Abstract
Long noncoding RNAs (lncRNAs) have gained widespread attention in recent years as a potentially new and crucial layer of biological regulation. lncRNAs of all kinds have been implicated in a range of developmental processes and diseases, but knowledge of the mechanisms by which they act is still surprisingly limited, and claims that almost the entirety of the mammalian genome is transcribed into functional noncoding transcripts remain controversial. At the same time, a small number of well-studied lncRNAs have given us important clues about the biology of these molecules, and a few key functional and mechanistic themes have begun to emerge, although the robustness of these models and classification schemes remains to be seen. Here, we review the current state of knowledge of the lncRNA field, discussing what is known about the genomic contexts, biological functions, and mechanisms of action of lncRNAs. We also reflect on how the recent interest in lncRNAs is deeply rooted in biology’s longstanding concern with the evolution and function of genomes.
Keywords: noncoding RNA (ncRNA), epigenetics, gene regulation, high-throughput sequencing, C-value enigma
THE past several years have witnessed a steep rise of interest in the study of lncRNAs. Almost on a weekly basis, it seems that a new lncRNA is found to be up- or downregulated in a particular disease, or a new class of noncoding transcripts is uncovered by a transcriptomic study, or a new article heralds a paradigm shift that lncRNAs will bring to our understanding of biology. Without a doubt, the advent of sensitive, high-throughput genomic technologies such as microarrays and next-generation sequencing (NGS) has resulted in an unprecedented ability to detect novel transcripts, the vast majority of which seem not to be derived from annotated protein-coding genes. Despite this explosion of data, however, surprisingly little is known about how lncRNAs function, how many different types of lncRNAs exist, or even whether most of them carry biological significance.
In this review, we focus on recently discovered lncRNAs of >200 nt, place these new discoveries in historical context, and outline areas where additional work is needed. The review does not cover “classic” ncRNAs such as ribosomal (r)RNAs, ribozymes, transfer (t)RNAs, small nuclear (sn)RNAs, small nucleolar (sno)RNAs, and telomere-associated RNAs (TERC, TERRA); nor does it cover small ncRNAs such as microRNAs (miRNAs), endogenous small interfering (endo-si)RNAs that participate in RNA interference (RNAi), and Piwi-associated (pi)RNAs. We refer readers to the many excellent recent reviews on these topics (Peculis 2000; Xiao et al. 2002; Henras et al. 2004; Okamura and Lai 2008; Kim et al. 2009; Feuerhahn et al. 2010; Blackburn and Collins 2011; Czech and Hannon 2011; Siomi et al. 2011). Although lncRNAs are found across many taxa, and many crucial discoveries have been made in plants, fungi, and invertebrates (Gelbart and Kuroda 2009; Au et al. 2011), we largely limit our discussion to mammalian examples, with occasional reference to lncRNAs of other taxa.
Historical Overview of the lncRNA Field: Where Did It Come From? What Is It? Where Is It Going?
The C-value enigma and junk DNA
While widespread attention on lncRNAs is a rather recent phenomenon, it fits into the broader historical interest in studying the size, evolution, and function of genomes. Since the 1950s, the C-value, or the amount of DNA in the haploid genome (i.e., genome size), has been known to show little correlation with organism size or developmental complexity (Mirsky and Ris 1951; Thomas 1971; Gall 1981). “Lower” animals such as the salamander can have a genome 15 times larger than that of “higher” animals like humans (Gall 1981). This “C-value paradox” (Thomas 1971) troubled scientists with a human-centric point of view: “Being a little chauvinistic toward our own species, we like to think that man is surely one of the most complicated species on earth and thus needs just about the maximum number of genes” (Comings 1972, p. 313).
The paradox was arguably “solved” with the discovery that much of the genome does not encode protein-coding genes. Based on both DNA–RNA hybridization experiments (Lewin 1980, Chap. 24) and calculations of mutational load in the genome (i.e., the cost to evolutionary fitness due to deleterious mutations, which depends on the mutation rate per gene and the number of genes) (Ohno 1972), it was determined in the 1970s that humans are unlikely to have >20,000–30,000 (protein-coding) genes, remarkably close to the current estimate based on the human genome sequence [unlike the overestimates of 50,000–100,000 from the early days of the Human Genome Project (Pertea and Salzberg 2010)]. The remaining noncoding space was termed “junk DNA” (Comings 1972; Ohno 1972) due to its overwhelming burden of transposons, pseudogenes, and simple repeats [in total accounting for 50–70% of the human genome (de Koning et al. 2011)].
Although the discrepancy in genome size may no longer be “paradoxical”, there remains a “C-value enigma” (Gregory 2001). Even among species that are morphologically similar and phylogenetically close [e.g., onion, garlic, and their relatives in the genus Allium (T. R. Gregory, personal communication)], genome size and thus, presumably, noncoding content can vary by four- to fivefold, even when counting only diploid species (Ricroch et al. 2005). Whole-genome sequencing in recent years suggests that noncoding content may correlate with organismic “complexity” (Taft et al. 2007), but such a conclusion must await a more extensive and varied sample of nonmammalian genome sequences.
Despite their status as “junk”, noncoding sequences received sustained interest during the period between the 1970s and the present. Early pioneers had the foresight to realize that “being junk doesn’t mean it is entirely useless” (Comings 1972, p. 316), and “it would be surprising if the host organism did not occasionally find some use for” a portion of these sequences (Orgel and Crick 1980, p. 606). Among a multitude of hypothesized functions are chromosomal pairing, genome integrity, gene regulation, messenger (m)RNA processing, and serving as a reservoir for evolutionary innovation (Britten and Davidson 1971; Yunis and Yasmineh 1971; Comings 1972; John and Miklos 1979; Lewin 1980, Chap. 17\x{2013}19; Orgel and Crick 1980; Lewin 1982). In light of recent discoveries, these early genomicists may not have been too far from the truth.
Pervasive transcription: Useful junk or transcriptional noise?
In the 1970s, hints had begun to emerge that more of the genome is transcribed than could be attributed to coding genes and various known RNAs such as rRNAs and tRNAs. These so-called “heterogeneous nuclear RNAs” (hnRNAs) are transcribed from repetitive and heterochromatic regions, as well as from upward of 20% of all nonrepetitive regions in mammalian genomes (>10-fold higher by the same measure than the amount of transcription into mRNA). It was also known that 50% of hnRNAs are restricted to the nucleus and do not contain coding sequences (Holmes et al. 1972; Pierpont and Yunis 1977; Lewin 1980, Chap. 25). Introns, discovered in 1977 (Berget et al. 1977; Chow et al. 1977), accounted for only a small part of the noncoding sequences. Shortly after, in the 1980s, snRNAs and snoRNAs became recognized as major players in post-transcriptional RNA processing.
The scale of “pervasive transcription,” however, was not fully appreciated until the arrival of whole-genome technologies in the late 1990s and early 2000s. From microarray hybridization and deep sequencing analyses, it is now estimated that as much as 70–90% of our genome is transcribed at some point during development (Okazaki et al. 2002; Rinn et al. 2003; Bertone et al. 2004; Ota et al. 2004; Carninci et al. 2005; Birney et al. 2007; Kapranov et al. 2010; Mercer et al. 2011; Djebali et al. 2012). However, it should be pointed out that the idea of pervasive transcription, while popular, has been challenged on a number of grounds, including the low cross-species conservation rate (Wang et al. 2004) and extremely low expression levels for many transcripts. Some recently identified transcripts are calculated to be present at as low as 0.0006 copies per cell (Mercer et al. 2011). There has also been criticism of technical limitations with tiling microarrays, including problems with false positives, low dynamic range, resolution, and low concordance between studies (Agarwal et al. 2010; van Bakel et al. 2010, 2011). Some high-throughput RNA sequencing (RNA-Seq) analyses suggest that much of pervasive transcription might be explained by alternative splicing and/or extensions of known protein-coding genes (He et al. 2008; Mortazavi et al. 2008; Sultan et al. 2008; van Bakel et al. 2010, 2011). Evidence supporting the existence of noncoding transcription in intergenic regions has come from correlations with chromatin signatures, such as DNase1 hypersensitivity; histone modifications like H3K9ac, H3K4me3, and H3K36me3; or binding of transcription factors (TFs) at the loci and dependence of expression levels on these TFs (Guttman et al. 2009, 2011; van Bakel et al. 2010; Encode Project Consortium 2012). These studies have revealed promising, novel, conserved lncRNAs, but the new transcripts number in only a few thousand—not enough to explain 70–90% of the genome.
The big question, then as now, is whether such transcriptional activities serve any biological function. Even as early as 1961, when Jacob and Monod first deduced the existence of mRNA and expounded the repressor–operator model of gene regulation, they speculated that the repressor could be an RNA molecule (Jacob and Monod 1961). In 1969, Britten and Davidson postulated a model for regulation of gene expression in eukaryotes where ncRNAs act as regulatory intermediaries to convey signals, received at sensory genetic elements, to receptor elements that affect coding gene production (although formally, the identity of the intermediaries could be proteins) (Britten and Davidson 1969). Some of the first cases of gene-specific regulatory roles of lncRNAs were uncovered in the early 1990s, with the discovery of lncRNAs involved in epigenetic regulation, such as H19 (Brannan et al. 1990) and Xist (Brockdorff et al. 1992; Brown et al. 1992). However, the idea of “transcriptional noise” (Hüttenhofer et al. 2005) continues to resonate in the field, with the argument that the proper null hypothesis is lack of function, and the burden of proof is on supporters to find function. Calculations for TF binding (Wunderlich and Mirny 2009) and RNA polymerase II (Pol II) initiation of transcription (Struhl 2007) in eukaryotes have shown that Pol II can initiate “nonspecifically” and that as much as 90% of Pol II transcription can be spurious. Transcription events also seem to have a tendency to spill over or “ripple” away from “legitimate” transcripts, potentially leading to leaky expression of neighboring regions (Ebisuya et al. 2008). How, then, should one approach the field of lncRNAs in 2013?
Genomic Contexts of lncRNAs
One way to find order in the plethora of reported lncRNAs is to classify them according to genomic location—i.e., from where in the genome these RNAs are transcribed, relative to well-established markers such as protein-coding genes (Figure 1). In this manner, lncRNAs can be grouped into five broad but mutually nonexclusive categories. The caveat to this approach, however, is that genomic context does not necessarily provide any information about lncRNA function or evolutionary origin, but it does serve as a convenient shorthand to organize these diverse species.
Figure 1.
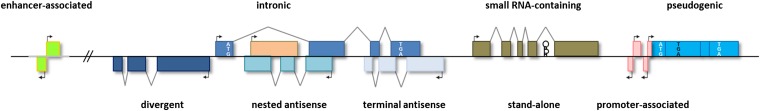
Genomic contexts of lncRNAs. lncRNAs may be stand-alone transcription units, or they may be transcribed from enhancers (eRNAs), promoters (TSSa-RNAs, uaRNAs, pasRNAs, and PROMPTs), or introns of other genes (in this case a protein-coding gene, with start codon ATG and stop codon TGA in white); from pseudogenes (shown here with a premature stop codon TGA in black); or antisense to other genes (NATs) with varying degrees of overlap, from none (divergent), to partial (terminal), to complete (nested). lncRNAs may also host one or more small RNAs (black hairpin) within their transcription units.
Stand-alone lncRNAs
These lncRNAs are distinct transcription units located in sequence space that do not overlap protein-coding genes. Some of these have been referred to as “lincRNAs” for “large intergenic (or intervening) noncoding RNAs” (Guttman et al. 2009; Cabili et al. 2011; Ulitsky et al. 2011). A large number were identified through chromatin signatures for actively transcribed genes (H3K4me3 at the promoter, H3K36me3 along the transcribed length). Many of the characterized ones are transcribed by RNA Pol II, polyadenylated, and spliced (usually with alternative isoforms, but with fewer exons than coding mRNAs) and have an average length of 1 kb. Known examples include Xist (Brockdorff et al. 1992; Brown et al. 1992), H19 (Brannan et al. 1990), HOTAIR (Rinn et al. 2007) and MALAT1 (Ji et al. 2003).
Natural antisense transcripts
Abundant transcription appears to occur opposite the sense DNA strand of annotated transcription units; as much as 70% of sense transcripts have reported antisense counterparts (Katayama et al. 2005; He et al. 2008; Faghihi and Wahlestedt 2009). The overlap between these sense–antisense (SAS) pairs can be complete, with either transcript nested within the other, but natural antisense transcripts (NATs) tend mostly to be enriched around the 5′ (promoter) or 3′ (terminator) ends of the sense transcript. There are a number of well-documented SAS pairs formed by two coding mRNAs, as well as dual lncRNA SAS pairs like Xist/Tsix, two RNAs that control X chromosome inactivation (Lee et al. 1999a). In addition, many imprinted regions contain coding/noncoding SAS pairs, such as Kcnq1/Kcnq1ot1 (Kanduri et al. 2006) and Igf2r/Air (Lyle et al. 2000). Fewer of the newly discovered NATs are spliced or polyadenylated when compared to mRNAs or stand-alone lncRNAs, and while the expression of SAS pairs is more intercorrelated (either positively or negatively) than expected by chance alone, whether most NATs have biological function remains to be seen.
Pseudogenes
These are the “relics” of genes that have lost their coding potential due to nonsense, frameshift, and other mutations (Balakirev and Ayala 2003; Pink et al. 2011). Many pseudogenes are products of tandem gene duplication or of mRNAs being carried along during retrotransposition, both of which create extra gene copies that are no longer under selective pressure. By some estimates, there might be as many pseudogenes as functional coding genes. The vast majority of pseudogenes are “dead”—i.e., they are no longer expressed and their genetic sequences drift at a neutral rate. However, a portion of pseudogenes are transcribed (estimates ranging from 2 to 20%) and sometimes have high levels of sequence conservation; a few rare examples have even been shown to be translated. Expressed pseudogenes may be intermediates on their way to complete pseudogenization (Harrison et al. 2005), or they may be dead pseudogenes that have been “resurrected” and acquired new functions (Bekpen et al. 2009). Some transcribed pseudogenes have been found to regulate gene expression (often of their ancestral coding genes) by epigenetic or post-transcriptional mechanisms. In fact, Xist is hypothesized to have evolved by the pseudogenization of the protein-coding gene Lnx3 and integration of various transposon-derived repeat elements (Duret et al. 2006; Elisaphenko et al. 2008).
Long intronic ncRNAs
Introns have long been known to harbor small ncRNAs such as snoRNAs and miRNAs. Recently, many long transcripts have been reported, by large-scale transcriptomic or computational analyses, to be encoded within the introns of annotated genes (Louro et al. 2009; Rearick et al. 2011). Many of these are observed to have differential expression patterns, respond to stimuli, or be misregulated in cancer, but only a few have been studied in detail to date (Guil et al. 2012). One example that has been implicated in plant vernalization is COLDAIR, which is located in the first intron of the flowering repressor locus FLC (Heo and Sung 2011).
Divergent transcripts, promoter-associated transcripts, and enhancer RNAs
Abundant short transcripts (ranging from 20 to 2500 nt) have been found to be produced from the vicinity of transcription start sites in both sense and antisense directions, corresponding to peaks of Pol II occupancy due to pausing (Buratowski 2008; Core et al. 2008; He et al. 2008; Preker et al. 2008; Seila et al. 2008). The shortest of these, called transcription start site-associated (TSSa-)RNAs, may be degradation products or processed from the longer upstream antisense (ua)RNAs or promoter upstream transcripts (PROMPTs). These heterogeneous transcripts are usually capped and polyadenylated, have low abundance (as little as 0.1 copy per cell), and are subject to rapid degradation by exosomes. It is currently unclear whether these are simply transcriptional by-products from nucleosome-free regions around promoters, whether the act of their of transcription helps maintain this environment of open chromatin, or whether the transcripts themselves play a regulatory role, especially since a subset called promoter-associated short (pas)RNAs has been found to interact with epigenetic factors such as Polycomb proteins (Kanhere et al. 2010). In addition to promoters, another class of genomic regulatory elements, the enhancers, has also been found to produce short (<2 kb) bidirectional transcripts [enhancer (e)RNAs], but these tend not to be processed, and as of yet no known biological function has been found (Kim et al. 2010; Wang et al. 2011a).
lncRNAs: A Functional Perspective
Before surveying molecular mechanisms, it is helpful to summarize what is currently known about lncRNA function. From a variety of screens and expression analyses, it is increasingly evident that changes in expression levels of many lncRNAs are correlated with developmental processes and disease states. Functional studies indicate important roles for several lncRNAs, but the majority of lncRNAs await further verification.
Regulation of allelic expression: X chromosome inactivation
Arguably the best-studied biological function for lncRNAs occurs in the epigenetic regulation of allelic expression, such as the processes of dosage compensation and genomic imprinting. The difference in X-linked gene dosage between XX females and XY males in therian mammals is compensated for by a mechanism known as X chromosome inactivation (XCI), in which one of the two X chromosomes in females is heterochromatinized and silenced (the inactive X, or Xi) such that only one X remains active and is expressed in each female cell (the active X, or Xa) (Lyon 1961; Lee 2011). In placental mammals there are two types of XCI: imprinted XCI in the fertilized embryo and extraembryonic tissues, where the paternal X is always inactivated (Takagi and Sasaki 1975; Okamoto et al. 2004), and random XCI, occurring in the inner cell mass (which becomes the embryo proper), where epigenetic marks have been erased after the embryo’s implantation into the uterus (Monk and Harper 1979; Mak et al. 2004). In random XCI, either the paternal or the maternal X is randomly chosen for inactivation, leading to a mosaic female (McMahon et al. 1983). XCI in placental mammals is largely controlled by a cluster of lncRNA loci known as the X-inactivation center (Xic) (Brown et al. 1991) (Figure 2). The 17-kb X (inactive)-specific transcript (Xist) is highly expressed from Xi during the onset of XCI, but not from Xa. Xist RNA then coats the X chromosome and forms an “Xist cloud” (Brown et al. 1992; Clemson et al. 1996), which acts as a scaffold for the recruitment of silencing factors such as Polycomb repressive complex 2 (PRC2) (Zhao et al. 2008).
Figure 2.
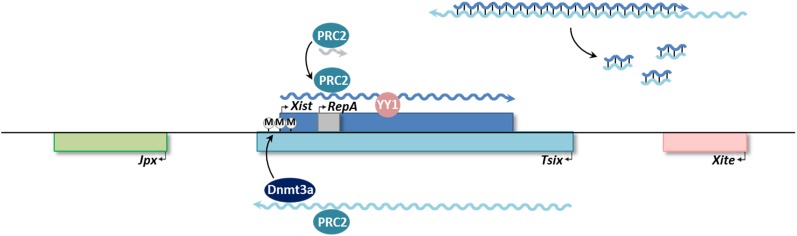
Noncoding loci at the Xic. On Xi, RepA is thought to recruit PRC2 to the Xist promoter to (paradoxically) upregulate transcription. PRC2 may then be loaded onto Xist, which remains tethered to its allele of origin via YY1 interactions with RNA and DNA. Meanwhile on Xa, Tsix expression is believed to repress Xist through a combination of several mechanisms: titrating away PRC2, preventing its proper docking onto RepA/Xist; recruiting Dnmt3a, laying down DNA methylation (Ⓜ) to silence the Xist promoter; and/or directly base pairing with Xist RNA, becoming a substrate for Dicer-dependent processing into small RNAs.
As it turns out, Xist itself is also regulated by other lncRNAs. Tsix, transcribed in the antisense orientation from a promoter downstream of Xist, is highly expressed before XCI initiates and then disappears from the future Xi while persisting on the presumptive Xa, thus exhibiting the reverse pattern as Xist expression (Lee et al. 1999a). Tsix has been demonstrated to coordinate X chromosome pairing to generate epigenetic asymmetry within the Xist locus (Bacher et al. 2006; Xu et al. 2006) and to downregulate Xist by a number of potential mechanisms (Sado et al. 2005; Sun et al. 2006; Ogawa et al. 2008; Zhao et al. 2008). Xist is also positively regulated in trans by the upstream Jpx RNA, although its mechanism of action is currently unclear (Chureau et al. 2002; Johnston et al. 2002; Tian et al. 2010).
Regulation of allelic expression: Imprinting
lncRNAs are also important in genomic imprinting, the process by which a gene is expressed monoallelically according to its parent of origin (Edwards and Ferguson-Smith 2007; Wan and Bartolomei 2008). As in XCI, imprinting is usually regulated by specific genomic loci called imprinting control regions, from which lncRNAs often emanate, similar to the Xic. Many imprinted clusters contain protein-coding genes and lncRNAs that are reciprocally expressed, such as Igf2r/Air (Lyle et al. 2000), Dlk1/Gtl2 (Schmidt et al. 2000; da Rocha et al. 2008), Nesp/Nespas/Gnas (Williamson et al. 2006), and the Beckwith–Wiedemann syndrome (BWS)-associated Kcnq1/Kcnq1ot1 (Lee et al. 1999b; Kanduri et al. 2006). Some of these lncRNAs may function by recruiting epigenetic factors, such as PRC2 and G9a, to control the imprinted expression of neighboring coding genes (Nagano et al. 2008; Pandey et al. 2008; Zhao et al. 2010). H19, another lncRNA from the BWS locus, was one of the first mammalian lncRNAs to be identified and is one of the most highly expressed transcripts in the embryo (Brannan et al. 1990; Bartolomei et al. 1991). It is reciprocally imprinted with the protein-coding gene Igf2. Although H19 does not seem to function as a lncRNA (Jones et al. 1998; Gabory et al. 2010), it serves as a miRNA precursor (Cai and Cullen 2007; Keniry et al. 2012).
Other roles in development
Beyond allelic regulation, the role of lncRNAs extends to other aspects of development, from the control of pluripotency to lineage specification. As explained above, the process of XCI is tightly coupled to early embryonic development, as well as to pluripotency in embryonic stem (ES) cells and induced pluripotent stem cells (Deuve and Avner 2011). Members of the core network of pluripotency transcription factors (e.g., Oct4, Sox2, and Nanog) have been shown to colocalize to Xist intron 1 (Navarro et al. 2008; Donohoe et al. 2009; Nesterova et al. 2011), and Oct4 has been demonstrated to regulate the expression of Tsix and Xite (an upstream regulator of Tsix also found in the Xic), in turn controlling X chromosome pairing and setting off the XCI cascade (Donohoe et al. 2009). More recently, Oct4 has been tied to expression of a series of pluripotency-associated lncRNAs (Sheikh Mohamed et al. 2010), some of which might serve as scaffolds for Sox2 and the PRC2 component Suz12 in regulating downstream targets (Ng et al. 2011). The expression of Oct4 may itself be regulated by a lncRNA antisense to one of its pseudogenes (Hawkins and Morris 2010).
lncRNAs are additionally implicated in processes later in animal development. The Hox genes encode homeodomain TFs that are crucial for anterior–posterior pattern formation in all bilaterian metazoans (Pearson et al. 2005). Hox genes are arranged in linear clusters along the chromosome, and mammals possess four paralogous clusters, HOXA, -B, -C, and -D. A number of lncRNAs are encoded within these clusters, including HOTAIR from HOXC, and HOTTIP and Mistral from HOXA (Rinn et al. 2007; Bertani et al. 2011; Wang et al. 2011b). These lncRNAs are proposed to regulate expression of Hox genes from either the host or a distant cluster. Neuronal development is another process where lncRNAs have been implicated. The noncoding Evf2 locus, which is associated with an enhancer located between two homeodomain TFs, Dlx5 and Dlx6, was shown by in vivo knockout experiments to regulate the development of GABAnergic interneurons in the postnatal mouse forebrain. It acts through both cis (transcription-based) repression of Dlx6 and trans (transcript-based) activation of Dlx5 (Bond et al. 2009). In addition, a number of lncRNAs might be necessary in the specification of neuronal vs. glial fate, perhaps through the recruitment of epigenetic complexes such as PRC2 (Ng et al. 2011).
Implications in cancer
lncRNAs have also been associated with disease, most notably cancer (Gutschner and Diederichs 2012). For example, a recent RNA-Seq study in prostate cancer tissues and cell lines uncovered a lncRNA, PCAT-1, that promotes cell proliferation and is a target of PRC2 regulation (while also possibly interacting with PRC2 itself) (Prensner et al. 2011). ANRIL, also upregulated in prostate cancer, is required for the repression of the tumor suppressors INK4a/p16 and INK4b/p15 (Yap et al. 2010; Kotake et al. 2011). HOTAIR overexpression is associated with poor prognosis in breast (Gupta et al. 2010), liver (Z. Yang et al. 2011), colorectal (Kogo et al. 2011), gastrointestinal (Niinuma et al. 2012), and pancreatic (Kim et al. 2012) cancers and is proposed to increase tumor invasiveness and metastasis (Gupta et al. 2010). MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), another lncRNA associated with various cancers and metastasis (Ji et al. 2003; Lin et al. 2011), is found to affect the transcriptional and post-transcriptional regulation of cytoskeletal and extracellular matrix genes (Tano et al. 2010). lincRNA-p21 (named for its vicinity to the CDKN1A/p21 locus) is upregulated by p53 upon DNA damage and implicated in downstream repressive effects of the p53 pathway, particularly on genes regulating apoptosis, possibly by directing the recruitment of hnRNP-K to its genomic targets (Huarte et al. 2010). Another DNA damage-responsive, p53-induced lncRNA that lies upstream of p21, PANDA (P21 associated ncRNA DNA damage activated), is also implicated in the repression of pro-apoptotic genes, such as FAS and BIK, by acting as a decoy for the transcription factor NF-YA. In some cancer types, p53 mutations have been found that maintain the protein’s ability to induce the PANDA pathway (and its antiapoptotic effects) while abolishing its ability to induce p21 and its promotion of cell-cycle arrest, thus leading to increased tumor cell survival (Hung et al. 2011). The above examples suggest that lncRNAs may be used as diagnostic markers or therapeutic targets in the treatment of cancer, but much work needs to be done before such applications become clinically practical.
Mechanisms of Action: Finding Pattern in Chaos
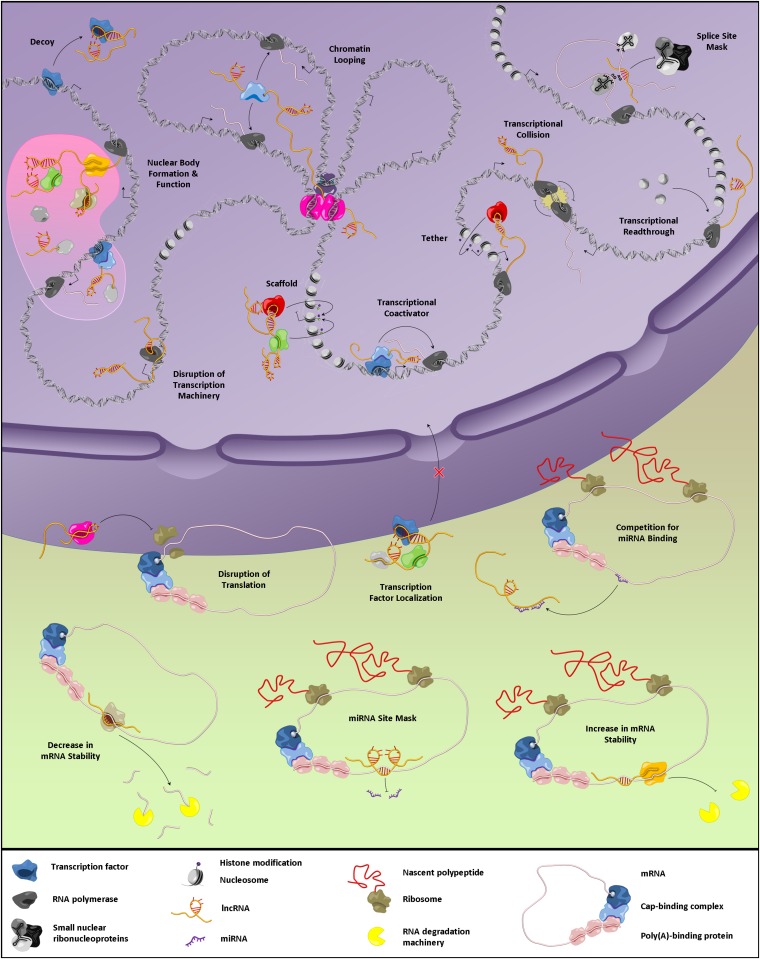
Although the vast majority of lncRNAs described in the literature have not yet been studied in mechanistic detail, the few that have provide clues regarding how lncRNAs might carry out their biological roles (Figure 3). However, it will be seen that many lncRNAs blur the lines between categories or employ more than one mechanism of action. The continued discovery of new lncRNAs and more thorough characterization of those already known will surely reveal additional themes.
Figure 3.
Mechanisms of lncRNA function. See text for detailed discussion.
lncRNAs in epigenetics: Recruiters, tethers, and scaffolds
A major recurrent theme in lncRNA biology is the ability to function in the recruitment of protein factors for regulation of chromatin states (Campos and Reiberg 2009). Members of this class of lncRNAs may function in cis, acting on linked genes in the vicinity of the RNA’s site of synthesis; or they might act in trans, regulating genes located in other, often distant domains or chromosomes. Large-scale and genome-wide studies of RNA–protein interactions have shown that chromatin-modifying complexes, such as PRC2, interact with a large number of lncRNAs (Khalil et al. 2009; Kanhere et al. 2010; Zhao et al. 2010; Guil et al. 2012). The Polycomb proteins, first discovered in Drosophila as regulators of homeotic gene expression during development (Schwartz and Pirrotta 2007), include a number of factors that bind or modify chromatin marks. These include Ezh2 in PRC2, which is a key H3K27 methyltransferase, and the Pc/Cbx family proteins in PRC1, chromodomain-containing proteins that can bind trimethylated H3K27 (Sparmann and Van Lohuizen 2006; Schwartz and Pirrotta 2007). The mechanisms by which the Polycomb complexes are recruited to specific genomic loci in mammals have been elusive, especially in the absence of definitive consensus binding sequences, unlike the well-defined Polycomb response elements (PREs) in fruit flies (Schwartz and Pirrotta 2007). However, observed interactions of Polycomb proteins with lncRNAs suggest that Polycomb recruitment in mammals might be directed by RNA.
HOTAIR in the HOXC cluster has been observed to repress transcription of HOXD in trans through interaction with PRC2 (Rinn et al. 2007), although the mechanism of trans-action has not been defined. Xist RNA and a related 1.6-kb transcript called RepA also recruit PRC2 (Zhao et al. 2008). The mechanism of action appears different from HOTAIR’s in that Xist/RepA targets PRC2 in cis to its site of synthesis to initiate XCI. RepA targets PRC2 to the Xist promoter and is associated with Xist upregulation; full-length Xist then binds and recruits PRC2 to the rest of the X chromosome. The RepA/Xist interaction with PRC2 may be blocked by the antisense Tsix transcript, which also binds PRC2 and may therefore competitively inhibit interaction with the sense transcripts (Zhao et al. 2008). Further investigation revealed that the RNA–protein complex loads onto a “nucleation center” within the first exon of Xist, a process that depends on another Polycomb group transcription factor, YY1 (homolog of the fly Pho protein that forms part of the PRE-binding complex), which is bound only to Xi and leads to Xi-specific recruitment of PRC2 (Jeon and Lee 2011). By cotranscriptionally tethering Xist RNA to the Xic, YY1 serves as a bridge between lncRNA and chromatin and explains the allele-specific binding of Xist to Xi in cis. Nevertheless, in post-XCI cells, Xist is capable of trans-silencing autosomal regions where nucleation centers have been ectopically introduced, bypassing the developmental programming that would normally block these centers in differentiated cells (Jeon and Lee 2011). Thus, a strict cis/trans distinction in describing lncRNA function may not be as feasible as once thought.
Other epigenetic complexes are likely to interact with lncRNAs as well, such as the H3K9 methyltransferase G9a, which interacts with the imprinted lncRNA Air (Nagano et al. 2008). In some cases, the RNA acts as a scaffold onto which multiple protein complexes can assemble, allowing the coordination of multiple layers of chromatin modifications. For example, Kcnq1ot1 has been hypothesized to recruit both PRC2 and G9a to the promoter of Kcnq1 (Pandey et al. 2008). ANRIL, located in the p15/INK4B-p16/INK4A-p14/ARF tumor suppressor gene cluster, interacts with both the PRC1 component Cbx7 and the PRC2 component Suz12 (Yap et al. 2010; Kotake et al. 2011). HOTAIR, in addition to PRC2, interacts with the LSD1/CoREST/REST complex that demethylates histone H3K4 to prevent gene activation (Tsai et al. 2010), thereby potentially synergizing the repressive effects. A number of other lncRNAs might dually bind both PRC2 and CoREST complexes (Khalil et al. 2009).
lncRNAs can also act by recruiting factors involved in gene activation. From the HOXA cluster, two lncRNAs, Mistral and HOTTIP, have been implicated in recruiting the MLL complex in cis (Bertani et al. 2011; Wang et al. 2011b). MLL, an H3K4 trimethylase, is a member of the Trithorax group of developmentally important gene-activating proteins that, like Polycomb, were discovered in flies (Schuettengruber et al. 2011). Using a technique called chromosome conformation capture, which assays long-range interactions among distant chromosomal regions in 3D space, it was found that multiple loci as far apart as 40 kb in the HOXA cluster are in close physical proximity, potentially enabling epigenetic factors such as MLL to coordinately regulate their expression. On the basis of knockdown analyses, both Mistral and HOTTIP are hypothesized to recruit MLL to distinct sets of genes that are sequentially proximal to the respective lncRNA locus, and Mistral may do so by facilitating formation of long-range chromosomal loops. This observation may lend support to the longstanding hypothesis that regulatory elements in the genome, such as enhancers, could assert their effects on distant gene loci via chromosomal looping (Dean 2011).
Beyond histone modifications, lncRNAs also influence epigenetic regulation through modulation of DNA methylation at CpG dinucleotides, which is crucial for the stable repression of genes (Law and Jacobsen 2010). During embryogenesis, methylation marks are first laid down on previously unmethylated DNA by the de novo methyltransferases Dnmt3a and -3b, and are later perpetuated through DNA replication by the maintenance methyltransferase Dnmt1. One way by which Tsix might function to repress Xist is to recruit Dnmt3a activity to methylate and silence the Xist promoter (Sado et al. 2006; Sun et al. 2006). Similarly, Kcnq1ot1 may recruit Dnmt1 (Mohammad et al. 2010).
lncRNA-directed methylation has also been implicated in the regulation of ribosomal (r)DNA. rDNA exists in the genome as tandemly repeated units, of which some always remain silenced by heterochromatic histone marks and DNA methylation (McStay and Grummt 2008). Each rDNA repeat unit encodes a polycistronic transcript comprising the various rRNAs, and each unit is separated by intergenic spacers (IGSs) that are transcribed by RNA Pol I (Mayer et al. 2006). The IGS transcripts have recently been shown to be processed into 150- to 300-nt fragments called promoter (p)RNAs, which may act as scaffolds to recruit poly(ADP-ribose)-polymerase-1 (PARP1) (Guetg et al. 2012), the ATP-dependent chromatin remodeling complex NoRC (Mayer et al. 2008), and the methyltransferase Dnmt3b (Schmitz et al. 2010). pRNA forms a conserved hairpin structure that binds both PARP1 and the TIP5 subunit of NoRC, leading to a conformation change in TIP5 that facilitates the recruitment of NoRC to the nucleolus, where rDNA is located (Mayer et al. 2008; Guetg et al. 2010). Particularly intriguing is that the recruitment of Dnmt3b by pRNA may be dependent on DNA:RNA triplexing, possibly via Hoogsteen base pairing, between the rDNA promoter and the 5′ end of pRNA (Schmitz et al. 2010). If proved, DNA:RNA triplex formation may be a general mechanism by which lncRNAs recruit trans factors to specific DNA loci.
While mechanisms of trans-action remain undefined, it seems clear that several features make lncRNAs excellent candidates for cis-acting molecular tethers (Lee 2009). Because lncRNAs are inherently attached to chromatin via DNA:RNA hybridization during transcription, and because they are generally transcribed from a single locus in the genome, they are poised to direct allele- and locus-specific control in cis. This is a feat that cannot be performed by protein TFs, which do not retain allelic or positional memory once translated in the cytoplasm and can recognize short DNA sequence motifs only a few nucleotides in length that occur thousands of times in the genome. Furthermore, the length of lncRNAs allows them to reach out and capture epigenetic complexes while tethered (either by Pol II or by bridging factors like YY1), and the unmasking of 3′-degradation signals upon transcriptional termination would limit the RNA’s half-life and prevent diffusion and action at ectopic sites. This cis-acting mechanism is somewhat reminiscent of the RNAi-based transcriptional gene silencing used by the fission yeast Schizosaccharomyces pombe in assembling centromeric heterochromatin (Cam et al. 2009; Moazed 2009).
lncRNAs in transcription: Decoys, coregulators, and Pol II inhibitors
In addition to regulating gene expression by recruiting epigenetic complexes, lncRNAs can directly affect the process of transcription. Some act as decoys for TFs, as in the case of PANDA sequestering NF-YA away from its pro-apoptotic target genes (Hung et al. 2011). Others compete for TF binding, such as the Gas5 RNA that, in addition to being a host for snoRNAs (Smith and Steitz 1998), binds the DNA-binding domain of nuclear glucocorticoid receptors and precludes contact with glucocorticoid response elements on genomic DNA (Kino et al. 2010). lncRNAs may even influence the cellular localization of TFs. The cytoplasmic NRON (noncoding repressor of NFAT) RNA was found to prevent NFAT from shuttling into the nucleus, possibly by interfering with NFAT’s interactions with the importin family of nuclear transport proteins (Willingham et al. 2005). NRON may also sequester the TF in a cytoplasmic ribonucleoprotein complex that contains kinases, further counteracting nuclear import as only unphosphorylated NFAT can localize to the nucleus (Liu et al. 2011; Sharma et al. 2011).
lncRNAs can also act as transcriptional coregulators. For example, SRA RNA serves as a coactivator for a number of nuclear steroid receptors (Lanz et al. 1999, 2002). These receptors in turn exert their function on downstream targets by recruiting additional transcription and epigenetic factors, including ATP-dependent chromatin remodeling complexes and histone acetyltransferases (HATs) (Gronemeyer et al. 2004). Interestingly, SRA is a bifunctional transcript, with one isoform functioning as ncRNA and another as mRNA that is translated into the protein SRAP (Kawashima et al. 2003; Chooniedass-Kothari et al. 2004), which may in turn antagonize the function of its noncoding counterpart (Hubé et al. 2011). Conversely, an example of a lncRNA corepressor can be found at the cyclin D1 (CCND1) locus, where a series of independently transcribed, variable-length lncRNAs called ncRNACCND1 are upregulated during stress (Klein and Assoian 2008; Wang et al. 2008). These transcripts may remain partially tethered to DNA as RNA:DNA hybrids and thereby recruit and allosterically modulate the protein TLS, which can then inhibit HAT activity of the coactivators p300/CBP. In this sense, ncRNACCND1 serve as both recruiters and effectors of TLS.
In addition to acting through TFs, lncRNAs can directly interfere with Pol II activity. One case is the inhibition of the major, coding transcript of dihydrofolate reductase (DHFR) (Schnell et al. 2004). In quiescent cells, an upstream minor promoter of DHFR produces a lncRNA that apparently disrupts formation of the transcription preinitiation complex at the major promoter. This is thought to occur through direct binding with the general transcription factor TFIIB, as well as possibly through DNA:RNA triplex formation at the major promoter (Martianov et al. 2007). lncRNAs transcribed from SINEs, an abundant class of retrotransposons that includes B2 in mice and Alu in humans, may also block transcription of heat-shock genes by binding Pol II to prevent formation of preinitiation complexes (Espinoza et al. 2004; Mariner et al. 2008; Yakovchuk et al. 2009).
lncRNAs and nuclear compartments
lncRNAs may also emerge as key regulators of nuclear compartments. The interior of the nucleus is a dynamic place consisting of multiple “nuclear bodies” that perform important functions (Mao et al. 2011b). Perhaps the best known of these is the nucleolus, the major site of rRNA transcription and ribosome assembly, and the role of pRNA in regulating the function of this compartment has been discussed above. The nucleolus, along with the perinucleolar compartment, might have roles beyond ribosome biogenesis though, as both Xist and Kcnq1ot1 have been shown to target Xi and the imprinted Kcnq1 domain, respectively, to the perinucleolar compartment to maintain silencing (Zhang et al. 2007; Pandey et al. 2008).
The structure and function of several other nuclear bodies likewise seem to involve RNA. NEAT1 (nuclear enriched abundant transcript 1) seeds the formation and maintains stability of paraspeckles, which are believed to participate in the nuclear retention of mRNAs that have undergone adenosine-to-inosine hyperediting (Chen and Carmichael 2009; Sunwoo et al. 2009). NEAT1 interacts with paraspeckle proteins such as p54/NONO and PSP (Chen and Carmichael 2009; Clemson et al. 2009; Sasaki et al. 2009), and recent evidence suggests that the recruitment of these proteins to form paraspeckles is a dynamic process that requires continuous transcription of NEAT1 (Mao et al. 2011a). Its neighbor, NEAT2 or MALAT1, has been shown to localize serine/arginine (SR) splicing factors to a compartment called nuclear speckles where they can be stored and modified by phosphorylation (Bernard et al. 2010). MALAT1 is associated with proper relocation of these splicing factors to sites of transcription, where splicing occurs, and thus may have a role in controlling alternative splicing of certain mRNA precursors (Tripathi et al. 2010). More recently, MALAT1 has been shown to interact with the PRC1 subunit Cbx4/Pc2 and participate in the shuttling of genes between nuclear compartments for silencing and activation. In the presence of extracellular growth signals, unmethylated Cbx4 binds MALAT1 and localizes its target genes, along with coactivating factors such as LSD1, to “transcription factories” called interchromatin granules that usually cluster around nuclear speckles; in the absence of signal, Cbx4 becomes methylated and instead binds another lncRNA TUG1, associates with corepressors such as Ezh2, and translocates to silencing compartments called Polycomb bodies (L. Yang et al. 2011). This example illustrates the complex interplay among cell-signaling pathways, chromatin-modifying factors, lncRNAs, and nuclear bodies in regulating gene expression.
A most puzzling fact, however, is that in spite of their abundance and association with some of the most prominent subnuclear structures, knockouts of neither NEAT1 nor MALAT1 have robust phenotypes (Nakagawa et al. 2011, 2012; Eissmann et al. 2012; Zhang et al. 2012). These recent observations add to the mystery surrounding lncRNAs and support the assertion that quantity and function do not necessarily correlate. They also underscore the importance of performing conventional knockout studies to test the function of newly discovered lncRNAs, a crucial test that an overwhelming number of lncRNAs have not been subjected to.
Transcript or act of transcription?
The above examples postulate that the mature lncRNA transcript is required for the regulatory mechanism. However, in some cases it may be the act of transcription, and not the transcript itself, that is important. For example, in the β-globin locus control region (Ling et al. 2004, 2005), polyadenylated transcripts of varying lengths are generated from the HS2 enhancer site. No biological function has been ascribed to these transcripts, but because HS2 is unable to activate globin expression when a transcriptional terminator is placed between it and the globin promoter, the phenomenon has been interpreted to be transcription rather than transcript dependent (Ling et al. 2004). In fact, intergenic transcription has been seen to occur throughout the globin locus (Ashe et al. 1997) and plays a role in establishing open chromatin domains (Bender et al. 2000; Gribnau et al. 2000) marked by permissive histone modifications such as H3K4me2/3 and H3 acetylation (Miles et al. 2007). A similar scenario has been observed in chicks. During the stimulation of lysozyme expression in macrophages by microbial lipopolysaccharide (LPS) (Lefevre et al. 2008), the onset of LPS induction first leads to the transcription of an upstream, antisense lncRNA (called LINoCR) and the rapid deposition of chromatin-opening histone marks (namely, H3S10 phosphorylation and H3K9 acetylation) in the region. LINoCR transcription seems to drive nucleosome repositioning away from an enhancer toward a silencer, leading to the eviction of CTCF and cohesin from the latter while allowing the binding of C/BEP at the former, ultimately resulting in the upregulation of lysozyme expression. It should be noted, however, that in these examples, the available evidence cannot definitively rule out involvement of the lncRNA transcript, but the act of transcription remains the most parsimonious explanation.
Better-established examples of noncoding genes functioning through the act of transcription can be found in fungi. In Saccharomyces cerevisiae, a key enzyme in serine synthesis, SER3, is repressed in the presence of serine by “transcriptional interference” (TI) from an upstream noncoding locus, SRG1, that extends past the SER3 promoter (Martens et al. 2004, 2005). Replacement of the SRG1 sequence with a coding sequence provided strong support for the idea that SER3 repression is dependent on the act of transcription alone (Martens et al. 2004). This effect was found to be mediated by the transcription elongation factors Spt6 and Spt16, which reassemble nucleosomes in transcribed regions in the wake of Pol II, inhibiting further transcription from downstream promoters (Hainer et al. 2011). An opposite mechanism is observed for the fission yeast S. pombe gluconeogenesis enzyme fbp1+, where the transcription of multiple upstream noncoding loci, in the event of glucose starvation, leads to the opening of chromatin at the fbp1+ promoter and removal of the TUP corepressors, allowing other transcription factors such as Rst2 to bind and induce fbp1+ expression (Hirota et al. 2008). Blocking transcription with an inserted terminator abrogates this induction, and reintroducing the ncRNA in trans fails to rescue the defect, again supporting a transcription-based mechanism.
The discovery of eRNAs (Kim et al. 2010) raises the possibility that similar transcription-based mechanisms may operate at mammalian enhancers, although at present, the biological function of these RNAs is under debate. Another group of lncRNAs with enhancer-like function, called ncRNA-a, seems to be associated with robust expression of coding genes within 3 kb of the ncRNA locus, but reporter-based assays support the idea that they function as mature transcripts and not through the act of transcription (Ørom et al. 2010).
One particular form of TI, called “transcriptional collision,” has been proposed as a possible mechanism of action for NATs, where the RNA polymerases on the two strands transcribing toward each other literally crash and stall. While this phenomenon has been observed in bacteria (Crampton et al. 2006) and budding yeast (Prescott and Proudfoot 2002), none has been reported in mammals. However, computational analyses of genes with NATs have found an anticorrelation between the length of overlap and the expression levels of SAS pairs (i.e., the greater the overlap is, the lower the expression of either transcript) (Osato et al. 2007), consistent with a transcriptional collision model in which convergent transcripts sterically hinder transcription of one another.
lncRNAs in post-transcriptional regulation: mRNA processing, stability, and translation
lncRNAs also act at various steps of mRNA processing and stability control. The case of MALAT1 affecting alternative splicing through interactions with splicing factors has already been mentioned above. Another lncRNA, Gomafu/MIAT, which localizes to a novel nuclear domain and has a neuron-restricted expression, may hinder spliceosome formation and affect the splicing of a subset of mRNAs by sequestering splicing factor 1 (SF1) (Sone et al. 2007; Tsuiji et al. 2011).
NATs may affect the alternative splicing of their overlapping transcripts by virtue of masking splice sites through base complementarity. Early observations that the ratio of splice isoforms for a number of mRNAs can be influenced by expression of overlapping antisense transcripts, and that the latter can form RNA duplexes with the mRNAs opposite them in vivo and/or inhibit splicing in vitro, led to speculation that NAT-mediated alternative splicing may be a common method of post-transcriptional regulation (Munroe 1988; Krystal et al. 1990; Khochbin et al. 1992; Yan et al. 2005; Beltran et al. 2008; Annilo et al. 2009). More recent genome-wide analysis has indeed revealed correlations between overlapping SAS pairs and alternative splicing (namely, enrichment of alternative exons in overlapping regions and increased number of alternative splice isoforms) (Morrissy et al. 2011). One notable case is the α-thyroid hormone receptor gene erbAα, which produces two mRNA isoforms and has a downstream, translated antisense transcript called RevErb (thus, not a lncRNA per se) whose 3′-untranslated region (UTR) overlaps the last splice acceptor site of the long erbAα isoform (Lazar et al. 1989). RevErb expression is correlated with lower levels of the long erbAα isoform without affecting its transcription or stability, and the antisense RNA has been implicated by both in vivo and in vitro experiments to repress splicing at the overlapping acceptor site, thus tilting the balance in favor of the short erbAα isoform (Lazar et al. 1990; Munroe and Lazar 1991; Hastings et al. 1997). Antisense transcripts can theoretically affect alternative polyadenylation site selection in a similar manner, as was shown for two convergent mRNAs encoded by the mouse polyomavirus (Gu et al. 2009).
Once in the cytoplasm, a transcript may be regulated in a signal-responsive manner by factors that alter its stability. More than 5% of human genes contain 50–150 nt of AU-rich elements (AREs) in their 3′-UTRs, which recruit RNA-binding proteins such as AUF1 and lead to destabilization of the transcript through deadenylation, decapping, and degradation via the action of cellular machineries such as the exosome (Barreau et al. 2006). A NAT produced from the 3′-UTR of iNOS (inducible nitric oxide synthase) has been found to interact with its sense counterpart and with HuR, an ARE-binding factor that increases the stability of ARE-containing transcripts (Matsui et al. 2008). This suggests a potential mechanism whereby a NAT helps stabilize its sense counterpart by aiding in the recruitment of stabilizing factors. In a more intricate example, the protein Staufen1 (STAU1), in complex with the nonsense-mediated decay factor Upf1, is involved in the regulated decay of ∼1% of coding transcripts, a process named “STAU1-mediated decay” (Kim et al. 2005, 2007). While STAU1 was found to bind its first identified target, ARF1, at a STAU1-binding site in the 3′-UTR of ARF1 that forms a double-stranded (ds)RNA stem structure, the same motif could not be found in other STAU1 targets. Instead, it was discovered that a subset of these mRNA targets contains an Alu element in its 3′-UTRs that can base pair with a group of cytoplasmic and polyadenylated lncRNAs, named half-STAU1-binding site RNAs (1/2-sbsRNAs), that contain complementary Alu elements necessary to form the dsRNA structure (Gong and Maquat 2011). Thus, in contrast to the NAT that serves to stabilize iNOS, these 1/2-sbsRNAs promote the decay of their target mRNAs by permitting recruitment of destabilizing factors.
lncRNAs may even exert their effects at the level of translational regulation. PU.1, an important TF involved in hematogenesis, has an overlapping NAT that was found to negatively influence PU.1 protein level but not polysome-bound PU.1 sense mRNA level, and the antisense RNA seems to compete with the sense transcript for binding to the translation initiation factor eIF4A (Ebralidze et al. 2008). While failure to detect siRNA-like small RNAs potentially rules out involvement of RNAi, whether this kind of antisense regulation of translation is a common alternative to RNAi-based mechanisms remains to be seen. Additionally, it has been reported that lincRNA-p21 associates with polysome-bound β-catenin and JunB mRNAs in a manner dependent on the general translation repressor Rck, leading to decreased polysome number, and thus translation, of these mRNAs (Yoon et al. 2012).
Intersection of the long and the small: lncRNAs as sources and sinks
It seemed inevitable that the long and small ncRNA worlds would eventually intertwine. Indeed, lncRNAs have been suggested to interfere with miRNA-mediated mRNA destabilization. For example, the antisense transcript of the Alzheimer-associated β-secretase-1 (BACE), known as BACE-AS, increases BACE mRNA stability (Faghihi et al. 2008), most likely by masking the binding sites for miR-485-5p (Faghihi et al. 2010).
Rather than competing for miRNA-binding sites, lncRNAs can compete for the miRNAs themselves. A number of mammalian pseudogenes, including PTENP1 and KRASP1 (Poliseno et al. 2010), and other lncRNAs (Wang et al. 2010; Cesana et al. 2011) have miRNA-binding sites in their 3′-UTRs and may therefore serve as “sponges” to sequester miRNAs away from their mRNA targets. First discovered in plants (Franco-Zorrilla et al. 2007), this phenomenon has been hypothesized to be part of a genome-wide fine-tuning regulatory network composed of miRNA “pseudotargets” (Seitz 2009) and/or bona fide targets called “competing endogenous RNAs” (ceRNAs) (Salmena et al. 2011). Changes in the expression level of one member of this network, and thus the amount of miRNAs bound up by it, would affect the overall accessible pool of shared miRNAs, leading to concordant changes in the transcript levels of other members of the network. For example, the developmentally regulated linc-MD1 is reported to influence the mRNA levels of miRNA-targeted muscle differentiation genes (Cesana et al. 2011). The task now is to determine whether this actually represents a new layer of post-transcriptional regulation directed by precise and signal-responsive changes in a ceRNA’s expression level or if this is simply an inevitable consequence of several mRNAs and ncRNAs being regulated by the same pool of miRNAs.
lncRNAs can themselves be host genes for small RNAs. H19 is host to miR-675 (Cai and Cullen 2007; Keniry et al. 2012); Gas5 gives rise to 10 highly conserved snoRNAs (Smith and Steitz 1998); and the imprinted Gtl2, anti-Rtl1, and Mirg RNAs are hosts to almost 50 miRNAs and 40 snoRNAs (da Rocha et al. 2008). Moreover, a large number of endo-siRNAs have been found in mammalian germ cells that are produced from the complementarily base-paired regions between coding and/or noncoding SAS pairs, between mRNAs and expressed pseudogenes, or from within pseudogenes containing inverted repeats (Tam et al. 2008; Watanabe et al. 2008). In XCI, it has been proposed that Tsix might repress Xist partly through an RNAi-related pathway, by base pairing to form an RNA duplex that yields Dicer-dependent small RNAs (Ogawa et al. 2008). Thus, observed biological outcomes of lncRNA knockout, knockdown, or overexpression studies could be consequences of the long transcript itself and/or the small RNAs therein.
Translated lncRNAs: Pervasive Translation or Gene Evolution in Progress?
A transcript is usually considered to act as ncRNA, rather than protein, if it lacks any substantial open reading frame (ORF) or fails to produce protein during in vitro translation experiments (Brannan et al. 1990; Brockdorff et al. 1992; Niazi and Valadkhan 2012). However, an interesting twist in the ncRNA debate occurred when a developmentally important Drosophila transcript previously thought to be noncoding, called tarsal-less or polished rice, was found to be a polycistronic mRNA that encodes multiple short (11–32 amino acids) but functional peptides, and this gene structure is conserved among insects (Galindo et al. 2007; Kondo et al. 2007, 2010). More recently, ribosome-profiling experiments in mouse ES cells revealed that the majority of lncRNAs are in fact translated into small peptides (Ingolia et al. 2011). Like the identification of new transcripts, however, just because an RNA is translated should not automatically be taken to mean that these peptides are functional, but could instead be “translational noise.” Moreover, it would be a false dichotomy to maintain that a transcript must either be coding or be noncoding—the example of SRA/SRAP demonstrates that an RNA could potentially function at multiple levels, both as a ncRNA performing a regulatory role and as an mRNA for protein synthesis (Kawashima et al. 2003; Chooniedass-Kothari et al. 2004).
Another interesting possible explanation is that these short translated ORFs are “protogenes” in the making. A number of studies in the past several years across multiple taxa, including primates, suggest that new protein-coding genes can arise de novo from previously nongenic sequences, by first becoming transcribed noncoding genes and then evolving into translated protein-coding genes (Tautz and Domazet-Lošo 2011). Recently, a survey of S. cerevisiae ORFs identified >1000 short (<400 nt) and poorly conserved (found only in the closest relatives of S. cerevisiae and with no sequence similarities to any known genes) ORFs that are nonetheless translated, as assayed by ribosome profiling, but only less than one-quarter show any sign of purifying selection (Carvunis et al. 2012). In other words, it may be common in our genome for a large number of protein-coding units to have been generated by random processes, most of which have no meaningful biological function and will be weeded out by natural selection, leaving the few that have acquired adaptive functions to become new bona fide genes. By the same token, it is plausible that most of the tens of thousands of RNA transcripts produced from our genome do not actually have active functions, but instead serve as a reservoir for evolution to tinker with and generate new tools (whether ncRNA or protein) for the species.
Conclusions
The foregoing discussion has provided a survey of the present state of knowledge regarding the locations, functions, and mechanisms of lncRNAs. A large fraction of the genome in many organisms is likely transcribed, but the characteristics and function of the overwhelming majority of these lncRNAs are currently not known. Some are nuclear, some cytoplasmic; some are highly expressed, some barely detectable. In the end, it may not be possible or meaningful to try to apply criteria such as stability, conservation, and expression level to find order in this chaos. High-turnover, poorly conserved, and low-abundance transcripts could still have essential functions. For instance, Kcnq1ot1 has a half-life of ∼1 hr (Clark et al. 2012), the lncRNAs in the Xic are found only in placental mammals and have low conservation even within this taxon (Davidow et al. 2007), and RepA exists at ∼5–10 copies per cell (Zhao et al. 2008). Yet even fully processed transcripts that interact with protein factors might not be essential (Ponting and Belgard 2010; van Bakel et al. 2010; Schorderet and Duboule 2011; Eissmann et al. 2012). Ultimately, the true test for function lies in the detailed, mechanistic dissection of the genetic pathways and cellular activities for each individual putative lncRNA (see Figure 4). We must also keep in mind that the genome may not be a streamlined, highly sculpted space honed by natural selection; it could instead be quite noisy, even wasteful, where genomic junk and evolutionary relics accumulate but may yet adopt useful functions eventually (Lynch 2007; Koonin and Wolf 2010). Thus, at present, it may be best to avoid blanket statements about structure, function, and mechanism, as indeed we have barely begun to scratch the surface of the lncRNA world. There is still plenty to learn and much work to be done. This is an exciting time for the study of RNA biology!
Figure 4.
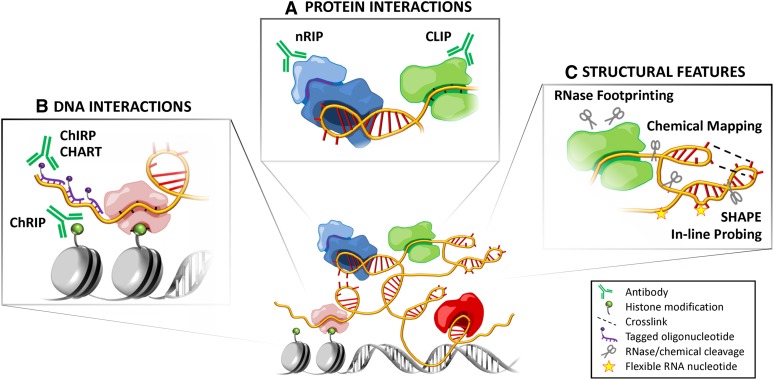
Methods for studying lncRNAs. (A) Protein interactions: Identifying protein partners of lncRNAs provides clues into their functional mechanisms and pathways. RNA-immunoprecipitation (RIP) techniques such as chemical-cross-linked RIP (Selth et al. 2009), native RIP (nRIP) (Zhao et al. 2008), and UV-crosslinked immunoprecipitation (CLIP) (Ule et al. 2005) use antibodies to pull down ribonucleoprotein complexes, from which the associated RNAs are isolated for analysis. Each variation has its own advantages and disadvantages: nRIP avoids cross-linking artifacts, whereas CLIP may be better at avoiding reassociation artifacts and can be used to identify protein-interacting regions (even nucleotides) of the RNA. These techniques are being combined with high-throughput sequencing (e.g., RIP-Seq, HITS-CLIP/CLIP-Seq) to identify lncRNA interactions with a whole host of protein factors (Licatalosi et al. 2008; Zhao et al. 2010), although further validation by mechanistic studies is required. (B) DNA interactions: Several techniques have been developed to identify the genomic targets of lncRNAs. Based on principles of both chromatin immunoprecipitation (ChIP) and RIP, chromatin RNA immunoprecipitation (ChRIP) can be used to identify RNAs associated with a particular chromatin mark (Pandey et al. 2008). On the other hand, techniques such as chromatin oligo-affinity precipitation (ChOP) (Mariner et al. 2008), chromatin isolation by RNA purification (ChIRP) (Chu et al. 2011), and capture hybridization of RNA targets (CHART) (Simon et al. 2011) use tagged complementary oligonucleotides to identify DNA loci that interact with an RNA of interest. (C) Structural features: ncRNAs form specific secondary (base pairing) and tertiary (three-dimensional) structures to carry out their functions. Such structures can be mapped using chemical reagents that cleave at specific nucleotides or attack solvent-exposed regions of the RNA backbone or by cross-linking three-dimensionally proximal regions of the RNA to reveal long-range intramolecular interactions (Weeks 2010). Ribonucleases with different cleavage specificities are also used to identify single- or double-stranded regions, as well as protein-protected regions (RNase footprinting). Other methods such as selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (Wilkinson et al. 2006) and in-line probing (Regulski and Breaker 2008) assess local nucleotide flexibility. SHAPE has since been adapted for high-throughput analysis (SHAPE-Seq) (Lucks et al. 2011), joining other techniques that rely on RNase digestion such as fragmentation sequencing (FragSeq) (Underwood et al. 2010) and parallel analysis of RNA structure (PARS) (Kertesz et al. 2010).
Acknowledgments
We thank members of the laboratory and T. R. Gregory for many helpful discussions, and we apologize to any of our colleagues whose work could not be cited here for space constraints. J.T.Y.K. was supported by a Postgraduate Scholarship from the Natural Sciences and Engineering Research Council of Canada, and J.T.L. is supported by the National Institutes of Health (R01-GM090278). J.T.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Agarwal A., Koppstein D., Razowsky J., Sboner A., Habegger L., et al. , 2010. Comparison and calibration of transcriptome data from RNA-Seq and tiling arrays. BMC Genomics 11: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annilo T., Kepp K., Laan M., 2009. Natural antisense transcript of natriuretic peptide precursor A (NPPA): structural organization and modulation of NPPA expression. BMC Mol. Biol. 10: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe H. L., Monks J., Wijgerde M., Fraser P., Proudfoot N. J., 1997. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 11: 2494–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au P. C., Zhu Q. H., Dennis E. S., Wang M. B., 2011. Long non-coding RNA-mediated mechanisms independent of the RNAi pathway in animals and plants. RNA Biol. 8: 404–414. [DOI] [PubMed] [Google Scholar]
- Bacher C. P., Guggiari M., Brors B., Augui S., Clerc P., et al. , 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8: 293–299. [DOI] [PubMed] [Google Scholar]
- Balakirev E. S., Ayala F. J., 2003. Pseudogenes: Are they “junk” or functional DNA? Annu. Rev. Genet. 37: 123–151. [DOI] [PubMed] [Google Scholar]
- Barreau C., Paillard L., Osborne H. B., 2006. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 33: 7138–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei M. S., Zemel S., Tilghman S. M., 1991. Parental imprinting of the mouse H19 gene. Nature 351: 153–155. [DOI] [PubMed] [Google Scholar]
- Bekpen C., Margues-Bonet T., Alkan C., Antonacci F., Leogrande M. B., et al. , 2009. Death and resurrection of the human IRGM gene. PLoS Genet. 5: e1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M., Puig I., Peña C., García J. M., Alvarez A. B., et al. , 2008. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 22: 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender M. A., Bulger M., Close J., Groudine M., 2000. Beta-globin gene switching and DNase I sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol. Cell 5: 387–393. [DOI] [PubMed] [Google Scholar]
- Berget S. M., Moore C., Sharp P. A., 1977. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA 74: 3171–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D., Prasanth K. V., Tripathi V., Colasse S., Nakamura T., et al. , 2010. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29: 3082–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani S., Sauer S., Bolotin E., Sauer F., 2011. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol. Cell 43: 1040–1046. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bertone P., Stolc V., Royce T. E., Rozowsky J. S., Urban A. E., et al. , 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246. [DOI] [PubMed] [Google Scholar]
- Birney E., Stamatoyannopoulos J. A., Dutta A., Guigó R., Gingeras T. R., et al. , 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H., Collins K., 2011. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 3: a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond A. M., Vangompel M. J., Sametsky E. A., Clark M. F., Savage J. C., et al. , 2009. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12: 1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan C. I., Dees E. C., Ingram R. S., Tilghman S. M., 1990. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 20: 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H., 1969. Gene regulations for higher cells: a theory. Science 165: 349–357. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H., 1971. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q. Rev. Biol. 46: 111–138. [DOI] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G. F., McCabe V. M., Norris D .P., et al. , 1992. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71551 526. [DOI] [PubMed] [Google Scholar]
- Brown C. J., Lafrenière R. G., Powers V. E., Sebastio G., Ballabio A., et al. , 1991. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 349: 82–84. [DOI] [PubMed] [Google Scholar]
- Brown C. J., Hendrich B. D., Rupert J. L., Lafrenière R. G., Xing Y., et al. , 1992. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542. [DOI] [PubMed] [Google Scholar]
- Buratowski S., 2008. Gene expression—Where to start? Science 322: 1804–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M. N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., et al. , 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25: 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Cullen B. R., 2007. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13: 313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H. P., Chen E. S., Grewal S. I., 2009. Transcriptional scaffolds for heterochromatin assembly. Cell 136: 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos E. I., Reiberg D., 2009. Histones: annotating chromatin. Annu. Rev. Genet. 43: 559–599. [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., et al. , 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- Carvunis A. R., Rolland T., Wapinski I., Calderwood M. A., Yildirim M. A., et al. , 2012. Proto-genes and de novo gene birth. Nature 487: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., et al. , 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. L., Carmichael G. G., 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell 35: 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooniedass-Kothari S., Emberley E., Hamedani M. K., Troup S., Wang X., et al. , 2004. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 566: 43–47. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Gelinas R. E., Broker T. R., Roberts R. J., 1977. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12: 1–8. [DOI] [PubMed] [Google Scholar]
- Chu C., Qu K., Zhong F. L., Artandi S. E., Chang H. Y., 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44: 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chureau C., Prissette M., Bourdet A., Barbe V., Cattolico L., et al. , 2002. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 12: 894–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. B., Johnston R. L., Inostroza-Ponta M., Fox A. H., Fortini E., et al. , 2012. Genome-wide analysis of long noncoding RNA stability. Genome Res. 22: 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C. M., McNeil J. A., Willard H. F., Lawrence J. B., 1996. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132: 259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C. M., Hutchinson J. N., Sara S. A., Ensminger A. W., Fox A. H., et al. , 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33: 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comings D. E., 1972. The genetic organization of chromosomes. Adv. Hum. Genet. 3: 237–431. [DOI] [PubMed] [Google Scholar]
- Core L. J., Waterfall J. J., Lis J. T., 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton N., Bonass W. A., Kirkham J., Rivetti C., Thomson N. H., 2006. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 34: 5416–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B., Hannon G. J., 2011. Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 12: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha S. T., Edwards C. A., Ito M., Ogata T., Ferguson-Smith A. C., 2008. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 24: 306–316. [DOI] [PubMed] [Google Scholar]
- Davidow L. S., Breen M., Duke S. E., Samollow P. B., McCarrey J. R., et al. , 2007. The search for a marsupial XIC reveals a break with vertebrate synteny. Chromosome Res. 15: 137–146. [DOI] [PubMed] [Google Scholar]
- Dean A., 2011. In the loop: long range chromatin interactions and gene regulation. Brief. Funct. Genomics 10: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning A. P., Gu W., Castoe T. A., Batzer M. A., Pollock D. D., 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7: E1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuve J. L., Avner P., 2011. The coupling of X-chromosome inactivation to pluripotency. Annu. Rev. Cell Dev. Biol. 27: 611–629. [DOI] [PubMed] [Google Scholar]
- Djebali S., Davis C. A., Merkel A., Dobin A., Lassmann T., et al. , 2012. Landscape of transcription in human cells. Nature 489: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe M. E., Silva S. S., Pinter S. F., Xu N., Lee J. T., 2009. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature 460: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Chureau C., Samain S., Weissenbach J., Avner P., 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312: 1653–1655. [DOI] [PubMed] [Google Scholar]
- Ebisuya M., Yamamoto T., Nakajima M., Nishida E., 2008. Ripples from neighbouring transcription. Nat. Cell Biol. 10: 1106–1113. [DOI] [PubMed] [Google Scholar]
- Ebralidze A. K., Guibal F. C., Steidl U., Zhang P., Lee S., et al. , 2008. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev. 22: 2085–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards C. A., Ferguson-Smith A. C., 2007. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 19: 281–289. [DOI] [PubMed] [Google Scholar]
- Eissmann M., Gutschner T., Hämmerle M., Günther S., Caudron-Herger M., et al. , 2012. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elisaphenko E. A., Kolesnikov N. N., Shevchenko A. I., Rogozin I. B., Nesterova T. B., 2008. A dual origin of the Xist gene from a protein-coding gene and a set of transposable elements. PLoS ONE 3: e2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium, 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza C. A., Allen T. A., Hieb A. R., Kugel J. F., Goodrich J. A., 2004. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 11: 822–829. [DOI] [PubMed] [Google Scholar]
- Faghihi M. A., Wahlestedt C., 2009. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 10: 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi M. A., Modarresi F., Khalil A. M., Wood D. E., Sahagan B. G., et al. , 2008. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14: 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi M. A., Zhang M., Huang J., Modarresi F., van der Brug M. P., et al. , 2010. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerhahn S., Iglesias N., Panza A., Porro A., Lingner J., 2010. TERRA biogenesis, turnover and implications for function. FEBS Lett. 584: 3812–3818. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Valli A., Todesco M., Mateos I., Puga M. I., et al. , 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gabory A., Jammes H., Dandolo L., 2010. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays 32: 473–480. [DOI] [PubMed] [Google Scholar]
- Galindo M. I., Puevo J. I., Fouix S., Bishop S. A., Couso J. P., 2007. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 5: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G., 1981. Chromosome structure and the C-value paradox. J. Cell Biol. 91: 3s–14s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart M. E., Kuroda M. I., 2009. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136: 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., Maquat L. E., 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory T. R., 2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. Camb. Philos. Soc. 76: 65–101. [DOI] [PubMed] [Google Scholar]
- Gribnau J., Diderich K., Pruzina S., Calzolari R., Fraser P., 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol. Cell 5: 377–386. [DOI] [PubMed] [Google Scholar]
- Gronemeyer H., Gustafsson J. A., Laudet V., 2004. Principles for modulation of the nuclear receptor superfamily. Nat. Rev. Drug Discov. 3: 950–964. [DOI] [PubMed] [Google Scholar]
- Gu R., Zhang Z., DeCerbo J. N., Carmichael G. G., 2009. Gene regulation by sense-antisense overlap of polyadenylation signals. RNA 15: 1154–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C., Lienemann P., Sirri V., Grummt I., Hernandez-Verdun D., et al. , 2010. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 29: 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C., Scheifele F., Rosenthal F., Hottiger M. O., Santoro R., 2012. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell 45: 790–800. [DOI] [PubMed] [Google Scholar]
- Guil S., Soler M., Portela A., Carrère J., Fonalleras E., et al. , 2012. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat. Struct. Mol. Biol. 19: 664–670. [DOI] [PubMed] [Google Scholar]
- Gupta R. A., Shah N., Wang K. C., Kim J., Horlings H. M., et al. , 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T., Diederichs S., 2012. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 9: 703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Amit I., Garber M., French C., Lin M. F., et al. , 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Donaghey J., Carey B. W., Garber M., Grenier J. K., et al. , 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Pruneski J. A., Mitchell R. D., Monteverde R. M., Martens J. A., 2011. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 25: 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P. M., Zheng D., Zhang Z., Carriero N., Gerstein M., 2005. Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 33: 2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M. L., Milcarek C., Martincic K., Peterson M. L., Munroe S. H., 1997. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res. 25: 4296–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins P. G., Morris K. V., 2010. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription 1: 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Vogelstein B., Velculescu V. E., Papadopoulos N., Kinzler K. W., 2008. The antisense transcriptomes of human cells. Science 322: 1855–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A. K., Dez C., Henry Y., 2004. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 14: 335–343. [DOI] [PubMed] [Google Scholar]
- Heo J. B., Sung S., 2011. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79. [DOI] [PubMed] [Google Scholar]
- Hirota K., Miyoshi T., Kugou K., Hoffman C. S., Shibata T., et al. , 2008. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456: 130–134. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Mayfield J. E., Sander G., Bonner J., 1972. Chromosomal RNA: its properties. Science 177: 72–74. [DOI] [PubMed] [Google Scholar]
- Huarte M., Guttman M., Feldser D., Garber M., Koziol M. J., et al. , 2010. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubé F., Velasco G., Rollin J., Furling D., Francastel C., 2011. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 39: 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T., Wang Y., Lin M. F., Koegel A. K., Kotake Y., et al. , 2011. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43: 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A., Schattner P., Polacek N., 2005. Non-coding RNAs: Hope or hype? Trends Genet. 21: 289–297. [DOI] [PubMed] [Google Scholar]
- Ingolia N. T., Lareau L. F., Weissman J. S., 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Monod J., 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3: 318–356. [DOI] [PubMed] [Google Scholar]
- Jeon Y., Lee J. T., 2011. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146: 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P., Diederichs S., Wang W., Böing S., Metzger R., et al. , 2003. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22: 8031–8041. [DOI] [PubMed] [Google Scholar]
- John B., Miklos G. L. G., 1979. Functional aspects of satellite DNA and heterochromatin. Int. Rev. Cytol. 58: 1–114. [DOI] [PubMed] [Google Scholar]
- Johnston C. M., Newall A. E., Brockdorff N., Nesterova T. B., 2002. Enox, a novel gene that maps 10 kb upstream of Xist and partially escapes X inactivation. Genomics 80: 236–244. [DOI] [PubMed] [Google Scholar]
- Jones B. K., Levorse J. M., Tilghman S. M., 1998. Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes Dev. 12: 2200–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C., Thakur N., Pandey R. R., 2006. The length of the transcript encoded from the Kcnq1ot1 antisense promoter determines the degree of silencing. EMBO J. 25: 2096–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A., Viiri K., Araújo C. C., Rasaiyaah J., Bouwman R. D., et al. , 2010. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell 38: 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., St Laurent G., Raz T., Ozsolak F., Reynolds C. P., et al. , 2010. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ’dark matter’ un-annotated RNA. BMC Biol. 8: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., 2005. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566. [DOI] [PubMed] [Google Scholar]
- Kawashima H., Takano H., Sugita S., Takahara Y., Sugimura K., et al. , 2003. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem. J. 369: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry A., Oxley D., Monnier P., Kyba M., Dandolo L., et al. , 2012. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 14: 859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M., Wan Y., Mazor E., Rinn J. L., Nutter R. C., et al. , 2010. Genome-wide measurement of RNA secondary structure in yeast. Nature 467: 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A. M., Guttman M., Huarte M., Garber M., Raj A., et al. , 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 106: 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khochbin S., Brocard M. P., Grunwald D., Lawrence J. J., 1992. Antisense RNA and p53 regulation in induced murine cell differentiation. Ann. N. Y. Acad. Sci. 28: 77–87. [DOI] [PubMed] [Google Scholar]
- Kim K., Jutooru I., Chadalapaka G., Johnson G., Frank J., et al. , 2012. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. DOI: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. K., Hemberg M., Gray J. M., Costa A. M., Bear D. M., et al. , 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V. N., Han J., Siomi M. C., 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10: 126–139. [DOI] [PubMed] [Google Scholar]
- Kim Y. K., Furic L., Desgroseillers L., Maquat L. E., 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195–208. [DOI] [PubMed] [Google Scholar]
- Kim Y. K., Furic L., Parisien M., Major F., Desgroseillers L., et al. , 2007. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 26: 2670–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T., Hurt D. E., Ichijo T., Nader N., Chrousos G. P., 2010. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E. A., Assoian R. K., 2008. Transcriptional regulation of the cyclin D1 gene at a glance. J. Cell Sci. 121: 3853–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., et al. , 2011. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71: 6320–6326. [DOI] [PubMed] [Google Scholar]
- Kondo T., Hashimoto Y., Kato K., Inagaki S., Hayashi S., et al. , 2007. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat. Cell Biol. 9: 660–665. [DOI] [PubMed] [Google Scholar]
- Kondo T., Plaza S., Zanet J., Benrabah E., Valenti P., et al. , 2010. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329: 336–339. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Wolf Y. I., 2010. Constraints and plasticity in genome and molecular-phenome evolution. Nat. Rev. Genet. 11: 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y., Nakagawa T., Kitagawa K., Suzuki S., Liu N., et al. , 2011. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30: 1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal G. W., Armstrong B. C., Battey J. F., 1990. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol. Cell. Biol. 10: 4180–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz R. B., McKenna N. J., Onate S. A., Albrecht U., Wong J., et al. , 1999. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97: 17–27. [DOI] [PubMed] [Google Scholar]
- Lanz R. B., Razani B., Goldberg A. D., O’Malley B. W., 2002. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA). Proc. Natl. Acad. Sci. USA 99: 16081–16086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J. A., Jacobsen S. E., 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar M. A., Hodin R. A., Darling D. S., Chin W. W., 1989. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. Mol. Cell. Biol. 9: 1128–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar M. A., Hodin R. A., Cardona G., Chin W. W., 1990. Gene expression from the c-erbA alpha/Rev-ErbA alpha genomic locus. Potential regulation of alternative splicing by opposite strand transcription. J. Biol. Chem. 265: 12859–12863. [PubMed] [Google Scholar]
- Lee J.T., 2009. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 23: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. T., 2011. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 12: 815–826. [DOI] [PubMed] [Google Scholar]
- Lee J. T., Davidow L. S., Warshawsky D., 1999a Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21: 400–404. [DOI] [PubMed] [Google Scholar]
- Lee M. P., DeBaun M. R., Mitsuya K., Galonek H. L., Brandenburg S., et al. , 1999b Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc. Natl. Acad. Sci. USA 96: 5203–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre P., Witham J., Lacroix C. E., Cockerill P. N., Bonifer C., 2008. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol. Cell 32: 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin B., 1980. Gene Expression, 2: Eucaryotic Chromosomes John Wiley & Sons, New York. [Google Scholar]
- Lewin R., 1982. Repeated DNA still in search of a function. Science 217: 621–623. [DOI] [PubMed] [Google Scholar]
- Licatalosi D. D., Mele A., Fak J. J., Ule J., Kavikci M., et al. , 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Roychowdhury-Saha M., Black C., Watt A. T., Marcusson E. G., et al. , 2011. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 585: 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J., Ainol L., Zhang L., Yu X., Pi W., et al. , 2004. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J. Biol. Chem. 279: 51704–51713. [DOI] [PubMed] [Google Scholar]
- Ling J., Baibakov B., Pi W., Emerson B. M., Tuan D., 2005. The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J. Mol. Biol. 350: 883–896. [DOI] [PubMed] [Google Scholar]
- Liu Z., Lee J., Krummeu S., Lu W., Cai H., et al. , 2011. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol. 12: 1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louro R., Smirnova A. S., Verjovski-Almeida S., 2009. Long intronic noncoding RNA transcription: Expression noise or expression choice? Genomics 93: 291–298. [DOI] [PubMed] [Google Scholar]
- Lucks J. B., Mortimer S. A., Trapnell C., Luo S., Aviran S., et al. , 2011. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc. Natl. Acad. Sci. USA 108: 11063–11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyle R., Watanabe D., te Vruchte D., Lerchner W., Smrzka O. W., et al. , 2000. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 25: 19–21. [DOI] [PubMed] [Google Scholar]
- Lynch M., 2007. The evolution of genetic networks by non-adaptive processes. Nat. Rev. Genet. 8: 803–813. [DOI] [PubMed] [Google Scholar]
- Lyon M. F., 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. [DOI] [PubMed] [Google Scholar]
- Mak W., Nesterova T. B., de Napoles M., Appanah R., Yamanaka S., et al. , 2004. Reactivation of the paternal X chromosome in early mouse embryos. Science 303: 666–669. [DOI] [PubMed] [Google Scholar]
- Mao Y. S., Sunwoo H., Zhang B., Spector D. L., 2011a Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13: 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y. S., Zhang B., Spector D. L., 2011b Biogenesis and function of nuclear bodies. Trends Genet. 27: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner P. D., Walters R. D., Espinoza C. A., Drullinger L. F., Wagner S. D., et al. , 2008. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell 29: 499–509. [DOI] [PubMed] [Google Scholar]
- Martens J. A., Laprade L., Winston F., 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574. [DOI] [PubMed] [Google Scholar]
- Martens J. A., Wu P. Y., Winston F., 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19: 2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov J., Ramadass A., Serra Barros A., Chow N., Akoulitchev A., 2007. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445: 666–670. [DOI] [PubMed] [Google Scholar]
- Matsui K., Nishizawa M., Ozaki T., Kimura T., Hashimoto I., et al. , 2008. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology 47: 686–697. [DOI] [PubMed] [Google Scholar]
- Mayer C., Schmitz K. M., Li J., Grummt I., Santoro R., 2006. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 22: 351–361. [DOI] [PubMed] [Google Scholar]
- Mayer C., Neubert M., Grummt I., 2008. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 9: 774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A., Fosten M., Monk M., 1983. X-chromosome inactivation mosaicism in the three germ layers and the germ line of the mouse embryo. J. Embryol. Exp. Morphol. 74: 207–220. [PubMed] [Google Scholar]
- McStay B., Grummt I., 2008. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24: 131–157. [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Gerhardt D. J., Dinger M. E., Crawford J., Trapnell C., et al. , 2011. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 30: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles J., Mitchell J. A., Chakalova L., Govenechea B., Osborne C. S., et al. , 2007. Intergenic transcription, cell-cycle and the developmentally regulated epigenetic profile of the human beta-globin locus. PLoS ONE 2: e630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky A. E., Ris H., 1951. The desoxyribonucleic acid content of animal cells and its evolutionary significance. J. Gen. Physiol. 34: 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F., Mondal T., Guseva N., Pandey G. K., Kanduri C., 2010. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 137: 2493–2499. [DOI] [PubMed] [Google Scholar]
- Monk M., Harper M. I., 1979. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature 281: 311–313. [DOI] [PubMed] [Google Scholar]
- Morrissy A. S., Malachi G., Marra M. A., 2011. Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res. 21: 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Munroe S. H., 1988. Antisense RNA inhibits splicing of pre-mRNA in vitro. EMBO J. 7: 2523–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe S. H., Lazar M. A., 1991. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J. Biol. Chem. 266: 22083–22086. [PubMed] [Google Scholar]
- Nagano T., Mitchell J. A., Sanz L. A., Pauler F. M., Ferguson-Smith A. C., et al. , 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720. [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Naganuma T., Shioi G., Hirose T., 2011. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 193: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Ip J. Y., Shioi G., Tripathi V., Zong X., et al. , 2012. Malat1 is not an essential component of nuclear speckles in mice. RNA 18: 1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P., Chambers I., Karwacki-Neisius V., Chureau C., Morey C., et al. , 2008. Molecular coupling of Xist regulation and pluripotency. Science 321: 1693–1695. [DOI] [PubMed] [Google Scholar]
- Nesterova T. B., Senner C. E., Schneider J., Alcayna-Stevens T., Tattermusch A., et al. , 2011. Pluripotency factor binding and Tsix expression act synergistically to repress Xist in undifferentiated embryonic stem cells. Epigenetics Chromatin 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. Y., Johnson R., Stanton L. W., 2011. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31: 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niazi F., Valadkhan S., 2012. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3′ UTRs. RNA 18: 825–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niinuma T., Suzuki H., Nojima M., Nosho K., Yamamoto H., et al. , 2012. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 72: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ogawa Y., Sun B. K., Lee J. T., 2008. Intersection of the RNA interference and X-inactivation pathways. Science 320: 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., 1972. So much “junk” DNA in our genome, pp. 366–370 in Evolution of Genetic Systems, edited by Smith H. H. Gordon & Breach, New York. [Google Scholar]
- Okamoto I., Otte A. P., Allis C. D., Reinberg D., Heard E., 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303: 644–649. [DOI] [PubMed] [Google Scholar]
- Okamura K., Lai E. C., 2008. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 9: 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., et al. , 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563–573. [DOI] [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. C., 1980. Selfish DNA: the ultimate parasite. Nature 284: 604–607. [DOI] [PubMed] [Google Scholar]
- Ørom U. A., Derrien T., Beringer M., Gumireddy K., Gardini A., et al. , 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato N., Suzuki Y., Ikeo K., Gojobori T., 2007. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics 176: 1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., et al. , 2004. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36: 40–45. [DOI] [PubMed] [Google Scholar]
- Pandey R. R., Mondal T., Mohammad F., Enroth S., Redrup L., et al. , 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32: 232–246. [DOI] [PubMed] [Google Scholar]
- Pearson J. C., Lemons D., McGinnis W., 2005. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6: 893–904. [DOI] [PubMed] [Google Scholar]
- Peculis B. A., 2000. RNA-binding proteins: if it looks like a sn(o)RNA…. Curr. Biol. 10: R916–R918. [DOI] [PubMed] [Google Scholar]
- Pertea M., Salzberg S. L., 2010. Between a chicken and a grape: estimating the number of human genes. Genome Biol. 11: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpont M. E., Yunis J. J., 1977. Localization of chromosomal RNA in humn G-banded metaphase chromosomes. Exp. Cell Res. 106: 303–308. [DOI] [PubMed] [Google Scholar]
- Pink R. C., Wicks K., Caley D. P., Punch E. K., Jacobs L., et al. , 2011. Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA 17: 792–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L., Salmena L., Zhang J., Carver B., Haveman W. J., et al. , 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Belgard T. G., 2010. Transcribed dark matter: Meaning or myth? Hum. Mol. Genet. 19: R162–R168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P., Nielsen J., Kammler S., Lykke-Adersen S., Christensen M. S., et al. , 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854. [DOI] [PubMed] [Google Scholar]
- Prensner J. R., Iyer M. K., Balbin O. A., Dhanasekaran S. M., Cao Q., et al. , 2011. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 29: 742–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott E. M., Proudfoot N. J., 2002. Transcriptional collision between convergent genes in budding yeast. Proc. Natl. Acad. Sci. USA 99: 8793–8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rearick D., Prakash A., McSweeny A., Shepard S. S., Fedorova L., et al. , 2011. Critical association of ncRNA with introns. Nucleic Acids Res. 39: 2357–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski E. E., Breaker R. R., 2008. In-line probing analysis of riboswitches. Methods Mol. Biol. 419: 53–67. [DOI] [PubMed] [Google Scholar]
- Ricroch A., Yockteng R., Brown S. C., Nadot S., 2005. Evolution of genome size across some cultivated Allium species. Genome 48: 511–520. [DOI] [PubMed] [Google Scholar]
- Rinn J. L., Euskirchen G., Bertone P., Martone R., Luscombe N. M., 2003. The transcriptional activity of human chromosome 22. Genes Dev. 17: 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., et al. , 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T., Hoki Y., Sasaki H., 2005. Tsix silences Xist through modification of chromatin structure. Dev. Cell 9: 159–165. [DOI] [PubMed] [Google Scholar]
- Sado T., Hoki Y., Sasaki H., 2006. Tsix defective in splicing is competent to establish Xist silencing. Development 133: 4925–4931. [DOI] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. P., 2011. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 146: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y. T., Ideue T., Sano M., Mituyama T., Hirose T., 2009. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 106: 2525–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. V., Matteson P. G., Jones B. K., Guan X. J., Tilghman S. M., 2000. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 14: 1997–2002. [PMC free article] [PubMed] [Google Scholar]
- Schmitz K. M., Mayer C., Postepska A., Grummt I., 2010. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24: 2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell J. R., Dyson H. J., Wright P. E., 2004. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu. Rev. Biophys. Biomol. Struct. 33: 119–140. [DOI] [PubMed] [Google Scholar]
- Schorderet P., Duboule D., 2011. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 7: e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Martinez A. M., Iovino N., Cavalli G., 2011. Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12: 799–814. [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Pirrotta V., 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8: 9–22. [DOI] [PubMed] [Google Scholar]
- Seila A. C., Calabrese J. M., Levine S. S., Yeo G. W., Rahl P. B., et al. , 2008. Divergent transcription from active promoters. Science 322: 1849–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H., 2009. Redefining microRNA targets. Curr. Biol. 19: 870–873. [DOI] [PubMed] [Google Scholar]
- Selth, L. A., C. Gilbert, and J. Q. Svejstrup, 2009 RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harb. Protoc. pdb.prot5234. [DOI] [PubMed]
- Sharma S., Findlay G. M., Bandukwala H. S., Oberdoerffer S., Baust B., et al. , 2011. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc. Natl. Acad. Sci. USA 108: 11381–11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh Mohamed J., Gaughwin P. M., Lim B., Robson P., Lipovich L., 2010. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 16: 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M. D., Wang C. I., Kharchenko P. V., West J. A., Chapman B. A., et al. , 2011. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA 108: 20497–20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M. C., Sato K., Pezic D., Aravin A. A., 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12: 246–258. [DOI] [PubMed] [Google Scholar]
- Smith C. M., Steitz J. A., 1998. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 18: 6897–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M., Hayashi T., Tarui H., Agata K., Takeichi M., et al. , 2007. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 120: 2498–2506. [DOI] [PubMed] [Google Scholar]
- Sparmann A., van Lohuizen M., 2006. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6: 846–856. [DOI] [PubMed] [Google Scholar]
- Struhl K., 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 14: 103–105. [DOI] [PubMed] [Google Scholar]
- Sultan M., Schulz M. H., Richard H., Magen A., Klingenhoff A., et al. , 2008. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960. [DOI] [PubMed] [Google Scholar]
- Sun B. K., Deaton A. M., Lee J. T., 2006. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell 21: 617–628. [DOI] [PubMed] [Google Scholar]
- Sunwoo H., Dinger M. E., Wilusz J. E., Amaral P. P., Mattick J. S., et al. , 2009. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19: 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft R. J., Pheasant M., Mattick J. S., 2007. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 29: 288–299. [DOI] [PubMed] [Google Scholar]
- Takagi N., Sasaki M., 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256: 640–642. [DOI] [PubMed] [Google Scholar]
- Tam O. H., Aravin A. A., Stein P., Girard A., Murchison E. P., et al. , 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano K., Mizuno R., Okada T., Rakwal R., Shibato J., et al. , 2010. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 584: 4575–4580. [DOI] [PubMed] [Google Scholar]
- Tautz D., Domazet-Lošo T., 2011. The evolutionary origin of orphan genes. Nat. Rev. Genet. 12: 692–702. [DOI] [PubMed] [Google Scholar]
- Thomas C. A., 1971. The genetic organization of chromosomes. Annu. Rev. Genet. 5: 237–256. [DOI] [PubMed] [Google Scholar]
- Tian D., Sun S., Lee J. T., 2010. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143: 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V., Ellis J. D., Shen Z., Song D. Y., Pan Q., et al. , 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39: 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. C., Manor O., Wan Y., Mosammaparast N., Wang J. K., et al. , 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuiji H., Yoshimoto R., Hasegawa Y., Furuno M., Yoshida M., et al. , 2011. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells 16: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J., Jensen K., Mele A., Darnell R. B., 2005. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37: 376–386. [DOI] [PubMed] [Google Scholar]
- Ulitsky I., Shkumatava A., Jan C. H., Sive H., Bartel D. P., 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood J. G., Uzilov A. V., Katzman S., Onodera C. S., Mainzer J. E., et al. , 2010. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods 7: 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H., Nislow C., Blencowe B. J., Hughes T. R., 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 8: e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H., Nislow C., Blencowe B. J., Hughes T. R., 2011. Response to “The reality of pervasive transcription.” PLoS Biol. 9: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L. B., Bartolomei M. S., 2008. Regulation of imprinting in clusters: noncoding RNAs vs. insulators. Adv. Genet. 61: 207–223. [DOI] [PubMed] [Google Scholar]
- Wang D., Garcia-Bassets I., Benner C., Li W., Su X., et al. , 2011a Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang J., Zheng H., Li J., Liu D., et al. , 2004. Mouse transcriptome: neutral evolution of ’non-coding’ complementary DNAs. Nature 431: 7010. [PubMed] [Google Scholar]
- Wang J., Liu X., Wu H., Ni P., Gu Z., et al. , 2010. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 38: 5366–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. C., Yang Y. W., Liu B., Sanyal A., Corces-Zimmerman R., et al. , 2011b A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Arai S., Song X., Reichart D., Du K., et al. , 2008. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454: 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., et al. , 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., 2010. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 20: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson K. A., Merino E. J., Weeks K. M., 2006. Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1: 1610–1616. [DOI] [PubMed] [Google Scholar]
- Williamson C. M., Turner M. D., Ball S. T., Nottingham W. T., Glenister P., et al. , 2006. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat. Genet. 38: 350–355. [DOI] [PubMed] [Google Scholar]
- Willingham A. T., Orth A. P., Batalov S., Peters E. C., Wen B. G., et al. , 2005. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–1573. [DOI] [PubMed] [Google Scholar]
- Wunderlich Z., Mirny L. A., 2009. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 25: 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Scott F., Fierke C. A., Engleke D. R., 2002. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 71: 165–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Tsai C. L., Lee J. T., 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science 311: 1149–1152. [DOI] [PubMed] [Google Scholar]
- Yakovchuk P., Goodrich J. A., Kugel J. F., 2009. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. USA 106: 5569–5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M. D., Hong C. C., Lai G. M., Cheng A. L., Lin Y. W., et al. , 2005. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum. Mol. Genet. 14: 1465–1474. [DOI] [PubMed] [Google Scholar]
- Yang L., Lin C., Liu W., Zhang J., Ohgi K. A., et al. , 2011. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Zhou L., Wu L. M., Lai M. C., Xie H. Y., et al. , 2011. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 18: 1243–1250. [DOI] [PubMed] [Google Scholar]
- Yap K. L., Li S., Muñoz-Cabello A. M., Raguz S., Zeng L., et al. , 2010. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. H., Abdelmohsen K., Srikantan S., Yang X., Martindale J. L., et al. , 2012. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis J. J., Yasmineh W. G., 1971. Heterochromatin, satellite DNA, and cell function. Science 174: 1200–1209. [DOI] [PubMed] [Google Scholar]
- Zhang B., Arun G., Mao Y. S., Lazar Z., Hung G., et al. , 2012. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. F., Huynh K. D., Lee J. T., 2007. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129: 693–706. [DOI] [PubMed] [Google Scholar]
- Zhao J., Sun B. K., Erwin J. A., Song J. J., Lee J. T., 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Ohsumi T. K., Kung J. T., Ogawa Y., Grau D. J., et al. , 2010. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40: 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]