Abstract
All living organisms require nutrient minerals for growth and have developed mechanisms to acquire, utilize, and store nutrient minerals effectively. In the aqueous cellular environment, these elements exist as charged ions that, together with protons and hydroxide ions, facilitate biochemical reactions and establish the electrochemical gradients across membranes that drive cellular processes such as transport and ATP synthesis. Metal ions serve as essential enzyme cofactors and perform both structural and signaling roles within cells. However, because these ions can also be toxic, cells have developed sophisticated homeostatic mechanisms to regulate their levels and avoid toxicity. Studies in Saccharomyces cerevisiae have characterized many of the gene products and processes responsible for acquiring, utilizing, storing, and regulating levels of these ions. Findings in this model organism have often allowed the corresponding machinery in humans to be identified and have provided insights into diseases that result from defects in ion homeostasis. This review summarizes our current understanding of how cation balance is achieved and modulated in baker’s yeast. Control of intracellular pH is discussed, as well as uptake, storage, and efflux mechanisms for the alkali metal cations, Na+ and K+, the divalent cations, Ca2+ and Mg2+, and the trace metal ions, Fe2+, Zn2+, Cu2+, and Mn2+. Signal transduction pathways that are regulated by pH and Ca2+ are reviewed, as well as the mechanisms that allow cells to maintain appropriate intracellular cation concentrations when challenged by extreme conditions, i.e., either limited availability or toxic levels in the environment.
IN addition to the major components of organic molecules, i.e., carbon, nitrogen, hydrogen, and oxygen, living organisms require multiple chemical elements, termed nutrient minerals, for growth. In the aqueous cellular environment, these elements exist as charged ions that, together with protons and hydroxide ions, facilitate biochemical reactions. Charged ions, which cannot diffuse across lipid bilayers, also provide the raw material to establish electrochemical gradients that drive cellular processes such as ATP synthesis. Potassium ions help balance negative charge inside cells and activate critical metabolic processes such as protein translation. Trace elements, such as zinc, copper, iron, and manganese, are critical determinants of protein structure and serve as essential enzyme cofactors. Calcium performs structural, enzymatic, and signaling roles within cells. All of these essential elements can also be toxic. Thus, cells must be able to acquire, utilize, and store nutrient minerals effectively, but have also developed sophisticated homeostatic mechanisms to regulate their levels and avoid toxicity. Genetic studies in yeast have identified key components responsible for acquiring, utilizing, storing, and regulating levels of these ions. Furthermore, because many of these proteins are highly conserved, yeast serves as an excellent model to identify the corresponding machinery in humans and understand diseases that result from defects in ion homeostasis. A genome-wide study measured levels of 13 elements in >4000 yeast deletion strains grown in rich medium to establish the yeast “ionome.” Relatively few mutations (212) were found to significantly perturb the ionome, revealing that robust mechanisms exist to compensate for loss of a single component of ion homeostasis (Eide et al. 2005). However, the vast majority of the 212 mutations identified altered the level of more than one element, and subsets of elements covaried, illustrating the cooperative nature of the regulatory networks that control intracellular ion levels. These studies also highlighted the critical role that intracellular organelles, particularly the vacuole and the mitochondria, play in ion regulation.
This chapter reviews our current understanding of how cation balance is achieved and regulated in baker’s yeast. Starting with monovalent cations and proceeding to divalent metal ions, the role of each cation is briefly reviewed, with particular emphasis on current knowledge of its uptake, storage, and efflux mechanisms. Where appropriate, roles for cation in signal transduction pathways are also discussed.
Maintenance of Intracellular pH
The concentration of protons (H+) in the cell, expressed as intracellular pH (pHi), dramatically influences every aspect of cellular biochemistry and must be carefully regulated both in the cytosol and in the lumen of intracellular organelles. In rapidly growing yeast cells, cytosolic pH is stable at 7.2 and changes little over a broad range of extracellular pH (Martinez-Munoz and Kane 2008; Orij et al. 2009). However, mounting evidence indicates that intracellular pH is regulated and can serve a signaling function, in particular, to report nutrient availability (reviewed in Orij et al. 2011). Different methods have been used to assess pHi in Saccharomyces cerevisiae including 31P NMR (Navon et al. 1979), pH sensitive dyes (Haworth et al. 1991), and more recently, expression of pHlourin, a ratiometric pH-sensitive fluorescent protein (Orij et al. 2009) that can be targeted to the cytosol or intracellular compartments to measure pH in living cells (Braun et al. 2010; Maresova et al. 2010). The concentration of protons in the cytosol is largely determined by the activity of two proton pumps: Pma1, which resides in the plasma membrane (PM), and a large protein complex, termed the V-ATPase, which acidifies the endomembrane system including the vacuole, Golgi, and endosomal compartments.
Pma1, the essential plasma membrane proton pump
Pma1, a P2-type ATPase, is made up of a single 100-kDa subunit that pumps one proton across the plasma membrane per ATP molecule hydrolyzed. This H+-ATPase is one of the most abundant cellular proteins, consuming at least 20% of cellular ATP. Pma1 is essential and rate limiting for growth; mutations that compromise its activity decrease both cytosolic pH and growth (McCusker et al. 1987; Portillo and Serrano 1989). Mutants with impaired Pma1 function are unable to grow in low pH media or in the presence of weak acids, reflecting their reduced ability to pump protons across the plasma membrane. They are also resistant to a variety of cationic drugs and ions, including the aminoglycoside, hygromycin B, because decreasing the proton-motive force across the plasma membrane leads to reduced cellular uptake of these compounds (Perlin et al. 1988). PMA2 encodes a closely related gene product that is normally present at a very low level, but can substitute for Pma1 when expressed from a strong promoter (Supply et al. 1993).
Regulation of Pma1
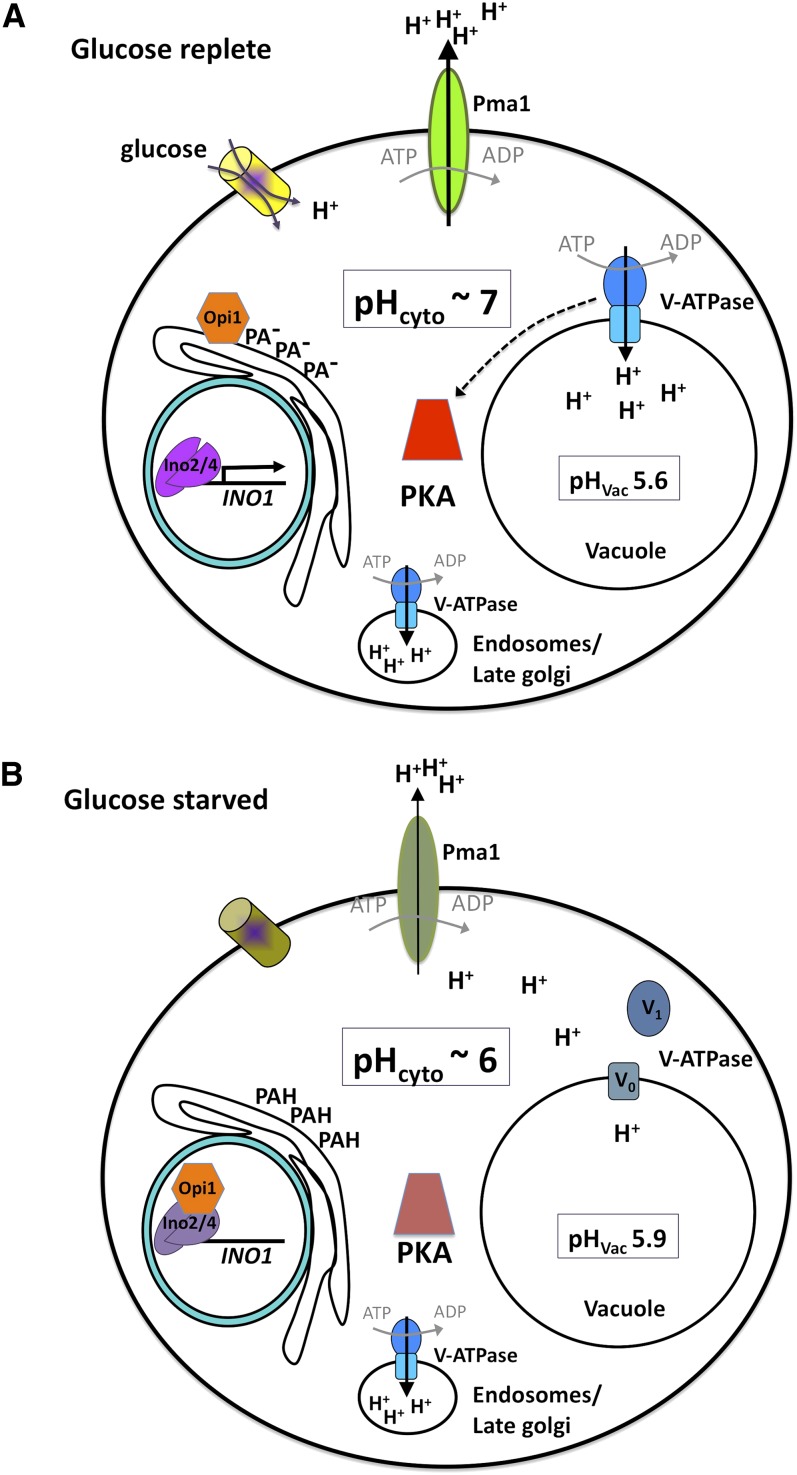
Pma1 is regulated by at least two distinct mechanisms: First, to maintain neutral pH in the cytosol, Pma1 is activated when intracellular pH drops. This has been most clearly shown in cells exposed to weak organic acids, which cross the PM in their protonated state and become deprotonated in the cell to lower pHi. Under these conditions, the Km of Pma1 is decreased through an unknown mechanism (Eraso and Gansedo 1987; Carmelo et al. 1997). A drop in internal pH also affects Pma1 activity indirectly by activating K+ uptake through plasma membrane transporters, Trk1/2; this offsets the electrogenic potential created by Pma1-mediated proton pumping and facilitates its activation (Yenush et al. 2005). Pma1 is also regulated post-translationally by phosphoryation, especially in response to nutrients. When glucose is added back to starved cells, pHi decreases transiently (30 sec), followed by activation of Pma1, which promotes recovery of pHi to pH 7.2 (Figure 1). Pma1 activation is mediated in part by phosphorylation of the autoinhibitory C-terminal tail of Pma1, which increases its Vmax, affinity for ATP, and pH optimum. Thr-912 of Pma1 is constitutively phosphorylated; a modification that is necessary but not sufficient for Pma1 activation (Portillo et al. 1991). Glucose addition results in further phosphorylation at Ser-911 to produce a tandemly phosphorylated, activated ATPase (Serrano 1983; Lecchi et al. 2007). Ptk2 and Hrk1, members of the Npr1 family of protein kinases, which regulates multiple membrane transporters, are thought to phosphorylate Pma1 (Goossens et al. 2000; Eraso et al. 2006), and it may be dephosphorylated by a PP1-type phosphatase, containing the Glc7 catalytic subunit (Williams-Hart et al. 2002). Other studies suggest that Ca2+-dependent signaling also contributes to activation of Pma1, by unknown mechanisms, in response to glucose addition (Tropia et al. 2006; Bouillet et al. 2012).
Figure 1.
During glucose starvation, changes in intracellular pH serve a signaling function. (A) In the presence of glucose, Pma1 and the V-ATPase are active; cytosolic pH is 7.0–7.2 and vacuolar pH is 5.6. PKA is active, and V-ATPase association is proposed to regulate its activity. Inositol synthesis is active due to sequestration of the Opi1 repressor on the surface of the ER. (B) In glucose-starved cells, cytosolic pH drops due to decreased activity of Pma1 and dissociation of the V-ATPase subcomplexes. Opi1 is released from the ER due to protonation of phosphatadic acid (PA); Op1 entry into the nucleus represses expression of INO1 by Ino2/4 transcriptonal activators. Bright colors in A indicate active proteins; dull colors in B indicate reduced activity. See text for details.
Vacuolar ATPase acidifies the vacuole and secretory organelles
The other major protein that impacts cytosolic pH is the vacuolar proton-translocating ATPase, or V-ATPase, which pumps protons into the vacuole. The V-ATPase helps maintain cytosolic pH by removing protons from the cytosol; treating cells with concanamycin A, a V-ATPase inhibitor, causes a rapid drop in intracellular pH (Martinez-Munoz and Kane 2008). The V-ATPase is a large highly conserved protein complex related to the mitochondrial F1/F0 ATPase. It is made up of 14 different subunits (some in multiple copies per enzyme) that are organized into two discrete subcomplexes: an integral membrane complex, V0, which contains the proton pore, and an associated V1 complex, which is responsible for ATP hydrolysis and is made up of peripheral membrane proteins (Kane 2006). In addition to these structural components, several additional proteins are required for assembly of the V0 subcomplex in the endoplasmic reticulum (ER) and its subsequent transport/exit from the ER, but are not present in the mature enzyme (Graham et al. 2000; Malkus et al. 2004; Davis-Kaplan et al. 2006; Ryan et al. 2008). There are two different forms of the V-ATPase in cells; one, containing the Vph1 subunit of the V0 complex, resides on the vacuolar membrane and is responsible for acidifying that compartment (Tarsio et al. 2011). The other contains Stv1 (similar to Vph1) instead of Vph1, and is transported to endosomes, the late Golgi, and secretory vesicles whose lumens are also acidic (Braun et al. 2010).
The major function of the V-ATPase is to maintain acidic pH in the vacuole, which is approximately 5.6 in growing cells (Martinez-Munoz and Kane 2008). This acidic pH is essential for enzymatic processes that take place in the vacuole, such as proteolysis, and creates a proton gradient across the vacuolar membrane that enables proton exchangers (i.e., Ca2+/H+, K+/H+, and Na+/H+) to transport their substrates into the vacuole. The V-ATPase is not essential for growth under standard laboratory conditions; however, vma− mutants, which lack V-ATPase enzyme function, have growth defects on both low (<3) and high (>7) pH media and are sensitive to a variety of cations, including Ca2+, due to decreased sequestration of ions in the vacuole. In cells lacking functional V-ATPase, the only mechanism for vacuole acidification is the uptake of protons from the media via endocytosis (Munn and Riezman 1994).
Regulation of V-ATPase activity
One mode of V-ATPase regulation is through the reversible dissociation/association of its two subcomplexes. This phenomenon was first demonstrated in response to glucose deprivation and readdition; during glucose starvation, when ATP is limited, the intact V1 subcomplex detaches from the integral membrane V0 complex (Kane 2011), causing inactivation of both portions of the V-ATPase and loss of ATP hydrolysis (Figure 1). This regulation is only observed for V-ATPase enzymes residing on the vacuolar membrane (Kawasaki-Nishi et al. 2001).
Association of the V1 and V0 in response to glucose readdition promotes vacuolar acidification and requires the RAVE complex (Smardon et al. 2002), PKA/Ras activity, and other factors (Smardon et al. 2002; Kane 2011). This rapid mode of V-ATPase regulation appears to be highly conserved, as V-ATPase integrity is regulated in many organisms including mammals (Kane 2011) and responds to additional signals including pH; decreasing intracellular pH is necessary and sufficient to trigger V-ATPase disassembly (Dechant et al. 2010), and increasing extracellular pH suppresses dissociation and stimulates V-ATPase activity (Diakov and Kane 2010). Furthermore, in cells grown in poor nutrient conditions, the V-ATPase pool is partially disassociated, suggesting that this regulatory mechanism fine tunes the amount of V-ATPase activity to fit different growth conditions (Kane 2011).
Coordination of Pma1 and V-ATPase
In normal cells, cytosolic pH is maintained by the combined efforts of Pma1 and the V-ATPase, and the activities of these proton pumps are coordinated at multiple levels. For example, in vma− mutants Pma1 is partially mislocalized to the vacuole and other compartments, which may help compensate for loss of the V-ATPase (Martinez-Munoz and Kane 2008). Other conditions that alter vacuolar pH, such as incubation with concamycin A or deletion of VPH1, cause a reduction in Pma1 activity without perturbing its localization; the mechanism that mediates this Pma1 modulation in response to changes in vacuolar pH is unknown (Martinez-Munoz and Kane 2008; Tarsio et al. 2011).
Effects of pH on transport across membranes
In addition to their effects on intracellular pH, the proton gradients created by Pma1 and the V-ATPase are major determinants of membrane potential (Δψ), which is the sum of all ionic gradients over a membrane. This electrochemical gradient is harnessed by protein transporters to drive the uptake and efflux of ions and nutrients across the membrane. The uptake of nutrients depends on maintenance of the plasma membrane proton gradient, as major nutrient permeases for glucose and other sugars as well as amino acids are H+ symporters. Furthermore, these proton-coupled transporters, together with the K+ transporters, Trk1 and Trk2, and the Na+/H+ antiporter, Nha1, influence cytosolic pH by consuming the proton gradient (Orij et al. 2011). Similarly, the electrochemical gradient across the vacuolar membrane powers transport into and out of the vacuole. Ca2+ accumulates in the vacuole via the Ca2+/H+ antiporter, Vcx1. Similarly, transport of heavy metals (Ycf1 and Zrc1), amino acids (Avt3, Avt4, and Avt6), and polyphosphate accumulation depend on the proton gradient across the vacuolar membrane (reviewed in Li and Kane 2009).
Do changes in intracellular pH serve a signaling function?
When cells are actively growing, homeostatic mechanisms maintain a relatively constant intracellular pH of 7.2 (Martinez-Munoz and Kane 2008; Orij et al. 2009). In contrast, glucose starvation causes a drop in cytosolic pH due to decreased Pma1 activity and V-ATPase disassembly (Figure 1). Readdition of glucose to starved cells causes a further transient (30 sec) intracellular acidification, followed by a rise in intracellular pH mediated by activation of Pma1 and association of the V-ATPase subcomplexes. Some observations suggest that these rapid changes in pHi may be a signal that couples nutrient status to downstream events. In particular, adding glucose back to starved cells results in rapid activation of PKA, which phosphorylates multiple targets and signals high glucose availability. PKA is activated by a spike of cAMP, produced when the small GTPase, Ras, activates adenylate cyclase (Cyr1). Recent studies suggest that the V-ATPase is required for this glucose-mediated stimulation of PKA and that cytosolic pH serves as a second messenger in this regulatory pathway (Dechant et al. 2010) (Figure 1). However, glucose acts through a complex network of signaling events, some of which, i.e., activation of Cyr 1 through negative regulation of Ras GAPs (Ira1 and Ira2), are independent of changes in intracellular pH (Thevelein and de Winde 1999).
Changes in intracellular pH are also thought to regulate phospholipid metabolism. During normal growth, when cytosolic pH is close to neutral, the Opi1 transcriptional repressor is tethered to the surface of the ER by its interactions with Scs2 and phosphatidic acid (PA), and as a result, inositol-regulated genes are active (Loewen et al. 2004) (Figure 1). However, when intracellular pH drops, as in glucose-starved cells, the phosphate head group of PA becomes protonated, decreasing its affinity for Opi1 and releasing the repressor from the ER. This allows Opi1 to enter the nucleus, where it inhibits expression of genes including INO1, which encodes the rate-limiting enzyme in inositol biosynthesis (Young et al. 2010). Thus, changes in intracellular pH couple nutrient availability to the production of key enzymes required for membrane biogenesis.
The Alkali Metals
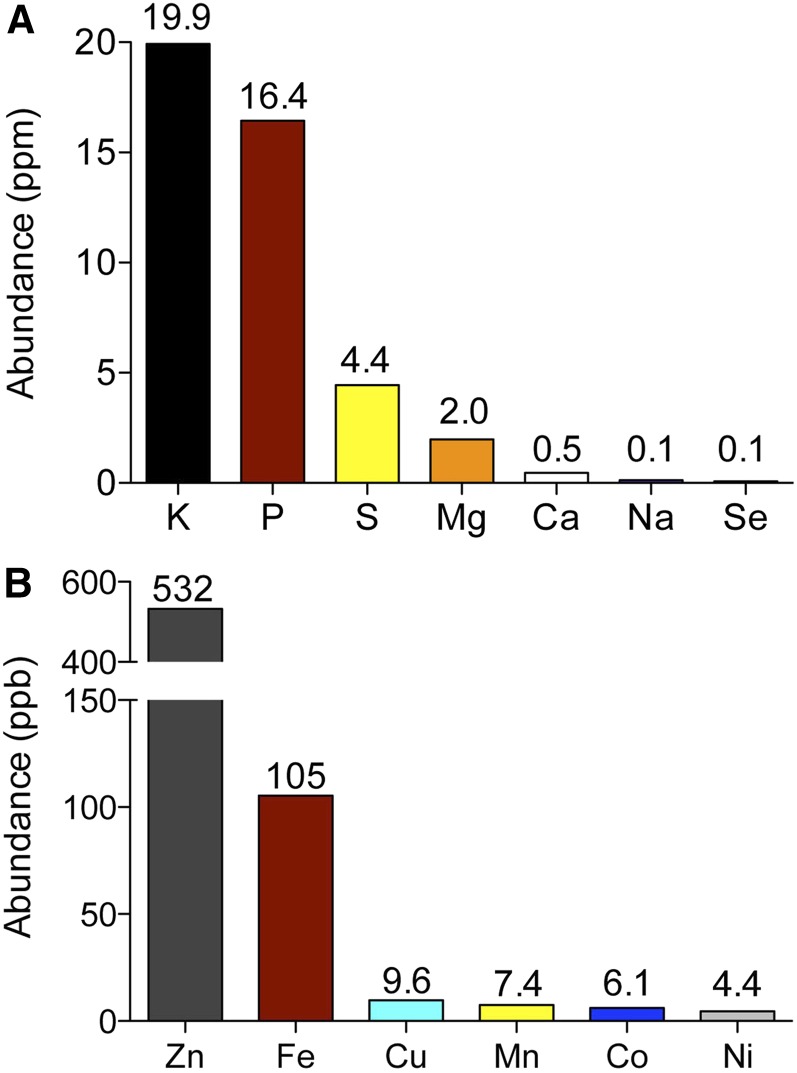
Sodium and potassium, together with lithium, rubidium, cesium, and francium, make up the family of alkali metals, which share similar properties and atomic structures and readily form monovalent cations. In cells, K+, plays many important physiological roles; it is required for negative charge compensation and activation of key metabolic processes, such as pyruvate synthesis and protein translation (Page and Di Cera 2006). In contrast, Na+ is toxic at high levels, in part because it readily substitutes for K+ (Page and Di Cera 2006). Thus, despite the much greater abundance of Na+ in the environment, yeast must maintain a high intracellular ratio of K+/Na+ (Figure 4A), and achieve this by selectively accumulating K+, while actively extruding Na+. K+ is critical for balancing charge across the plasma membrane, and thus contributes to maintaining both intracellular pH and membrane potential. K+ is continually taken up and extruded by cells, and both processes are important; membrane potential increases when potassium influx is crippled and decreases in cells defective for K+ efflux (Madrid et al. 1998; Kinclova-Zimmermannova et al. 2006); furthermore, K+ efflux systems are regulated in response to changes in the membrane potential (Zahradka and Sychrová 2012). K+ transport is also closely coordinated with H+-ATPase activity, and Pma1 is activated when K+ uptake increases and also under K+ starvation conditions (Seto-Young and Perlin 1991; Kahm et al. 2012).
Figure 4.
Abundance of common elements in S. cerevisiae. Strain BY4741 was grown in rich (YPD) medium and ionic content was measured using inductively coupled plasma-atomic emission spectroscopy. (A) Data expressed as parts per million (ppm). (B) Abundance of metals, with data expressed as parts per billion (ppb). Data from Eide et al. (2005).
K+ entry into cells
Under ideal growth conditions, cellular K+ content is 200–300 mM (Figure 4A); however, yeast can grow over a wide range of external K+ concentrations (from 10 μM to 2.5 M), and must maintain a minimal amount of internal K+ (∼30 mM) to survive (Arino et al. 2010). Gene products responsible for high affinity K+ uptake, the closely related Trk1 and Trk2, were identified by screening for mutants unable to grow under K+-limiting conditions (Gaber et al. 1988; Ko et al. 1990; Ko and Gaber 1991) (Figure 2). Of these, Trk1 is the dominant determinant of K+ influx, due to its higher expression level. However, trk1Δ trk2Δ cells require ∼10-fold higher levels of K+ supplementation for normal growth than trk1Δ, demonstrating the contribution of Trk2 to K+ uptake. Trk1 and Trk2 are large plasma membrane proteins, 1235 and 889 amino acids long, respectively, each containing four M1-P-M2 sequence motifs, where M1 and M2 denote hydrophobic domains that are connected by an α-helical P segment. By analogy with the crystal structure of KcsA, a K+ channel from Streptomyces lividans, each protein is proposed to fold into a symmetric array of four repeating pairs of membrane-spanning domains that contain a central K+-conducting pore (Durell and Guy 1999). Each Trk1 or Trk2 monomer is further suggested to associate into a homotetramer. Trk1/2-mediated transport displays high affinity for K+ and Rb+, and high velocity (Vmax = 30 nmol/mg cells/min) that is driven by the negative electrochemical potential established by the H+-APTase (Rodriguez-Navarro and Ramos 1984). Each transporter has two binding sites for cations, and normally cotransports two identical (K+) ions, in contrast to related transporters in plants that cotransport one K+ and one Na+ ion (Haro and Rodriguez-Navarro 2002). Surprisingly, these proteins also mediate efflux of anions, including halides (I−, Br−, and Cl−) and nonhalide chaotropic anions (SCN− and NO3−), an activity that is not coupled to K+ transport (Kuroda et al. 2004; Rivetta et al. 2011). The physiological relevance of this efflux activity is not well understood, but may allow cells to balance charges created by Pma1-mediated proton extrusion (Rivetta et al. 2011).
Figure 2.
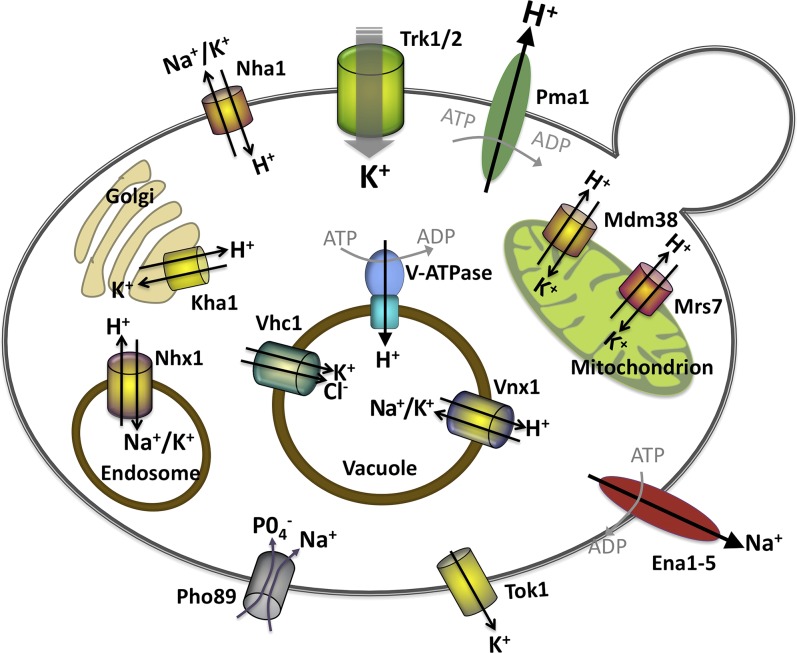
The major transporters responsible for uptake, efflux, and intracellular transport of alkali metal ions in S. cerevisiae. Note that the V-ATPase also resides in endosomes and late Golgi, but was omitted from the figure due to space constraints.
The activity and/or stability of Trk1/2 are altered by several protein kinases and phosphatases. The functionally redundant kinases, Hal4 and Hal5, are required to stabilize Trk1/2 at the plasma membrane, especially under conditions of low extracellular [K+] (Perez-Valle et al. 2007). Furthermore cells lacking calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, display decreased K+ uptake, which is due in part to a defect in HAL5 expression, which is regulated by Crz1 (see below) and potentially to direct regulation of the transporters as well (Mendoza et al. 1994; Casado et al. 2010). Sky1, an SR protein kinase, so named for its role in regulating SR-type splicing factors, may also regulate Trk1/2 and/or additional components of K+ homeostasis (Erez and Kahana 2002; Forment et al. 2002). Finally, Trk1 physically associates with the Ppz1 phosphatase in membrane microdomains, and increased phosphorylation of Trk1 in ppz1ppz2 mutants suggests that it is a substrate for these phosphatases. Hal3 negatively regulates Ppz1 and associates with it in a pH-dependent manner, thus leading to the proposal that the Hal3–Ppz1 interaction allows intracellular levels of H+ and K+ to be adjusted coordinately through regulation of Trk1 activity (Yenush et al. 2002, 2005).
In addition to high affinity K+/Rb+ transport, yeast display a low-affinity mode, which is likely also mediated by Trk1/2 (Arino et al. 2010). However, trk1trk2 cells display a very low affinity K+ transport activity, with a Km in the millimolar range, for which the responsible protein(s) has not been identified (Madrid et al. 1998). NSC1 (for nonspecific cation channel), an ion conductance identified using electrophysiological methods whose molecular identity is unknown, mediates K+ currents that are blocked by Ca2+ and other divalent cations in trk1trk2 cells, and could be responsible for the low affinity K+ transport observed (Bihler et al. 1998; Madrid et al. 1998). Thus, additional K+ transporters remain to be identified in yeast.
K+ efflux
Nha1 and Ena1, discussed below, play primary roles in Na+ transport, but also efflux K+. Tok1, an outwardly rectifying K+ channel in the plasma membrane that is activated by membrane depolarization, is the only K+-specific efflux mechanism in yeast cells (Gustin et al. 1986; Bertl et al. 1993; Ketchum et al. 1995; Zhou et al. 1995) (Figure 2). Tok1 is a 691-amino-acid-long integral membrane protein that contains two pore domains, each of which conducts K+. It is the founding member of a conserved family of K+ channels (2P) that contain two tandem pore domains (Enyedi and Czirjak 2010). The cytosolic carboxy terminal tail of Tok1 participates in its regulation and gating by preventing channel closure (Loukin and Saimi 2002). Despite extensive electrophysiological characterization of this channel, its physiological significance is somewhat unclear. Tok1-mediated release of cellular K+ at low membrane potential should boost the electrochemical potential across the plasma membrane. In fact, deletion of TOK1 does lead to depolarization, and its overexpression hyperpolarizes cells (Maresova et al. 2006), but tok1 mutants display no growth defects, changes in K+ content, or transport activity. This channel is the target of the K1 viral killer toxin, which kills yeast by binding to and activating Tok1 (Ahmed et al. 1999).
Maintenance of intracellular K+ levels
Although many components of K+ transport in yeast have been identified, a greater understanding of the ways in which these transport systems work together is needed. A computational model was recently developed to explain how cells maintain a minimal concentration of intracellular K+ under K+ starvation conditions. Surprisingly, these investigations suggest that increased levels of Pma1 activity and bicarbonate ion, rather than regulation of Trk1/2 or Nha1, are critical under these conditions (Kahm et al. 2012). Intracellular CO2, produced in many enzymatic reactions, is converted by carbonic anhydrase (encoded by NCE103) to carbonic acid, which dissociates into bicarbonate (HCO3−) and protons. Protons are pumped out of the cell by Pma1; however, HCO3− accumulates inside the cell and becomes a negative-charge sink that increases K+ retention inside the cell. Experimental studies confirm that NCE103 expression increases under conditions of K+ starvation (Kahm et al. 2012) and also show that while cells maintain a minimal intracellular K+ concentration, the amount of K+ in a yeast cell varies as a function of extracellular K+ concentration and is an example of nonperfect adaptation (Kahm et al. 2012).
Na+ entry into cells
Yeast cells do not actively accumulate Na+; cellular levels of this ion are low under standard growth conditions, but rise when cells are challenged with a high Na+ environment. For wild-type yeast, Na+ is a competitive inhibitor of K+ transport and has a lower affinity than K+ for Trk1 (Haro and Rodriguez-Navarro 2002). In high Na+ environments, Trk1 is thought to transport some Na+, although it is also modified under these conditions, through an unknown mechanism, to become more selective for K+ (Mendoza et al. 1994). Cells lacking high affinity K+ transport (trk1trk2) have a higher Na+ content than wild-type cells (Gomez et al. 1996), because the remaining, nonspecific cation transporters, such as NSC1, mediate influx of both K+ and Na+. In general, yeast transport mechanisms are biased toward minimizing Na+ uptake and promoting its efflux. However, somewhat surprisingly, S. cerevisiae also depend on Na+ for phosphate assimilation by Pho89, a plasma protein containing 12 predicted membrane-spanning domains that cotransports Na+ and inorganic phosphate (Figure 2). Pho89p has a Km for phosphate of 0.5 μM, is highly specific for Na+, and is maximally active at alkaline pH (9.5) (Martinez and Persson 1998). It is the only nutrient transporter in yeast known to require Na+ for its activity. Expression of Pho89 is induced by phosphate limitation and by high pH; it is one of two high-affinity phosphate transporters, the other of which, Pho84, mediates proton-coupled phosphate transport and is active at low pH (Pattison-Granberg and Persson 2000).
Na+ efflux
Two different mechanisms promote efflux of Na+, Li−, and to a lesser extent, K+; the Ena P-type ATPases and the Nha1 antiporter both promote yeast growth in the presence of toxic cations (Figure 2). Chromosome IV of S. cerevisiae contains a group of tandemly repeated genes, ENA1–5, each of which encodes a highly related (>97% identical in amino acid sequence) P-type ATPase of the fungal-specific P2D subtype (Palmgren and Nissen 2011). These gene products localize to the plasma membrane, where they use ATP hydrolysis to extrude Na+, Li+, and possibly K+ (Haro et al. 1991; Benito et al. 1997). The size of the ENA cluster varies; most laboratory strains contain three to five genes, with more Na+- and Li+-tolerant strains containing larger arrays (Arino et al. 2010). CEN.PK strains, which contain a single atypical gene (ENA6), and deletion mutants lacking the entire gene cluster are extremely sensitive to Na+ and Li+ (Haro et al. 1991; Daran-Lapujade et al. 2009) and display growth defects under alkaline conditions (Haro et al. 1991). Expression of ENA1 is induced under these growth conditions by a constellation of signaling pathways including the osmotic stress-responsive HOG pathway, the Ca2+-dependent calcineurin/Crz1 pathway that promotes survival under a variety of stress conditions, the Rim101 pathway, which modulates tolerance to NaCl and high pH as well as regulating meiotic gene transcription, and nutrient-dependent pathways regulated by Snf1 and TOR (reviewed in Arino et al. 2010). At high pH, Ena-mediated Na+ and K+ efflux become essential, most likely because the Na+, K+/H+ antiporter, Nha1 cannot function under these conditions (Haro et al. 1991).
Nha1 is a plasma membrane proton antiporter that mediates extrusion of Na+ and K+ with similar affinities and promotes tolerance to alkali cations at acidic pH (Bañuelos et al. 1998; Ohgaki et al. 2005). This protein functions as a dimer and is electrogenic, exchanging multiple protons for each molecule of Na+ or K+ transported (Mitsui et al. 2005; Ohgaki et al. 2005). Nha1 is 985 amino acids in length; its overall structure is similar to other Na+/H+ exchangers (NHE) with a predicted short N-terminal cytosolic domain, followed by 12 membrane-spanning segments and a very long (551 amino acids) C-terminal cytosolic domain. This C-terminal domain contains six regions that are conserved in other fungal NHEs, and includes sequences required for its targeting to the plasma membrane, activity, substrate specificity, and regulation, such as its interaction with 14-3-3 proteins (Kinclova et al. 2001; Simon et al. 2001; Mitsui et al. 2004; Zahradka et al. 2012). In addition to its role in Na+ detoxification, Nha1-mediated K+ efflux contributes to the constitutive K+ uptake and efflux that is used to regulate the membrane potential of yeast cells (Zahradka and Sychrová 2012). It also promotes rapid adaptations to alkaline and osmotic stress (Bañuelos et al. 1998; Kinclova et al. 2001; Proft and Struhl 2004), and may contribute to cell-cycle regulation, as Nha1 overexpression suppresses the G1 arrest of a phosphatase-deficient (sit4hal3) strain (Simon et al. 2001, 2003).
Alkali metal cation transport in intracellular compartments
Mitochondria:
K+/H+ exchange (KHE) is carried out by mitochondria, and mutations that compromise this exchange alter mitochondrial K+ content, cause defects in respiratory chain assembly, and disrupt mitochondrial morphology and volume homeostasis (Nowikovsky et al. 2012). In S. cerevisiae, Mdm38 and Mrs7, members of the LETM1 protein family, are required for KHE (Figure 2); however, LETM1 proteins contain a single trans-membrane domain, suggesting that they modulate transport rather than directly conducting ion exchange. In support of this hypothesis, both Mdm38 and Mrs7 are components of larger protein complexes (Nowikovsky et al. 2012). Human LETM1 family members contain EF hand calcium-binding domains that are not present in the yeast proteins, and Letm1 has been proposed to participate in mitochondrial Ca2+ uptake, although this issue is controversial. Human LETM1 is associated with Wolf-Hirschhorn syndrome (WHS), a pleiotropic neurological disorder, which is caused by a partial deletion of chromosome 4; loss of Letm1 occurs in a subset of WHS cases and is linked to the occurrence of seizures (Nowikovsky et al. 2012).
Vacuole, Golgi, and endosomes:
Yeast cells accumulate Na+ and K+ in the vacuole and other organelles of the secretory system, i.e., the Golgi and endosomes, via proton-coupled antiport, with the V-type ATPase generating the proton gradient that drives accumulation of the alkali cation. Vnx1, which was identified using reverse genetics, mediates the majority of this transport in the vacuole (Cagnac et al. 2007) (Figure 2). Vnx1 encodes a 102-kDa protein with 13 predicted membrane-spanning domains and is a member of the calcium exchanger (CAX) superfamily, although it lacks several sequence motifs thought to mediate Ca2+ binding and does not transport Ca2+. Vnx1 localizes to the vacuole, and vnx1Δ yeast display increased sensitivity to NaCl and to the positively charged aminoglycoside, hygromycin B (Cagnac et al. 2007). Vacuolar vesicles isolated from vnx1Δ cells are greatly deficient in Na+ and K+/H+ exchange activity (Cagnac et al. 2007, 2010). Surprisingly, the vacuolar Ca2+/H+ exchanger, Vcx1 was shown to mediate a small amount of residual vacuolar K+/H+ exchange observed in vnx1Δ cells (Cagnac et al. 2010).
Kha1 encodes a Golgi-localized, 97-kDa protein with 12 predicted transmembrane domains that is related to bacterial K+ and Na+ transporters (Maresova and Sychrová 2005). The transport activity of Kha1 has not been examined biochemically; however, phenotypic characterization suggests that it carries out K+/H+ transport. kha1Δ strains display sensitivity to hygromycin B and a growth defect under alkaline conditions that can be suppressed by addition of K+ to the media (Maresova and Sychrová 2005). Furthermore, studies of a mammalian chloride channel, CIC-2, showed that this channel was able to substitute for a yeast chloride channel, Gef1, only in cells that also overexpressed Kha1. This suggests that increased levels of Kha1 improves CIC-2 function by bringing the pH of the yeast Golgi closer to the CIC-2 optimum (Flis et al. 2005). Thus, Kha1 is thought to play roles in both K+ and pH homeostasis by carrying out K+/H+ exchange.
Nhx1 encodes an endosomal Na+/H+ antiporter that is a member of the NHE superfamily and localizes mainly to the late endosome, as well as other secretory organelles (Nass and Rao 1998; Kojima et al. 2012) (Figure 2). A role for Nhx1 in sequestration of Na+ and K+ into secretory organelles has been demonstrated, for which it utilizes the proton gradient established by the V-type ATPase (Nass et al. 1997; Brett et al. 2005). nxh1Δ cells display growth defects in high salt or low pH conditions, and a decrease in intracellular pH (Nass et al. 1997; Brett et al. 2005). However, Nhx1 was also identified in a screen for mutants with vacuolar protein sorting defects (Bowers et al. 2000), and its role in determining the pH of endosomal compartments is critical to protein trafficking, where it may regulate the fusion of these compartments (Qiu and Fratti 2010; Kallay et al. 2011; Kojima et al. 2012).
Proton-independent transport may also contribute to K+ accumulation in the vacuole. Vhc1, a member of the cation-Cl− cotransporter (CCC) family of transporters, is thought to function as a vacuolar K+-Cl− cotransporter (Figure 2) (Andre and Scherens 1995; Petrezselyova et al. 2012). Deletion of VHC1 improves growth of a trk1Δ trk2Δ strain under low potassium conditions, but does not alter cytosolic pH, suggesting that this protein sequesters K+ in the vacuole without transporting protons (Petrezselyova et al. 2012). Like the related K+-Cl− cotransporter, NKCC1, in humans, Vhc1 contributes to cellular volume control. When exposed to hyperosmotic shock, vacuoles in vhc1Δ cells shrink less than those in wild-type cells, and the mutants show reduced survival (Petrezselyova et al. 2012). Although Vhc1 transport activity has not been demonstrated directly, this physiological characterization is consistent with the proposal that it mediates K+-Cl− cotransport into the vacuole.
Divalent Cations: Ca2+ and Mg2+
Ca2+ ion homeostasis and regulation
Ca2+ is ubiquitous in the environment and serves a variety of signaling and structural functions in all eukaryotes. This cation is required for growth of S. cerevisae. Yeast undergo G2 arrest in growth medium that has been depleted of metal ions, but resume cell cycle progression upon readdition of Zn2+, together with either Ca2+ or Mn2+ (Loukin and Kung 1995). Calcium is abundant in yeast cells (Figure 4A). However, for Ca2+ to serve its signaling function, a low cytosolic concentration of this ion (50–200 nM) must be maintained (Miseta et al. 1999); this is achieved by Ca2+ pumps and exchangers that actively sequester Ca2+ in intracellular compartments. Under specific conditions, such as exposure to environmental stress or mating pheromone, cytosolic Ca2+ transiently increases and activates downstream events. These signals are generated by Ca2+ entry through plasma membrane ion channels and/or release from intracellular organelles. The primary Ca2+ store in yeast cells is the vacuole, where >90% of all cellular Ca2+ resides in association with polyphosphate (total concentration, 2 mM) (Dunn et al. 1994). Intracellular Ca2+ stores in yeast differ significantly from those in mammalian cells: Yeast mitochondria do not accumulate Ca2+ and do not express the Ca2+ uniporter found in their mammalian counterparts (Carafoli et al. 1970; Baughman et al. 2011; De Stefani et al. 2011). Also, the endoplasmic reticulum is not a significant Ca2+ store in yeast cells, containing 10 μM Ca2+, compared to 250–600 μM observed in the ER of mammalian cells (Demaurex and Frieden 2003). However, there are direct parallels between the main components of Ca2+ homeostasis in yeast cells and cardiac myocytes (described in Cui et al. 2009b).
Ca2+ entry into yeast cells
Yeast cells possess both high and low affinity mechanisms for Ca2+ influx across the plasma membrane. The high affinity, low capacity, Ca2+ system (HACS) requires at least three proteins: Mid1, Cch1, and Ecm7 (Cunningham 2011). Mid1 and Cch1 are trans-membrane proteins that interact with each other (Iida et al. 1994; Fischer et al. 1997; Paidhungat and Garrett 1997; Locke et al. 2000). Mid1 has four predicted trans-membrane domains and is broadly conserved in yeast and fungi. Cch1 is a large protein that is related to the alpha subunit of mammalian voltage-gated calcium channels (VGCCs) and is partially inhibited by the L-type VGCC blockers nifedipine and verapamil (Teng et al. 2008). However, key residues that are required for voltage sensing in mammalian channels are lacking in Cch1 (Teng et al. 2008). Ecm7, the third protein proposed to be a component of the channel, is related to gamma subunits of VGCCs and is a member of the claudin superfamily of proteins. In cells lacking Mid1, Ecm7 is destabilized, suggesting that it associates with Mid1, although this interaction has yet to be demonstrated (Martin et al. 2011). Cells lacking either Mid1 or Cch1 or both have similar phenotypes, such as low Ca2+ uptake activity and loss of viability during prolonged exposure to mating pheromone (Iida et al. 1994; Fischer et al. 1997; Paidhungat and Garrett 1997), consistent with their contributing to a single functional complex. The electrophysiological properties of this Ca2+ channel and its mechanism of activation are unclear. Expression of Mid1 in mammalian cells resulted in a new stretch-activated Ca2+ channel activity, and the endogenous yeast channel may be similarly mechanosensitive. Studies of the analogous channel in the fungal pathogen, Cryptococcus neoformans, indicate that the channel is activated by the depletion of intracellular Ca2+ stores (Hong et al. 2010). Several physiological conditions that activate influx through HACS have been identified, although the molecular details of this regulation have not been worked out (see below).
A second, less well-defined Ca2+ influx activity in yeast has been termed LACS for low affinity Ca2+ system. LACS is activated when cells grown in rich medium are exposed to high concentrations of pheromone. Genome-wide screens for mutants defective in LACS identified components of the polarisome (Bni1, Spa2, and Pea2), a protein complex that localizes to the tip of the mating projection and is involved in organizing and polymerizing actin (Howell and Lew 2012), as well as other actin-binding proteins (Rvs167) and gene products required for cell fusion during mating (Fig1, Fus1, and Fus2) (Muller et al. 2003, 2009). Pheromone induces expression of Fig1, a member of the claudin superfamily of proteins that contains four predicted transmembrane domains (Erdman et al. 1998), and localizes to sites of cell fusion during mating (Aguilar et al. 2007). Cells lacking Fig1 fail to induce LACS and have fusion defects during mating that can be suppressed by addition of Ca2+ to the medium. This suggests that Ca2+ influx through LACS is intimately coupled to the cell fusion process and may represent a Ca2+-dependent wound repair pathway that is triggered by membrane holes induced during the fusion process (Engel and Walter 2008).
In addition to HACS and LACS, yeast cells possess additional Ca2+ influx pathways that have not yet been characterized at the molecular level. Mathematical modeling of Ca2+ transients predicts the presence of additional influx pathways that are subject to rapid feedback inhibition by Ca2+ (Cui et al. 2009a). Subsequent experimental analyses indicated that these additional influx pathways are inhibited by Mg2+ (Cui et al. 2009a), which is consistent with observations of rapid Ca2+ influx in cells grown in Mg2+-deficient medium (Wiesenberger et al. 2007). Ca2+ influx is also stimulated when glucose is added back to glucose-starved cells, and this pathway requires the Gpr1 plasma membrane receptor coupled to Gpa2, a Gα subunit, and phospholipase C (see below). For cells grown in minimal medium, Mid1/Cch1 mediates this Ca2+ influx. However, cells grown in rich medium show a large Ca2+ signal upon glucose addition that is retained in mutants lacking Mid1, Cch1, or Fig1. This new activity, termed glucose-induced calcium (GIC) system, displays unique characteristics, including sensitivity to magnesium, gadolinium, and nifedipine and resistance to the related L-type VGCC inhibitor, verapamil (Groppi et al. 2011). Thus, GIC likely represents one of the previously identified Mg2+-sensitive influx pathways.
Ca2+ sequestration in intracellular compartments
ER/Golgi/secretory pathway:
As mentioned, the ER-lumenal Ca2+ concentration is significantly lower in yeast than in mammalian cells. However, the secretory pathway contains many Ca2+-dependent enzymes, and the lumen of the ER and other secretory organelles contains a much higher concentration of Ca2+ than the cytosol (∼10 μM in the ER lumen compared to 125 nM in the cytosol). This Ca2+ gradient is achieved primarily by the activity of Pmr1, a conserved P-type ATPase that pumps Ca2+ and Mn2+, whose human homolog is hSPCA1 (for secretory pathway Ca2+-ATPase 1). Mutations in the gene that encodes hSPCA1 result in a blistering skin disorder termed Hailey-Hailey disease (HHD) (Hu et al. 2000; Sudbrak et al. 2000). Both pmr1Δ yeast and human HHD cells have secretory vesicles and organelles that are deficient in Ca2+ and Mn2+, which results in defects in the folding, modification, and sorting of proteins that pass through the secretory pathway (Antebi and Fink 1992; Durr et al. 1998; Hu et al. 2000; Sudbrak et al. 2000). Both proteins localize primarily to the Golgi (Burk et al. 1989; Antebi and Fink 1992); however, pmr1Δ yeast are also significantly depleted for Ca2+ in the ER. Thus, the minor pool of Pmr1 passing through the ER plays a major role in concentrating Ca2+ and Mn2+ in this organelle (Strayle et al. 1999). In contrast, ER-localized SERCA-type Ca2+-ATPases fulfill this role in mammalian cells.
Spf1/Cod1, an ER-localized P-type ATPase that is a member of the evolutionarily conserved, but poorly understood p5 subfamily of P-type ATPases (Palmgren and Nissen 2011), also contributes to Ca2+ homeostasis in the ER and Golgi. spf1Δ and pmr1Δ exhibit similar defects; however, double mutants spf1Δ pmr1Δ are more severely compromised for Ca2+/Mn2+-dependent glycosylation events, and constitutively induce the unfolded protein response (Cronin et al. 2002; Vashist et al. 2002). Thus, although the substrate it transports remains unidentified, Spf1/Cod1 clearly overlaps functionally with Pmr1 to maintain proper Ca2+ and Mn2+ homeostasis in the ER and Golgi of yeast cells.
Vacuole:
Yeast cells contain one or more vacuoles: dynamic, acidic, storage organelles akin to mammalian lysosomes and plant tonoplasts, that undergo fusion and fission during the cell cycle and in response to environmental stimuli (Baars et al. 2007). The vacuole is the primary Ca2+ storage organelle in yeast, containing >90% of total cellular Ca2+, and the primary mechanism for clearing Ca2+ from the yeast cytosol is its rapid delivery to the vacuole. This vacuolar Ca2+ sequestration is carried out by the combined action of Vcx1, a H+/Ca2+ exchanger and member of the type II calcium exchanger family (CAX) and Pmc1, a P-type Ca2+-ATPase.
Vcx1 utilizes the steep proton gradient across the vacuolar membrane (generated by the V-type H+-ATPase) to couple Ca2+ entry into the vacuole with H+ efflux into the cytosol (Cunningham and Fink 1996; Pozos et al. 1996; Pozos 1998). When wild-type yeast are exposed to a sudden increase in extracellular Ca2+, cytosolic Ca2+ levels rise, peaking within seconds, and then sharply decrease within 30 sec to restore a low steady state level of cytosolic Ca2+ (Miseta et al. 1999). Vcx1 is primarily responsible for this rapid Ca2+ flux into the vacuole, and vcx1Δ mutants display a greater increase and significantly slower decrease in cytosolic Ca2+ under these same conditions. However, vcx1Δ cells do not exhibit lower amounts of steady state vacuolar Ca2+ and these cells are as tolerant as wild type to growth on media containing high levels of Ca2+ (Cunningham and Fink 1996; Pozos et al. 1996; Pozos 1998).
In contrast, pmc1Δ cells display Ca2+ flux dynamics that are identical to wild-type cells (Miseta et al. 1999), but exhibit Ca2+-sensitive growth and reduced vacuolar content (Cunningham and Fink 1994); both phenotypes are exacerbated in pmc1Δ vcx1Δ cells (Cunningham and Fink 1996; Pozos et al. 1996; Pozos 1998). Thus, Vcx1 and Pmc1 work together to sequester Ca2+ into the yeast vacuole. Vcx1 rapidly handles a large volume of Ca2+ and plays the dominant role in short-term Ca2+ dynamics, and the slower, ATP-dependent action of Pmc1 establishes the extremely steep Ca2+ concentration gradient across the vacuolar membrane that exists in wild-type cells. A mathematical model, developed to explain the key characteristics of yeast Ca2+ transients, predicted the existence of an additional regulatory element, i.e., rapid Ca2+-dependent negative feedback of Ca2+ influx pathways, which has not yet been demonstrated experimentally (Cui et al. 2009a). Calcineurin, the Ca2+/calmodulin-dependent phosphatase regulates Ca2+ homeostasis, but on a slower time scale. Calcineurin increases expression of PMC1 and PMR1 by activating the Crz1 transcription factor, and negatively regulates Vcx1 activity through an undetermined mechanism (Cunningham and Fink 1994; Cunningham and Fink 1996; Matheos et al. 1997; Stathopoulos and Cyert 1997; Pittman et al. 2004) (see below).
Ca2+ release from intracellular compartments
Vacuolar Ca2+ release mediated by Yvc1:
Ca2+ stored in the vacuole can be released into the cytosol through the Yvc1 Ca2+ channel, also called TRPY1 (Figure 3). The Yvc1 channel was first identified by electrophysiological studies of ion conductances in isolated yeast vacuoles (Bertl and Slayman 1990; Saimi et al. 1992), which showed that it was cation selective and inhibited by vacuolar Ca2+ (mM) or low pH (<5). Many years later, the gene product responsible for this conductance was identified as the product of the Yvc1 gene, which is a member of the large family of TRP channels (transient receptor potential), whose mammalian members are responsible for sensory phenomenon such as detection of heat, cold, and pain and are associated with diseases including retinal degeneration and polycystic kidney disease (Palmer et al. 2001; Nilius and Owsianik 2011). Yeast strains lacking Yvc1 display no growth defects, but overexpression of the gene confers Ca2+ sensitivity, suggesting a role for this protein in Ca2+ homeostasis (Denis and Cyert 2002). Indeed, Yvc1 mediates vacuolar Ca2+ release when yeast are exposed to osmotic shock (Denis and Cyert 2002), a condition also reported to trigger Ca2+ influx through Mid1/Cch1 (Matsumoto et al. 2002). Further studies showed that the Yvc1-encoded channel is mechanosensitive, suggesting that vacuolar shrinkage during osmotic shock may directly activate the channel (Zhou et al. 2003). Yvc1 is also thought to contribute to Ca2+ transients observed after glucose readdition to starved cells (Bouillet et al. 2012), although the mechanism of its activation is unknown.
Figure 3.
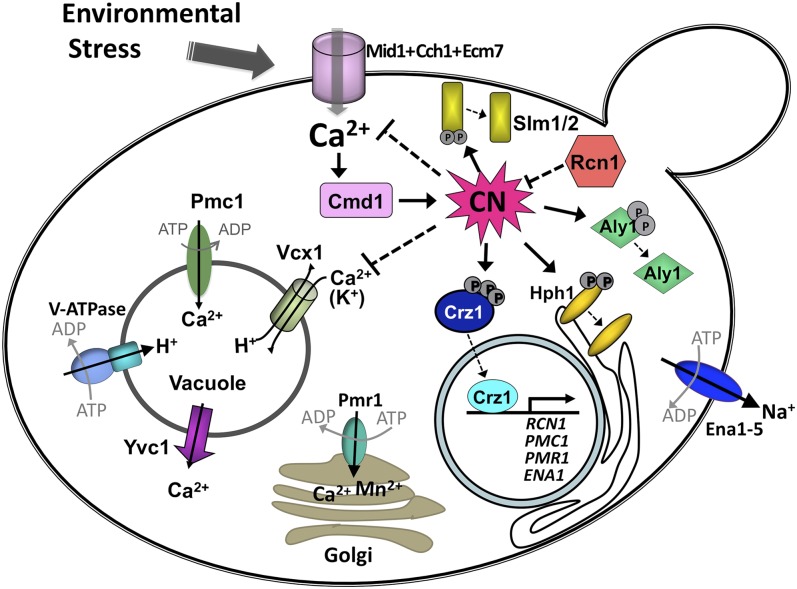
Environmental stress induces Ca2+ influx into S. cerevisiae via the Mid1/Cch1 high affinity Ca2+ channel and activation of the calcineurin phosphatase (CN), which dephosphorylates substrates Crz1, Aly1, Slm1/2, and Hph1. See text for details. Note that the V-ATPase also resides in endosomes and late Golgi, but was omitted from the figure due to space constraints.
Ca2+-dependent signaling pathways
Responses to stress:
Ca2+ is widely used in prokaryotic and eukaryotic cells as a second messenger that triggers downstream signaling events. Ca2+ signals are extremely dynamic and are controlled both temporally and spatially to confer specific cellular responses (Clapham 2007). In yeast, calmodulin is a major target of Ca2+ which, in its Ca2+-bound form, binds to and activates protein kinases (Cmk1 and Cmk2) and calcineurin, the conserved Ca2+/calmodulin-dependent phosphatase (Cyert 2001) (Figure 3). These kinases and phosphatases are often stimulated by the same physiological conditions (Moser et al. 1996; Dudgeon et al. 2008), however only calcineurin-dependent pathways, which are better understood, will be described here. Calcineurin is a heterodimer composed of a catalytic and a regulatory subunit (encoded by CNA1, CNA2, and CNB1, respectively). Calcineurin deficiency can by induced by mutation (cna1Δ cna2Δ or cnb1Δ) or by incubating yeast with the calcineurin inhibitors, cyclosporine or FK506 (Foor et al. 1992). Calcineurin is not essential for growth under typical laboratory conditions, i.e., when [Ca2+]cyto is low and the enzyme exhibits little activity. However, exposure of yeast to any of a number of environmental stress conditions, including high pH, cell wall damage, and high concentrations of cations (Mn2+, Na+, and Li+) activates calcineurin, and the phosphatase is required for cell survival under these conditions (Cyert 2003; Garcia et al. 2004; Viladevall et al. 2004; Zakrzewska et al. 2005) (Figure 3). This Ca2+-mediated stress response is conserved in pathogenic fungi, where it promotes survival in the host and is required for pathogenesis (Bastidas et al. 2008). Calcineurin dephosphorylates a range of protein targets in yeast, including the Crz1 transcription factor, which rapidly translocates from the cytosol to the nucleus upon dephosphorylation to express genes encoding cell wall biosynthetic enzymes (Fks2), ion pumps (Pmc1, Pmr1, and Ena1), signaling enzymes (Cmk2), components of vesicle trafficking (Gyp7 and Ypt53), and regulators of calcineurin (Rcn1) (Yoshimoto et al. 2002; Cyert 2003; Heath et al. 2004; Viladevall et al. 2004; Bultynck et al. 2006). In addition to Crz1, calcineurin dephosphorylates proteins involved in ER translocation (Hph1) (Pina et al. 2011), TORC2 effectors (Slm1 and Slm2) (Bultynck et al. 2006; Berchtold et al. 2012; Niles et al. 2012), its regulator (Rcn1) (Hilioti et al. 2004), and as mentioned, may regulate K+ transporters (Mendoza et al. 1994) (Figure 3). In response to excess amino acids, calcineurin also activates endocytosis of the Dip5 amino acid transporter by dephosphorylating the α-arrestin trafficking adaptor, Aly1/Art6 (A. F. O’Donnell, unpublished results). Calcineurin-dependent signaling is activated by the influx of extracellular Ca2+, rather than Ca2+ release from intracellular stores. In some cases, such as the responses to alkaline and ER stress, Ca2+ influx is mediated by the Mid1/Cch1 plasma membrane channel (Bonilla et al. 2002; Viladevall et al. 2004); however, for many calcineurin-activating conditions, the effect of mid1 or cch1 mutations on signaling has not been tested. Although the mechanism of Mid1/Cch1 activation has not yet been clearly defined, the channel is suggested to be mechanosensitive (Kanzaki et al. 1999), which could explain the link between environmental stress and calcineurin activation. Crz1/calcineurin signaling is active when protein secretion is compromised, i.e., in pmr1Δ cells, in many mutants with defects in vesicle-mediated transport, and in cells experiencing ER stress caused by the accumulation of unfolded proteins (Locke et al. 2000; Bonilla et al. 2002; Martin et al. 2011). All of these conditions cause alterations in cell wall composition, and as yeast cells are under constant turgor pressure, could result in activation of Ca2+ influx through a mechanosensitive ion channel in the plasma membrane. Furthermore, many conditions that activate the protein kinase C-regulated cell wall integrity pathway, which responds to cell wall damage caused by chemical agents, heat, mutation in cell wall biosynthetic enzymes, or hypotonic shock, also activate calcineurin-mediated stress responses (Levin 2011). During heat stress, for example, both Ca2+/calcineurin and the cell wall integrity pathway are activated and work together to regulate expression of a cell wall biosynthetic enzyme, Fks2 (Zhao et al. 1998a). Thus, plasma membrane/cell wall stretch may activate the cell wall integrity pathway through its upstream sensors, Wsc1-3, Mid2, and Mtl1 (Levin 2011), while simultaneously causing Ca2+ influx via activation of Mid1/Cch1. Surprisingly, Ca2+ influx stimulated by hypotonic shock requires Cch1 but not Mid1 (Groppi 2011).
Response to mating factor:
Exposure of haploid MATa yeast cells to α-factor mating pheromone activates a MAPK signaling pathway to induce cell-cycle arrest and morphological changes, such as cell polarization and production of cell surface agglutinins, that promote interaction and, ultimately, fusion with a mating partner (Dohlman and Slessareva 2006). Changes in Ca2+ ion accumulation also play an important signaling role in this process, as cells incubated with pheromone for extensive periods die unless Ca2+ is present in the growth medium (Iida et al. 1990; Cunningham 2011). Indeed, exposure to mating factor stimulates the Mid1/Cch1 or HACS Ca2+ channel, inducing a rise in [Ca2+]cyto that activates Ca2+/calmodulin-dependent phosphatases, Cna1/2, and kinases, Cmk1/2. Cells lacking any of the gene products in this signaling pathway behave like cells deprived of extracellular Ca2+ and lose viability during prolonged exposure to pheromone (Iida et al. 1994; Moser et al. 1996; Fischer et al. 1997; Paidhungat and Garrett 1997; Withee et al. 1997). Calcineurin activates gene expression in pheromone-treated cells through regulation of Crz1, and like calcineurin mutants, crz1Δ cells exhibit decreased survival when incubated with pheromone (Stathopoulos and Cyert 1997; Yoshimoto et al. 2002). The extensive cell wall remodeling that occurs as cells extend mating projections may cause cell wall damage, and thus explain the requirement for both calcineurin and PKC-mediated stress responses for survival under these conditions (Levin 2011). Once Ca2+ influx is activated by pheromone, calcineurin negatively regulates this pathway by inhibiting HACS-mediated Ca2+ influx, possibly by dephosphorylating Cch1 (Locke et al. 2000). Ca2+ also seems to facilitate cell fusion during mating. At higher pheromone concentrations than those required for HACS activation, Ca2+ influx through LACS is stimulated, and transcription of Fig1, which encodes a component of LACS is induced (Erdman et al. 1998). Mutants that disrupt LACS, including fig1Δ, have defects in cell fusion during mating that can be alleviated by addition of high extracellular Ca2+ (Muller et al. 2003; Aguilar et al. 2007).
Response to nutrients:
Yeast respond to the presence of glucose, sucrose, and other sugars using a variety of intracellular signals including cAMP and pHi (as discussed earlier) as well as Ca2+ (Gancedo 2008). Addition of glucose to yeast cells deprived of sugar results in a rapid influx of extracellular Ca2+ (Nakajima-Shimada et al. 1991) that requires glucose phosphorylation and Gpr1 (Tisi et al. 2002), a plasma membrane protein that is a member of a large class of G protein-coupled receptors containing seven membrane-spanning domains (Xue et al. 2008). Although such proteins typically interact with a heterotrimeric GTP-binding, or G protein, composed of α, β, and γ subunits, Gpr1 interacts with and activates the Gα-related Gpa2, in the absence of a traditional β or γ subunit (Gancedo 2008). Sugars, i.e., glucose or sucrose, likely bind to Gpr1 (although this has not been directly demonstrated), causing activation of Gpa2, which activates adenylcyclase to produce cAMP (Kraakman et al. 1999; Lemaire et al. 2004; Peeters et al. 2006) and Plc1, a phospholipase C-type γ that cleaves PI4,5P2 to create diacylglycerol and inositol-(1,4,5)-trisphosphate (IP3) (Ansari et al. 1999; Tisi et al. 2002). It is IP3 that is required for Ca2+ influx, which occurs through two different influx pathways: Mid1/Cch1 and GIC (discussed earlier) (Tisi et al. 2004). The mechanism by which IP3 stimulates these influx pathways has not yet been determined; however, the Ca2+ signal generated may regulate Pma1 (Tropia et al. 2006; Bouillet et al. 2012) and is sufficient to activate calcineurin/Crz1 signaling, suggesting a role for this pathway, which up-regulates the expression of several genes encoding carbohydrate-metabolizing enzymes, in the glucose response (Ruiz et al. 2008; Groppi et al. 2011).
Regulation of cell cycle and morphogenesis:
An early approach to studying the role of Ca2+ in yeast was the identification of Ca2+-sensitive mutants in which V-ATPase activity was compromised and a budding-defective mutant, cls4, which is an allele of Cdc24, the essential guanine nucleotide exchange factor for Cdc42, a GTPase that is critical for yeast cell polarity and budding (Ohya et al. 1986a,b; Howell and Lew 2012). The mechanism by which Ca2+ prevents budding in cls4 cells is still not understood; however, recent work identified additional Ca2+-sensitive mutants and showed multiple effects of this ion on the morphology of both mutant and wild-type yeast cells (Ohnuki et al. 2007). This suggests that our knowledge of Ca2+-dependent pathways regulating morphogenesis is quite deficient, perhaps due to regulatory redundancy. For example, study of zds1 mutants, which lack a regulatory subunit of the PP2A phosphatase (Rossio and Yoshida 2011), uncovered Ca2+ and calcineurin-dependent regulation of the morphogenesis checkpoint, which controls entry into mitosis (Miyakawa and Mizunuma 2007; Howell and Lew 2012).
Mg2+ ion homeostasis and regulation
Magnesium is abundant in the environment, serves as an essential cofactor for many cellular enzymes, and is required for cell growth and proliferation (Wolf and Trapani 2008). Yeast cells actively accumulate this ion and regulate its concentration in the cytoplasm and intracellular organelles. Although Mg2+ is stored in the vacuole and mitochondria, when grown under Mg2+-deficient conditions, yeast cells eventually stop dividing when the cellular Mg2+ content drops below ∼15 nmol/106 cells (Beeler et al. 1997; Pisat et al. 2009). Four yeast proteins that contribute to Mg2+ homoeostasis belong to the CorA or MIT (metal ion transporter) superfamily of Mg2+ transporters that are found in prokaryotes and eukaryotes and function as oligomers (Maguire 2006). Mrs2, a member of one subfamily, is responsible for Mg2+ uptake into mitochondria (Schindl et al. 2007), and Alr1 and Alr2, closely related proteins that belong to a distinct branch of CorA-related proteins, mediate Mg2+ uptake across the plasma membrane. Alr1-deficient cells display reduced Mg2+ content and a growth defect that can be suppressed by high extracellular Mg2+ or overexpression of Alr2, which is normally present at low levels (Lim et al. 2011). A third member of the MIT superfamily, Mnr2, localizes to the vacuolar membrane and is proposed to function in utilization of Mg2+ stores. The vacuole accumulates Mg2+, and this is thought to occur via a yet-to-be-identified Mg2+/H+ exchanger (Borrelly et al. 2001). mnr2 mutants exhibit increased cellular Mg2+ content together with a Mg2+-suppressible growth defect indicative of low cytoplasmic [Mg2+]. Wild-type cells grown under Mg2+-replete conditions are able to survive for several generations in the absence of extracellular Mg2+, and this requires Mnr2-mediated release of Mg2+ from the vacuole (Pisat et al. 2009). Thus, Mg2+-limitation results in utilization of intracellular Mg2+ stores, as well as stimulation of Mg2+ uptake through Alr1, whose activity may be directly regulated by this ion (Lim et al. 2011).
Biological Roles of Transition Metal Ions
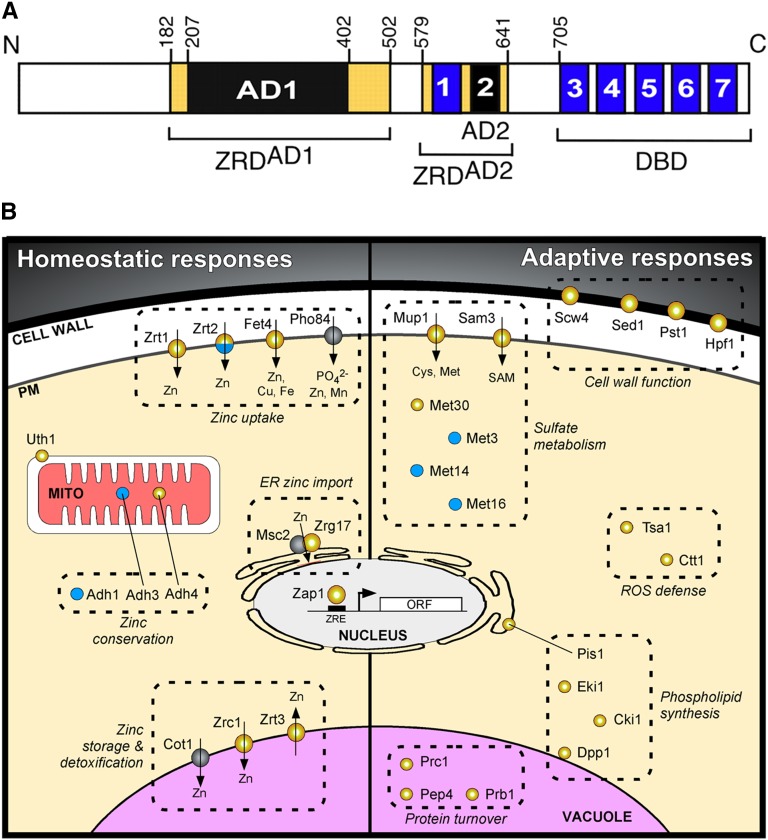
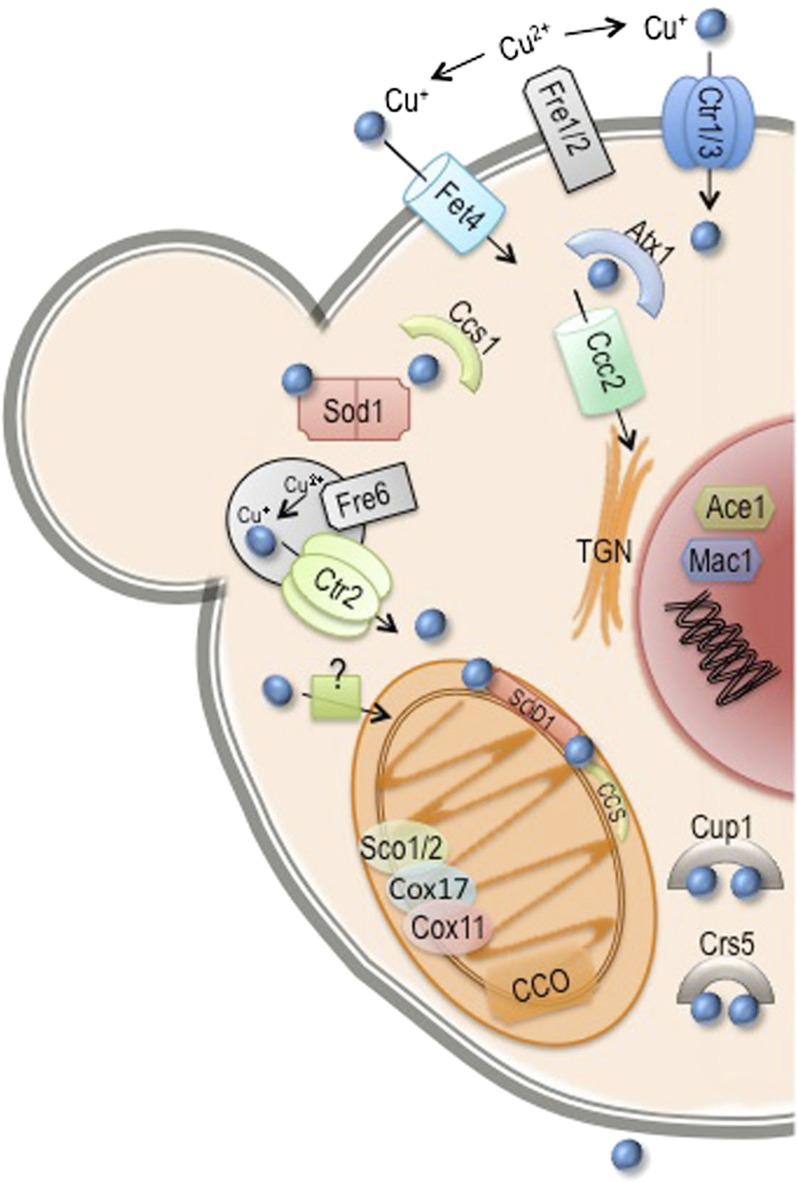
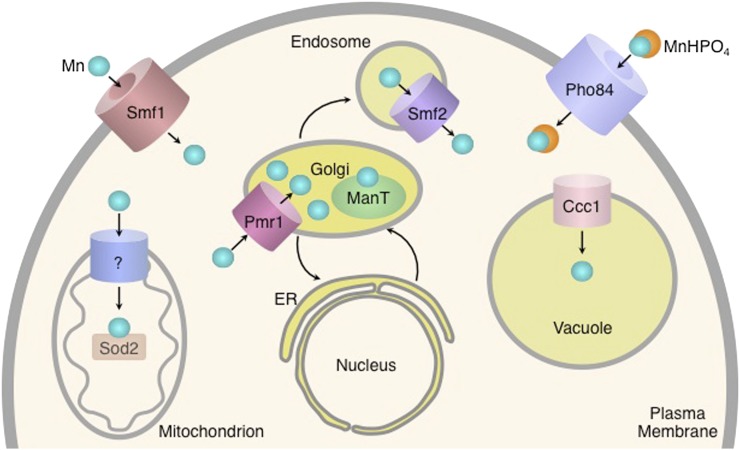
Eukaryotic cells contain hundreds of proteins that are functionally dependent on bound transition metal cofactors. These metalloproteins perform critical functions in virtually every cellular process. Proteins lacking their metal cofactors are typically inactive; therefore, metal ions constitute essential nutrients for all organisms. The exact number of metalloproteins in the yeast proteome is not known, as the presence of metal centers in known proteins continues to be discovered (Cvetkovic et al. 2010). An estimate of the prevalence of metalloproteins can be obtained by examining the metal content of enzymes for which three-dimensional structures have been solved. Using this approach, 9% of proteins contain zinc, 8% contain iron, 6% contain manganese, and 1% contains copper (Andreini et al. 2008). While the presence of a metal ion in a crystal structure strongly suggests that a metal ion is present in the protein in vivo, the identity of the metal ion in the solved structure may not correspond to that of the metal ion present in the endogenous protein. Bioinformatic approaches based on the number of known metal binding domains identified in sequenced genomes have also been used to estimate the prevalence of metalloproteins. This approach indicates that zinc metalloproteins are the most abundant and comprise ∼10% of the yeast proteome (Andreini et al. 2009). Iron proteins containing mononuclear and diiron centers, iron-sulfur clusters, and heme cofactors are also predicted to be very abundant, comprising ∼2% of the yeast proteome. Copper and manganese are considered trace nutrients, with cells containing many fewer copper and manganese metalloproteins and far lower levels of these elements. Nickel, cobalt, molybdenum, and cadmium are present in trace amounts in yeast but are not known to be incorporated into metalloenzymes and are not considered nutrient metals (Bleackley and Macgillivray 2011; Zhang and Gladyshev 2011). The abundance of transition metals and other ions in yeast cells grown in rich medium has been measured and roughly parallels the estimates of metalloprotein abundance (Figure 4) (Eide et al. 2005).
This reliance on metal ions exacts a toll on cells. Iron, manganese, and copper are redox-active metals that occupy multiple valence states in biological systems. This propensity to readily pick up or lose electrons renders these metals extremely useful in enzymatic reactions involving the transfer of electrons; however, the presence of reduced iron or copper in cells that also contain oxygen can lead to the formation of reactive oxygen species and consequent oxidative damage to cellular components. Metals can also cause toxicity by binding to noncognate sites in metalloproteins (Waldron et al. 2009). This binding of the “wrong” metal ion can preclude the binding of the correct metal ion and typically inactivates metalloenzymes. For these reasons, cellular systems involved in the uptake and utilization of metal ions are precisely regulated according to the availability of and the cellular requirement for the ion.
Uptake systems for individual metals are homeostatically regulated in S. cerevisiae, with expression of uptake systems at high levels during periods of metal scarcity and at low levels when metals are abundant (reviewed in Philpott and Protchenko 2008; Eide 2009; Reddi et al. 2009a; Nevitt et al. 2012). Yeast cells are also capable of adjusting metal uptake to accommodate different metabolic states. For example, respiratory growth is accompanied by expansion of mitochondria rich in respiratory complexes that contain numerous iron and copper cofactors (Stevens 1977). Respiratory growth is also accompanied by increases in iron and copper uptake and expansion of intracellular pools of these metals. Conversely, anaerobic growth is associated with down-regulation of oxygen-requiring processes and is associated with large reductions in iron uptake (Hassett et al. 1998a).
Here we will discuss the major nutrient metals: iron, zinc, copper, and manganese. For each metal we first discuss the regulation of systems involved in uptake, utilization, and detoxification, and then the functions of the protein components of these systems and the metabolic adaptations associated with scarcity or excess of metals.
Iron
Transcriptional control through Aft1 and Aft2
Iron homeostasis is largely achieved through the transcriptional regulation of genes involved in iron uptake. These genes are controlled by the major iron-dependent transcriptional activator, Aft1 (Yamaguchi-Iwai et al. 1995, 1996; Shakoury-Elizeh et al. 2004), and, to a lesser extent, by its paralog, Aft2 (Blaiseau et al. 2001; Rutherford et al. 2001, 2003; Courel et al. 2005), which bind DNA through a consensus upstream activation sequence (PyPuCACCC). Aft1 is constitutively expressed and shuttles in and out of the nucleus (Yamaguchi-Iwai et al. 2002). When cytosolic iron levels fall, Aft1 accumulates in the nucleus, where it binds DNA and activates transcription. Elevated intracellular iron levels trigger the nuclear export and inactivation of Aft1, a process that requires several proteins involved in the assembly and transport of iron–sulfur clusters (ISCs) (Chen et al. 2004; Rutherford et al. 2005).
ISCs are inorganic cofactors of iron and sulfide, frequently in the form of a simple 2Fe–2S cluster or a cubane 4Fe–4S cluster, which are directly coordinated by cysteine or histidine residues in recipient proteins (reviewed in Lill and Muhlenhoff 2008). In yeast, ISCs are assembled in the mitochondrial matrix and in the cytosol. Cells lacking Yfh1 (a mitochondrial iron carrier and component of the ISC assembly machinery) (Chen et al. 2004), Grx5 (the mitochondrial monothiol glutaredoxin) (Belli et al. 2004), or glutathione (Rutherford et al. 2005) exhibit impaired mitochondrial ISC assembly as well as defects in iron-mediated inactivation of Aft1. An as yet undefined product of the mitochondrial ISC machinery is exported to the cytosol, where it interacts with proteins of the cytosolic ISC assembly machinery to deliver ISCs to target enzymes of the cytosol and nucleus (Kispal et al. 1999; Lange et al. 2000; Li et al. 2001a). Most of the cytosolic ISC machinery is not required for the inactivation of Aft1, however. Instead, a complex consisting of monothiol glutaredoxins, Grx3 or Grx4, a BolA-like protein, Fra2, and a third protein, Fra1, is required for iron sensing through Aft1 (Ojeda et al. 2006; Pujol-Carrion et al. 2006; Kumanovics et al. 2008; Li et al. 2009).
While the exact mechanism of iron-induced inactivation of Aft1 is not yet clear, several observations, considered together, suggest a possible model. Grx3/4, Fra2, and Fra1 can be isolated in a complex with Aft1 in vivo. Grx3 and Fra2 form a heterodimer that, along with glutathione, contains a bridging 2Fe–2S cluster in vitro. Aft1 undergoes an iron-induced dimerization when it is inactivated, and Aft1 contains a conserved Cys-Xaa-Cys motif that is required for both iron-dependent inactivation and dimerization (Ueta et al. 2007). Together, these observations suggest a model in which the 2Fe–2S cluster bound to a Grx3/4-Fra2 heterodimer could be transferred to the Cys-Xaa-Cys motifs in Aft1 to promote the formation of a transcriptionally inactive Aft1 homodimer. Thus, ISC-bound Grx3/4-Fra2 heterodimers may constitute the iron signal that leads to Aft1 inactivation.
Remarkably, only Saccharomyces and related species of yeast rely on Aft1-like transcription factors to control iron homeostasis. Other fungal species, such as Schizosaccharomyces pombe, Candida albicans, and Aspergillus spp. rely on iron-regulated transcriptional repressors of the GATA and CCAAT-box binding families (Labbe et al. 2007; Schrettl and Haas 2011). The evolutionary forces that led to this diversity of iron regulators are not clear; however, the monothiol glutaredoxins also appear to have a conserved role as iron sensors in S. pombe (Jbel et al. 2011).
The Aft1/Aft2 regulon
Aft1 and Aft2 activate the transcription of a set of genes involved in the uptake of iron from the environment, the mobilization of iron from sites of intracellular storage, and the adaptation to an iron-limited metabolism (Philpott and Protchenko 2008). Table 1 contains a list of the Aft1/2 target genes along with their function and cellular location (see also Figure 5). The majority of these genes are involved either directly or indirectly in the uptake of iron at the plasma membrane. Most of the iron in the aerobic extracellular milieu is present as poorly soluble ferric oxyhydroxides. To solve this bioavailability problem, unicellular organisms enhance the solubility of iron through (1) acidification of the environment, (2) reduction of ferric iron to the more soluble ferrous form, and (3) secretion of soluble iron-chelating molecules. S. cerevisiae relies on all three strategies to varying extents. Many unicellular organisms and some plants synthesize and secrete siderophores, which are a heterogeneous group of organic compounds that bind ferric iron with extremely high affinity and specificity. These compounds are secreted in their iron-free form to the extracellular environment, where they bind and solublize ferric iron. The iron–siderophore complexes can then be taken up by specific transport systems. While S. cerevisiae does not synthesize or secrete siderophores, it can take up and utilize the iron bound to siderophores secreted by a variety of other species.
Table 1. Genes activated by Aft1 and Aft2.
| ORF name | Gene name | Location | Function |
|---|---|---|---|
| Uptake of iron at the cell surface | |||
| YDR534C | FIT1 | Cell wall | Siderophore binding/uptake |
| YOR382W | FIT2 | Cell wall | Siderophore binding/uptake |
| YOR383C | FIT3 | Cell wall | Siderophore binding/uptake |
| YLR214W | FRE1 | Plasma membrane | Metalloreductase |
| YKL220C | FRE2 | Plasma membrane | Metalloreductase |
| YOR381W | FRE3 | Plasma membrane | Siderophore reductase |
| YNR060W | FRE4 | Unknown | Putative reductase |
| YMR058W | FET3 | Plasma membrane | Multicopper oxidase, Fe(II) uptake |
| YER145C | FTR1 | Plasma membrane | Permease, Fe(II) uptake |
| YNL259C | ATX1 | Cytosol | Cu chaperone, deliver Cu to Ccc2p |
| YDR270W | CCC2 | Post-Golgi vesicle | Cu transport into vesicles |
| YHL040C | ARN1 | Endosome, plasma membrane | Ferrichrome transport |
| YHL047C | ARN2/TAF1 | Unknown | TAFC transport |
| YEL065W | ARN3/SIT1 | Endosome, plasma membrane | Hydroxamate siderophore transport |
| YOL158C | ARN4/ENB1 | Plasma membrane | Enterobactin transport |
| Efflux of iron from vacuole to cytosol | |||
| YLL051C | FRE6 | Vacuole | Metalloreductase |
| YLR034C | SMF3a | Vacuole | Fe(II) transport |
| YFL041W | FET5 | Vacuole | Multicopper oxidase, Fe(II) transport |
| YBR207W | FTH1 | Vacuole | Permease, Fe(II) transport |
| Other transporters | |||
| YGR065C | VHT1 | Plasma membrane | Biotin transporter |
| YOR316C | COT1 | Vacuole | Zn, Co storage/detoxification |
| YKR052C | MRS4a | Mitochondria | Mitochondrial iron import |
| YOR384W | FRE5 | Mitochondria | Putative reductase |
| Metabolic adaptation to low iron | |||
| YLR205C | HMX1 | Endoplasmic reticulum | Heme oxygenase |
| YLR136C | CTH2/TIS11 | Cytosol | mRNA degradation |
Predominately regulated by Aft2p. All others predominately regulated by Aft1p.
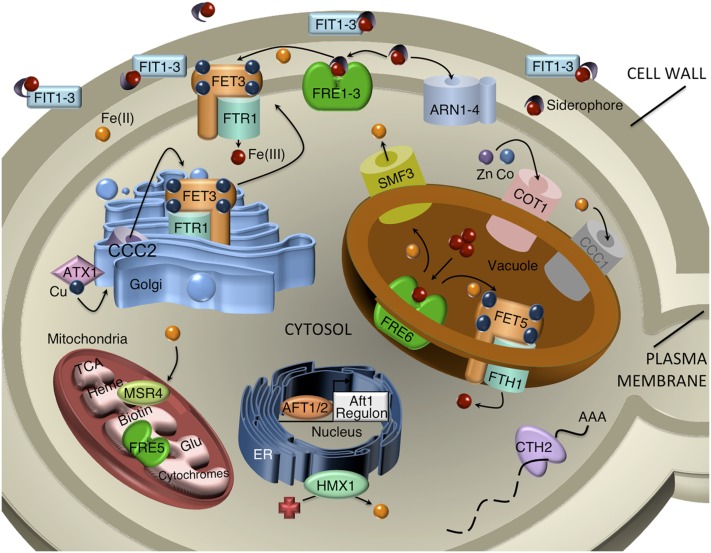
Figure 5.
Iron homeostasis in S. cerevisiae. Protein products of Aft1- and Aft2-regulated genes are shown in their respective subcellular locations. Red spheres are Fe(III); orange spheres are Fe(II). Ccc1 and mitochondrial proteins involved in the heme synthesis, the TCA cycle, biotin synthesis, and glutamate synthesis are down-regulated by iron deficiency. Reproduced with permission (Philpott and Smith 2013).
Before any iron compound can be taken up by a yeast cell, the iron must first traverse the cell wall. The yeast cell wall is a dynamic structure and contains a layer of mannoproteins covalently attached to a latticework of glucans and chitin. The most highly expressed genes of the Aft1 regulon are three cell wall mannoproteins, Fit1, Fit2, and Fit3 (Protchenko et al. 2001). These proteins enhance the retention of siderophore in the cell wall and increase the uptake of siderophore–iron at the cell surface, although the precise mechanisms by which these proteins function is not known.
Reductive uptake of iron at the cell surface
Aft1 controls the expression of two genetically separate systems of iron uptake, one that requires the external reduction of ferric iron to ferrous before uptake (the reductive system) and one that takes up intact ferric–siderophore chelates (the nonreductive system). Reductive uptake of iron is a two-step process in which ferric salts and ferric chelates are first reduced to the ferrous form by members of the FRE family of metalloreductases, then the ferrous ion is transferred to the cytosol by a high-affinity, ferrous-specific transport complex (reviewed in Philpott 2006). FRE1 and FRE2 encode flavocytochromes that constitute the majority (>90%) of the cell surface reductase activity (Dancis et al. 1990, 1992; Georgatsou and Alexandraki 1994; Lesuisse and Labbe 1994; Hassett and Kosman 1995; Georgatsou et al. 1997). They are required for growth on media containing low concentrations of ferric salts and can catalyze the reduction of ferric–siderophore chelates (Yun et al. 2001). Because siderophores have low affinity for ferrous iron, reduction of the ferric–siderophore chelate results in the release of ferrous iron, which can be taken up by ferrous-specific transporters. Additional family members (Fre3 and Fre4) have weak activity against ferric–siderophore chelates. Fre5 has been detected in purified mitochondria (Sickmann et al. 2003), where its function is unknown, and Fre6 is expressed in the vacuole (see below) (Rees and Thiele 2007; Singh et al. 2007). The FRE reductases exhibit specificity for oxidized forms of both iron and copper and can enzymatically reduce a variety of nonmetallic compounds that act as one-electron acceptors (Lesuisse et al. 1987).
Reduced iron is taken up via a high-affinity transport complex that consists of a copper-dependent ferrous oxidase (Fet3) (Askwith et al. 1994) and an iron permease (Ftr1) (Stearman et al. 1996). Fet3 oxidizes ferrous iron to ferric before transferring the ferric iron directly to Ftr1 for transport across the plasma membrane (de Silva et al. 1995; de Silva et al. 1997; Hassett et al. 1998a,b). Why the cell couples the oxidation of iron to its transport is not known, but the coupled process may permit the transport complex to distinguish between ferrous iron and other transition metals. Because Fet3 requires four copper ions for activity and because oxygen is a cosubstrate for the enzyme (Hassett et al. 1998b), iron uptake through the Fet3–Ftr1 complex is both a copper- and oxygen-dependent process (Yuan et al. 1995). Two genes activated by Aft1 under iron-deficient conditions are dedicated to the post-translational delivery of copper to Fet3. CCC2 encodes a P-type ATPase that transports copper ions from the cytosol to the lumen of post-Golgi vesicles (Yuan et al. 1995), where the copper is inserted into Fet3 (Dancis et al. 1994b). ATX1 encodes a copper chaperone that binds cytosolic copper and delivers it to the Ccc2 transporter (Lin et al. 1997).
When iron is abundant, Aft1 is inactive, the high-affinity uptake systems are not expressed, and yeast rely on broad specificity metal transporters for iron uptake. Yeast express three members of the Nramp family of divalent metal transporters, Smf1, Smf2, and Smf3 (Portnoy et al. 2000). Smf1 and Smf2 are primarily manganese transporters, but they can efficiently transport ferrous iron, as well, and cells overexpressing Smf1 accumulate higher levels of intracellular iron (Cohen et al. 2000). Fet3 cannot function under anaerobic conditions and cells grown in reduced oxygen induce the expression of a low-affinity, low-specificity ferrous iron transporter, Fet4 (Dix et al. 1994; Hassett et al. 1998a; Jensen and Culotta 2002). Fet4 also exhibits transport activity for zinc, copper, and cadmium (Hassett et al. 2000; Portnoy et al. 2001; Waters and Eide 2002), but may be the primary system for iron uptake in hypoxic environments.
Nonreductive uptake of siderophore–iron chelates
Even though S. cerevisiae does not synthesize or secrete siderophores, this yeast expresses four siderophore–iron transporters of the ARN/SIT family that specifically take up siderophore–iron chelates produced by other species of fungi and bacteria (Lesuisse et al. 1998; Heymann et al. 1999, 2000; Yun et al. 2000a,b). These transporters are members of the major facilitator superfamily, have 14 predicted membrane-spanning domains, and are thought to be energized by proton symport. Each transporter exhibits specificity for a subset of fungal or bacterial siderophores (Table 2). Two of these transporters, Arn1 and Arn3/Sit1, are regulated post-translationally through their trafficking through the late secretory pathway.
Table 2. Specificity and kinetics of ARN Transporters.
| Transporter | Siderophore substrates | Km for transport (μM) |
|---|---|---|
| Arn1 | Ferrichromesa | 0.9 |
| Coprogen | ||
| (Triacetylfusarinine C)b | ||
| Arn2/Taf1 | Triacetylfusarinine C | 1.6 |
| Arn3/Sit1 | Ferrioxamine B | 0.5 |
| Ferrichromesa | 2.3 | |
| Coprogen | ||
| (Triacetylfusarinine C)b | ||
| Arn4/Enb1 | Enterobactin | 1.9 |
Arn1 and Arn3 exhibit specificity for multiple members of the ferrichrome family of siderophores. Km determined for ferrichrome.
Arn1 and Arn3 exhibit a trace amount of transport activity for triacetylfusarinine C.
Under conditions of iron deficiency, Aft1 directs the transcription of the ARN/SIT family of siderophore transporters and after translation the proteins proceed through the secretory pathway to the trans-Golgi network (TGN). When siderophore substrates for Arn1 and Arn3 are not available outside the cell, the transporters at the TGN are sorted into vesicles destined for degradation in the vacuolar lumen, bypassing the plasma membrane (Kim et al. 2002; Froissard et al. 2007). This sorting process requires recognition by clathrin adaptor proteins at the TGN, followed by ubiquitination and sorting into luminal vesicles of the multivesicular body, which are then degraded in the lumen of the vacuole (Kim et al. 2007; Erpapazoglou et al. 2008; Deng et al. 2009). When siderophore substrates for Arn1 or Arn3 are present outside the cell, even in very low concentrations, the substrates entering the endosome through fluid phase endocytosis bind to a receptor domain on the transporter, which redirects the transporter into vesicles destined for the plasma membrane (Moore et al. 2003; Kim et al. 2005; Froissard et al. 2007; Erpapazoglou et al. 2008; Deng et al. 2009). Thus the Arn1 and Arn3 transporters are not expressed on the surface of yeast unless the cells are both iron deficient and the specific siderophore substrates are available for uptake.
Mobilization of vacuolar iron stores
When yeast are grown in media containing adequate amounts of iron, iron transported into the cytosol that is not immediately required for metabolic purposes is stored in the vacuole. CCC1 encodes an iron and manganese transporter that localizes to the vacuolar membrane and is required for the transfer of iron from the cytosol to the vacuole (Lapinskas et al. 1996; Chen and Kaplan 2000; Li et al. 2001b). CCC1 is actively transcribed under conditions of iron excess by the transcription factor Yap5 (Li et al. 2008). Cells lacking Ccc1 are sensitive to the toxic effects of iron. When cells are grown in iron-poor medium, Ccc1 expression is turned off and Aft1 and Aft2 direct the transcription of a set of genes that effect the transfer of vacuolar iron back to the cytosol. These genes largely duplicate on the vacuolar membrane the reductive iron uptake system of the plasma membrane. Fre6, a paralog of the plasma membrane ferric and cupric reductases (Martins et al. 1998), is expressed exclusively on the vacuolar membrane where it is required for the reduction of vacuolar iron and copper (Rees and Thiele 2007; Singh et al. 2007). Reduced vacuolar iron is transported into the cytosol via the Fet5/Fth1 oxidase/permease complex that is homologous to the plasma membrane Fet3/Ftr1 complex (Spizzo et al. 1997; Urbanowski and Piper 1999). Smf3 is a vacuolar iron transporter of the Nramp family that is transcriptionally activated by Aft2 in response to iron deficiency (Portnoy et al. 2002; Courel et al. 2005). Both of these ferrous-specific transporters contribute to the mobilization of stored iron when cells are iron deficient.
Other transporters
Iron-deficient yeast activate the expression of a low-specificity transporter, Cot1, that mediates vacuolar accumulation of zinc and cobalt (Conklin et al. 1992; MaDiarmid et al. 2000; Shakoury-Elizeh et al. 2004). Because iron-deficient cells are sensitive to the toxic effects of other metals, activation of this transporter suggests that the sequestration of other metals in the vacuole is an adaptive response during iron deficiency (Li and Kaplan 1998). Iron-deficient cells also activate the expression of Fre5, a metalloreductase localized to mitochondrial membranes (Sickmann et al. 2003), and Mrs4, a protein of the mitochondrial carrier family that, along with Mrs3, is involved in the uptake of ferrous iron (and possibly copper) across the mitochondrial inner membrane (Foury and Roganti 2002; Muhlenhoff et al. 2003b; Froschauer et al. 2009). As the mitochondrial intermembrane space is a relatively oxidizing environment, Fre5 may serve to supply Mrs3 and Mrs4 with reduced iron for uptake. Iron deficiency also triggers the expression of Vht1, the high-affinity biotin transporter, and Bio5, a transporter of biotin biosynthetic intermediates (Phalip et al. 1999; Stolz et al. 1999; Belli et al. 2004; Shakoury-Elizeh et al. 2004). Although yeast are formally auxotrophic for biotin, they express enzymes for the terminal three steps of the biosynthetic pathway and can synthesize biotin from intermediates. The ultimate step in biotin synthesis is catalyzed by Bio2, an Fe–S protein (Ugulava et al. 2001), and the entire synthetic pathway is transcriptionally down-regulated during iron deficiency. This shift from biosynthesis to uptake of biotin during iron deficiency allows yeast to conserve iron, a pattern that is replicated in other Fe–S- and heme-dependent pathways.
Metabolic adaptation to iron deficiency
Budding yeast can survive in environments that contain very low or very high concentrations of iron. Survival in low iron environments involves metabolic adaptations that reduce the cell’s dependence on iron-containing proteins. Metabolic adaptation to iron deficiency includes shifting from iron-dependent biosynthetic pathways to iron-independent pathways. As mentioned above, iron-deficient yeast shut down the iron-requiring biosynthetic pathway for biotin and up-regulate uptake systems for biotin and biotin precursors. Another example of the shift to iron-independent pathways is glutamate synthesis. Yeast express two genetically separate systems for the synthesis of glutamate. One requires the glutamate dehydrogenases Gdh1 or Gdh3. The other requires glutamine synthetase, Gln1, and glutamate synthase, Glt1, an iron-sulfur cluster enzyme. GLT1 is transcriptionally shut off in iron-deficient yeast while GDH3 transcripts increase, thereby shifting glutamate synthesis to an iron-independent pathway (Shakoury-Elizeh et al. 2004). Similarly, yeast alter carbon source utilization according to the availability of iron. Growth on nonfermentable carbon sources is accompanied by an expansion of mitochondria containing iron-rich respiratory complexes, and yeast cannot grow on media containing nonfermentable carbon sources that are also low in iron. Yeast grown on glucose media metabolize glucose largely through fermentation, with only a small amount of respiration. When yeast are shifted to iron-poor media, levels of transcripts involved in respiration fall and metabolite analysis indicates increased flux through the glycolytic pathway. These studies suggest that iron-deficient yeast shut down respiration, thereby allowing the cell to reuse the iron scavenged from respiratory complexes for other metabolic purposes (Shakoury-Elizeh et al. 2010). These metabolic adaptations occur through multiple mechanisms. Some are transcriptional and depend on Aft1 and Aft2 or the heme-dependent activator Hap1 or Hap4. Some occur post-transcriptionally and depend on the mRNA-destabilizing proteins Cth1 and Cth2.
Under conditions of iron deficiency, Aft1 activates the transcription of HMX1, which encodes the yeast heme oxygenase (Protchenko and Philpott 2003). Hmx1 degrades heme, releasing the heme iron for other metabolic uses. Growth in iron-poor medium is associated with large reductions in cellular heme levels. This loss of heme is due, in part, to the activity of Hmx1, in part to less iron availability for insertion into heme, and in part to lower expression levels of heme biosynthetic enzymes (Lesuisse et al. 2003). In addition to its role as an enzyme cofactor, heme is also a regulatory molecule that is required for the activation of the transcription factor Hap1 (Guarante 1992; Kwast et al. 1998; Lee and Zhang 2009). Hap1 activates the transcription of many genes involved in respiration and aerobic growth. These genes are down-regulated in iron-deficient cells due, in part, to loss of heme-dependent activation of Hap1. Many proteins involved in the tricarboxylic acid cycle and respiration require heme and Fe–S cluster cofactors; thus, down-regulation of these pathways reduces expression of proteins that may ultimately lack required cofactors and allows the limited iron that may have been used in these pathways to be redirected to other pathways.
CTH2 and, to a lesser extent, CTH1 are Aft1 targets that are actively transcribed under iron deficiency and are involved in mediating the metabolic adaptation to iron deficiency (Puig et al. 2005, 2008). Cth1 and Cth2 are mRNA-binding proteins that recognize and bind to AU-rich sequences in the 3′-UTR of a number of transcripts that are down-regulated during iron deficiency. Binding of Cth1 or Cth2 to the AU-rich elements leads to destabilization and degradation of the transcripts. Many of the transcripts that are putative targets of Cth1 or Cth2 encode proteins that either contain iron cofactors or function in pathways with other iron-dependent enzymes. These pathways include cellular respiration, heme and Fe–S cluster biosynthesis, iron homeostasis, fatty acid and ergosterol metabolism, and amino acid metabolism. Strains lacking both Cth1 and Cth2 exhibit slow growth on iron-poor medium, indicating that the reduced expression of the set of Cth1 and Cth2 targets offers an adaptive advantage to iron-deficient yeast. Although the amount of mRNA degradation that can be attributed to Cth1 or Cth2 is relatively small, usually twofold or less, the net effect represents a significant “iron savings” for the cell because of the relatively large number of genes targeted.
Cellular iron utilization
Cellular iron is used for the synthesis of heme and ISCs or it can be directly coordinated by enzymes in the form of mononuclear or diiron centers. In the cytosol, the monothiol glutaredoxins, Grx3 and Grx4, have roles in iron delivery that go beyond the regulation of Aft1 (Ojeda et al. 2006; Pujol-Carrion et al. 2006). Grx3 can form a homodimer that coordinates a bridging 2Fe–2S cluster in vitro and Grx3/4 that is affinity purified from yeast also contains bound iron (Li et al. 2009). Yeast strains lacking Grx3 and Grx4 exhibit defects in heme and ISC synthesis as well as defects in the metallation and activity of ribonucleotide reductase, a cytosolic diiron enzyme (Muhlenhoff et al. 2010). These observations suggest that Grx3/4 is involved in the delivery of iron to mitochondria, the cytosolic ISC assembly complex, and to at least one cytosolic diiron protein. Direct physical interactions between Grx3/4 and these iron recipients have not yet been demonstrated.
Iron is imported from the cytosol into the mitochondrial matrix for the synthesis of heme and Fe–S clusters. The mitochondrial carrier proteins Mrs3 and Mrs4 facilitate the transport of iron across the mitochondrial inner membrane, but additional pathways of mitochondrial iron import exist, as strains deleted for both MRS3 and MRS4 can maintain heme and Fe–S cluster synthesis when grown in iron-rich media (Foury and Roganti 2002; Muhlenhoff et al. 2003b). The mitochondrial pyrimidine exchanger Rim2, also a member of the mitochondrial carrier family, was identified in a screen for mutations synthetically lethal with a mrs3Δmrs4Δ strain (Yoon et al. 2011). Rim2 contributes to mitochondrial iron utilization, but appears not to directly facilitate the uptake of iron.
Heme biosynthesis is a highly conserved eight-step pathway with individual steps occurring in both the mitochondria and cytosol (reviewed in Labbe-Bois and Labbe 1989). The initial step of heme synthesis occurs in the mitochondrial matrix where glycine and succinyl-coA condense to form δ-aminolevulinic acid (ALA). ALA is exported to the cytosol, where the next five steps occur, then protoporphyrinogen IX moves from the cytosol to the mitochondria, where it is converted to protoporphyrin IX. The final step in heme synthesis is the insertion of iron into protoporphyrin IX, which is catalyzed by ferrocheletase and occurs in the mitochondrial matrix. Heme is then distributed to proteins located in the mitochondria, cytosol, and organelles throughout the cell. Proteins involved in the intracellular trafficking of heme have not been identified.
ISC assembly is a highly conserved process in both prokaryotes and eukaryotes (reviewed in Lill and Muhlenhoff 2008). While protein complexes devoted to the assembly of ISCs are located in both the mitochondria and cytosol, products of mitochondrial ISC synthesis are required for cytosolic ISC assembly. The core ISC biosynthetic complex consists of the cysteine desulfurase Nfs1 (Kispal et al. 1999), which supplies sulfide, an accessory protein Isd11 (Adam et al. 2006; Wiedemann et al. 2006), the yeast frataxin homolog Yfh1, which supplies iron (Chen et al. 2002; Gerber et al. 2003; Muhlenhoff et al. 2003a; Ramazzotti et al. 2004) and allosterically activates Nfs1 (Tsai and Barondeau 2010), and Isu1 or Isu2, which serves as a scaffold for assembly (Muhlenhoff et al. 2003a). Additional components provide reducing equivalents, and protein chaperones, along with a monothiol glutaredoxin, facilitate the transfer of nascent ISCs to recipient proteins. Some product of the ISC assembly complex is exported to the cytosolic ISC assembly complex. This product has not been molecularly characterized, but it must include sulfide, as there is no alternative source of sulfide in the cytosol. The mitochondrial ABC transporter Atm1 was shown to be required for cytosolic ISC assembly, presumably by exporting a product of the mitochondrial ISC assembly complex (Kispal et al. 1999). However, a more recent study found that Atm1 was not involved in cytosolic Fe–S cluster assembly in yeast (Bedekovics et al. 2011). A sulfhydryl oxidase, Erv1, located in the intermembrane space (Lange et al. 2001), and mitochondrial glutathione are also required for export of the unknown product.
Assembly of ISCs in the cytosol requires a genetically and biochemically distinct set of proteins from that of the mitochondria (reviewed in Lill 2009). Cytosolic ISCs appear to be assembled in a two-step manner. The first step involves the transient assembly of a pair of bridging 4Fe–4S clusters on a scaffold composed of the P-loop NTPases Cfd1 and Nbp35 arranged in a heterotetramer (Roy et al. 2003; Hausmann et al. 2005; Netz et al. 2007). An electron transfer complex consisting of the flavoprotein Tah18 and the Fe–S protein Dre2 are also required for this early step of ISC assembly (Zhang et al. 2008; Netz et al. 2010). Clusters are then transferred to a complex consisting of Nar1, similar to iron-only hydrogenases of bacteria, and Cia1, a WD40 repeat protein (Balk et al. 2004, 2005). From this complex ISCs are then transferred to recipient proteins in the cytosol and nucleus.
Zinc
Based on the detection of known zinc-binding domains in the S. cerevisiae proteome, budding yeast are estimated to contain ∼476 different zinc proteins (Andreini et al. 2006). Zinc can have a catalytic function, as in the active site of an enzyme, or a structural function, as in a zinc finger. The use of zinc as a catalytic cofactor is widespread. Of the zinc-dependent enzymes, more than half are hydrolases (Andreini and Bertini 2011). Structural zinc-binding domains are very common in trancriptional regulators and represent approximately one-third of all zinc proteins in eukaryotes. By far, the most common structural zinc-binding domains are zinc fingers of the Cys4 or Cys2His2 type (Andreini et al. 2006).
Transcriptional regulation through Zap1
Zinc homeostasis is largely achieved through transcriptional control of gene expression (Eide 2009). Zap1 is the zinc-sensing transcription factor that controls the response to zinc deficiency in yeast (Zhao and Eide 1997). Under conditions of zinc deficiency, Zap1 activates the transcription of ∼80 genes, while also repressing the transcription of a small number (Lyons et al. 2000; Yuan 2000; Higgins et al. 2003; De Nicola et al. 2007; Wu et al. 2008). Zap1 target genes are largely involved in zinc homeostasis and in the metabolic adaptation to zinc deficiency (Figure 6). In zinc-replete cells, Zap1 is inactivated. The protein contains both a DNA-binding domain and multiple transcriptional activation domains, all of which bind zinc (Figure 6A). The DNA binding domain is located in the carboxy-terminal third of the protein and consists of five zinc fingers of the C2H2 type, each of which is required for full DNA binding activity (Bird et al. 2000a,b; Evans-Galea et al. 2003). The high-affinity, structural zinc-binding sites formed by the zinc finger motifs are occupied by zinc ions whether cells are zinc deficient or zinc replete. However, in zinc-replete cells, Zap1 does not bind DNA, which is likely due to a post-translational modification that interferes with DNA binding (Frey et al. 2011). In zinc-deficient cells, Zap1 binds as a monomer to a palindromic consensus DNA sequence, ACCTTNAAGGT, known as a zinc-responsive element (ZRE) (Zhao et al. 1998b). Considerable variation from this sequence has been observed in functional Zap1 binding sites, but Zap1-regulated promoters typically contain one or a few recognizable ZREs. The ZAP1 gene itself contains a ZRE and is activated by Zap1, thereby increasing the amount of Zap1 protein during zinc deficiency. Some Zap1 targets are transcribed during mild zinc deficiency, while others are activated only when zinc deficiency becomes severe. ZREs with the highest binding affinity for Zap1 tend to appear in genes activated by mild iron deficiency, which suggests that Zap1 occupies lower-affinity sites only when the Zap1 levels rise, as they do in severe zinc deficiency. The autoactivation of ZAP1 is thus a mechanism by which cells can achieve a graded response to differing levels of zinc deficiency (Wu et al. 2008).
Figure 6.
Zinc homeostasis. (A) Schematic of Zap1. Regions representing zinc fingers are filled and numbered. Activation domains (AD) in black are embedded within zinc-regulatory domains (ZRD) in orange. Reproduced with permission (Frey and Eide 2011). (B) Transcriptional response to zinc deficiency. Proteins involved in zinc homeostasis are on the left, proteins involved in metabolic adaptation to zinc deficiency on the right. Yellow spheres indicate up-regulation, blue spheres indicate down-regulation, gray spheres indicate no zinc regulation. Subcellular localizations are shown. PM, plasma membrane; MITO, mitochondria; ORF, open reading frame. Reproduced with permission (Eide 2009).
Zap1’s two activation domains, known as AD1 and AD2, function independently in zinc sensing and activation (Bird et al. 2000b). AD1 is part of a larger zinc-regulatory domain located in the amino-terminal half of the protein and it contains two clusters of potential zinc-binding residues at either end of the domain (Herbig et al. 2005). Zinc binding in this domain has been demonstrated in vitro, and mutation of potential zinc-binding residues results in constitutive activity of AD1. Thus, AD1 likely senses zinc through direct binding of the metal, which results in loss of transcriptional activation.
AD2 of Zap1 consists of two C2H2-type zinc fingers located in the middle of the protein. Neither of these zinc fingers is involved in DNA binding, but together they can confer zinc-regulated transcriptional activation (Bird et al. 2003). The zinc-binding sites within AD2 are of lower affinity than those of most C2H2 zinc fingers, as would be appropriate for a zinc sensor. In vitro studies indicate that zinc binding by the two zinc fingers promotes a stable interaction between the fingers (Wang et al. 2006). Presumably this conformation of AD2 prevents the recruitment of transcriptional coactivators.
Most Zap1 target genes can be fully activated by a mutated Zap1 containing only AD1 (Frey and Eide 2011). Although some genes respond to both AD1 and AD2, only a few genes require AD2 for full induction. AD2 appears to be more important for Zap1 activity when zinc deficiency is combined with other environmental stresses.
Transporters regulated by Zap1
The primary response to zinc deficiency is the Zap1-mediated expression of zinc transporters that serve to transfer zinc from the extracellular milieu and the vacuolar lumen to the cytosol. Zrt1 and Zrt2 are Zn-specific transporters of the ZIP family, a large, ancient family of zinc transporters with representatives in most species of prokaryotes and eukaryotes. Zrt1 provides essentially all of the high-affinity zinc uptake activity (Zhao and Eide 1996a); deletion of ZRT1 results in loss of high-affinity zinc uptake and slow growth in zinc-limited medium. Zrt2 exhibits a moderate degree of sequence identity (44%) with Zrt1 and functions as a low-affinity zinc transporter (Zhao and Eide 1996b). Strains deleted for both ZRT1 and ZRT2 can grow on media containing moderately reduced levels of zinc, indicating the presence of additional low-affinity zinc-uptake systems. Fet4 is a low-affinity, broad-specificity metal transporter, the expression of which is induced by Zap1 under zinc-limiting conditions (Waters and Eide 2002). Expression of Fet4 in a zrt1Δzrt2Δ strain was associated with improved growth in low zinc medium. Yeast grown in a very zinc-restricted environment depend on Zrt1 for uptake, while Zrt2 and Fet4 provide uptake activity when zinc levels are normal or mildly reduced.
Surprisingly, Zrt2 is repressed by Zap1 under conditions of severe zinc deficiency (Bird et al. 2004). This repression is mediated through a lower-affinity ZRE located upstream of ZRT2. Transcriptional activation of ZRT2 is mediated by Zap1 binding to high-affinity ZREs located upstream of the TATA box. Severe zinc deficiency increases the levels of Zap1 through autoactivation at the ZAP1 promoter. In the setting of these higher levels, Zap1 also binds to a lower affinity ZRE located near the transcription start site of ZRT2, thereby inhibiting transcription. Thus, under conditions where zinc levels are too low for Zrt2 to function, expression is repressed.
As is the case with iron, yeast grown in zinc-replete meda store excess zinc in the vacuole, both to prevent toxicity and to meet metabolic need during zinc deficiency. Zinc storage is mediated by two unrelated transporters located on the vacuolar membrane, Cot1 and Zrc1 (Kamizono et al. 1989; MacDiarmid et al. 2000). Under conditions of zinc deficiency, vacuolar zinc stores are mobilized through the activity of Zrt3, a third yeast member of the ZIP family of transporters that is expressed on vacuolar membranes. Strains deleted for ZRT3 exhibit high levels of intracelluar zinc, which is not effectively mobilized for use during zinc deficiency (MacDiarmid et al. 2000).
Although Zrc1 is involved in zinc storage when yeast are grown in zinc-replete media, somewhat counterintuitively, this transporter is also induced by Zap1 during zinc deficiency. An explanation for this expression pattern came from studies examining the response of zinc-deficient yeast to a sudden increase in extracellular zinc (MacDiarmid et al. 2003). Because zinc-deficient yeast express uptake systems at high levels, acute exposure of these yeast to high concentrations of zinc results in a sudden influx of zinc to the cytosol. The presence of Zrc1 on the vacuole exerts a protective effect for the yeast exposed to this influx of zinc, as zinc-deficient strains lacking ZRC1 exhibited increased sensitivity to zinc.
Eukaryotic cells have a requirement for zinc within the lumen of the ER. This requirement is due to the presence of several zinc metalloenzymes (metalloproteases, protein chaperones, and a transferase of glycosylphosphatidylinositol anchor synthesis) in the lumen of the ER (Ellis et al. 2004, 2005). Zinc is transferred from the cytosol to the ER lumen by the activity of a heterodimeric transporter composed of Msc2 and Zrg17, both of which are members of the cation diffusion facilitator family of efflux pumps. Although both peptide components of this pump are required for zinc transport, only one, Zrg17, is zinc-regulated directly through Zap1 (Wu et al. 2011).
Adaptation to zinc deficiency
Zap1 also controls the expression of several genes involved in the adaptation to zinc deficiency. One of the most highly zinc-regulated genes is ADH1 (Lyons et al. 2000), which encodes a very abundant, zinc-binding alcohol dehydrogenase (Leskovac et al. 2002). Adh1 is repressed by Zap1 in zinc-limited cells. This repression is mediated by the Zap1-dependent transcription of a noncoding RNA, termed ZRR1, from a site just upstream of the ADH1 promoter (Bird et al. 2006). Transcription of ZRR1 leads to the displacement of Rap1, the transcriptional activator for ADH1, from the promoter, thus inhibiting ADH1 transcription. A mitochondrially targeted paralog of Adh1, Adh3, is regulated in a similar manner. Zinc deficiency also induces the Zap1-mediated transcription of ADH4, which encodes another zinc-dependent alcohol dehydrogenase (Drewke and Ciriacy 1988). The adaptive advantage of the shift from Adh1 and Adh3 to Adh4 during zinc limitation is not entirely clear, but likely involves the overall conservation of zinc. Structural studies of Adh1 indicate that it binds two zinc ions per monomer. Adh4 is closely related to the iron-dependent alcohol dehydrogenase of Zymomonas mobilis, which binds a single metal ion per monomer (Neale et al. 1986).
Zinc deficiency is accompanied by significant changes in the lipid composition of the cell, namely a large increase in phophatidylinositol (PI) and a large decrease in phosphatidylethanolamine (Iwanyshyn et al. 2004). The major phospholipids of the cell are primarily synthesized in a stepwise manner from the precursor cytodine diphosphate (CDP)–diacylglycerol (Henry et al. 2012). A secondary pathway, termed the Kennedy pathway, can also supply phospholipids from the precursors ethanolamine and choline. Under zinc limitation, Zap1 increases the transcription of PIS1, the PI synthase, PAH1, the phosphatidate (PA) phosphatase, and EKI1 and CKI1, two genes of the Kennedy pathway. Increased PI synthase leads to greater synthesis of PI, and increased PA phosphatase leads to depletion of PA levels (Soto-Cardalda et al. 2012). Reduced PA triggers the release of the transcriptional repressor Opi1, which results in down-regulation of multiple genes of the CDP–diacylglycerol pathway. Thus zinc deficiency leads to a shift in phospholipid synthesis to the secondary Kennedy pathway and to a major change in the phospholipid composition of the cell. The adaptive value of these changes is not known, but one could speculate that the activity or trafficking of the zinc transporters could be affected. Alternatively, reliance on the Kennedy pathway would reduce the requirement for S-adenosyl methionine (possibly limiting in zinc deficiency, see below), which is consumed in the synthesis of phospholipids via the CDP–diacylglycerol pathway.
Zinc deficiency in yeast is also associated with a decrease in sulfate assimilation and reduced levels of free intracellular cysteine and methionine, which can again be traced to transcriptional events under the control of Zap1 (Wu et al. 2009). Diminished sulfate assimilation is due to transcriptional repression of MET3, MET14, and MET16, the three genes required for the stepwise reduction of intracellular sulfate to sulfite, which is used in the synthesis of sulfur-containing amino acids. These genes are under the control of Met4, a transcriptional activator that is, in turn, post-translationally regulated through ubiquitin-mediated degradation (Rouillon et al. 2000; Smothers et al. 2000). Met30, an F-box protein and component of the SCF(Met30) ubiquitin ligase complex, targets Met4 for degradation. In zinc-deficient cells, Zap1 activates the transcription of MET30, which leads to the degradation of Met4 and reduced expression of genes of sulfate assimilation. Permeases for the uptake of S-adenosyl methionine (Sam3) and cysteine and methionine (Mup1) are also transcriptionally activated by Zap1. Although the reduced availability of cysteine and methionine that results from these regulatory changes would appear to be maladaptive, this regulatory pattern partially protects cells from the oxidative stress associated with zinc deficiency. The source of the increased oxidative stress that occurs with zinc deficiency is not known, but cells grown in zinc-limited medium exhibit increased sensitivity to exogenous H2O2 and produce increased amounts of reactive oxygen species (Wu et al. 2007, 2009). The relationship between zinc deficiency, sulfate assimilation, and oxidative stress may be traced to intracellular pools of NADPH. Sulfate assimilation involves the consumption of relatively large amounts of reducing equivalents in the form of NADPH. Zinc deficiency triggers the Zap1-mediated transcription of TSA1, which encodes the major peroxiredoxin of yeast and protects cells against oxidative damage. The down-regulation of sulfate assimilation in zinc deficiency would make more NADPH available for the regeneration of reduced peroxiredoxin and glutathione in the setting of oxidative stress (Eide 2009).
Copper
Transcriptional regulation through Mac1 and Ace1
Although copper ions and copper proteins are present in yeast cells at far lower levels than iron or zinc, copper proteins play a role in multiple critical cellular processes. Similar to iron, copper ions can occupy multiple valence states in cells and can thus participate in redox reactions that generate toxic reactive oxygen species. Similar to zinc, copper ions have an affinity for thiolate ligands and can thus compete with zinc or iron-sulfur clusters for cysteine-rich metal binding sites. Yeast cells therefore tightly regulate copper uptake and utilization. This regulation is largely accomplished at the level of transcription by a pair of reciprocally regulated, copper-sensing transcriptional activators, Mac1 and Ace1 (also called Cup2) (Figure 7).
Figure 7.
Copper homeostasis. Proteins involved in the regulation, uptake, distribution, and utilization of copper. Subcellular localizations are indicated. Reproduced with permission (Nevitt et al. 2012).
Mac1 is a constitutively expressed transcriptional activator that is stably localized to the nucleus in both copper-deficient and copper-replete cells (Jungmann et al. 1993; Graden and Winge 1997; Jensen and Winge 1998; Serpe et al. 1999; Keller et al. 2005). Under conditions of copper deficiency, Mac1 binds to the consensus sequence TTTGC(T/G)C(A/G) located in the promoters of the Mac1-regulated genes (Labbe et al. 1997; Yamaguchi-Iwai et al. 1997). These promoter elements are present in multiple copies as either direct or inverted repeats, and Mac1 binds as a dimer to these cis-acting elements (Joshi et al. 1999). When intracellular copper levels rise, Mac1 becomes transcriptionally inactive and dissociates from promoter elements. Structurally, Mac1 contains an amino-terminal DNA binding domain and a carboxy-terminal activation domain that also contains two cysteine-rich, copper-binding motifs (Jensen et al. 1998; Brown et al. 2002). These two motifs can bind up to eight copper ions as a copper-thiolate cluster and are thought to function as direct copper sensors. In the presence of copper, Mac1 inactivation is accomplished through an intramolecular interaction between the amino-terminal DNA binding domain and the first of the two cysteine-rich motifs, which results in the loss of both DNA binding and transactivation (Graden and Winge 1997; Labbe et al. 1997; Jensen and Winge 1998). Copper binding in the second cysteine-rich motif also plays a role in the loss of transactivation (Keller et al. 2005). Mac1 is phosphorylated in vivo and treatment with phosphatase disrupts DNA binding in vitro (Heredia et al. 2001). The copper-dependent regulation of Mac1 suggests that cells contain a nuclear pool of copper that can bind to and inactivate Mac1. The molecular form of this copper pool and whether it is coordinated by a copper-binding metallochaperone is not known.
Yeast are protected from the toxic effects of excess copper through the activity of Ace1 (Thiele 1988). When cells are exposed to excess copper, this transcription factor activates a group of genes involved in the detoxification of copper. Ace1 is also thought to be a direct sensor of nuclear copper levels and has some structural features similar to those of Mac1. Ace1 contains an amino-terminal DNA-binding domain that is also the site of regulatory copper binding (Buchman et al. 1989; Huibregtse et al. 1989; Szczypka and Thiele 1989). Both Mac1 and Ace1 contain a single zinc-binding domain that is involved in DNA minor groove interactions (Thorvaldsen et al. 1994; Farrell et al. 1996). Ace1 contains four pairs of cysteine residues that bind four atoms of copper in a polycopper cluster that is structurally similar to those of Mac1 (Dameron et al. 1991; Graden et al. 1996; Brown et al. 2002). Copper-binding by Ace1 induces a conformational change in the protein that allows it to bind to and activate transcription at specific copper regulatory sites with the sequence TTX2GCTG (Huibregtse et al. 1989; Evans et al. 1990; Pachkov et al. 2007). Ace1 binds DNA as a monomer and is present in the nucleus at both high and low copper levels. Affinity measurements for the copper binding domains of both Ace1 and Mac1 indicate that Mac1 has a higher affinity for copper (Kd = 9.7 × 10−20) than does Ace1 (Kd = 4.7 × 10−18). These extremely high binding affinities would indicate that the copper binding sites of both sensors are fully occupied at concentrations equating to less than a single free copper ion in the cell. However, these measurements are obtained under equilibrium conditions in which the metal fully dissociates from the protein and is fully hydrated in aqueous solution. In living cells, metal ions are surrounded by metal buffers, consisting of small molecules and proteins that constitute labile ligands. In this situation, metal exhange between ligands of the buffer and ligands of the sensors is likely to occur with much more labile kinetics. These measured affinities should be regarded as reference values, which suggest that, as intracellular copper levels rise, Mac1 is inactivated by copper binding at concentrations below those that activate Ace1 by copper binding (Wegner et al. 2011) and well below one fully hydrated (“free”) copper ion per cell.
Copper transport systems
Similar to iron, extracellular copper is largely present in the oxidized state, Cu(II), and must undergo reduction to Cu(I) before uptake. Under conditions of copper scarcity, Mac1 activates the transcription of two members of the FRE family of metalloreductases, FRE1 and FRE7 (Yamaguchi-Iwai et al. 1997; Martins et al. 1998; Georgatsou and Alexandraki 1999). Fre1 catalyzes the majority of Cu(II) reduction at the plasma membrane while Fre2 catalyzes a lesser amount (Dancis et al. 1992; Georgatsou and Alexandraki 1994; Georgatsou et al. 1997). Overexpression studies of Fre7 suggest it also functions as a metalloreductase on the plasma membrane (Rees and Thiele 2007). Mac1 also activates the transcription of three high-affinity copper transporters. Two of these, Ctr1 and Ctr3, are expressed on the plasma membrane and mediate the uptake of reduced copper (Dancis et al. 1994a,b; Knight et al. 1996). A third Ctr paralog, Ctr2, is expressed on the vacuolar membrane and, along with Fre6, mediates the reduction and transfer of copper from the lumen of the vacuole to the cytosol (Portnoy et al. 2001; Rees and Thiele 2007). Ctr-type transporters contain extramembranous copper-binding domains at both the amino and carboxy termini. They also contain three central membrane-spanning domains, with copper-binding motifs in the second transmembrane domain (Puig et al. 2002; Xiao et al. 2004). Structural studies of human Ctr1 indicate that the functional unit is a trimer, with an externally oriented amino terminus and an internally oriented carboxy terminus that cap a central, funnel-shaped pore (Aller and Unger 2006; De Feo et al. 2009). Ctr1 has been proposed to function by a series of sequential copper exchange reactions between the copper-binding sites that are coupled to sequential changes in the conformation of the transporter. In copper-replete media, high-affinity copper transport is shut off, Ctr1 is degraded (Ooi et al. 1996), and copper uptake is mediated by low-affinity, broad-specificity transporters, such as Fet4 and Smf1 (Cohen et al. 2000; Portnoy et al. 2001). Mac1 also activates the transcription of IRC7, a beta-lyase involved in the production of thiols (Roncoroni et al. 2011). Its role in the response to copper deficiency is not yet clear.
Intracellular copper is present at exceedingly low concentrations (Rae et al. 1999), and yeast rely on cytosolic copper-binding proteins called copper chaperones for the transfer of copper from import proteins (such as Ctr1) to enzymes (such as Cu,Zn superoxide dismutase, Sod1, and the copper exporter, Ccc1). It is unclear whether copper chaperones pick up their metal ligands directly or indirectly from copper transporters, although rapid exchange of copper ligands between copper-binding domains of transporters and chaperones has been demonstrated in vitro (Xiao et al. 2004). Copper chaperones deliver copper to target enzymes via a metal-enhanced, protein–protein interaction that facilitates a ligand exchange reaction for the copper ion. Ccs1 is the chaperone for Sod1 (Culotta et al. 1997) and Atx1 is required for the delivery of copper to Ccc2 (Lin et al. 1997; Pufahl et al. 1997). Ccc2 is a P-type ATPase that couples the hydrolysis of ATP to the transport of Cu(I) from the cytosol to the lumen of post-Golgi vesicles (Yuan et al. 1995). Luminal copper binds to and activates the multicopper oxidase Fet3 (Askwith et al. 1994), which is required for high-affinity iron uptake. Thus, iron uptake is a copper-dependent process in yeast.
Copper detoxification and utilization
When extracellular copper concentrations rise, yeast are protected from the influx of copper to the cytosol by the expression of copper-binding proteins called metallothioneins. Ace1 directs the transcription of two metallothionein genes, CUP1 (often present in multiple copies) and CRS5, which encode cysteine-rich proteins that bind and detoxify cytosolic copper (Butt et al. 1984; Winge et al. 1985; Culotta et al. 1994). Ace1 also activates the transcription of Sod1 under conditions of copper excess (Gralla et al. 1991). Sod1 is expressed at basal levels in copper-deficient cells. Increased expression under conditions of copper excess may offer cells additional protection against oxidative damage in the presence of this redox-active metal. Sod1 and its chaperone, Ccs1, are primarily located in the cytosol, but a small amount of both are also located in the mitochondrial intermembrane space (Sturtz et al. 2001) and in the nucleus (Wood and Thiele 2009). Surprisingly, both the nuclear localization and enzymatic activity of Sod1 are required for the Mac1 response to low copper.
Mitochondrial copper utilization
Copper plays an essential role in respiration through its activation of cytochrome c oxidase (CcO). CcO is a multisubunit enzyme (also known as complex IV), located in the mitochondrial inner membrane, which catalyzes the final step of the electron transport chain of cellular respiration. Cox1 (subunit 1) contains a single copper ion bound in a heterobimetallic active site (termed the CuB site) and Cox2 (subunit 2) contains two copper ions bound in a dinuclear center termed the CuA site. In addition to the copper ions, CcO contains two heme a cofactors plus zinc, magnesium, and sodium ions. Assembly of functional CcO is a complex process and requires the assistance of copper chaperones specific for CcO, namely Cox17, Cox11, and Sco1 (Carr and Winge 2003; Robinson and Winge 2010). Mitochondrial copper is largely present in the matrix associated with a low molecular weight ligand and must be transferred to the intermembrane space for incorporation into CcO (Cobine et al. 2004). Cox17 is a soluble protein with localization to both the cytosol and the mitochondrial intermembrane space (Beers et al. 1997). Yeast lacking functional Cox17 exhibit respiratory and CcO deficiency that can be rescued with high concentrations of copper (Glerum et al. 1996a). Cox17 binds three copper ions per monomer of protein and likely binds copper after it translocates to the intermembrane space, where it can specifically interact with and donate copper to Sco1 and Cox11 (Horng et al. 2004).
Sco1 is a mitochondrial copper-binding protein that is required for the formation of the CuA site in Cox2. Sco1 was initially linked to Cox17 when it was isolated as a high-copy suppressor of the respiratory defect of a Cox17 mutant (Glerum et al. 1996b). Cells lacking Sco1 exhibit respiratory deficiency and defects in CcO biogenesis (Schulze and Rodel 1988; Krummeck and Rodel 1990). Sco1 is anchored in the mitochondrial inner membrane by a single amino-terminal transmembrane helix. The carboxy-terminal domain extends into the intermembrane space and binds a single Cu(I) ion (Rentzsch et al. 1999; Nittis et al. 2001). Interactions between Sco1 and Cox2 are likely mediated by electrostatic interactions on a surface distinct from the copper-binding site (Rigby et al. 2008).
Formation of the CuB site in Cox1 requires Cox11, another copper-binding protein anchored in the mitochondrial inner membrane. Although yeast lacking Cox11 also exhibit respiratory deficiency and reduced levels of CcO (Tzagoloff et al. 1990), the specific function of Cox11 came from studies in the aerobic bacterium, Rhodobacter sphaeroides (Hiser et al. 2000). CcO isolated from strains lacking Cox11 contained both heme centers and the CuA site, but lacked the spectroscopic evidence for a CuB site. The CuB site of CcO is buried in the inner membrane in the mature enzyme, and copper is thought to be inserted into Cox1 via a channel-like cavity that is transiently occupied by Cu-bound Cox11 (Khalimonchuk et al. 2007).
Manganese
Manganese is a redox-active metal that can occupy multiple valence states in biological systems. Similar to iron, Mn(II) and Mn(III) [and to a lesser extent, Mn(IV)] are frequently found in biological systems. In vitro, Mn(II) can substitute for a number of divalent cations (such as Mg2+ and Ca2+) in the activation of enzymes; thus the endogenous, in vivo metal ligand for a protein is not always clear. It may differ in different species or depend on the relative concentrations of cations in the cellular compartment where the enzyme is located. Manganese-dependent enzymes include oxidoreductases, dehydrogenases, transferases, and hydrolases (Crowley et al. 2000). In yeast, particular attention has been given to the manganese-containing superoxide dismutase (Sod2) of the mitochondria and the manganese-dependent sugar transferases of the secretory pathway. Similar to other nutrient metals, uptake of manganese is homeostatically regulated. Unlike other nutrient metals, the regulation of manganese uptake appears to occur exclusively through post-translational mechanisms.
Uptake of manganese through Smf1 and Smf2
As is the case with other nutrient metals, under conditions of manganese deficiency, yeast express high-affinity manganese transporters (Figure 8). Two homologous transporters of the Nramp family, Smf1 and Smf2, account for the majority of the high-affinity transport of manganese into the cytosol (Supek et al. 1996; Liu et al. 1997; Cohen et al. 2000). Both transporters exhibit proton-coupled metal transport when expressed in xenopus oocytes (Chen et al. 1999), and, similar to other members of the Nramp family, can transport Mn(II), Fe(II), Cu(II), and Zn(II), as well as other divalent metal ions (Chen et al. 1999; Cohen et al. 2000). In cells grown in media containing sufficient concentrations of manganese, both Smf1 and Smf2 have relatively short half-lives and the transporters are sorted directly from the Golgi compartment to the vacuole for degradation (Liu et al. 1997; Liu and Culotta 1999; Portnoy et al. 2000, 2002). When manganese levels are low, both proteins become relatively more stable, and Smf1 is predominately expressed on the plasma membrane while Smf2 is predominately localized to endomembranes that likely correspond to the Golgi or endosomal compartment. Deletion of either SMF1 or SMF2 leads to slow growth in metal-depleted medium (Supek et al. 1996; Cohen et al. 2000), but the relative contributions of these transporters to cellular manganese homeostasis are not equal. In rich medium, a strain lacking SMF1 exhibits nearly wild-type levels of intracellular manganese, while a strain lacking SMF2 exhibits a marked reduction in intracellular manganese as well as a loss of Mn-SOD activity and defects in glycosylation (Luk and Culotta 2001). Elevated intracellular levels of manganese can protect cells from oxidative stress, and Smf1 is required for the intracellular accumulation of manganese when high levels of exogenous manganese are used to rescue the oxidative sensitivity of a sod1Δ strain (Reddi et al. 2009b).
Figure 8.
Manganese homeostasis. Proteins involved in the transport and utilization of manganese are shown. Subcellular localization of proteins observed under manganese-deficient conditions is depicted.
The phenotypic differences between the smf1Δ and the smf2Δ strains may be due to the distinct subcellular localizations of Smf1 and Smf2. Overall manganese uptake in yeast is relatively low compared to iron or zinc uptake, and only a small number of transporters on the cell surface would be required to account for the measured levels of uptake (Stimpson et al. 2006). In contrast, levels of manganese within the lumen of the secretory pathway are relatively high due to the activity of the Golgi-localized, Ca2+/Mn2+ efflux pump, Pmr1 (see below) (Lapinskas et al. 1995; Durr et al. 1998). Recovery of intraluminal manganese by Smf2 in the late secretory pathway may represent the majority of manganese influx to the cytosol.
The activities of Smf1 and Smf2 are controlled post-translationally by both intracellular trafficking and regulated degradation pathways. Yeast grown in manganese-sufficient medium rapidly degrade newly synthesized Smf1 and Smf2. This degradation occurs by sorting Smfs from the late Golgi compartment into the multivesicular body of the late secretory pathway with subsequent degradation in the vacuole. Similar to other transporters, degradation of Smf1 and Smf2 is dependent on ubiquitination by the ubiquitin ligase Rsp5 (Sullivan et al. 2007), but also requires several adaptor proteins that are involved in recruiting Rsp5 to the transporters. These include Bsd2 and Tre1/Tre2 (Liu et al. 1997; Liu and Culotta 1999; Stimpson et al. 2006). In yeast grown in manganese-depleted medium, Smf1 and Smf2 escape recognition by Bsd2 and Tre1/Tre2 and are stably expressed on the plasma membrane and endosomes, respectively. Mechanistically, it is not clear how Smf1 and Smf2 escape detection or what controls their sorting to different membranes. Intraluminal manganese appears to be required for recognition by Bsd2 and Tre1/Tre2, however, because a pmr1Δ strain that fails to pump manganese into the lumen of Golgi vesicles also exhibits impaired degradation of Smf1 in manganese-sufficient medium (Jensen et al. 2009). These observations have led to a model in which it is the conformational changes associated with transport activity that lead to recruitment of Rsp5 and subsequent degradation (Stimpson et al. 2006). Under conditions of manganese toxicity, Smf1 degradation becomes independent of Bsd2 and Tre1/2 and is only partially dependent on Rsp5. Elevated cytosolic levels of manganese trigger this pathway of degradation and the presence of manganese in the Golgi lumen is no longer required (Jensen et al. 2009).
Phosphate-dependent manganese transport
When yeast are grown in medium containing elevated levels of manganese, uptake can occur through the high-affinity phosphate transporter, Pho84. The transport substrate of Pho84 is a complex of phosphate anion and divalent metal cation (Fristedt et al. 1999). In standard laboratory media, magnesium concentrations are relatively high, and the substrate for Pho84 is likely a MgHPO4 complex. However, when extracellular manganese levels are high, Pho84 can also transport MnHPO4, and strains lacking PHO84 exhibit resistance to manganese toxicity (Jensen et al. 2003). Pho84 is strongly regulated at the transcriptional level by intracellular phosphate (Johnston and Carlson 1992), but no evidence exists for regulation by manganese. Thus, uptake of manganese through Pho84 can significantly contribute to manganese toxicity.
Transport of manganese into the secretory pathway
Yeast require substantial concentrations of manganese in Golgi compartments to correctly mannosylate proteins of the secretory pathway that have been modified with O- and N-glucosylation in the endoplasmic reticulum. Yeast mannosyl transferases, similar to the oligosaccharide transferases of bacteria, plants, and animals, are dependent on bound manganese cofactors for activity. Full activation of these enzymes in vitro requires manganese at >1 mM concentrations (Crowley et al. 2000). It is inferred from these observations that the intraluminal concentration of manganese must be substantial and similar to the concentration of Ca2+ (1–2 mM) (Pezzati et al. 1997). A single transporter in yeast, Pmr1, catalyzes the ATP-dependent transfer of both Ca2+ and Mn2+ into the lumen of the Golgi (Rudolph et al. 1989; Antebi and Fink 1992; Lapinskas et al. 1995; Durr et al. 1998) (see also Divalent Cations: Ca2+ and Mg2+, Ca2+ sequestration in intracellular compartments, above). Strains lacking Pmr1 exhibit multiple phenotypes attributable to defects in Golgi and endoplasmic reticulum function. While the protein sorting defects are largely attributable to luminal Ca2+ deficiency, the glycosylation defects are rescued only by manganese supplementation. Strains lacking Pmr1 also exhibit increased intracellular manganese accumulation.
Yeast can also store manganese in the vacuolar lumen via the efflux pump Ccc1. Although primarily an iron transporter, overexpression of Ccc1 is associated with increased vacuolar manganese and can partially suppress the Mn2+ hypersensitivity of a pmr1Δ strain (Lapinskas et al. 1996; Li et al. 2001b).
Unanswered Questions
While considerable progress has been made in dissecting the molecular details of ion homeostasis in S. cerevisiae, several areas remain mysterious. Several activities that mediate transport of the alkali metal ions, Na+ and K+, as well as the cation Ca2+, have been described using electrophysiology, but remain to be identified at the molecular level. Similarly, the regulation of transporters, including the Trk1/2 potassium transporters and the Ca2+ channel made up of Mid1, Cch1, and Ecm7, must be characterized at the molecular level. Many components of signal transduction pathways that are regulated by these ions must be elucidated, and methods for directly identifying and analyzing changes in intracellular ions, particularly Ca2+, within yeast cells should be further developed, as these tools have been invaluable for studying Ca2+-dependent signaling in mammalian cells. Recent advancements in the design and use of genetically encoded calcium sensors offer hope that such methods may be successfully applied to the study of Ca2+-dependent signaling in yeast (Mehta and Zhang 2011).
Metal trafficking within the cell is another significant area that needs exploration. All of the nutrient metals are required for activation of mitochondrial proteins, but only some of the transporters that control the uptake and efflux of iron in that organelle have been identified. The regulatory mechanisms that control the amount of metal taken up by mitochondria also remain to be identified. Copper, and possibly other metals, is present in mitochondria as a complex with a low-molecular-weight ligand that is uncharacterized. Whether mitochondrial metal chaperones in addition to the identified copper chaperones are required for protein metallation there has not been determined.
Heme and some product of the mitochondrial ISC machinery must exit the mitochondria and carrier proteins and/or molecules are likely involved, but how this occurs is unknown (in the case of heme) or only beginning to be understood (in the case of ISCs). With the exception of Atx1 and Ccs1, the existence or identity of cytosolic metal chaperones for the metallation of iron, zinc, or manganese proteins is unknown. ISCs are very likely involved in signaling cellular iron status to the Aft1 and Aft2 regulators, but the molecular details of regulation have not been described. Copper must access the nucleus in some form to bind to the Mac1 and Ace1 regulators, but how this occurs is not known. The number of proteins known to comprise the yeast metalloproteome continues to expand, as does our appreciation of the importance of nutrient metals in the function of cells. Studies of cat regulation in S. cerevisiae have already provided many insights into these processes in larger eukaryotes, and will likely continue to uncover conserved components of these fundamental cellular networks.
Acknowledgments
The authors would like to thank the anonymous reviewers for their helpful comments. M.S.C. is funded the National Institutes of Health (NIH) grant R01GM48729 from the National Institute of General Medicine. C.C.P. is supported by the NIH Intramural Research Program of the National Institue of Diabetes and Digestive and Kidney Disease.
Footnotes
Communicating editor: J. Thorner
Literature Cited
- Adam A. C., Bornhovd C., Prokisch H., Neupert W., Hell K., 2006. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 25: 174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar P. S., Engel A., Walter P., 2007. The plasma membrane proteins Prm1 and Fig1 ascertain fidelity of membrane fusion during yeast mating. Mol. Biol. Cell 18: 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A., Sesti F., Ilan N., Shih T. M., Sturley S. L., et al. , 1999. A molecular target for viral killer toxin: TOK1 potassium channels. Cell 99: 283–291. [DOI] [PubMed] [Google Scholar]
- Aller S. G., Unger V. M., 2006. Projection structure of the human copper transporter CTR1 at 6-A resolution reveals a compact trimer with a novel channel-like architecture. Proc. Natl. Acad. Sci. USA 103: 3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre B., Scherens B., 1995. The yeast YBR235w gene encodes a homolog of the mammalian electroneutral Na(+)-(K+)-C1- cotransporter family. Biochem. Biophys. Res. Commun. 217: 150–153. [DOI] [PubMed] [Google Scholar]
- Andreini C., Bertini I., 2011. A bioinformatics view of zinc enzymes. J. Inorg. Biochem. 111: 150–156. [DOI] [PubMed] [Google Scholar]
- Andreini C., Banci L., Bertini I., Rosato A., 2006. Zinc through the three domains of life. J. Proteome Res. 5: 3173–3178. [DOI] [PubMed] [Google Scholar]
- Andreini C., Bertini I., Cavallaro G., Holliday G. L., Thornton J. M., 2008. Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorg. Chem. 13: 1205–1218. [DOI] [PubMed] [Google Scholar]
- Andreini C., Bertini I., Rosato A., 2009. Metalloproteomes: a bioinformatic approach. Acc. Chem. Res. 42: 1471–1479. [DOI] [PubMed] [Google Scholar]
- Ansari K., Martin S., Farkasovsky M., Ehbrecht I. M., Kuntzel H., 1999. Phospholipase C binds to the receptor-like GPR1 protein and controls pseudohyphal differentiation in Saccharomyces cerevisiae. J. Biol. Chem. 274: 30052–30058. [DOI] [PubMed] [Google Scholar]
- Antebi A., Fink G. R., 1992. The yeast Ca2+-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell 3: 633–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arino J., Ramos J., Sychrová H., 2010. Alkali metal cation transport and homeostasis in yeasts. Microbiol. Mol. Biol. Rev. 74: 95–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askwith C., Eide D., Van Ho A., Bernard P. S., Li L., et al. , 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76: 403–410. [DOI] [PubMed] [Google Scholar]
- Baars T. L., Petri S., Peters C., Mayer A., 2007. Role of the V-ATPase in regulation of the vacuolar fission-fusion equilibrium. Mol. Biol. Cell 18: 3873–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J., Pierik A. J., Netz D. J., Muhlenhoff U., Lill R., 2004. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23: 2105–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J., Aguilar Netz D. J., Tepper K., Pierik A. J., Lill R., 2005. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol. Cell. Biol. 25: 10833–10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañuelos M. A., Sychrová H., Bleykasten-Grosshans C., Souciet J. L., Potier S., 1998. The Nha1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology 144(Pt 10): 2749–2758. [DOI] [PubMed] [Google Scholar]
- Bastidas R. J., Reedy J. L., Morales-Johansson H., Heitman J., Cardenas M. E., 2008. Signaling cascades as drug targets in model and pathogenic fungi. Curr. Opin. Investig. Drugs 9: 856–864. [PMC free article] [PubMed] [Google Scholar]
- Baughman J. M., Perocchi F., Girgis H. S., Plovanich M., Belcher-Timme C. A., et al. , 2011. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476: 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedekovics T., Li H., Gajdos G. B., Isaya G., 2011. Leucine biosynthesis regulates cytoplasmic iron-sulfur enzyme biogenesis in an Atm1p-independent manner. J. Biol. Chem. 286: 40878–40888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler T., Bruce K., Dunn T., 1997. Regulation of cellular Mg2+ by Saccharomyces cerevisiae. Biochim. Biophys. Acta 1323: 310–318. [DOI] [PubMed] [Google Scholar]
- Beers J., Glerum D. M., Tzagoloff A., 1997. Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J. Biol. Chem. 272: 33191–33196. [DOI] [PubMed] [Google Scholar]
- Belli G., Molina M. M., Garcia-Martinez J., Perez-Ortin J. E., Herrero E., 2004. Saccharomyces cerevisiae glutaredoxin 5-deficient cells subjected to continuous oxidizing conditions are affected in the expression of specific sets of genes. J. Biol. Chem. 279: 12386–12395. [DOI] [PubMed] [Google Scholar]
- Benito B., Quintero F. J., Rodriguez-Navarro A., 1997. Overexpression of the sodium ATPase of Saccharomyces cerevisiae: conditions for phosphorylation from ATP and Pi. Biochim. Biophys. Acta 1328: 214–226. [DOI] [PubMed] [Google Scholar]
- Berchtold D., Piccolis M., Chiaruttini N., Riezman I., Riezman H., et al. , 2012. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat. Cell Biol. 14: 542–547. [DOI] [PubMed] [Google Scholar]
- Bertl A., Slayman C. L., 1990. Cation-selective channels in the vacuolar membrane of Saccharomyces: dependence on calcium, redox state, and voltage. Proc. Natl. Acad. Sci. USA 87: 7824–7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertl A., Slayman C. L., Gradmann D., 1993. Gating and conductance in an outward-rectifying K+ channel from the plasma membrane of Saccharomyces cerevisiae. J. Membr. Biol. 132: 183–199. [DOI] [PubMed] [Google Scholar]
- Bihler H., Slayman C. L., Bertl A., 1998. NSC1: a novel high-current inward rectifier for cations in the plasma membrane of Saccharomyces cerevisiae. FEBS Lett. 432: 59–64. [DOI] [PubMed] [Google Scholar]
- Bird A., Evans-Galea M. V., Blankman E., Zhao H., Luo H., et al. , 2000a Mapping the DNA binding domain of the Zap1 zinc-responsive transcriptional activator. J. Biol. Chem. 275: 16160–16166. [DOI] [PubMed] [Google Scholar]
- Bird A. J., Zhao H., Luo H., Jensen L. T., Srinivasan C., et al. , 2000b A dual role for zinc fingers in both DNA binding and zinc sensing by the Zap1 transcriptional activator. EMBO J. 19: 3704–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. J., McCall K., Kramer M., Blankman E., Winge D. R., et al. , 2003. Zinc fingers can act as Zn2+ sensors to regulate transcriptional activation domain function. EMBO J. 22: 5137–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. J., Blankman E., Stillman D. J., Eide D. J., Winge D. R., 2004. The Zap1 transcriptional activator also acts as a repressor by binding downstream of the TATA box in ZRT2. EMBO J. 23: 1123–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. J., Gordon M., Eide D. J., Winge D. R., 2006. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 25: 5726–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaiseau P. L., Lesuisse E., Camadro J. M., 2001. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J. Biol. Chem. 276: 34221–34226. [DOI] [PubMed] [Google Scholar]
- Bleackley M. R., Macgillivray R. T., 2011. Transition metal homeostasis: from yeast to human disease. Biometals 24: 785–809. [DOI] [PubMed] [Google Scholar]
- Bonilla M., Nastase K. K., Cunningham K. W., 2002. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 21: 2343–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelly G., Boyer J. C., Touraine B., Szponarski W., Rambier M., et al. , 2001. The yeast mutant vps5Delta affected in the recycling of Golgi membrane proteins displays an enhanced vacuolar Mg2+/H+ exchange activity. Proc. Natl. Acad. Sci. USA 98: 9660–9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet L. E., Cardoso A. S., Perovano E., Pereira R. R., Ribeiro E. M., et al. , 2012. The involvement of calcium carriers and of the vacuole in the glucose-induced calcium signaling and activation of the plasma membrane H(+)-ATPase in Saccharomyces cerevisiae cells. Cell Calcium 51: 72–81. [DOI] [PubMed] [Google Scholar]
- Bowers K., Levi B. P., Patel F. I., Stevens T. H., 2000. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 11: 4277–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N. A., Morgan B., Dick T. P., Schwappach B., 2010. The yeast CLC protein counteracts vesicular acidification during iron starvation. J. Cell Sci. 123: 2342–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett C. L., Tukaye D. N., Mukherjee S., Rao R., 2005. The yeast endosomal Na+K+/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol. Biol. Cell 16: 1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. R., Keller G. L., Pickering I. J., Harris H. H., George G. N., et al. , 2002. Structures of the cuprous-thiolate clusters of the Mac1 and Ace1 transcriptional activators. Biochemistry 41: 6469–6476. [DOI] [PubMed] [Google Scholar]
- Buchman C., Skroch P., Welch J., Fogel S., Karin M., 1989. The CUP2 gene product, regulator of yeast metallothionein expression, is a copper-activated DNA-binding protein. Mol. Cell. Biol. 9: 4091–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultynck G., Heath V. L., Majeed A. P., Galan J. M., Haguenauer-Tsapis R., et al. , 2006. Slm1 and slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol. Cell. Biol. 26: 4729–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk S. E., Lytton J., MacLennan D. H., Shull G. E., 1989. cDNA cloning, functional expression, and mRNA tissue distribution of a third organellar Ca2+ pump. J. Biol. Chem. 264: 18561–18568. [PubMed] [Google Scholar]
- Butt T. R., Sternberg E. J., Gorman J. A., Clark P., Hamer D., et al. , 1984. Copper metallothionein of yeast, structure of the gene, and regulation of expression. Proc. Natl. Acad. Sci. USA 81: 3332–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnac O., Leterrier M., Yeager M., Blumwald E., 2007. Identification and characterization of Vnx1p, a novel type of vacuolar monovalent cation/H+ antiporter of Saccharomyces cerevisiae. J. Biol. Chem. 282: 24284–24293. [DOI] [PubMed] [Google Scholar]
- Cagnac O., Aranda-Sicilia M. N., Leterrier M., Rodriguez-Rosales M. P., Venema K., 2010. Vacuolar cation/H+ antiporters of Saccharomyces cerevisiae. J. Biol. Chem. 285: 33914–33922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafoli E., Balcavage W. X., Lehninger A. L., Mattoon J. R., 1970. Ca2+ metabolism in yeast cells and mitochondria. Biochim. Biophys. Acta 205: 18–26. [DOI] [PubMed] [Google Scholar]
- Carmelo V., Santos H., Sa-Correia I., 1997. Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1325: 63–70. [DOI] [PubMed] [Google Scholar]
- Carr H. S., Winge D. R., 2003. Assembly of cytochrome c oxidase within the mitochondrion. Acc. Chem. Res. 36: 309–316. [DOI] [PubMed] [Google Scholar]
- Casado C., Yenush L., Melero C., Ruiz Mdel C., Serrano R., et al. , 2010. Regulation of Trk-dependent potassium transport by the calcineurin pathway involves the Hal5 kinase. FEBS Lett. 584: 2415–2420. [DOI] [PubMed] [Google Scholar]
- Chen O. S., Kaplan J., 2000. CCC1 suppresses mitochondrial damage in the yeast model of Friedreich’s ataxia by limiting mitochondrial iron accumulation. J. Biol. Chem. 275: 7626–7632. [DOI] [PubMed] [Google Scholar]
- Chen O. S., Hemenway S., Kaplan J., 2002. Inhibition of Fe-S cluster biosynthesis decreases mitochondrial iron export: evidence that Yfh1p affects Fe-S cluster synthesis. Proc. Natl. Acad. Sci. USA 99: 12321–12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen O. S., Crisp R. J., Valachovic M., Bard M., Winge D. R., et al. , 2004. Transcription of the yeast iron regulon does not respond directly to iron but rather to iron-sulfur cluster biosynthesis. J. Biol. Chem. 279: 29513–29518. [DOI] [PubMed] [Google Scholar]
- Chen X. Z., Peng J. B., Cohen A., Nelson H., Nelson N., et al. , 1999. Yeast SMF1 mediates H(+)-coupled iron uptake with concomitant uncoupled cation currents. J. Biol. Chem. 274: 35089–35094. [DOI] [PubMed] [Google Scholar]
- Clapham D. E., 2007. Calcium signaling. Cell 131: 1047–1058. [DOI] [PubMed] [Google Scholar]
- Cobine P. A., Ojeda L. D., Rigby K. M., Winge D. R., 2004. Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J. Biol. Chem. 279: 14447–14455. [DOI] [PubMed] [Google Scholar]
- Cohen A., Nelson H., Nelson N., 2000. The family of SMF metal ion transporters in yeast cells. J. Biol. Chem. 275: 33388–33394. [DOI] [PubMed] [Google Scholar]
- Conklin D. S., McMaster J. A., Culbertson M. R., Kung C., 1992. COT1, a gene involved in cobalt accumulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 3678–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courel M., Lallet S., Camadro J. M., Blaiseau P. L., 2005. Direct activation of genes involved in intracellular iron use by the yeast iron-responsive transcription factor Aft2 without its paralog Aft1. Mol. Cell. Biol. 25: 6760–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin S. R., Rao R., Hampton R. Y., 2002. Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J. Cell Biol. 157: 1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley J. D., Traynor D. A., Weatherburn D. C., 2000. Enzymes and proteins containing manganese: an overview. Met. Ions Biol. Syst. 37: 209–278. [PubMed] [Google Scholar]
- Cui J., Kaandorp J. A., Ositelu O. O., Beaudry V., Knight A., et al. , 2009a Simulating calcium influx and free calcium concentrations in yeast. Cell Calcium 45: 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Kaandorp J. A., Sloot P. M., Lloyd C. M., Filatov M. V., 2009b Calcium homeostasis and signaling in yeast cells and cardiac myocytes. FEMS Yeast Res. 9: 1137–1147. [DOI] [PubMed] [Google Scholar]
- Culotta V. C., Howard W. R., Liu X. F., 1994. CRS5 encodes a metallothionein-like protein in Saccharomyces cerevisiae. J. Biol. Chem. 269: 25295–25302. [PubMed] [Google Scholar]
- Culotta V. C., Klomp L. W., Strain J., Casareno R. L., Krems B., et al. , 1997. The copper chaperone for superoxide dismutase. J. Biol. Chem. 272: 23469–23472. [DOI] [PubMed] [Google Scholar]
- Cunningham K., Fink G. R., 1996. Calcineurin inhibits VCX1-Dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2226–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K. W., 2011. Acidic calcium stores of Saccharomyces cerevisiae. Cell Calcium 50: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K. W., Fink G. R., 1994. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 124: 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic A., Menon A. L., Thorgersen M. P., Scott J. W., Poole F. L., 2nd, et al. , 2010. Microbial metalloproteomes are largely uncharacterized. Nature 466: 779–782. [DOI] [PubMed] [Google Scholar]
- Cyert M. S., 2001. Genetic analysis of calmodulin and its targets in Saccharomyces cerevisiae. Annu. Rev. Genet. 35: 647–672. [DOI] [PubMed] [Google Scholar]
- Cyert M. S., 2003. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311: 1143–1150. [DOI] [PubMed] [Google Scholar]
- Dameron C. T., Winge D. R., George G. N., Sansone M., Hu S., et al. , 1991. A copper-thiolate polynuclear cluster in the ACE1 transcription factor. Proc. Natl. Acad. Sci. USA 88: 6127–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancis A., Klausner R. D., Hinnebusch A. G., Barriocanal J. G., 1990. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 2294–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancis A., Roman D. G., Anderson G. J., Hinnebusch A. G., Klausner R. D., 1992. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc. Natl. Acad. Sci. USA 89: 3869–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancis A., Haile D., Yuan D. S., Klausner R. D., 1994a The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 269: 25660–25667. [PubMed] [Google Scholar]
- Dancis A., Yuan D. S., Haile D., Askwith C., Elde D., et al. , 1994b Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell 76: 393–402. [DOI] [PubMed] [Google Scholar]
- Daran-Lapujade P., Daran J. M., Luttik M. A., Almering M. J., Pronk J. T., et al. , 2009. An atypical PMR2 locus is responsible for hypersensitivity to sodium and lithium cations in the laboratory strain Saccharomyces cerevisiae CEN.PK113–7D. FEMS Yeast Res. 9: 789–792. [DOI] [PubMed] [Google Scholar]
- Davis-Kaplan S. R., Compton M. A., Flannery A. R., Ward D. M., Kaplan J., et al. , 2006. PKR1 encodes an assembly factor for the yeast V-type ATPase. J. Biol. Chem. 281: 32025–32035. [DOI] [PubMed] [Google Scholar]
- De Feo C. J., Aller S. G., Siluvai G. S., Blackburn N. J., Unger V. M., 2009. Three-dimensional structure of the human copper transporter hCTR1. Proc. Natl. Acad. Sci. USA 106: 4237–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nicola R., Hazelwood L. A., De Hulster E. A., Walsh M. C., Knijnenburg T. A., et al. , 2007. Physiological and transcriptional responses of Saccharomyces cerevisiae to zinc limitation in chemostat cultures. Appl. Environ. Microbiol. 73: 7680–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva D. M., Askwith C. C., Eide D., Kaplan J., 1995. The FET3 gene product required for high affinity iron transport in yeast is a cell surface ferroxidase. J. Biol. Chem. 270: 1098–1101. [DOI] [PubMed] [Google Scholar]
- de Silva D., Davis-Kaplan S., Fergestad J., Kaplan J., 1997. Purification and characterization of Fet3 protein, a yeast homologue of ceruloplasmin. J. Biol. Chem. 272: 14208–14213. [DOI] [PubMed] [Google Scholar]
- De Stefani D., Raffaello A., Teardo E., Szabo I., Rizzuto R., 2011. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476: 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant R., Binda M., Lee S. S., Pelet S., Winderickx J., et al. , 2010. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 29: 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaurex N., Frieden M., 2003. Measurements of the free luminal ER Ca(2+) concentration with targeted “cameleon” fluorescent proteins. Cell Calcium 34: 109–119. [DOI] [PubMed] [Google Scholar]
- Deng Y., Watson H., Guo Y., Au W.-C., Shakoury-Elizeh M., et al. , 2009. Gga2 mediates sequential ubiquitin-independent and -dependent steps in the trafficking of Arn1 from the trans-Golgi network to the vacuole. J. Biol. Chem. 284: 23830–23841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis V., Cyert M. S., 2002. Internal Ca2+ release in yeast is triggered by hypertonic shock and mediated by a TRP channel homologue. J. Cell Biol. 156: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakov T. T., Kane P. M., 2010. Regulation of vacuolar proton-translocating ATPase activity and assembly by extracellular pH. J. Biol. Chem. 285: 23771–23778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix D. R., Bridgham J. T., Broderius M. A., Byersdorfer C. A., Eide D. J., 1994. The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J. Biol. Chem. 269: 26092–26099. [PubMed] [Google Scholar]
- Dohlman H. G., Slessareva J. E., 2006. Pheromone signaling pathways in yeast. Sci. STKE 2006: cm6. [DOI] [PubMed] [Google Scholar]
- Drewke C., Ciriacy M., 1988. Overexpression, purification and properties of alcohol dehydrogenase IV from Saccharomyces cerevisiae. Biochim. Biophys. Acta 950: 54–60. [DOI] [PubMed] [Google Scholar]
- Dudgeon D. D., Zhang N., Ositelu O. O., Kim H., Cunningham K. W., 2008. Nonapoptotic death of Saccharomyces cerevisiae cells that is stimulated by Hsp90 and inhibited by calcineurin and Cmk2 in response to endoplasmic reticulum stresses. Eukaryot. Cell 7: 2037–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T., Gable K., Beeler T., 1994. Regulation of cellular Ca2+ by yeast vacuoles. J. Biol. Chem. 269: 7273–7278. [PubMed] [Google Scholar]
- Durell S. R., Guy H. R., 1999. Structural models of the KtrB, TrkH, and Trk1,2 symporters based on the structure of the KcsA K(+) channel. Biophys. J. 77: 789–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr G., Strayle J., Plemper R., Elbs S., Klee S. K., et al. , 1998. The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Mol. Biol. Cell 9: 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D. J., 2009. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 284: 18565–18569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D. J., Clark S., Nair T. M., Gehl M., Gribskov M., et al. , 2005. Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol. 6: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C. D., Wang F., MacDiarmid C. W., Clark S., Lyons T., et al. , 2004. Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J. Cell Biol. 166: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C. D., MacDiarmid C. W., Eide D. J., 2005. Heteromeric protein complexes mediate zinc transport into the secretory pathway of eukaryotic cells. J. Biol. Chem. 280: 28811–28818. [DOI] [PubMed] [Google Scholar]
- Engel A., Walter P., 2008. Membrane lysis during biological membrane fusion: collateral damage by misregulated fusion machines. J. Cell Biol. 183: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi P., Czirjak G., 2010. Molecular background of leak K+ currents: two-pore domain potassium channels. Physiol. Rev. 90: 559–605. [DOI] [PubMed] [Google Scholar]
- Eraso P., Gansedo C., 1987. Activation of yeast plasma membrane ATPase by acid pH during growth. FEBS Lett. 224: 187–192. [DOI] [PubMed] [Google Scholar]
- Eraso P., Mazon M. J., Portillo F., 2006. Yeast protein kinase Ptk2 localizes at the plasma membrane and phosphorylates in vitro the C-terminal peptide of the H+-ATPase. Biochim. Biophys. Acta 1758: 164–170. [DOI] [PubMed] [Google Scholar]
- Erdman S., Lin L., Malczynski M., Snyder M., 1998. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140: 461–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez O., Kahana C., 2002. Deletions of SKY1 or PTK2 in the Saccharomyces cerevisiae trk1Deltatrk2Delta mutant cells exert dual effect on ion homeostasis. Biochem. Biophys. Res. Commun. 295: 1142–1149. [DOI] [PubMed] [Google Scholar]
- Erpapazoglou Z., Froissard M., Nondier I., Lesuisse E., Haguenauer-Tsapis R., et al. , 2008. Substrate- and ubiquitin-dependent trafficking of the yeast siderophore transporter Sit1. Traffic 9: 1372–1391. [DOI] [PubMed] [Google Scholar]
- Evans C. F., Engelke D. R., Thiele D. J., 1990. ACE1 transcription factor produced in Escherichia coli binds multiple regions within yeast metallothionein upstream activation sequences. Mol. Cell. Biol. 10: 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans-Galea M. V., Blankman E., Myszka D. G., Bird A. J., Eide D. J., et al. , 2003. Two of the five zinc fingers in the Zap1 transcription factor DNA binding domain dominate site-specific DNA binding. Biochemistry 42: 1053–1061. [DOI] [PubMed] [Google Scholar]
- Farrell R. A., Thorvaldsen J. L., Winge D. R., 1996. Identification of the Zn(II) site in the copper-responsive yeast transcription factor, AMT1: a conserved Zn module. Biochemistry 35: 1571–1580. [DOI] [PubMed] [Google Scholar]
- Fischer M., Schnell N., Chattaway J., Davies P., Dixon G., et al. , 1997. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Lett. 419: 259–262. [DOI] [PubMed] [Google Scholar]
- Flis K., Hinzpeter A., Edelman A., Kurlandzka A., 2005. The functioning of mammalian ClC-2 chloride channel in Saccharomyces cerevisiae cells requires an increased level of Kha1p. Biochem. J. 390: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foor F., Parent S. A., Morin N., Dahl A. M., Ramadan N., et al. , 1992. Calcineurin mediates inhibition by FK506 and cyclosporin of recovery from alpha-factor arrest in yeast. Nature 360: 682–684. [DOI] [PubMed] [Google Scholar]
- Forment J., Mulet J. M., Vicente O., Serrano R., 2002. The yeast SR protein kinase Sky1p modulates salt tolerance, membrane potential and the Trk1,2 potassium transporter. Biochim. Biophys. Acta 1565: 36–40. [DOI] [PubMed] [Google Scholar]
- Foury F., Roganti T., 2002. Deletion of the mitochondrial carrier genes MRS3 and MRS4 suppresses mitochondrial iron accumulation in a yeast frataxin-deficient strain. J. Biol. Chem. 277: 24475–24483. [DOI] [PubMed] [Google Scholar]
- Frey A. G., Eide D. J., 2011. Roles of two activation domains in Zap1 in the response to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 286: 6844–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A. G., Bird A. J., Evans-Galea M. V., Blankman E., Winge D. R., et al. , 2011. Zinc-regulated DNA binding of the yeast Zap1 zinc-responsive activator. PLoS ONE 6: e22535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt U., van Der Rest M., Poolman B., Konings W. N., Persson B. L., 1999. Studies of cytochrome c oxidase-driven H(+)-coupled phosphate transport catalyzed by the Saccharomyces cerevisiae Pho84 permease in coreconstituted vesicles. Biochemistry 38: 16010–16015. [DOI] [PubMed] [Google Scholar]
- Froissard M., Belgareh-Touze N., Dias M., Buisson N., Camadro J. M., et al. , 2007. Trafficking of Siderophore Transporters in Saccharomyces cerevisiae and Intracellular Fate of Ferrioxamine B Conjugates. Traffic 8: 1601–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froschauer E. M., Schweyen R. J., Wiesenberger G., 2009. The yeast mitochondrial carrier proteins Mrs3p/Mrs4p mediate iron transport across the inner mitochondrial membrane. Biochim. Biophys. Acta 1788: 1044–1050. [DOI] [PubMed] [Google Scholar]
- Gaber R. F., Styles C. A., Fink G. R., 1988. TRK1 encodes a plasma membrane protein required for high-affinity potassium transport in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 2848–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J. M., 2008. The early steps of glucose signalling in yeast. FEMS Microbiol. Rev. 32: 673–704. [DOI] [PubMed] [Google Scholar]
- Garcia R., Bermejo C., Grau C., Perez R., Rodriguez-Pena J. M., et al. , 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279: 15183–15195. [DOI] [PubMed] [Google Scholar]
- Georgatsou E., Alexandraki D., 1994. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 3065–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgatsou E., Alexandraki D., 1999. Regulated expression of the Saccharomyces cerevisiae Fre1p/Fre2p Fe/Cu reductase related genes. Yeast 15: 573–584. [DOI] [PubMed] [Google Scholar]
- Georgatsou E., Mavrogiannis L. A., Fragiadakis G. S., Alexandraki D., 1997. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 272: 13786–13792. [DOI] [PubMed] [Google Scholar]
- Gerber J., Muhlenhoff U., Lill R., 2003. An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 4: 906–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glerum D. M., Shtanko A., Tzagoloff A., 1996a Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 271: 14504–14509. [DOI] [PubMed] [Google Scholar]
- Glerum D. M., Shtanko A., Tzagoloff A., 1996b SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 271: 20531–20535. [DOI] [PubMed] [Google Scholar]
- Gomez M. J., Luyten K., Ramos J., 1996. The capacity to transport potassium influences sodium tolerance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 135: 157–160. [DOI] [PubMed] [Google Scholar]
- Goossens A., de La Fuente N., Forment J., Serrano R., Portillo F., 2000. Regulation of yeast H(+)-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters. Mol. Cell. Biol. 20: 7654–7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graden J. A., Winge D. R., 1997. Copper-mediated repression of the activation domain in the yeast Mac1p transcription factor. Proc. Natl. Acad. Sci. USA 94: 5550–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graden J. A., Posewitz M. C., Simon J. R., George G. N., Pickering I. J., et al. , 1996. Presence of a copper(I)-thiolate regulatory domain in the copper-activated transcription factor Amt1. Biochemistry 35: 14583–14589. [DOI] [PubMed] [Google Scholar]
- Graham L. A., Powell B., Stevens T. H., 2000. Composition and assembly of the yeast vacuolar H(+)-ATPase complex. J. Exp. Biol. 203: 61–70. [DOI] [PubMed] [Google Scholar]
- Gralla E. B., Thiele D. J., Silar P., Valentine J. S., 1991. ACE1, a copper-dependent transcription factor, activates expression of the yeast copper, zinc superoxide dismutase gene. Proc. Natl. Acad. Sci. USA 88: 8558–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppi S., 2011. Glucose and osmotic stress-dependent calcium signalling in Saccharomyces cerevisiae: evidences for novel transporter systems and calcineurin involvement, Ph.D. Thesis, University of Milano-Bicocca, Milan, Italy. [Google Scholar]
- Groppi S., Belotti F., Brandao R. L., Martegani E., Tisi R., 2011. Glucose-induced calcium influx in budding yeast involves a novel calcium transport system and can activate calcineurin. Cell Calcium 49: 376–386. [DOI] [PubMed] [Google Scholar]
- Guarante L., 1992. Messenger RNA transcription and its control in Saccharomyces cerevisiae, pp. 49–98 in The Molecular and Cellular Biology of the Yeast Saccaromyces: Gene Expression, edited by Jones E., Pringle J., and Broach J. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Gustin M. C., Martinac B., Saimi Y., Culbertson M. R., Kung C., 1986. Ion channels in yeast. Science 233: 1195–1197. [DOI] [PubMed] [Google Scholar]
- Haro R., Rodriguez-Navarro A., 2002. Molecular analysis of the mechanism of potassium uptake through the TRK1 transporter of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1564: 114–122. [DOI] [PubMed] [Google Scholar]
- Haro R., Graciadebles B., Rodriguez-Navarro A., 1991. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 291: 189–191. [DOI] [PubMed] [Google Scholar]
- Hassett R., Kosman D. J., 1995. Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J. Biol. Chem. 270: 128–134. [DOI] [PubMed] [Google Scholar]
- Hassett R., Dix D. R., Eide D. J., Kosman D. J., 2000. The Fe(II) permease Fet4p functions as a low affinity copper transporter and supports normal copper trafficking in Saccharomyces cerevisiae. Biochem. J. 351(Pt 2): 477–484. [PMC free article] [PubMed] [Google Scholar]
- Hassett R. F., Romeo A. M., Kosman D. J., 1998a Regulation of high affinity iron uptake in the yeast Saccharomyces cerevisiae. Role of dioxygen and Fe. J. Biol. Chem. 273: 7628–7636. [DOI] [PubMed] [Google Scholar]
- Hassett R. F., Yuan D. S., Kosman D. J., 1998b Spectral and kinetic properties of the Fet3 protein from Saccharomyces cerevisiae, a multinuclear copper ferroxidase enzyme. J. Biol. Chem. 273: 23274–23282. [DOI] [PubMed] [Google Scholar]
- Hausmann A., Aguilar Netz D. J., Balk J., Pierik A. J., Muhlenhoff U., et al. , 2005. The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. USA 102: 3266–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth R. S., Lemire B. D., Crandall D., Cragoe E. J., Jr., Fliegel L., 1991. Characterisation of proton fluxes across the cytoplasmic membrane of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1098: 79–89. [PubMed] [Google Scholar]
- Heath V. L., Shaw S. L., Roy S., Cyert M. S., 2004. Hph1p and Hph2p, novel components of calcineurin-mediated stress responses in Saccharomyces cerevisiae. Eukaryot. Cell 3: 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S. A., Kohlwein S. D., Carman G. M., 2012. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190: 317–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig A., Bird A. J., Swierczek S., McCall K., Mooney M., et al. , 2005. Zap1 activation domain 1 and its role in controlling gene expression in response to cellular zinc status. Mol. Microbiol. 57: 834–846. [DOI] [PubMed] [Google Scholar]
- Heredia J., Crooks M., Zhu Z., 2001. Phosphorylation and Cu+ coordination-dependent DNA binding of the transcription factor Mac1p in the regulation of copper transport. J. Biol. Chem. 276: 8793–8797. [DOI] [PubMed] [Google Scholar]
- Heymann P., Ernst J. F., Winkelmann G., 1999. Identification of a fungal triacetylfusarinine C siderophore transport gene (TAF1) in Saccharomyces cerevisiae as a member of the major facilitator superfamily. Biometals 12: 301–306. [DOI] [PubMed] [Google Scholar]
- Heymann P., Ernst J. F., Winkelmann G., 2000. Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 186: 221–227. [DOI] [PubMed] [Google Scholar]
- Higgins V. J., Rogers P. J., Dawes I. W., 2003. Application of genome-wide expression analysis to identify molecular markers useful in monitoring industrial fermentations. Appl. Environ. Microbiol. 69: 7535–7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilioti Z., Gallagher D. A., Low-Nam S. T., Ramaswamy P., Gajer P., et al. , 2004. GSK-3 kinases enhance calcineurin signaling by phosphorylation of RCNs. Genes Dev. 18: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiser L., Di Valentin M., Hamer A. G., Hosler J. P., 2000. Cox11p is required for stable formation of the Cu(B) and magnesium centers of cytochrome c oxidase. J. Biol. Chem. 275: 619–623. [DOI] [PubMed] [Google Scholar]
- Hong M. P., Vu K., Bautos J., Gelli A., 2010. Cch1 restores intracellular Ca2+ in fungal cells during endoplasmic reticulum stress. J. Biol. Chem. 285: 10951–10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng Y. C., Cobine P. A., Maxfield A. B., Carr H. S., Winge D. R., 2004. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J. Biol. Chem. 279: 35334–35340. [DOI] [PubMed] [Google Scholar]
- Howell A. S., Lew D. J., 2012. Morphogenesis and the cell cycle. Genetics 190: 51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Bonifas J. M., Beech J., Bench G., Shigihara T., et al. , 2000. Mutations in ATP2C1, encoding a calcium pump, cause Hailey-Hailey disease. Nat. Genet. 24: 61–65. [DOI] [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R., Thiele D. J., 1989. Copper-induced binding of cellular factors to yeast metallothionein upstream activation sequences. Proc. Natl. Acad. Sci. USA 86: 65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida H., Yagawa Y., Anraku Y., 1990. Essential role for induced Ca2+influx followed by [Ca2+]i rise in maintaining viability of yeast cells late in the mating pheromone response pathway. J. Biol. Chem. 265: 13391–13399. [PubMed] [Google Scholar]
- Iida H., Nakamura H., Ono T., Okumura M., Anraku Y., 1994. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and Mating. Mol. Cell. Biol. 14: 8259–8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanyshyn W. M., Han G. S., Carman G. M., 2004. Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc. J. Biol. Chem. 279: 21976–21983. [DOI] [PubMed] [Google Scholar]
- Jbel M., Mercier A., Labbe S., 2011. Grx4 monothiol glutaredoxin is required for iron limitation-dependent inhibition of Fep1. Eukaryot. Cell 10: 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L. T., Ajua-Alemanji M., Culotta V. C., 2003. The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J. Biol. Chem. 278: 42036–42040. [DOI] [PubMed] [Google Scholar]
- Jensen L. T., Culotta V. C., 2002. Regulation of Saccharomyces cerevisiae FET4 by oxygen and iron. J. Mol. Biol. 318: 251–260. [DOI] [PubMed] [Google Scholar]
- Jensen L. T., Winge D. R., 1998. Identification of a copper-induced intramolecular interaction in the transcription factor Mac1 from Saccharomyces cerevisiae. EMBO J. 17: 5400–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L. T., Posewitz M. C., Srinivasan C., Winge D. R., 1998. Mapping of the DNA binding domain of the copper-responsive transcription factor Mac1 from Saccharomyces cerevisiae. J. Biol. Chem. 273: 23805–23811. [DOI] [PubMed] [Google Scholar]
- Jensen L. T., Carroll M. C., Hall M. D., Harvey C. J., Beese S. E., et al. , 2009. Down-regulation of a manganese transporter in the face of metal toxicity. Mol. Biol. Cell 20: 2810–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, M., and M. Carlson, 1992 Regulation of Carbon and Phosphate Utilization, pp. 193–281 in The Molecular and Cellular Biology of the Yeast Saccharomyces, edited by E. W. Jones, J. R. Pringle, and J. R. Broach. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Joshi A., Serpe M., Kosman D. J., 1999. Evidence for (Mac1p)2.DNA ternary complex formation in Mac1p-dependent transactivation at the CTR1 promoter. J. Biol. Chem. 274: 218–226. [DOI] [PubMed] [Google Scholar]
- Jungmann J., Reins H. A., Lee J., Romeo A., Hassett R., et al. , 1993. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 12: 5051–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahm M., Navarrete C., Llopis-Torregrosa V., Herrera R., Barreto L., et al. , 2012. Potassium starvation in yeast: mechanisms of homeostasis revealed by mathematical modeling. PLOS Comput. Biol. 8: e1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallay L. M., Brett C. L., Tukaye D. N., Wemmer M. A., Chyou A., et al. , 2011. Endosomal Na+ (K+)/H+ exchanger Nhx1/Vps44 functions independently and downstream of multivesicular body formation. J. Biol. Chem. 286: 44067–44077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamizono A., Nishizawa M., Teranishi Y., Murata K., Kimura A., 1989. Identification of a gene conferring resistance to zinc and cadmium ions in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 219: 161–167. [DOI] [PubMed] [Google Scholar]
- Kane P. M., 2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70: 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane P. M., 2012. Targeting reversible disassembly as a mechanism of controlling V-ATPase activity. Curr. Protein Pept. Sci. 13: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki M., Nagasawa M., Kojima I., Sato C., Naruse K., et al. , 1999. Molecular identification of a eukaryotic, stretch-activated nonselective cation channel. Science 285: 882–886. [DOI] [PubMed] [Google Scholar]
- Kawasaki-Nishi S., Nishi T., Forgac M., 2001. Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation. J. Biol. Chem. 276: 17941–17948. [DOI] [PubMed] [Google Scholar]
- Keller G., Bird A., Winge D. R., 2005. Independent metalloregulation of Ace1 and Mac1 in Saccharomyces cerevisiae. Eukaryot. Cell 4: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchum K. A., Joiner W. J., Sellers A. J., Kaczmarek L. K., Goldstein S. A., 1995. A new family of outwardly rectifying potassium channel proteins with two pore domains in tandem. Nature 376: 690–695. [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O., Bird A., Winge D. R., 2007. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 282: 17442–17449. [DOI] [PubMed] [Google Scholar]
- Kim Y., Yun C. W., Philpott C. C., 2002. Ferrichrome induces endosome to plasma membrane cycling of the ferrichrome transporter, Arn1p, in Saccharomyces cerevisiae. EMBO J. 21: 3632–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lampert S. M., Philpott C. C., 2005. A receptor domain controls the intracellular sorting of the ferrichrome transporter, ARN1. EMBO J. 24: 952–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Deng Y., Philpott C. C., 2007. GGA2- and ubiquitin-dependent trafficking of Arn1, the ferrichrome transporter of Saccharomyces cerevisiae. Mol. Biol. Cell 18: 1790–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinclova O., Ramos J., Potier S., Sychrová H., 2001. Functional study of the Saccharomyces cerevisiae Nha1p C-terminus. Mol. Microbiol. 40: 656–668. [DOI] [PubMed] [Google Scholar]
- Kinclova-Zimmermannova O., Gaskova D., Sychrová H., 2006. The Na+,K+/H+ -antiporter Nha1 influences the plasma membrane potential of Saccharomyces cerevisiae. FEMS Yeast Res. 6: 792–800. [DOI] [PubMed] [Google Scholar]
- Kispal G., Csere P., Prohl C., Lill R., 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18: 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S. A., Labbe S., Kwon L. F., Kosman D. J., Thiele D. J., 1996. A widespread transposable element masks expression of a yeast copper transport gene. Genes Dev. 10: 1917–1929. [DOI] [PubMed] [Google Scholar]
- Ko C. H., Buckley A. M., Gaber R. F., 1990. TRK2 is required for low affinity K+ transport in Saccharomyces cerevisiae. Genetics 125: 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko C. H., Gaber R. F., 1991. TRK1 and TRK2 encode structurally related K+ transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 4266–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima A., Toshima J. Y., Kanno C., Kawata C., Toshima J., 2012. Localization and functional requirement of yeast Na+/H+ exchanger, Nhx1p, in the endocytic and protein recycling pathway. Biochim. Biophys. Acta 1823: 534–543. [DOI] [PubMed] [Google Scholar]
- Kraakman L., Lemaire K., Ma P., Teunissen A. W., Donaton M. C., et al. , 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32: 1002–1012. [DOI] [PubMed] [Google Scholar]
- Krummeck G., Rodel G., 1990. Yeast SCO1 protein is required for a post-translational step in the accumulation of mitochondrial cytochrome c oxidase subunits I and II. Curr. Genet. 18: 13–15. [DOI] [PubMed] [Google Scholar]
- Kumanovics A., Chen O. S., Li L., Bagley D., Adkins E. M., et al. , 2008. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 283: 10276–10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda T., Bihler H., Bashi E., Slayman C. L., Rivetta A., 2004. Chloride channel function in the yeast TRK-potassium transporters. J. Membr. Biol. 198: 177–192. [DOI] [PubMed] [Google Scholar]
- Kwast K. E., Burke P. V., Poyton R. O., 1998. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 201: 1177–1195. [DOI] [PubMed] [Google Scholar]
- Labbe S., Zhu Z., Thiele D. J., 1997. Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway. J. Biol. Chem. 272: 15951–15958. [DOI] [PubMed] [Google Scholar]
- Labbe S., Pelletier B., Mercier A., 2007. Iron homeostasis in the fission yeast Schizosaccharomyces pombe. Biometals 20: 523–537. [DOI] [PubMed] [Google Scholar]
- Labbe-Bois, R., and P. Labbe, 1989 Tetrapyrrole and Heme Biosynthesis in the Yeast Saccharomyces cerevisiae, pp. 235–285 in Biosynthesis of Heme and Chlorophylls, edited by H. A. Dailey. McGraw Hill, New York. [Google Scholar]
- Lange H., Kaut A., Kispal G., Lill R., 2000. A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc. Natl. Acad. Sci. USA 97: 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., Lisowsky T., Gerber J., Muhlenhoff U., Kispal G., et al. , 2001. An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep. 2: 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinskas P. J., Cunningham K. W., Liu X. F., Fink G. R., Culotta V. C., 1995. Mutations in PMR1 suppress oxidative damage in yeast cells lacking superoxide dismutase. Mol. Cell. Biol. 15: 1382–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinskas P. J., Lin S. J., Culotta V. C., 1996. The role of the Saccharomyces cerevisiae CCC1 gene in the homeostasis of manganese ions. Mol. Microbiol. 21: 519–528. [DOI] [PubMed] [Google Scholar]
- Lecchi S., Nelson C. J., Allen K. E., Swaney D. L., Thompson K. L., et al. , 2007. Tandem phosphorylation of Ser-911 and Thr-912 at the C terminus of yeast plasma membrane H+-ATPase leads to glucose-dependent activation. J. Biol. Chem. 282: 35471–35481. [DOI] [PubMed] [Google Scholar]
- Lee H. C., Zhang L., 2009. A unique mechanism of chaperone action: heme regulation of Hap1 activity involves separate control of repression and activation. Protein Pept. Lett. 16: 642–649. [DOI] [PubMed] [Google Scholar]
- Lemaire K., Van de Velde S., Van Dijck P., Thevelein J. M., 2004. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16: 293–299. [DOI] [PubMed] [Google Scholar]
- Leskovac V., Trivic S., Pericin D., 2002. The three zinc-containing alcohol dehydrogenases from baker’s yeast, Saccharomyces cerevisiae. FEMS Yeast Res. 2: 481–494. [DOI] [PubMed] [Google Scholar]
- Lesuisse E., Labbe P., 1994. Reductive iron assimilation in Saccharomyces cerevisiae, pp. 149–178 in Metal Ions in Fungi, edited by G. Winkelmann, Winge D. R. Marcel Dekker, New York. [Google Scholar]
- Lesuisse E., Raguzzi F., Crichton R. R., 1987. Iron uptake by the yeast Saccharomyces cerevisiae: involvement of a reduction step. J. Gen. Microbiol. 133: 3229–3236. [DOI] [PubMed] [Google Scholar]
- Lesuisse E., Simon-Casteras M., Labbe P., 1998. Siderophore-mediated iron uptake in Saccharomyces cerevisiae: the SIT1 gene encodes a ferrioxamine B permease that belongs to the major facilitator superfamily. Microbiology 144: 3455–3462. [DOI] [PubMed] [Google Scholar]
- Lesuisse E., Santos R., Matzanke B. F., Knight S. A., Camadro J. M., et al. , 2003. Iron use for haeme synthesis is under control of the yeast frataxin homologue (Yfh1). Hum. Mol. Genet. 12: 879–889. [DOI] [PubMed] [Google Scholar]
- Levin D. E., 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189: 1145–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Mapolelo D. T., Dingra N. N., Naik S. G., Lees N. S., et al. , 2009. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 48: 9569–9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Saxena S., Pain D., Dancis A., 2001a Adrenodoxin reductase homolog (Arh1p) of yeast mitochondria required for iron homeostasis. J. Biol. Chem. 276: 1503–1509. [DOI] [PubMed] [Google Scholar]
- Li L., Kaplan J., 1998. Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J. Biol. Chem. 273: 22181–22187. [DOI] [PubMed] [Google Scholar]
- Li L., Chen O. S., McVey Ward D., Kaplan J., 2001b CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 276: 29515–29519. [DOI] [PubMed] [Google Scholar]
- Li L., Bagley D., Ward D. M., Kaplan J., 2008. Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol. Cell. Biol. 28: 1326–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. C., Kane P. M., 2009. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim. Biophys. Acta 1793: 650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., 2009. Function and biogenesis of iron-sulphur proteins. Nature 460: 831–838. [DOI] [PubMed] [Google Scholar]
- Lill R., Muhlenhoff U., 2008. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77: 669–700. [DOI] [PubMed] [Google Scholar]
- Lim P. H., Pisat N. P., Gadhia N., Pandey A., Donovan F. X., et al. , 2011. Regulation of Alr1 Mg transporter activity by intracellular magnesium. PLoS ONE 6: e20896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. J., Pufahl R. A., Dancis A., O’Halloran T. V., Culotta V. C., 1997. A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J. Biol. Chem. 272: 9215–9220. [PubMed] [Google Scholar]
- Liu X. F., Culotta V. C., 1999. Post-translation control of Nramp metal transport in yeast. Role of metal ions and the BSD2 gene. J. Biol. Chem. 274: 4863–4868. [DOI] [PubMed] [Google Scholar]
- Liu X. F., Supek F., Nelson N., Culotta V. C., 1997. Negative control of heavy metal uptake by the Saccharomyces cerevisiae BSD2 gene. J. Biol. Chem. 272: 11763–11769. [DOI] [PubMed] [Google Scholar]
- Locke E. G., Bonilla M., Liang L., Takita Y., Cunningham K. W., 2000. A homolog of voltage-gated Ca(2+) channels stimulated by depletion of secretory Ca(2+) in yeast. Mol. Cell. Biol. 20: 6686–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., et al. , 2004. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304: 1644–1647. [DOI] [PubMed] [Google Scholar]
- Loukin S., Kung C., 1995. Manganese effectively supports yeast cell-cycle progression in place of calcium. J. Cell Biol. 131: 1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukin S. H., Saimi Y., 2002. Carboxyl tail prevents yeast K(+) channel closure: proposal of an integrated model of TOK1 gating. Biophys. J. 82: 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk E. E., Culotta V. C., 2001. Manganese superoxide dismutase in Saccharomyces cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transporter, Smf2p. J. Biol. Chem. 276: 47556–47562. [DOI] [PubMed] [Google Scholar]
- Lyons T. J., Gasch A. P., Gaither L. A., Botstein D., Brown P. O., et al. , 2000. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 97: 7957–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid C. W., Gaither L. A., Eide D., 2000. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19: 2845–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid C. W., Milanick M. A., Eide D. J., 2003. Induction of the ZRC1 metal tolerance gene in zinc-limited yeast confers resistance to zinc shock. J. Biol. Chem. 278: 15065–15072. [DOI] [PubMed] [Google Scholar]
- Madrid R., Gomez M. J., Ramos J., Rodriguez-Navarro A., 1998. Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J. Biol. Chem. 273: 14838–14844. [DOI] [PubMed] [Google Scholar]
- Maguire M. E., 2006. Magnesium transporters: properties, regulation and structure. Front. Biosci. 11: 3149–3163. [DOI] [PubMed] [Google Scholar]
- Malkus P., Graham L. A., Stevens T. H., Schekman R., 2004. Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol. Biol. Cell 15: 5075–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresova L., Sychrová H., 2005. Physiological characterization of Saccharomyces cerevisiae kha1 deletion mutants. Mol. Microbiol. 55: 588–600. [DOI] [PubMed] [Google Scholar]
- Maresova L., Urbankova E., Gaskova D., Sychrová H., 2006. Measurements of plasma membrane potential changes in Saccharomyces cerevisiae cells reveal the importance of the Tok1 channel in membrane potential maintenance. FEMS Yeast Res. 6: 1039–1046. [DOI] [PubMed] [Google Scholar]
- Maresova L., Hoskova B., Urbankova E., Chaloupka R., Sychrová H., 2010. New applications of pHluorin–measuring intracellular pH of prototrophic yeasts and determining changes in the buffering capacity of strains with affected potassium homeostasis. Yeast 27: 317–325. [DOI] [PubMed] [Google Scholar]
- Martin D. C., Kim H., Mackin N. A., Maldonado-Baez L., Evangelista C. C., Jr, et al. , 2011. New regulators of a high affinity Ca2+ influx system revealed through a genome-wide screen in yeast. J. Biol. Chem. 286: 10744–10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P., Persson B. L., 1998. Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol. Gen. Genet. 258: 628–638. [DOI] [PubMed] [Google Scholar]
- Martinez-Munoz G. A., Kane P., 2008. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 283: 20309–20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins L. J., Jensen L. T., Simon J. R., Keller G. L., Winge D. R., et al. , 1998. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. [published erratum appears in J Biol Chem 1998 Nov 6;273(45):30056] J. Biol. Chem. 273: 23716–23721. [DOI] [PubMed] [Google Scholar]
- Matheos D., Kingsbury T., Ahsan U., Cunningham K., 1997. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 11: 3445–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T. K., Ellsmore A. J., Cessna S. G., Low P. S., Pardo J. M., et al. , 2002. An osmotically induced cytosolic Ca2+ transient activates calcineurin signaling to mediate ion homeostasis and salt tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 277: 33075–33080. [DOI] [PubMed] [Google Scholar]
- McCusker J. H., Perlin D., Haber J. E., 1987. Pleiotropic plasma membrane ATPase mutations of Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 4082–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S., Zhang J., 2011. Reporting from the field: genetically encoded fluorescent reporters uncover signaling dynamics in living biological systems. Annu. Rev. Biochem. 80: 375–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza I., Rubio F., Rodriguez-Navarro A., Pardo J. M., 1994. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 269: 8792–8796. [PubMed] [Google Scholar]
- Miseta A., Kellermayer R., Aiello D. P., Fu L., Bedwell D. M., 1999. The vacuolar Ca2+/H+ exchanger Vcx1p/Hum1p tightly controls cytosolic Ca2+ levels in S. cerevisiae. FEBS Lett. 451: 132–136. [DOI] [PubMed] [Google Scholar]
- Mitsui K., Kamauchi S., Nakamura N., Inoue H., Kanazawa H., 2004. A conserved domain in the tail region of the Saccharomyces cerevisiae Na+/H+ antiporter (Nha1p) plays important roles in localization and salinity-resistant cell-growth. J. Biochem. 135: 139–148. [DOI] [PubMed] [Google Scholar]
- Mitsui K., Yasui H., Nakamura N., Kanazawa H., 2005. Oligomerization of the Saccharomyces cerevisiae Na+/H+ antiporter Nha1p: implications for its antiporter activity. Biochim. Biophys. Acta 1720: 125–136. [DOI] [PubMed] [Google Scholar]
- Miyakawa T., Mizunuma M., 2007. Physiological roles of calcineurin in Saccharomyces cerevisiae with special emphasis on its roles in G2/M cell-cycle regulation. Biosci. Biotechnol. Biochem. 71: 633–645. [DOI] [PubMed] [Google Scholar]
- Moore R. E., Kim Y., Philpott C. C., 2003. The mechanism of ferrichrome transport through Arn1p and its metabolism in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100: 5664–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M. J., Geiser J. R., Davis T. N., 1996. Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol. Cell. Biol. 16: 4824–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff U., Gerber J., Richhardt N., Lill R., 2003a Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 22: 4815–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff U., Stadler J. A., Richhardt N., Seubert A., Eickhorst T., et al. , 2003b A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J. Biol. Chem. 278: 40612–40620. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U., Molik S., Godoy J. R., Uzarska M. A., Richter N., et al. , 2010. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 12: 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E. M., Mackin N. A., Erdman S. E., Cunningham K. W., 2003. Fig1p facilitates Ca2+ influx and cell fusion during mating of Saccharomyces cerevisiae. J. Biol. Chem. 278: 38461–38469. [DOI] [PubMed] [Google Scholar]
- Muller M. R., Sasaki Y., Stevanovic I., Lamperti E. D., Ghosh S., et al. , 2009. Requirement for balanced Ca/NFAT signaling in hematopoietic and embryonic development. Proc. Natl. Acad. Sci. USA 106: 7034–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn A. L., Riezman H., 1994. Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J. Cell Biol. 127: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima-Shimada J., Iida H., Tsuji F. I., Anraku Y., 1991. Monitoring of intracellular calcium in Saccharomyces cerevisiae with an apoaequorin cDNA expression system. Proc. Natl. Acad. Sci. USA 88: 6878–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass R., Rao R., 1998. Novel localization of a Na+/H+ exchanger in a late endosomal compartment of yeast. J. Biol. Chem. 273: 21054–21060. [DOI] [PubMed] [Google Scholar]
- Nass R., Cunningham K. W., Rao R., 1997. Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase. J. Biol. Chem. 272: 26145–26152. [DOI] [PubMed] [Google Scholar]
- Navon G., Shulman R. G., Yamane T., Eccleshall T. R., Lam K. B., et al. , 1979. Phosphorus-31 nuclear magnetic resonance studies of wild-type and glycolytic pathway mutants of Saccharomyces cerevisiae. Biochemistry 18: 4487–4499. [DOI] [PubMed] [Google Scholar]
- Neale A. D., Scopes R. K., Kelly J. M., Wettenhall R. E., 1986. The two alcohol dehydrogenases of Zymomonas mobilis. Purification by differential dye ligand chromatography, molecular characterisation and physiological roles. Eur. J. Biochem. 154: 119–124. [DOI] [PubMed] [Google Scholar]
- Netz D. J., Pierik A. J., Stumpfig M., Muhlenhoff U., Lill R., 2007. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 3: 278–286. [DOI] [PubMed] [Google Scholar]
- Netz D. J., Stumpfig M., Dore C., Muhlenhoff U., Pierik A. J., et al. , 2010. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 6: 758–765. [DOI] [PubMed] [Google Scholar]
- Nevitt T., Ohrvik H., Thiele D. J., 2012. Charting the travels of copper in eukaryotes from yeast to mammals. Biochim. Biophys. Acta 1823: 1580–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles B. J., Mogri H., Hill A., Vlahakis A., Powers T., 2012. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc. Natl. Acad. Sci. USA 109: 1536–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B., Owsianik G., 2011. The transient receptor potential family of ion channels. Genome Biol. 12: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nittis T., George G. N., Winge D. R., 2001. Yeast Sco1, a protein essential for cytochrome c oxidase function is a Cu(I)-binding protein. J. Biol. Chem. 276: 42520–42526. [DOI] [PubMed] [Google Scholar]
- Nowikovsky K., Pozzan T., Rizzuto R., Scorrano L., Bernardi P., 2012. Perspectives on: SGP symposium on mitochondrial physiology and medicine: the pathophysiology of LETM1. J. Gen. Physiol. 139: 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki R., Nakamura N., Mitsui K., Kanazawa H., 2005. Characterization of the ion transport activity of the budding yeast Na+/H+ antiporter, Nha1p, using isolated secretory vesicles. Biochim. Biophys. Acta 1712: 185–196. [DOI] [PubMed] [Google Scholar]
- Ohnuki S., Nogami S., Kanai H., Hirata D., Nakatani Y., et al. , 2007. Diversity of Ca2+-induced morphology revealed by morphological phenotyping of Ca2+-sensitive mutants of Saccharomyces cerevisiae. Eukaryot. Cell 6: 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya Y., Miyamoto S., Ohsumi Y., Anraku Y., 1986a Calcium-sensitive cls4 mutant of Sacchormyces cerevisiae with a defect in bud formation. J. Bacteriol. 165: 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya Y., Ohsumi Y., Anraku Y., 1986b Isolation and characterization of Ca2+-sensitive mutants of Saccharomyces cerevisiae. J. Gen. Microbiol. 132: 979–988. [DOI] [PubMed] [Google Scholar]
- Ojeda L., Keller G., Muhlenhoff U., Rutherford J. C., Lill R., et al. , 2006. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 281: 17661–17669. [DOI] [PubMed] [Google Scholar]
- Ooi C. E., Rabinovich E., Dancis A., Bonifacino J. S., Klausner R. D., 1996. Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctr1p in the apparent absence of endocytosis. EMBO J. 15: 3515–3523. [PMC free article] [PubMed] [Google Scholar]
- Orij R., Brul S., Smits G. J., 2011. Intracellular pH is a tightly controlled signal in yeast. Biochim. Biophys. Acta 1810: 933–944. [DOI] [PubMed] [Google Scholar]
- Orij R., Postmus J., Ter Beek A., Brul S., Smits G. J., 2009. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 155: 268–278. [DOI] [PubMed] [Google Scholar]
- Pachkov M., Erb I., Molina N., van Nimwegen E., 2007. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 35: D127–D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M. J., Di Cera E., 2006. Role of Na+ and K+ in enzyme function. Physiol. Rev. 86: 1049–1092. [DOI] [PubMed] [Google Scholar]
- Paidhungat M., Garrett S., 1997. A homolog of mammalian, voltage-gated calcium channels mediates yeast pheromone-stimulated Ca2+ uptake and exacerbates the cdc1(Ts) growth defect. Mol. Cell. Biol. 17: 6339–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C. P., Zhou X. L., Lin J., Loukin S. H., Kung C., et al. , 2001. A TRP homolog in Saccharomyces cerevisiae forms an intracellular Ca(2+)-permeable channel in the yeast vacuolar membrane. Proc. Natl. Acad. Sci. USA 98: 7801–7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren M. G., Nissen P., 2011. P-type ATPases. Annu Rev Biophys 40: 243–266. [DOI] [PubMed] [Google Scholar]
- Pattison-Granberg J., Persson B. L., 2000. Regulation of cation-coupled high-affinity phosphate uptake in the yeast Saccharomyces cerevisiae. J. Bacteriol. 182: 5017–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters T., Louwet W., Geladé R., Nauwelaers D., Thevelein J. M., et al. , 2006. Kelch-repeat proteins interacting with the Galpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc. Natl. Acad. Sci. USA 103: 13034–13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Valle J., Jenkins H., Merchan S., Montiel V., Ramos J., et al. , 2007. Key role for intracellular K+ and protein kinases Sat4/Hal4 and Hal5 in the plasma membrane stabilization of yeast nutrient transporters. Mol. Cell. Biol. 27: 5725–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin D., Brown C., Haber J. E., 1988. Membrane potential defect in hygromycin B-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem. 263: 18118–18122. [PubMed] [Google Scholar]
- Petrezselyova S., Kinclova-Zimmermannova O., Sychrová H., 2012. Vhc1, a novel transporter belonging to the family of electroneutral cation-Cl(-) cotransporters, participates in the regulation of cation content and morphology of Saccharomyces cerevisiae vacuoles. Biochim. Biophys. Acta 1828: 623–631. [DOI] [PubMed] [Google Scholar]
- Pezzati R., Bossi M., Podini P., Meldolesi J., Grohovaz F., 1997. High-resolution calcium mapping of the endoplasmic reticulum-Golgi-exocytic membrane system. Electron energy loss imaging analysis of quick frozen-freeze dried PC12 cells. Mol. Biol. Cell 8: 1501–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalip V., Kuhn I., Lemoine Y., Jeltsch J. M., 1999. Characterization of the biotin biosynthesis pathway in Saccharomyces cerevisiae and evidence for a cluster containing BIO5, a novel gene involved in vitamer uptake. Gene 232: 43–51. [DOI] [PubMed] [Google Scholar]
- Philpott C., Smith P., 2013. The iron starvation response in Saccharomyces cerevisiae, Metals in Cells, edited by R.Scott, Culotta V. C., The Encyclopedia of Inorganic Biochemistry John Wiley & Sons, New York. [Google Scholar]
- Philpott C. C., 2006. Iron uptake in fungi: a system for every source. Biochim. Biophys. Acta 1763: 636–645. [DOI] [PubMed] [Google Scholar]
- Philpott C. C., Protchenko O., 2008. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 7: 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina F. J., O’Donnell A. F., Pagant S., Piao H. L., Miller J. P., et al. , 2011. Hph1 and Hph2 are novel components of the Sec63/Sec62 posttranslational translocation complex that aid in vacuolar proton ATPase biogenesis. Eukaryot. Cell 10: 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisat N. P., Pandey A., MacDiarmid C. W., 2009. MNR2 regulates intracellular magnesium storage in Saccharomyces cerevisiae. Genetics 183: 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman J. K., Cheng N. H., Shigaki T., Kunta M., Hirschi K. D., 2004. Functional dependence on calcineurin by variants of the Saccharomyces cerevisiae vacuolar Ca2+/H+ exchanger Vcx1p. Mol. Microbiol. 54: 1104–1116. [DOI] [PubMed] [Google Scholar]
- Portillo F., Serrano R., 1989. Growth control strength and active site of yeast plasma membrane ATPase studied by site-directed mutagenesis. Eur. J. Biochem. 186: 501–507. [DOI] [PubMed] [Google Scholar]
- Portillo F., Eraso P., Serrano R., 1991. Analysis of the regulatory domain of yeast plasma membrane H+-ATPase by directed mutagenesis and intragenic suppression. FEBS Lett. 287: 71–74. [DOI] [PubMed] [Google Scholar]
- Portnoy M. E., Liu X. F., Culotta V. C., 2000. Saccharomyces cerevisiae expresses three functionally distinct homologues of the nramp family of metal transporters. Mol. Cell. Biol. 20: 7893–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy M. E., Schmidt P. J., Rogers R. S., Culotta V. C., 2001. Metal transporters that contribute copper to metallochaperones in Saccharomyces cerevisiae. Mol. Genet. Genomics 265: 873–882. [DOI] [PubMed] [Google Scholar]
- Portnoy M. E., Jensen L. T., Culotta V. C., 2002. The distinct methods by which manganese and iron regulate the Nramp transporters in yeast. Biochem. J. 362: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozos T. C., 1998. HUM1, a Novel Yeast Gene, Is Required for Vacuolar Calcium/Proton Exchange in S. cerevisiae, p. 130. Stanford University, Stanford, CA. [Google Scholar]
- Pozos T. C., Sekler I., Cyert M. S., 1996. The product of HUM1, a novel yeast gene, is required for vacuolar Ca2+/H+ exchange and is related to mammalian Na+/Ca2+ exchangers. Mol. Cell. Biol. 16: 3730–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M., Struhl K., 2004. MAP kinase-mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell 118: 351–361. [DOI] [PubMed] [Google Scholar]
- Protchenko O., Ferea T., Rashford J., Tiedeman J., Brown P. O., et al. , 2001. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J. Biol. Chem. 276: 49244–49250. [DOI] [PubMed] [Google Scholar]
- Protchenko O., Philpott C. C., 2003. Regulation of intracellular heme levels by HMX1, a homologue of heme oxygenase, in Saccharomyces cerevisiae. J. Biol. Chem. 278: 36582–36587. [DOI] [PubMed] [Google Scholar]
- Pufahl R. A., Singer C. P., Peariso K. L., Lin S. J., Schmidt P. J., et al. , 1997. Metal ion chaperone function of the soluble Cu(I) receptor Atx1. [see comments] Science 278: 853–856. [DOI] [PubMed] [Google Scholar]
- Puig S., Lee J., Lau M., Thiele D. J., 2002. Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J. Biol. Chem. 277: 26021–26030. [DOI] [PubMed] [Google Scholar]
- Puig S., Askeland E., Thiele D. J., 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120: 99–110. [DOI] [PubMed] [Google Scholar]
- Puig S., Vergara S. V., Thiele D. J., 2008. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 7: 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol-Carrion N., Belli G., Herrero E., Nogues A., de la Torre-Ruiz M. A., 2006. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J. Cell Sci. 119: 4554–4564. [DOI] [PubMed] [Google Scholar]
- Qiu Q. S., Fratti R. A., 2010. The Na+/H+ exchanger Nhx1p regulates the initiation of Saccharomyces cerevisiae vacuole fusion. J. Cell Sci. 123: 3266–3275. [DOI] [PubMed] [Google Scholar]
- Rae T. D., Schmidt P. J., Pufahl R. A., Culotta V. C., O’Halloran T. V., 1999. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284: 805–808. [DOI] [PubMed] [Google Scholar]
- Ramazzotti A., Vanmansart V., Foury F., 2004. Mitochondrial functional interactions between frataxin and Isu1p, the iron-sulfur cluster scaffold protein, in Saccharomyces cerevisiae. FEBS Lett. 557: 215–220. [DOI] [PubMed] [Google Scholar]
- Reddi A. R., Jensen L. T., Culotta V. C., 2009a Manganese homeostasis in Saccharomyces cerevisiae. Chem. Rev. 109: 4722–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi A. R., Jensen L. T., Naranuntarat A., Rosenfeld L., Leung E., et al. , 2009b The overlapping roles of manganese and Cu/Zn SOD in oxidative stress protection. Free Radic. Biol. Med. 46: 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E. M., Thiele D. J., 2007. Identification of a vacuole-associated metalloreductase and its role in Ctr2-mediated intracellular copper mobilization. J. Biol. Chem. 282: 21629–21638. [DOI] [PubMed] [Google Scholar]
- Rentzsch A., Krummeck-Weiss G., Hofer A., Bartuschka A., Ostermann K., et al. , 1999. Mitochondrial copper metabolism in yeast: mutational analysis of Sco1p involved in the biogenesis of cytochrome c oxidase. Curr. Genet. 35: 103–108. [DOI] [PubMed] [Google Scholar]
- Rigby K., Cobine P. A., Khalimonchuk O., Winge D. R., 2008. Mapping the functional interaction of Sco1 and Cox2 in cytochrome oxidase biogenesis. J. Biol. Chem. 283: 15015–15022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivetta A., Kuroda T., Slayman C., 2011. Anion currents in yeast K+ transporters (TRK) characterize a structural homologue of ligand-gated ion channels. Pflugers Arch. 462: 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N. J., Winge D. R., 2010. Copper metallochaperones. Annu. Rev. Biochem. 79: 537–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro A., Ramos J., 1984. Dual system for potassium transport in Saccharomyces cerevisiae. J. Bacteriol. 159: 940–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncoroni M., Santiago M., Hooks D. O., Moroney S., Harsch M. J., et al. , 2011. The yeast IRC7 gene encodes a beta-lyase responsible for production of the varietal thiol 4-mercapto-4-methylpentan-2-one in wine. Food Microbiol. 28: 926–935. [DOI] [PubMed] [Google Scholar]
- Rossio V., Yoshida S., 2011. Spatial regulation of Cdc55–PP2A by Zds1/Zds2 controls mitotic entry and mitotic exit in budding yeast. J. Cell Biol. 193: 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon A., Barbey R., Patton E. E., Tyers M., Thomas D., 2000. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30)complex. EMBO J. 19: 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Solodovnikova N., Nicholson T., Antholine W., Walden W. E., 2003. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22: 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph H. K., Antebi A., Fink G. R., Buckley C. M., Dorman T. E., et al. , 1989. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell 58: 133–145. [DOI] [PubMed] [Google Scholar]
- Ruiz A., Serrano R., Arino J., 2008. Direct regulation of genes involved in glucose utilization by the calcium/calcineurin pathway. J. Biol. Chem. 283: 13923–13933. [DOI] [PubMed] [Google Scholar]
- Rutherford J. C., Jaron S., Ray E., Brown P. O., Winge D. R., 2001. A second iron-regulatory system in yeast independent of Aft1p. Proc. Natl. Acad. Sci. USA 98: 14322–14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford J. C., Jaron S., Winge D. R., 2003. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 19: 19. [DOI] [PubMed] [Google Scholar]
- Rutherford J. C., Ojeda L., Balk J., Muhlenhoff U., Lill R., et al. , 2005. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 280: 10135–10140. [DOI] [PubMed] [Google Scholar]
- Ryan M., Graham L. A., Stevens T. H., 2008. Voa1p functions in V-ATPase assembly in the yeast endoplasmic reticulum. Mol. Biol. Cell 19: 5131–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saimi Y., Martinac B., Delcour A. H., Minorsky P. V., Gustin M. C., et al. , 1992. Patch clamp studies of microbial ion channels. Methods Enzymol. 207: 681–691. [DOI] [PubMed] [Google Scholar]
- Schindl R., Weghuber J., Romanin C., Schweyen R. J., 2007. Mrs2p forms a high conductance Mg2+ selective channel in mitochondria. Biophys. J. 93: 3872–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Haas H., 2011. Iron homeostasis–Achilles’ heel of Aspergillus fumigatus? Curr. Opin. Microbiol. 14: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze M., Rodel G., 1988. SCO1, a yeast nuclear gene essential for accumulation of mitochondrial cytochrome c oxidase subunit II. Mol. Gen. Genet. 211: 492–498. [DOI] [PubMed] [Google Scholar]
- Serpe M., Joshi A., Kosman D. J., 1999. Structure-function analysis of the protein-binding domains of Mac1p, a copper-dependent transcriptional activator of copper uptake in Saccharomyces cerevisiae. J. Biol. Chem. 274: 29211–29219. [DOI] [PubMed] [Google Scholar]
- Serrano R., 1983. In vivo glucose activation of the yeast plasma membrane ATPase. FEBS Lett. 156: 11–14. [DOI] [PubMed] [Google Scholar]
- Seto-Young D., Perlin D. S., 1991. Effect of membrane voltage on the plasma membrane H(+)-ATPase of Saccharomyces cerevisiae. J. Biol. Chem. 266: 1383–1389. [PubMed] [Google Scholar]
- Shakoury-Elizeh M., Tiedeman J., Rashford J., Ferea T., Demeter J., et al. , 2004. Transcriptional Remodeling in Response to Iron Deprivation in Saccharomyces cerevisiae. Mol. Biol. Cell 15: 1233–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakoury-Elizeh M., Protchenko O., Berger A., Cox J., Gable K., et al. , 2010. Metabolic response to iron deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 285: 14823–14833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., et al. , 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 100: 13207–13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon E., Barcelo A., Arino J., 2003. Mutagenesis analysis of the yeast Nha1 Na+/H+ antiporter carboxy-terminal tail reveals residues required for function in cell cycle. FEBS Lett. 545: 239–245. [DOI] [PubMed] [Google Scholar]
- Simon E., Clotet J., Calero F., Ramos J., Arino J., 2001. A screening for high copy suppressors of the sit4 hal3 synthetically lethal phenotype reveals a role for the yeast Nha1 antiporter in cell cycle regulation. J. Biol. Chem. 276: 29740–29747. [DOI] [PubMed] [Google Scholar]
- Singh A., Kaur N., Kosman D. J., 2007. The metalloreductase Fre6p in Fe-efflux from the yeast vacuole. J. Biol. Chem. 282: 28619–28626. [DOI] [PubMed] [Google Scholar]
- Smardon A. M., Tarsio M., Kane P. M., 2002. The RAVE complex is essential for stable assembly of the yeast V-ATPase. J. Biol. Chem. 277: 13831–13839. [DOI] [PubMed] [Google Scholar]
- Smothers D. B., Kozubowski L., Dixon C., Goebl M. G., Mathias N., 2000. The abundance of Met30p limits SCF(Met30p) complex activity and is regulated by methionine availability. Mol. Cell. Biol. 20: 7845–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Cardalda A., Fakas S., Pascual F., Choi H. S., Carman G. M., 2012. Phosphatidate phosphatase plays role in zinc-mediated regulation of phospholipid synthesis in yeast. J. Biol. Chem. 287: 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizzo T., Byersdorfer C., Duesterhoeft S., Eide D., 1997. The yeast FET5 gene encodes a FET3-related multicopper oxidase implicated in iron transport. Mol. Gen. Genet. 256: 547–556. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A. M., Cyert M. S., 1997. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11: 3432–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearman R., Yuan D. S., Yamaguchi-Iwai Y., Klausner R. D., Dancis A., 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. [see comments] Science 271: 1552–1557. [DOI] [PubMed] [Google Scholar]
- Stevens B., 1977. Variation in number and volume of mitochondria in yeast according to growth-conditions: study based on serial sectioning and computer graphics reconstitution. Biol. Cell. 28: 37–56. [Google Scholar]
- Stimpson H. E., Lewis M. J., Pelham H. R., 2006. Transferrin receptor-like proteins control the degradation of a yeast metal transporter. EMBO J. 25: 662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz J., Hoja U., Meier S., Sauer N., Schweizer E., 1999. Identification of the plasma membrane H+-biotin symporter of Saccharomyces cerevisiae by rescue of a fatty acid-auxotrophic mutant. J. Biol. Chem. 274: 18741–18746. [DOI] [PubMed] [Google Scholar]
- Strayle J., Pozzan T., Rudolph H. K., 1999. Steady-state free Ca(2+) in the yeast endoplasmic reticulum reaches only 10 microM and is mainly controlled by the secretory pathway pump pmr1. EMBO J. 18: 4733–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtz L. A., Diekert K., Jensen L. T., Lill R., Culotta V. C., 2001. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 276: 38084–38089. [DOI] [PubMed] [Google Scholar]
- Sudbrak R., Brown J., Dobson-Stone C., Carter S., Ramser J., et al. , 2000. Hailey-Hailey disease is caused by mutations in ATP2C1 encoding a novel Ca(2+) pump. Hum. Mol. Genet. 9: 1131–1140. [DOI] [PubMed] [Google Scholar]
- Sullivan J. A., Lewis M. J., Nikko E., Pelham H. R., 2007. Multiple interactions drive adaptor-mediated recruitment of the ubiquitin ligase rsp5 to membrane proteins in vivo and in vitro. Mol. Biol. Cell 18: 2429–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Supekova L., Nelson H., Nelson N., 1996. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc. Natl. Acad. Sci. USA 93: 5105–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P., Wach A., Thines-Sempoux D., Goffeau A., 1993. Proliferation of intracellular structures upon overexpression of the PMA2 ATPase in Saccharomyces cerevisiae. J. Biol. Chem. 268: 19744–19752. [PubMed] [Google Scholar]
- Szczypka M. S., Thiele D. J., 1989. A cysteine-rich nuclear protein activates yeast metallothionein gene transcription. Mol. Cell. Biol. 9: 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsio M., Zheng H., Smardon A. M., Martinez-Munoz G. A., Kane P. M., 2011. Consequences of loss of Vph1 protein-containing vacuolar ATPases (V-ATPases) for overall cellular pH homeostasis. J. Biol. Chem. 286: 28089–28096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng J., Goto R., Iida K., Kojima I., Iida H., 2008. Ion-channel blocker sensitivity of voltage-gated calcium-channel homologue Cch1 in Saccharomyces cerevisiae. Microbiology 154: 3775–3781. [DOI] [PubMed] [Google Scholar]
- Thevelein J. M., de Winde J. H., 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33: 904–918. [DOI] [PubMed] [Google Scholar]
- Thiele D. J., 1988. ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol. Cell. Biol. 8: 2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen J. L., Sewell A. K., Tanner A. M., Peltier J. M., Pickering I. J., et al. , 1994. Mixed Cu+ and Zn2+ coordination in the DNA-binding domain of the AMT1 transcription factor from Candida glabrata. Biochemistry 33: 9566–9577. [DOI] [PubMed] [Google Scholar]
- Tisi R., Baldassa S., Belotti F., Martegani E., 2002. Phospholipase C is required for glucose-induced calcium influx in budding yeast. FEBS Lett. 520: 133–138. [DOI] [PubMed] [Google Scholar]
- Tisi R., Belotti F., Wera S., Winderickx J., Thevelein J. M., et al. , 2004. Evidence for inositol triphosphate as a second messenger for glucose-induced calcium signalling in budding yeast. Curr. Genet. 45: 83–89. [DOI] [PubMed] [Google Scholar]
- Tropia M. J., Cardoso A. S., Tisi R., Fietto L. G., Fietto J. L., et al. , 2006. Calcium signaling and sugar-induced activation of plasma membrane H(+)-ATPase in Saccharomyces cerevisiae cells. Biochem. Biophys. Res. Commun. 343: 1234–1243. [DOI] [PubMed] [Google Scholar]
- Tsai C. L., Barondeau D. P., 2010. Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry 49: 9132–9139. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Capitanio N., Nobrega M. P., Gatti D., 1990. Cytochrome oxidase assembly in yeast requires the product of COX11, a homolog of the P. denitrificans protein encoded by ORF3. EMBO J. 9: 2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta R., Fujiwara N., Iwai K., Yamaguchi-Iwai Y., 2007. Mechanism underlying the iron-dependent nuclear export of the iron-responsive transcription factor Aft1p in Saccharomyces cerevisiae. Mol. Biol. Cell 18: 2980–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugulava N. B., Gibney B. R., Jarrett J. T., 2001. Biotin synthase contains two distinct iron-sulfur cluster binding sites: chemical and spectroelectrochemical analysis of iron-sulfur cluster interconversions. Biochemistry 40: 8343–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski J. L., Piper R. C., 1999. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 274: 38061–38070. [DOI] [PubMed] [Google Scholar]
- Vashist S., Frank C. G., Jakob C. A., Ng D. T., 2002. Two distinctly localized p-type ATPases collaborate to maintain organelle homeostasis required for glycoprotein processing and quality control. Mol. Biol. Cell 13: 3955–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladevall L., Serrano R., Ruiz A., Domenech G., Giraldo J., et al. , 2004. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 279: 43614–43624. [DOI] [PubMed] [Google Scholar]
- Waldron K. J., Rutherford J. C., Ford D., Robinson N. J., 2009. Metalloproteins and metal sensing. Nature 460: 823–830. [DOI] [PubMed] [Google Scholar]
- Wang Z., Feng L. S., Matskevich V., Venkataraman K., Parasuram P., et al. , 2006. Solution structure of a Zap1 zinc-responsive domain provides insights into metalloregulatory transcriptional repression in Saccharomyces cerevisiae. J. Mol. Biol. 357: 1167–1183. [DOI] [PubMed] [Google Scholar]
- Waters B. M., Eide D. J., 2002. Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J. Biol. Chem. 277: 33749–33757. [DOI] [PubMed] [Google Scholar]
- Wegner S. V., Sun F., Hernandez N., He C., 2011. The tightly regulated copper window in yeast. Chem. Commun. (Camb.) 47: 2571–2573. [DOI] [PubMed] [Google Scholar]
- Wiedemann N., Urzica E., Guiard B., Muller H., Lohaus C., et al. , 2006. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 25: 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenberger G., Steinleitner K., Malli R., Graier W. F., Vormann J., et al. , 2007. Mg2+ deprivation elicits rapid Ca2+ uptake and activates Ca2+/calcineurin signaling in Saccharomyces cerevisiae. Eukaryot. Cell 6: 592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Hart T., Wu X., Tatchell K., 2002. Protein phosphatase type 1 regulates ion homeostasis in Saccharomyces cerevisiae. Genetics 160: 1423–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge D. R., Nielson K. B., Gray W. R., Hamer D. H., 1985. Yeast metallothionein. Sequence and metal-binding properties. J. Biol. Chem. 260: 14464–14470. [PubMed] [Google Scholar]
- Withee J. L., Mulholland J., Jeng R., Cyert M. S., 1997. An essential role of the pheromone-dependent Ca2+ signal is to activate yeast calcineurin. Mol. Biol. Cell 8: 263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F. I., Trapani V., 2008. Cell (patho)physiology of magnesium. Clin. Sci. (Lond.) 114: 27–35. [DOI] [PubMed] [Google Scholar]
- Wood L. K., Thiele D. J., 2009. Transcriptional activation in yeast in response to copper deficiency involves copper-zinc superoxide dismutase. J. Biol. Chem. 284: 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Y., Bird A. J., Winge D. R., Eide D. J., 2007. Regulation of the yeast TSA1 peroxiredoxin by ZAP1 is an adaptive response to the oxidative stress of zinc deficiency. J. Biol. Chem. 282: 2184–2195. [DOI] [PubMed] [Google Scholar]
- Wu C. Y., Bird A. J., Chung L. M., Newton M. A., Winge D. R., et al. , 2008. Differential control of Zap1-regulated genes in response to zinc deficiency in Saccharomyces cerevisiae. BMC Genomics 9: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Y., Roje S., Sandoval F. J., Bird A. J., Winge D. R., et al. , 2009. Repression of sulfate assimilation is an adaptive response of yeast to the oxidative stress of zinc deficiency. J. Biol. Chem. 284: 27544–27556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. H., Frey A. G., Eide D. J., 2011. Transcriptional regulation of the Zrg17 zinc transporter of the yeast secretory pathway. Biochem. J. 435: 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Loughlin F., George G. N., Howlett G. J., Wedd A. G., 2004. C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking. J. Am. Chem. Soc. 126: 3081–3090. [DOI] [PubMed] [Google Scholar]
- Xue C., Hsueh Y. P., Heitman J., 2008. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol. Rev. 32: 1010–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y., Dancis A., Klausner R. D., 1995. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 14: 1231–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y., Stearman R., Dancis A., Klausner R. D., 1996. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 15: 3377–3384. [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y., Serpe M., Haile D., Yang W., Kosman D. J., et al. , 1997. Homeostatic regulation of copper uptake in yeast via direct binding of MAC1 protein to upstream regulatory sequences of FRE1 and CTR1. J. Biol. Chem. 272: 17711–17718. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y., Ueta R., Fukunaka A., Sasaki R., 2002. Subcellular localization of Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J. Biol. Chem. 277: 18914–18918. [DOI] [PubMed] [Google Scholar]
- Yenush L., Mulet J. M., Arino J., Serrano R., 2002. The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity and cell cycle progression. EMBO J. 21: 920–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenush L., Merchan S., Holmes J., Serrano R., 2005. pH-Responsive, posttranslational regulation of the Trk1 potassium transporter by the type 1-related Ppz1 phosphatase. Mol. Cell. Biol. 25: 8683–8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H., Zhang Y., Pain J., Lyver E. R., Lesuisse E., et al. , 2011. Rim2, a pyrimidine nucleotide exchanger, is needed for iron utilization in mitochondria. Biochem. J. 440: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto H., Saltsman K., Gasch A. P., Li H. X., Ogawa N., et al. , 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277: 31079–31088. [DOI] [PubMed] [Google Scholar]
- Young B. P., Shin J. J., Orij R., Chao J. T., Li S. C., et al. , 2010. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 329: 1085–1088. [DOI] [PubMed] [Google Scholar]
- Yuan D. S., 2000. Zinc-regulated genes in Saccharomyces cerevisiae revealed by transposon tagging. Genetics 156: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D. S., Stearman R., Dancis A., Dunn T., Beeler T., et al. , 1995. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc. Natl. Acad. Sci. USA 92: 2632–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C. W., Ferea T., Rashford J., Ardon O., Brown P. O., et al. , 2000a Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae. Evidence for two pathways of iron uptake. J. Biol. Chem. 275: 10709–10715. [DOI] [PubMed] [Google Scholar]
- Yun C. W., Tiedeman J. S., Moore R. E., Philpott C. C., 2000b Siderophore-iron uptake in saccharomyces cerevisiae. Identification of ferrichrome and fusarinine transporters. J. Biol. Chem. 275: 16354–16359. [DOI] [PubMed] [Google Scholar]
- Yun C. W., Bauler M., Moore R. E., Klebba P. E., Philpott C. C., 2001. The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J. Biol. Chem. 276: 10218–10223. [DOI] [PubMed] [Google Scholar]
- Zahradka J., Sychrová H., 2012. Plasma-membrane hyperpolarization diminishes the cation efflux via Nha1 antiporter and Ena ATPase under potassium-limiting conditions. FEMS Yeast Res. 12: 439–446. [DOI] [PubMed] [Google Scholar]
- Zahradka J., van Heusden G. P., Sychrová H., 2012. Yeast 14–3-3 proteins participate in the regulation of cell cation homeostasis via interaction with Nha1 alkali-metal-cation/proton antiporter. Biochim. Biophys. Acta 1820: 849–858. [DOI] [PubMed] [Google Scholar]
- Zakrzewska A., Boorsma A., Brul S., Hellingwerf K. J., Klis F. M., 2005. Transcriptional response of Saccharomyces cerevisiae to the plasma membrane-perturbing compound chitosan. Eukaryot. Cell 4: 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gladyshev V. N., 2011. Comparative genomics of trace element dependence in biology. J. Biol. Chem. 286: 23623–23629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lyver E. R., Nakamaru-Ogiso E., Yoon H., Amutha B., et al. , 2008. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol. Cell. Biol. 28: 5569–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Jung U. S., Garrett-Engele P., Roe T., Cyert M. S., et al. , 1998a Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 18: 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Eide D., 1996a The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 93: 2454–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Eide D., 1996b The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271: 23203–23210. [DOI] [PubMed] [Google Scholar]
- Zhao H., Eide D. J., 1997. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 5044–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Butler E., Rodgers J., Spizzo T., Duesterhoeft S., et al. , 1998b Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem. 273: 28713–28720. [DOI] [PubMed] [Google Scholar]
- Zhou X. L., Vaillant B., Loukin S. H., Kung C., Saimi Y., 1995. YKC1 encodes the depolarization-activated K+ channel in the plasma membrane of yeast. FEBS Lett. 373: 170–176. [DOI] [PubMed] [Google Scholar]
- Zhou X. L., Batiza A. F., Loukin S. H., Palmer C. P., Kung C., et al. , 2003. The transient receptor potential channel on the yeast vacuole is mechanosensitive. Proc. Natl. Acad. Sci. USA 100: 7105–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]