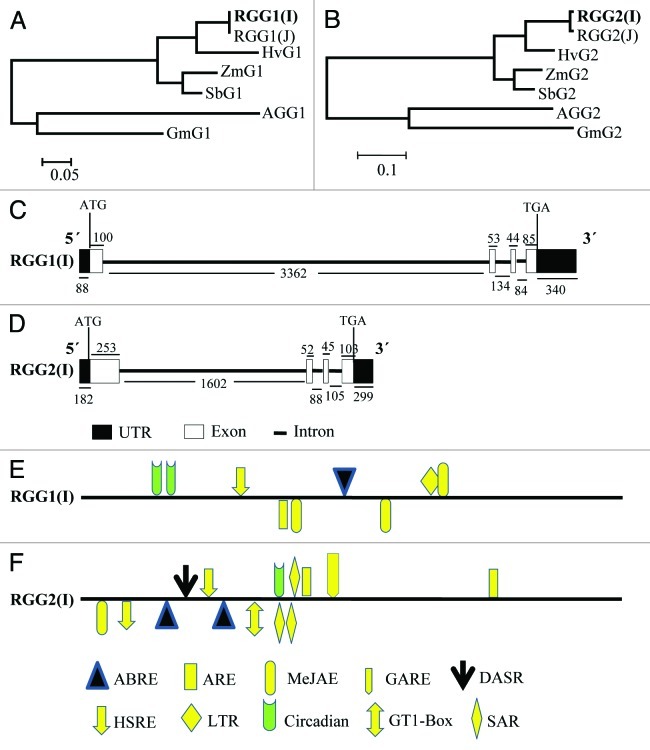
The phylogenetic trees constructed for gamma subunits of G-protein clustered all monocots together (Fig. 2A and B). The hypothetical proteins from barley and sorghum were considered in study due to high homology in Blastn with RGG(I) and were found to be putative Gγ subunits as they possess conserved motifs of G-protein gamma 1 and 2 subunits.
Figure 2. In silico analysis of RGG1(I) and RGG2(I). (A--B) Dendrogram showing evolutionary relationship of RGG1(I) (A) and RGG2(I) (B) with related proteins. The evolutionary history was inferred using the Neighbor-Joining method and evolutionary distances were computed using the Poisson correction method. These phylogenetic analyses were conducted in MEGA5. (C--D) The schematic representation of genomic organization (exon–intron organization) of the genomic sequence of RGG1(I) (C) and RGG2(I) (D) genes. Closed boxes represent exons, and lines between closed boxes represent introns. The dark boxes represent the UTRs. The position of ATG and TAA are marked. The numbers below the lines and the above boxes indicate the sizes (bp) of introns, UTR and exons, respectively. (E--F) Stress-responsive cis-elements and phytohormones responsive elements in the 2 kb 5′-upstream regions of RGG1(I) (E) and RGG2(I) (F). The lines represent 5′-upstream regions of RGG(I) genes. The elements located in the positive strand are above the lines, while those in the reverse strand are indicated below the line. ABRE, abscisic acid responsive element; ARE, auxin responsive factor (TGA-box; MeJAE, methyl jasmonate responsive element; GARE, gibberellic acid-responsive element; DASR, defense and stress responsive element; GT1-Box; SAR, salicylic acid responsive element; HSRE, heat stress responsive element; LTR, Low temperature responsive element.
Genomic organization of RGG1(I) and RGG2(I)
Alignment of the genomic sequence of RGG1(I) and RGG2(I) with their respective cDNA sequence identified four exons (100, 53, 44 and 88 bp) and three introns (3326, 134 and 84 bp) in RGG1(I) and in RGG2(I) (253, 52, 45 and 103 bp) and (1602, 88 and 105 bp) as well (Fig. 2C and D). The numbers of the exons and introns were found to be similar between RGG1(I) and RGG2(I), but their sizes were different (Fig. 2C and D).
In silico analysis of promoters of RGG1(I) and RGG2(I)
The distribution of regulatory cis-elements in 2.0 kb upstream promoter region of RGG1(I) and RGG2(I) were also analyzed and shown in Figure 2E and F, respectively. Stress-responsive cis-regulatory elements selected in the present study included are defense, stress responsive element, salt-induced responsive element (GT-1 motif), heat stress responsive element (HSRE), low temperature responsive element (LTR) and phytohormones responsive cis-regulatory elements, like abscisic acid responsive element (ABRE),26 auxin response factor (TGA-box),27,28 methyl jasmonate responsive element (MeJAE), salicylic acid responsive element (SAR), gibberellic acid-responsive element (GARE),29 and auxin response factor (ARF).
The results showed that RGG1(I) gene contained putative ABRE, HSRE, LTR, ARE, MeJAE and circadian cis-regulatory elements in their promoter regions (Fig. 2E). Whereas, RGG2(I), besides containing ABRE, HSRE, LTR, ARE, MeJAE and circadian, also had a GT-1 motif, which plays a role in pathogen- and salt-induced SCaM-4 gene expression,30 salicylic acid responsive element (SAR) and gibberellic acid-responsive element (GARE) in its 5′ upstream region genomic sequence (Fig. 2F). RGG1(I) contained LTR element (Fig. 2E) that was not present in RGG2(I) (Fig. 2F).
Transcript profile of RGG1(I) and RGG2(I) by quantitative real time PCR
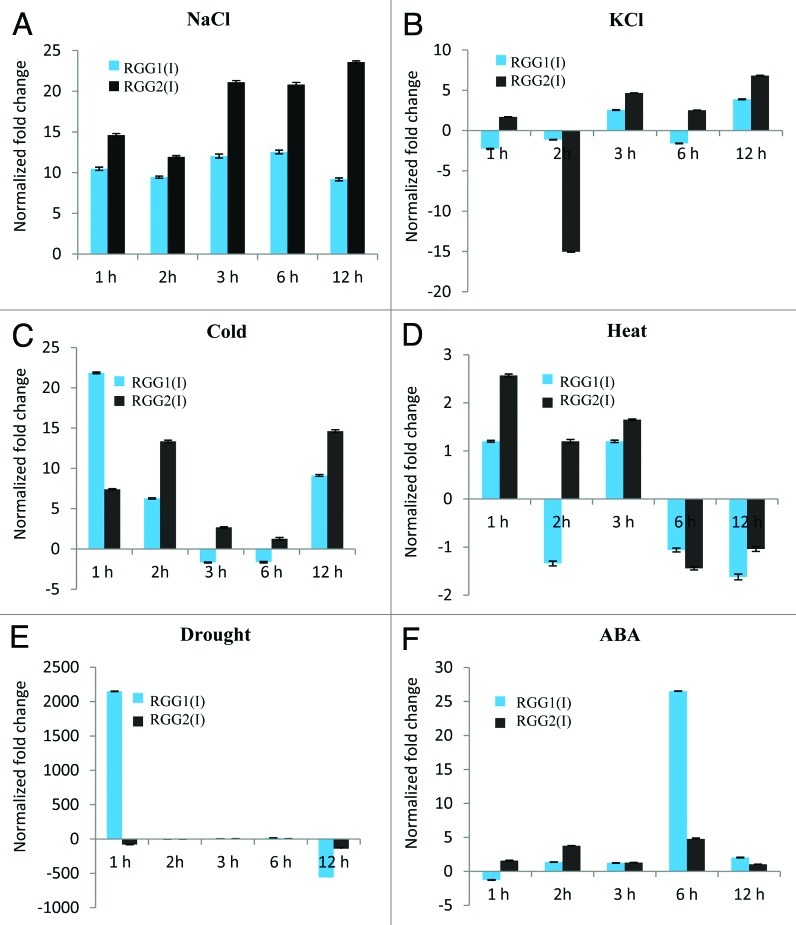
The 200 mM NaCl treatment induced the elevated expression of RGG1(I) and RGG2(I) by more than 10-fold at as early as 1 h. This elevation was maintained in case of RGG1(I) up to the observation period of 12 h while, RGG2(I) showed increased expression up to 23-fold at 12 h (Fig. 3A). Thus, it appears an early as well as prolong and strong response against NaCl. However, the same effect was not observed with 200 mM KCl treatment (Fig. 3B) suggesting that increased transcript levels of RGG1(I) and RGG2(I) was due to the high level of Na+. Exposure to cold stress caused expression of RGG1 and RGG 2 to increase by ca. 12- and 7-fold, respectively, for short time duration as soon as by 1h and subsequently, RGG1(I) expression decreased drastically (ca. 7-fold at 2 h) before maintaining the transient level (Fig. 3C). However, expression of RGG2(I) showed a rhythmic response by increasing ca. 13-fold at 2 h followed by decrease to basal level at 6 h and increased by ca. 14-fold at 12 h. On the other hand, expression of RGG1(I) and RGG2(I) under heat stress showed no significant change during observation period (Fig. 3D). During the drought-stress condition, expression of RGG1(I) rapidly increased up to 2150-fold by 1 h and decreased down to 560-fold at 12 h (Fig. 3E). On the other hand, RGG2(I) transcript level decreased 79-fold initially at 1h, after that it increased at 3 and 6 h before decreasing again at 12 h (141-fold) (Fig. 3E). Expression profile of RGG1 and RGG2, under 100 µM ABA treatment appeared as late response. In this case significant increase in expression of the RGG1(I) was observed at 6 h (26-fold) that decreased to 2.9-fold after 12 h (Fig. 3F). Whereas, RGG2(I) increased significantly by ca. 4 and 5-fold at 2 h and 6 h respectively, under ABA treatment (Fig. 3F).
Figure 3. Quantitative real-time PCR analyses showing expression profile of RGG1(I) and RGG2(I) in total RNA isolated from three-week-old rice seedling leaf blades samples collected at different time intervals, treated under different abiotic stress conditions (A) 200 mM NaCl; (B) 200 mM KCl; (C) Cold (4°C); (D) Heat (42°C); (E) drought condition and (F) 100 μM ABA. Error bars are SD.
Discussion
Rice is the most important staple crop that is cultivated worldwide. In the many rice-growing areas, there are frequent drought, salinity, extreme temperature, oxidative stress, heavy metal and many more abiotic stresses to impede rice growth and production. It promotes to elucidate the mechanisms of plant tolerance or resistance to a variety of stresses and improve the ability of crops to sustain against stresses. Heterotrimeric G-protein complex and related G-protein coupled receptor(s) are reported to play an important role in abiotic stresses.13 Heterotrimeric G-proteins consist of α, β and γ-subunits.31 A number of reports are available regarding the functions of heterotrimeric G-proteins in higher plants.32 Comprehensive analysis of plant G-proteins that integrate molecular, genetic and biochemical characterization and their roles in regulating specific signal transduction pathways is limited to α and β subunits of Arabidopsis and rice.3 Studies with mutants lacking α and β subunits have revealed their roles in transmission of external stimuli.33,34 However, in higher plants, very little information is available regarding function of G-protein γ subunits in abiotic stress signaling.
The γ subunit of heterotrimeric G-proteins of Indica rice seems to be an ortholog of the Gγ subunit of yeast and mammals. It can be judged from the length of the amino acid sequence and the presence of the important motifs. The RGG(I) proteins contain all the conserved features found in canonical Gγ. Both, RGG1(I) and RGG2(I) contain DPLL motif, which serves as an important hydrophobic contact to Gβ21. The RGG1(I) contains prenyl-group binding site (CAAX box) at its C-terminus. Prenylation is a post-translational lipid modification, which promotes protein-membrane and protein-protein interactions.35 It is also necessary for normal control of abscisic acid signaling and other fundamental processes.36 The increased expression level of RGG1(I) in presence of ABA at 6 h, suggest its role in ABA signaling pathway by activating downstream effectors due to presence of CAAX box. However, comparatively little increase in RGG2(I) expression under ABA treatment may be due to absence of prenyl-group-binding site (CAAX box) at its C-terminus. N-myristoylation is a co-translational or post-translational covalent modification of protein that can promote its association with membrane lipid. It is essential for the proper functioning of proteins in regulation of signaling pathways and involved in adaptation to high salt stress in plants.37 It is supported by the presence of N-myrisitoylation sites in RGG(I) subunits.
Although sequences of introns and UTRs are not the part of protein coding regions but they might have critical roles in gene expression regulation and evolution. The number and location of their introns and exons are similar and this conserved gene structure might lead to similar expression pattern RGG(I) genes. This is quite evident from their transcript profile under different abiotic stress conditions. It can be speculated that the structure of exon and intron might affect the expression RGG(I) of genes.38
Presence of stress responsive cis-regulatory elements in the promoter regions of RGG(I) genes can be well correlated with their transcript profile under different abiotic stresses. It strongly suggests their possible active role in regulation of abiotic stress signaling pathway. The increased expression levels of the RGG(I) subunits under high NaCl concentration suggest its possible role in adaptation to high salt stress. This increase in presence of high NaCl is unlike to high KCl as high K+/Na+ concentration is a requisite in view of plant nutrition.39 Moreover, G-protein α subunit mediated heat-stress signaling have been reported in pea13 and no considerable change in transcript profile of RGG(I) in this study suggests that 42°C temperature is either not a stress condition or heat-stress signaling is independent of RGG(I). Stress responsive genes are known to be expressed either through an ABA-dependent or ABA-independent pathway.40 This study suggests that gamma subunit of rice G-protein follow the ABA-dependent pathway. The presence of stress responsive cis elements indicate that some transcription factors may bind to these elements and activate the RGG(I) genes transcription under the stress conditions.
A generic signal transduction pathway starts with signal perception, followed by the generation of second messengers, which modulates intracellular Ca2+ levels, often initiating a protein phosphorylation cascade that finally targets proteins directly involved in cellular protection controlling specific sets of stress-regulated genes.41 Since the phosphorylation sites are present in RGG(I), they seems to be involved in cold and drought condition. Recently, ten Gγ genes have been found in the soybean genome and reported to have interesting expression profiles across different developmental stages.24
This research identifies the active participation of Gγ subunits in stress response, though its role in stress tolerance needs to be studied in detail. Taken together, the observations reported in this study present a first direct evidence for the regulation of transcript of Gγ1 and Gγ2 in response to abiotic stress. These studies could also provide new insight into the novel function of Gγ subunits of rice G-protein in abiotic stress response, thus suggesting a previously un-described molecule for manipulating stress tolerance in plants. These findings also provide an excellent starting point to investigate the potential roles of other subunits of rice G-proteins in plant stress tolerance. Overall, this study will contribute to our better understanding of G-proteins signaling under stress conditions in higher plants.
Materials and Methods
Plant material and stress treatment
Rice (Oryza sativa cv Indica group Swarna) seeds were grown in vermiculite in transgenic house under 16/8 h day light condition. For abiotic stress treatments the three week old seedlings were transferred to salt solutions (prepared in 1 × MS medium) in magenta boxes (200 mM NaCl, 200 mM KCl), abscisic acid (100 μM ABA) at room temperature for defined time intervals. For cold (4⁰C) and heat (42⁰C) treatment, seedlings in 1 × MS medium were kept in incubators at defined temperatures. Uprooted seedlings were kept on blotting paper for the mentioned period to mimic drought conditions. Young leaf blades of the stress treated seedlings were harvested at different time intervals (viz. 1 h, 2 h, 3 h, 6 h and 12 h). Seedlings grown in 1 × MS medium at room temperature were taken as a control. After sampling, the leaf blades (10 seedlings per treatment) were snap frozen in liquid nitrogen and stored at -80°C until use.
Isolation of RNA and cDNA preparation
Total RNA was isolated from 100 mg of stress treated and non-treated rice leaf blades, with TriZOL LS reagent (Invitrogen Life Technologies USA). DNaseI treatment was given to remove the contaminating genomic DNA. The total RNA obtained was used as template for cDNA synthesis. The first strand cDNA was synthesized from 5 μg of total RNA using Superscript II Reverse Transcriptase (Invitrogen Life Technologies USA) using oligo(dT)18 primer according to the manufacturer’s instructions. Experiments were performed thrice, independently.
Cloning of rice Gγ genes
For cloning, of the G-protein γ1 subunit [RGG1(I)], the primer pair 5′-CTCGAGCATATGCAGGCCGGAGGAGGA-3′ (Oligo-1, forward) and 5′-GAATTCTCACAAAAACCAGCATTTGCAT-3′ (Oligo-2, reverse), and for G-protein γ2 subunit [RGG2(I)], the primer pair 5′-CTCGAGCATATGAGGGGGGAGGCGAAC-3′ (Oligo-3, forward) and 5′-GAATTCCTAGGAAAAATCTGAGCCTTTG-3′ (Oligo-4, reverse) were used in PCR. The PCR reactions, using first strand cDNAs from Indica rice as template and respective primer pairs (Ta = 62°C), amplified DNA amplicon of 282 bp and 453 bp for RGG1(I) and RGG2(I) subunits, respectively. The full-length rice Gγ genes were cloned into the pGEMT easy vector (Promega). The putative recombinant colonies of E. coli DH5α, showing desired amplification were used for isolation of plasmid DNA using QIAprep Spin Miniprep kit (Qiagen) following manufacturer’s instructions. The plasmid DNA was confirmed for the gene insertion by restriction digestion using with NdeI and EcoRI enzymes. The putative positive colonies were subjected to nucleotide sequencing and thus obtained sequences were submitted to GenBank as Accession numbers GU111573 and GU066806 for RGG1(I) and RGG2(I), respectively.
In silico analysis of RGG1(I) and RGG2(I) proteins
The deduced amino acid sequences of RGG1 and RGG2 of Indica rice were compared with each other and with respective subunits of Japonica rice, maize, barley, Sorghum, Arabidopsis and soybean by multiple amino acid sequence alignment using ClustalW 2.0 program (www.ebi.ac.uk/clustalw).42 The pair wise amino acid sequence identity between RGG1(I) and RGG2(I), and with respective subunits of Japonica rice, maize, barley, Sorghum, Arabidopsis and soybean was calculated using ClustalW2 (EMBL-EBI). The ClustalW aligned amino acid sequences of RGG1(I) and RGG2(I) subunits, Japonica rice, maize, barley, Sorghum, Arabidopsis and soybean was used to infer the evolutionary relationship among them using the neighbor-joining method. The evolutionary distances were computed using the Poisson correction method43 and are in the units of the number of amino acid substitutions per site. These phylogenetic analyses were performed using MEGA5.44 The functional motifs, patterns and biologically significant sites in RGG1(I) and RGG2(I) amino acid sequence were located by ExPASy Proteomics Server ScanPro site (www.expasy.org/tools/scanprosite/).
In silico analysis of promoters and gene sequence of RGG1(I) and RGG2(I)
In order to analyze the putative cis-elements in the promoters, we searched 2.0 kb genomic sequence upstream of the translation initiation codon of RGG1(I) and RGG2(I) genes on cis-element database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).45 BLAST search in rice genome annotation project (http://rice.plantbiology.msu.edu/) was used to identify RGG(I) genomic DNA sequences including 5′ and 3′-UTR, exon and intron sequences.
Quantitative real-time PCR
The transcript profile of RGG1(I) and RGG2(I) under different stress conditions in leaf blades were determined by quantitative real time PCR. qPCR reactions were performed on StepOne Real Time PCR system (Applied Biosystems). Using Power SyberGreen PCR master mix (Applied BioSystems), a 20 μl reaction mixture containing 10 pM of each gene specific primer pair (α-tubulin forward 5′-GGTGGAGGTGATGATGCTTT-3′ and reverse 5′-ACCACGGGCAAAGTTGTTAG-3′; RGG1(I) forward 5′-CAAGAAGCTCGAGCAAGAGG-3′ and reverse 5′-CGGACCTTCAAACCATCTGT-3′; and RGG2(I) forward 5′-TGCAGGATGAACTGAACGAG-3′ and reverse 5′-GGATGCCCACCATTTGTTAC-3′) and 1 μl of stress treatment specific cDNA was prepared. PCR reaction conditions were as, one cycle of 10 min for 95°C for initial denaturation followed by 40 cycles of 15s at 95°C, 20s at 59°C and 30s at 72°C. Optical data were collected after every cycle. PCR products were melted by gradually increasing the temperature from 55–95°C in 0.5°C increment at every step. Rice α-tubulin gene was used as internal reference.46 The qPCR reactions were repeated thrice for each treatment. Relative gene expressions using the average CT values following Livaks’ method47 were calculated.
Acknowledgments
Work on signal transduction and plant stress signaling in N.T.’s laboratory is partially supported by Department of Science and Technology (DST), Government of India.
Glossary
Abbreviations:
- ABA
abscisic acid
- Gγ
G-protein γ subunits
- GPCR
G-protein coupled receptor
- RGG(I)
γ subunit of Indica rice
- ZmG
γ subunit of maize
- GmG
γ subunit of soybean
- SbG
γ subunit of Sorghum
- HvG
γ subunit of barley
- AGG
γ subunit of Arabidopsis
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/20356
References
- 1.Morris AJ, Malbon CC. Physiological regulation of G protein-linked signaling. Physiol Rev. 1999;79:1373–430. doi: 10.1152/physrev.1999.79.4.1373. [DOI] [PubMed] [Google Scholar]
- 2.Jones AM, Assmann SM. Plants: the latest model system for G-protein research. EMBO Rep. 2004;5:572–8. doi: 10.1038/sj.embor.7400174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perfus-Barbeoch L, Jones AM, Assmann SM. Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr Opin Plant Biol. 2004;7:719–31. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Tuteja N, Sopory SK. Plant signaling in stress: G-protein coupled receptors, heterotrimeric G-proteins and signal coupling via phospholipases. Plant Signal Behav. 2008;3:79–86. doi: 10.4161/psb.3.2.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trewavas AJ, Malho R. Signal perception and transduction: the origin of the phenotype. Plant Cell. 1997;9:1181–95. doi: 10.1105/tpc.9.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamm HE. The many faces of G protein signaling. J Biol Chem. 1998;273:669–72. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- 7.Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, et al. Insights into G protein structure, function, and regulation. Endocr Rev. 2003;24:765–81. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- 8.Offermanns S. G-proteins as transducers in transmembrane signalling. Prog Biophys Mol Biol. 2003;83:101–30. doi: 10.1016/S0079-6107(03)00052-X. [DOI] [PubMed] [Google Scholar]
- 9.Jones JC, Duffy JW, Machius M, Temple BR, Dohlman HG, Jones AM. The crystal structure of a self-activating G protein alpha subunit reveals its distinct mechanism of signal initiation. Sci Signal. 2011;4:ra8. doi: 10.1126/scisignal.2001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trusov Y, Zhang W, Assmann SM, Botella JR., Jr Ggamma1 + Ggamma2 not equal to Gbeta: heterotrimeric G protein Ggamma-deficient mutants do not recapitulate all phenotypes of Gbeta-deficient mutants. Plant Physiol. 2008;147:636–49. doi: 10.1104/pp.108.117655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma H, Yanofsky MF, Meyerowitz EM. Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1990;87:3821–5. doi: 10.1073/pnas.87.10.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa A, Tsubouchi H, Iwasaki Y, Asahi T. Molecular cloning and characterization of a cDNA for the alpha subunit of a G protein from rice. Plant Cell Physiol. 1995;36:353–9. doi: 10.1093/oxfordjournals.pcp.a078767. [DOI] [PubMed] [Google Scholar]
- 13.Misra S, Wu Y, Venkataraman G, Sopory SK, Tuteja N. Heterotrimeric G-protein complex and G-protein-coupled receptor from a legume (Pisum sativum): role in salinity and heat stress and cross-talk with phospholipase C. Plant J. 2007;51:656–69. doi: 10.1111/j.1365-313X.2007.03169.x. [DOI] [PubMed] [Google Scholar]
- 14.Assmann SM. Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell. 2002;14(Suppl):S355–73. doi: 10.1105/tpc.001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss CA, Garnaat CW, Mukai K, Hu Y, Ma H. Isolation of cDNAs encoding guanine nucleotide-binding protein beta-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1) Proc Natl Acad Sci U S A. 1994;91:9554–8. doi: 10.1073/pnas.91.20.9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaul S, Koo HL, Jenkins J, Rizzo M, Rooney T, Tallon LJ, Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 17.Mason MG, Botella JR. Isolation of a novel G-protein γ-subunit from Arabidopsis thaliana and its interaction with Gbeta. Biochim Biophys Acta. 2001;1520:147–53. doi: 10.1016/s0167-4781(01)00262-7. [DOI] [PubMed] [Google Scholar]
- 18.Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, et al. An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K⁺-channel regulation and morphological development in Arabidopsis thaliana. Plant J. 2011;67:840–51. doi: 10.1111/j.1365-313X.2011.04638.x. [DOI] [PubMed] [Google Scholar]
- 19.Marsh JF, 3rd, Kaufman LS. Cloning and characterisation of PGA1 and PGA2: two G protein alpha-subunits from pea that promote growth in the yeast Saccharomyces cerevisiae. Plant J. 1999;19:237–47. doi: 10.1046/j.1365-313X.1999.00516.x. [DOI] [PubMed] [Google Scholar]
- 20.Bisht NC, Jez JM, Pandey S. An elaborate heterotrimeric G-protein family from soybean expands the diversity of plant G-protein networks. New Phytol. 2010;190:35–48. doi: 10.1111/j.1469-8137.2010.03581.x. [DOI] [PubMed] [Google Scholar]
- 21.Temple BR, Jones AM. The plant heterotrimeric G-protein complex. Annu Rev Plant Biol. 2007;58:249–66. doi: 10.1146/annurev.arplant.58.032806.103827. [DOI] [PubMed] [Google Scholar]
- 22.Chen JG. Heterotrimeric G-proteins in plant development. Front Biosci. 2008;13:3321–33. doi: 10.2741/2928. [DOI] [PubMed] [Google Scholar]
- 23.Pandey S. More (G-proteins) please! Identification of an elaborate network of G-proteins in soybean. Plant Signal Behav. 2011;6:780–2. doi: 10.4161/psb.6.6.15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choudhury SR, Bisht NC, Thompson R, Todorov O, Pandey S. Conventional and novel Gγ protein families constitute the heterotrimeric G-protein signaling network in soybean. PLoS One. 2011;6:e23361. doi: 10.1371/journal.pone.0023361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klopffleisch K, Phan N, Augustin K, Bayne RS, Booker KS, Botella JR, et al. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol Syst Biol. 2011;7:532–8. doi: 10.1038/msb.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Busk PK, Pagès M. Regulation of abscisic acid-induced transcription. Plant Mol Biol. 1998;37:425–35. doi: 10.1023/A:1006058700720. [DOI] [PubMed] [Google Scholar]
- 27.Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–19. doi: 10.1046/j.1365-313X.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- 28.Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, et al. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell. 2005;17:1387–96. doi: 10.1105/tpc.105.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutoh K, Yamauchi D. Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J. 2003;34:635–45. doi: 10.1046/j.1365-313X.2003.01753.x. [DOI] [PubMed] [Google Scholar]
- 30.Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004;135:2150–61. doi: 10.1104/pp.104.041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason MG, Botella JR. Completing the heterotrimer: isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proc Natl Acad Sci U S A. 2000;97:14784–8. doi: 10.1073/pnas.97.26.14784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato C, Mizutani T, Tamaki H, Kumagai H, Kamiya T, Hirobe A, et al. Characterization of heterotrimeric G protein complexes in rice plasma membrane. Plant J. 2004;38:320–31. doi: 10.1111/j.1365-313X.2004.02046.x. [DOI] [PubMed] [Google Scholar]
- 33.Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, et al. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci U S A. 1999;96:7575–80. doi: 10.1073/pnas.96.13.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lease KA, Wen J, Li J, Doke JT, Liscum E, Walker JC. A mutant Arabidopsis heterotrimeric G-protein β subunit affects leaf, flower, and fruit development. Plant Cell. 2001;13:2631–41. doi: 10.1105/tpc.010315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–69. doi: 10.1146/annurev.bi.65.070196.001325. [My paper] [DOI] [PubMed] [Google Scholar]
- 36.Crowell DN, Huizinga DH. Protein isoprenylation: the fat of the matter. Trends Plant Sci. 2009;14:163–70. doi: 10.1016/j.tplants.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 37.de Jonge HR, Hogema B, Tilly BC. Protein N-myristoylation: critical role in apoptosis and salt tolerance. Sci STKE. 2000;2000:pe1. doi: 10.1126/stke.2000.63.pe1. [DOI] [PubMed] [Google Scholar]
- 38.Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- 39.Tuteja N. Mechanisms of high salinity tolerance in plants. Methods Enzymol. 2007;428:419–38. doi: 10.1016/S0076-6879(07)28024-3. b. [DOI] [PubMed] [Google Scholar]
- 40.Tuteja N. Abscisic Acid and abiotic stress signaling. Plant Signal Behav. 2007;2:135–8. doi: 10.4161/psb.2.3.4156. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14(Suppl):S165–83. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. Edited in Evolving Genes and Proteins by V. Bryson and H.J. Vogel. Academic Press, New York, 1965; 97-166. [Google Scholar]
- 44.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–7. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baisakh N, Subudhi PK, Varadwaj P. Primary responses to salt stress in a halophyte, smooth cordgrass (Spartina alterniflora Loisel.) Funct Integr Genomics. 2008;8:287–300. doi: 10.1007/s10142-008-0075-x. [DOI] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]