Abstract
Diploid sexual reproduction involves segregation of allelic pairs, ensuring equal representation of genotypes in the gamete pool. Some genes, however, are able to “cheat” the system by promoting their own transmission. The Segregation distorter (Sd) locus in Drosophila melanogaster males is one of the best-studied examples of this type of phenomenon. In this system the presence of Sd on one copy of chromosome 2 results in dysfunction of the non–Sd-bearing (Sd+) sperm and almost exclusive transmission of Sd to the next generation. The mechanism by which Sd wreaks such selective havoc has remained elusive. However, its effect requires a target locus on chromosome 2 known as Responder (Rsp). The Rsp locus comprises repeated copies of a satellite DNA sequence and Rsp copy number correlates with sensitivity to Sd. Under distorting conditions during spermatogenesis, nuclei with chromosomes containing greater than several hundred Rsp repeats fail to condense chromatin and are eliminated. Recently, Rsp sequences were found as small RNAs in association with Argonaute family proteins Aubergine (Aub) and Argonaute3 (AGO3). These proteins are involved in a germline-specific RNAi mechanism known as the Piwi-interacting RNA (piRNA) pathway, which specifically suppresses transposon activation in the germline. Here, we evaluate the role of piRNAs in segregation distortion by testing the effects of mutations to piRNA pathway components on distortion. Further, we specifically targeted mutations to the aub locus of a Segregation Distorter (SD) chromosome, using ends-out homologous recombination. The data herein demonstrate that mutations to piRNA pathway components act as enhancers of SD.
Keywords: meiotic drive, Piwi-interacting RNA (piRNA), Segregation Distorter (SD), PIWIs, RNAi
EVOLUTION of sexual organisms relies on the faithful segregation and transmission of alleles from one generation to the next, allowing unbiased exposure of these alleles to natural selection. Nevertheless, nature contains multiple examples of genes that violate this basic tenet of Mendelian inheritance and act selfishly to ensure their own propagation (Lyttle 1991). One such phenomenon, known as meiotic drive, occurs when one of two alleles alters the gametic ratio to enhance its own representation in the next generation, violating Mendel’s first law (Sandler and Novitski 1957). First discovered more than 50 years ago, Segregation Distorter (SD) in Drosophila melanogaster is one of the best-studied examples of this type of “selfish” genetic behavior (Sandler et al. 1959; Temin et al. 1991; Kusano et al. 2003).
Segregation Distorter chromosomes contain a dominant gain-of-function mutation that strongly favors the transmission of the SD chromosome from [SD/SD+] heterozygous males by causing dysfunction of wild-type (SD+) sperm (Sandler et al. 1959; Sandler and Hiraizumi 1960b; Hartl et al. 1967; Tokuyasu et al. 1977; Temin et al. 1991). While the SD/SD+ male transmits the SD chromosome to as many as 99% of his progeny, the gametes of heterozygous females show normal Mendelian segregation (Sandler and Hiraizumi 1959; Sandler et al. 1959; Burt and Trivers 2008; Larracuente and Presgraves 2012). The Segregation distorter (Sd) locus was mapped to the proximal euchromatin of chromosome 2L and identified as a truncated Ran GTPase Activating Protein (RanGAP) (Hartl 1974; McLean et al. 1994; Merrill et al. 1999). Formed by a tandem duplication event, this C-terminally truncated (shortened by 243 aa) version remains enzymatically active, but lacks part of a nuclear export signal (NES) and a sumoylation site required for docking at the nuclear pore (McLean et al. 1994; Merrill et al. 1999).
While wild-type RanGAP localizes to the cytoplasm, Sd-RanGAP is retained in the nucleus due to its truncated NES, potentially altering the GTP gradient required for Ran-mediated nuclear transport (Gorlich and Mattaj 1996; Gorlich and Kutay 1999b; Merrill et al. 1999; Kalab et al. 2002). In the proper genetic background, nuclear localization of even wild-type RanGAP is sufficient to cause distortion, suggesting that nuclear enzymatic activity of RanGAP causes abnormal nuclear retention of Ran cargo and sperm dysfunction in SD (Kusano et al. 2001, 2002).
The relative strength of Segregation Distorter chromosomes is dependent upon several modifiers distributed along the second chromosome (Figure 1A). The best studied of these, Enhancer of SD [E(SD)], is located in the h35 heterochromatic region of chromosome 2L (Ganetzky 1977; Brittnacher and Ganetzky 1984). The presence of the E(SD) locus not only strongly enhances drive, but also two doses of E(SD) result in the accumulation of wild-type RanGAP in the nucleus and low levels of distortion even in the absence of Sd (Temin 1991; Kusano et al. 2002).
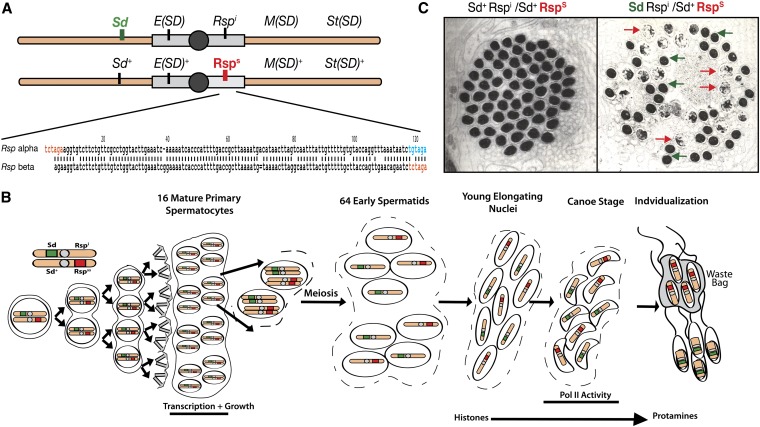
Figure 1.
Components of segregation distortion. (A) Top, schematic of D. melanogaster second chromosomes showing the relative locations of components of the Segregation Distorter system. Sd, E(Sd), Rspi, M(SD), and St(SD) all contribute to the drive phenotype while E(Sd)+, Rsps, M(SD)+, and St(SD)+ are wild-type loci and are normally found on nondistorting SD+ chromosomes. Bottom, an alignment of the α- and β-halves of the canonical Responder repeat sequence. The ∼120-bp α- and β-sequences are ∼87% homologous. In orange are the XbaI restriction sites that flank the 240-bp repeat. In blue is a mutated XbaI site that is found in at the 3′ end of the α-repeat. (B) An overview of SD in spermatogenesis: A germline stem cell (GSC) divides mitotically to produce a cyst of 16 mature primary spermatocytes. The spermatocytes enter a period of growth and increased transcription before entering meiosis to become 64 haploid early spermatids. The spermatids mature and condense, exchanging histones for protamines. Rsp-bearing nuclei fail to properly condense. The waste bag removes the uncondensed, Rsp-bearing spermatids during individualization and only Sd-bearing spermatids become mature sperm. (C) Wild-type and dysfunctional chromatin condensation. Left, an electron microscopy image of a cyst of 64 condensing spermatids from a wild-type male. Right, the same stage in the testis of the genotype Sd Rspi/Sd+ Rsps. Approximately half the nuclei fail to condense. (Reprinted from Tokuyasu et al. 1977 with permission from Elsevier.)
The other two loci, Stabilizer of SD [St(SD)] and Modifier of SD [M(SD)] are located on 2R. Both enhance drive of an SD chromosome, although their mechanisms of action are unknown (Sandler and Hiraizumi 1960a; Hiraizumi et al. 1980; Temin et al. 1991).
The sperm dysfunction observed in segregation distortion also depends on the allelic state of the target locus known as Responder (Rsp) (Hartl 1973, 1974). Rsp alleles range from completely insensitive (Rspi), which shows normal segregation in the presence of SD, to supersensitive (Rspss), which is almost completely eliminated in the presence of SD (Ganetzky 1977; Hiraizumi et al. 1980; Temin and Marthas 1984; Lyttle et al. 1986). Located in the heterochromatin of chromosome 2R, Responder is composed of an array of 120-bp satellite repeats. Repeat copy number correlates with sensitivity to SD (Wu et al. 1988; Pimpinelli and Dimitri 1989; Houtchens and Lyttle 2003). Rspss chromosomes are estimated to have several thousand repeats while the Rsps chromosome is estimated to have ∼700 copies (Wu et al. 1988). No completely insensitive SD+ chromosomes have been isolated from nature; however, a Rspi allele was generated by X-ray ablation of the Rsps locus (Ganetzky 1977). This Rspi allele is reported to have <20 remaining copies of Rsp (Wu et al. 1988). The loss of Rsps-containing nuclei occurs during the final stage of spermatogenesis when these nuclei fail to properly condense their chromatin, ensuring that the gametes produced carry the SD chromosome (Figure 1, B and C) (Tokuyasu et al. 1977).
The earliest models for the molecular mechanism of SD involved direct interaction between the Sd protein product and the Rsp locus (Hartl 1973). However, following the molecular characterization of these loci, this model appears unlikely. Later it was proposed that the mislocalization of Sd-RanGAP results in a defect in nuclear transport that prevents proper chromatin condensation by upsetting the balance of factors required for the transition from histones to protamines (a sperm-specific histone variant) (Kusano et al. 2001, 2003). In this model the mechanism that specifically targets only the Rsp-bearing sperm for destruction remains unclear. Understanding the function of the Rsp repeat in SD may be the key to unlocking the molecular mechanism of this phenomenon.
Several recent pieces of data have suggested a possible interaction between the Rsp repeat array and the germline-specific small RNA-based silencing system, known as the Piwi-interacting RNA (piRNA) pathway. This pathway is facilitated by a distinct subset of Argonaute family RNA slicer proteins in Drosophila, known as the PIWI clade (Carmell et al. 2002). These proteins, Piwi, Aubergine (Aub), and Argonaut3 (AGO3), are specifically expressed in gonads, where they utilize post-transcriptional gene silencing to ensure that transposons and other repetitive elements remain quiescent during gameteogenesis (Harris and MacDonald 2001; Brennecke et al. 2007; Gunawardane et al. 2007).
The piRNA pathway acts through a long single-stranded antisense RNA precursor that is cleaved to produce short sequences complementary to transposons or other targets. The Argonaute proteins use these short RNAs as guides to make an endonucleolytic cut in targeted transposon mRNAs, thus preventing transposition during gameteogenesis (Aravin et al. 2004, 2007; Brennecke et al. 2007; Gunawardane et al. 2007). This mechanism is distinct from both siRNA and miRNA generation in that it is germline specific; involves only Piwi family Argonautes; is Dicer independent; produces an unconventional length of small RNA (23–31 nt); and has a specific enrichment for noncoding, repetitive, and transposon-derived small RNAs (Brennecke et al. 2007; Gunawardane et al. 2007; Klattenhoff and Theurkauf 2008; Khurana and Theurkauf 2010; Senti and Brennecke 2010).
Given the role of the piRNA pathway in specifically suppressing repetitive elements in the germline, we asked whether the Responder array of satellite repeats could be a target of the piRNA pathway. In support of this hypothesis, Rsp sequence has been found as piRNA associated with both Aub and AGO3 in Drosophila testes (Nagao et al. 2010). Further, two other well-studied meiotic drive systems, the Stellate system in D. melanogaster and the Winters system in D. simulans, are thought to involve small RNA-based silencing mechanisms (Aravin et al. 2001, 2004; Tao et al. 2007a,b).
To evaluate the role of the piRNA pathway in SD, we utilized mutations of several components of the piRNA pathway and determined whether these mutations influenced the severity of the drive phenotype of SD. In addition, we have used ends-out homologous recombination to introduce mutations into the aubergine locus on an SD chromosome and assayed the effects of this mutation on distortion. These studies reveal that mutations to both aubergine and piwi act as enhancers of distortion, suggesting a model whereby the normal function of the piRNA pathway acts, in part, to prevent the altered transmission ratios that characterize SD.
Materials and Methods
Genetic stocks
The Drosophila stocks were maintained at room temperature on cornmeal molasses food. A complete list of stocks used in this experiment can be found in Supporting Information, Table S1.
K-tests
Segregation ratios were measured as described previously (McLean et al. 1994). k is the proportion of SD-bearing progeny as a fraction of the total progeny (Dunn 1953; Ganetzky 1977). For each cross 20–30 males aged <4 days were individually crossed with two RspS cn bw virgin females. Crosses were allowed to brood for 4 days at 25° and then passed to new food. After an additional 4 days of brooding, the parents were discarded. The progeny classes were counted on days 14, 18, and 22 from the initial cross as in McLean et al. (1994). To correct for viability differences between second chromosomes reciprocal crosses were carried out as previously described (Ganetzky 1977; McLean et al. 1994). Single SD/SD+ females were crossed to two Rsps cn bw males. The progeny counts were used to calculate the viability factor, W, where W = [SD+ progeny/SD progeny]. The corrected k value, kc, is then kc = [SD progeny/(SD progeny + SD+ progeny/W)] (Figure 2B). Significance was calculated using a two-tailed Z-test with a Bonferroni correction.
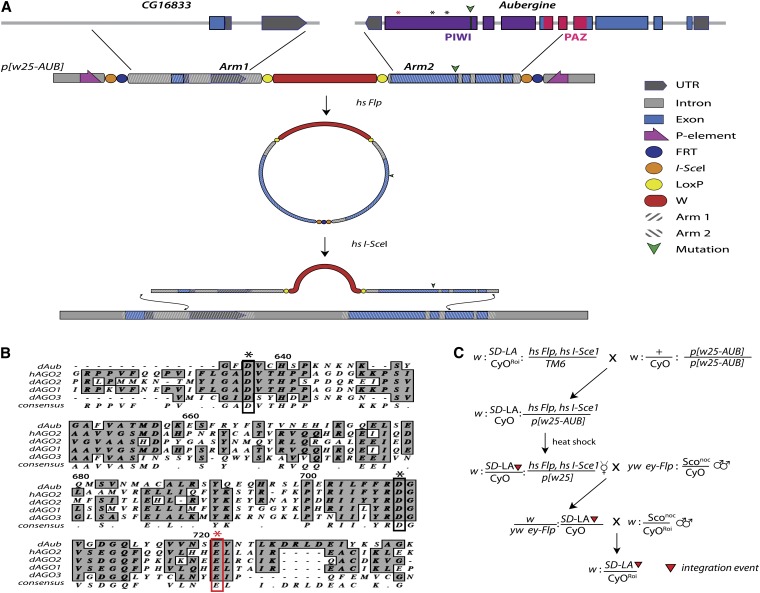
Figure 2.
Targeting of the aub locus by ends-out homologous recombination (HR). (A) Top, Genomic organization of the aub locus and nearby CG16833. Green triangle indicates the location of the Q622* mutation of aubHN and asterisks show the catalytic residues required for nuclease function. The red asterisk is the E721 residue mutated in this study. Aub exons 5–9 were cloned from cDNA into arm2 of the p[W25.2] cloning vector. Arm1 contains the 5′-UTR and intronic sequences from CG16833. This construct was integrated randomly into the genome, using standard transgenic methods. Mobilization and linearization were achieved by heat activation of Flp-recombinase and restriction enzyme I-SceI (hs Flp, hs I-SceI). The linear intermediate is able to recombine at the endogenous locus, using native DNA repair machinery, incorporating engineered mutations into the genome. (B) Alignment of human and Drosophila Argonaute protein sequences showing the location of the catalytic residues (asterisks). (C) Crossing scheme for specific targeting of the SD-Los Arrenos (SD-LA) chromosome by HR. Flies containing the transgenic p[w25-AUB] construct on the third chromosome were crossed to doubly balanced lines containing both SD-LA and the hs-Flp, hs I-SceI chromosome. Embryos from this cross were heat-shocked to allow mobilization and linearization of the p[w25-AUB] construct. Following eclosion, white- or mosaic-eyed virgins were crossed to ey-Flp males. The progeny were screened for red-eyed females that were then crossed to balancer stocks in preparation for PCR validation as described in Staber et al. (2011).
Generation of recombinant chromosomes
Recombinant chromosomes of the genotype aub+Rspmt were generated by allowing exchange between the aub mutant (aubmt) chromosomes and the Rspi16 chromosome. Females of the genotype aubmt Rspmt/aub+ Rspi were crossed with CyO/Sconoc balancer stocks to isolate potential recombinant second chromosomes. Forty isolated second chromosomes were screened by Southern blot for the presence of Rsp repeats matching the parental aub mutant chromosome.
The aub loci of aub+Rspmt recombinants from aubHN, aubQC, and aubAWE chromosomes were verified as wild type (aub+) by sequence analysis. The aubN11 chromosome contains a 110-bp deletion in the aub locus. The presence or absence of this mutation was determined by PCR product size. To generate an aubN11 Rsps chromosome, males from the line N11-35, previously identified as aubN11 Rspi, were crossed with the standard aub+ Rsps cn bw chromosome and recombinants were isolated and assayed as described above.
Southern blotting
For Southern analysis genomic DNA was prepared from ∼30 adult flies, using the Maxwell 16 DNA purification system (no. AS1030; Promega, Madison, WI), eluted in 500 μl of DNase-free water (no. 10977-015; GIBCO, Grand Island, NY), and treated with 20 μg RNAse A (no. AB-0548; Fisher Scientific, Pittsburgh). Samples were then concentrated by centrifugation at 8000 × g for 5 min in Microcon Ultracel YM-50 columns (no. 42416; Millipore, Billerica, MA). The final sample concentration was measured using the dsDNA HS assay kit with the Quibit 2.0 fluorometer (no. Q32854; Life Technologies), and 500 ng of DNA per sample was digested for 1.5 hr at 37° with XbaI, 100 μg/ml bovine serum albumin, and NEB Buffer 4 (20 mM Tris-acetate, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol, pH 7.9) (no. R0145L; New England Biolabs, Beverly, MA). Digested samples were run overnight on a 1.5% agarose gel (SeaKem LE Agarose; Lonza, Basel, Switzerland) in 0.5× TBE at 55 mV. The gel was washed for 30 min in denaturation solution (1.5 M NaCl, 0.5 M NaOH), rinsed with dH2O, and then washed for 30 min in neutralization solution (1.5 M NaCl, 0.5 M Tris-HCl, pH 7.5) with gentle shaking. The DNA was transferred overnight to a Hybond-N+ membrane (no. RPN203B; GE Healthcare) via capillary action, according to the protocol in Molecular Cloning: A Laboratory Manual (Sambrook et al. 1989). Following transfer, DNA was fixed to the membrane by soaking in 0.4 M NaOH for 2 min. The membrane was prehybridized in ECL Gold hybridization buffer (no RPN3006; GE Healthcare) in a Hybaid hybridization oven (Analytical Instruments) at 42° for 30 min.
The Rsp probe was generated by PCR amplification of a 238-bp sequence from the canonical Rsp repeat cloned into the pBluescript KS+ (Stratagene, La Jolla, CA) vector. The vector-specific primers, T3 and T7, were used to amplify the probe (Table S2). Product was run on a 0.5% agarose gel (SeaKem LE Agarose; Lonza) and the bands were cut and gel purified using the Wizard SV Gel and PCR Clean-Up System (no. A9282; Promega). Sample concentration was measured with the dsDNA HS Quibit fluorometer system (Life Technologies) and labeled using the ECL direct labeling kit according to manufacturer’s instructions (no. RPN3005; GE Healthcare). Labeled Rsp and 100-bp ladder probes (no N3231L; New England Biolabs) were added to blots at concentrations of 20 ng/ml and 10 ng/ml, respectively, in hybridization buffer and incubated overnight at 42°. Following incubation, blots were washed two times for 20 min each with ECL Primary wash buffer (0.4% SDS and 0.5× SSC) at 42° and then removed from the hybridization tubes and washed two more times at room temperature on a shaker with 2× SSC. The signal was generated using ECL detection reagents according to manufacturer’s instructions (no. RPN3004; GE Healthcare) and detected using the Kodak Image Station 4000R.
Homologous recombination
We performed ends-out homologous recombination, using a similar methodology to that reported previously (Staber et al. 2011). Briefly, we utilized the ends-out targeting vector p[w25.2] that contains the white+ selectable eye color minigene flanked by LoxP sites for subsequent removal by Cre-recombinase (Figure 2A). Homology arms were cloned and sequenced in pTOPO (Life Technologies) and then shuttled into the multiple cloning sites of the vector to generate p[w25-AUB], which was then introduced into the Drosophila genome by standard transgenic methods (Genetic Services).
The cloning strategy is as follows, where all genomic coordinates are given by the D. melanogaster draft assembly, BDGP Release 5, with release 5.12 annotation provided by FlyBase at the UCSC Genome Browser. Arm1 is the 5′ arm of p[w25-AUB], which contains intronic sequence as well as the 5′-UTR of CG168333 (Figure 2C) and was generated by PCR amplification to incorporate cloning sites as follows: BsiWI-Chr2L:10,995,293–10,997,756-AscI. The last six exons of aub are contained in the 3′ arm of p[w25-AUB] (Arm2), which was generated by PCR amplification and incorporated the following cloning sites: Acc65I-chr2L:10,997,757–11,000,338-NotI.
Mutations were introduced into Arm2 in pTOPO, using the Quik-change XL II kit (Agilent Technologies, La Jolla, CA). Once verified, the mutated Arm2 was liberated from pTOPO and ligated into p(w25.2), using the Acc65I and NotI cloning sites. A full list of cloning, mutagenic, and sequencing primers can be found in Table S2.
Targeting was performed to generate multiple independent targeting events that incorporate or exclude engineered mutations. To isolate targeting events to an SD chromosome, targeting was conducted in flies of the genotype w: SD-Los Arrenos/Cyocnbw Roi: hsFlp, hsSce-I/p[w25-AUB]. White or mosaic-eyed females were collected from the heat-shocked vials and then crossed with yw ey-Flp: nocsco/CyO males and only red-eyed female progeny were selected for additional validation (Figure 2C). Targeted alleles were validated by amplification, using primers outside the region of targeting and primers specific to the w+ minigene. All targeted alleles were sequenced to verify no unintended mutations were introduced.
Results
piRNA pathway mutants are genetic enhancers of SD
To test for genetic epistasis between mutations in piRNA pathway components and SD, we crossed females carrying an SD chromosome with males heterozygous for one of several piRNA pathway mutations including aub, piwi, zuc, and squ. These loci are located on chromosome 2L (Schupbach and Wieschaus 1991; Tweedie et al. 2009). Therefore, each mutant chromosome has an associated array of Rsp repeats located on chromosome 2R (Pimpinelli and Dimitri 1989). Distortion is dependent in part on the number of Rsp repeats present, making it necessary to assay the relative number of repeats on these chromosomes (Temin and Marthas 1984; Wu et al. 1988). The standard Rsps, Rspss, and Rspi chromosomes each exhibit a characteristic banding pattern when digested with the XbaI restriction enzyme and probed on Southern blots with labeled Rsp sequence (Figure 3A) (Wu et al. 1988). This pattern results from the presence of an XbaI digest site at the end of the 240-bp repeat (Figure 1B). However, divergence between Rsp repeats results in mutations that disrupt the restriction site in some sequences (Wu et al. 1988; Cabot et al. 1993; Houtchens and Lyttle 2003). These alterations result in larger band sizes increasing in intervals of 240 bp (Figure 3A).
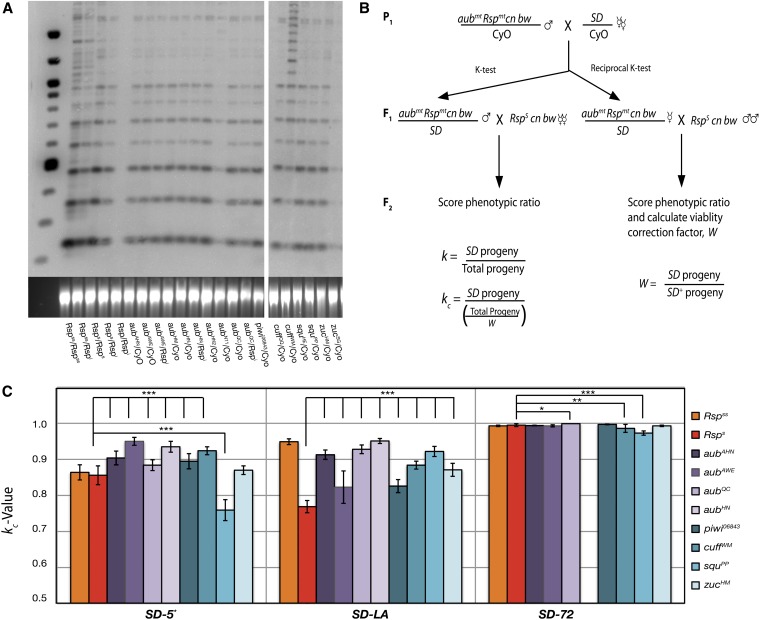
Figure 3.
Mutations in aub and piwi are genetic enhancers of distortion. (A) A representative genomic Southern blot for Rsp sequences of indicated genotypes. All lanes contain 500 ng of XbaI-digested genomic DNA. The aubAHN, aubAWE , aubHM, aubHN, aubHN2, aubQC, piwi06843, cuffQQ, squPP, and zucHM chromosomes all show repeat intensity and banding identical to a single copy of the standard Rsps chromosome (Rsps/Rspi). The CyO chromosome has no detectable Rsp repeats. (B) Crosses and calculations for k tests. Chromosomes with aub mutations were crossed to SD stocks. F1 males were then backcrossed to lines carrying recessive eye-color markers cn and bw to allow scoring of progeny classes by eye color. Reciprocal crosses were carried out to correct for viability differences between the chromosomes. W, k, and kc were calculated as indicated. (C) piRNA pathway mutants were crossed to three different SD chromosomes and kc was determined for each. Each bar represents the total kc value for the total progeny of 20–40 individual males. All five aub mutations tested as well as a piwi mutation showed a significant increase in drive over that of the standard Rsps chromosome (P < 0.0001). SquPP showed a significant reduction in drive when paired with either SD-72 or SD-5* (P < 0.0001). All genotypes gave kc values close to 1.0 for the strong driver SD-72. However, aubQC shows a significant increase in kc. Flies of the genotype SD-72/aubHN were nonviable and therefore could not be tested. Significance was calculated using a two-tailed Z-test (*P < 0.01, **P < 0.001, ***P < 0.0001).
To determine the Rsp repeat status of the mutants used in this study, these chromosomes were compared to the standard Rspss, Rsps, and Rspi chromosomes, using an XbaI digest followed by Southern blotting with Rsp probe. Strikingly, many second chromosomes mutant for piRNA pathway genes including aubAHN, aubAWE, aubHM, aubHN, aubQC, zucHM, squPP, and piwi06843 displayed a banding pattern identical to that of the standard Rsps cn bw chromosome (Figure 3A). These mutant chromosomes also contain the same recessive markers as the canonical Rsps chromosome, strongly suggesting the original mutagenesis was done in the Rsps background. We therefore predict that if the mutations do not affect distortion, SD should eliminate these mutant chromosomes at a rate equivalent to that of a Rsps chromosome.
To test the sensitivity of the piRNA pathway mutant chromosomes to SD we crossed flies carrying an SD+ chromosome bearing a mutant allele with several well-characterized SD chromosomes. Individual F1 males heterozygous for the SD+ aub mutant chromosome and the SD chromosome were then backcrossed with females that contained the same recessive eye color mutations (cnbw) that mark the mutant chromosomes (Figure 3B). The resulting F2 progeny were scored by eye color to determine kc, the fraction of progeny carrying the SD chromosome corrected for viability (Dunn 1953; Sandler and Novitski 1957; McLean et al. 1994).
Interestingly, when crossed into weak SD backgrounds, all of the aub alleles as well as a single piwi mutant chromosome showed a significant increase in distortion over the Rsps control with equivalent levels of repeats (Figure 3C) (two-tailed Z-test, P < 0.0001). In fact, many of these chromosomes were lost at levels equivalent to that seen with the Rspss chromosome. This result is surprising given that the Rspss is estimated to contain nearly 10-fold the number of repeats found in the Rsps background (Wu et al. 1988). When tested against a strongly distorting SD chromosome (SD-72), all of the kc values were very close to 1.0, making detecting changes in kc difficult (Figure 3C). The aubQC chromosome did, however, show a slight enhancement of drive (K = 0.999, P < 0.005) compared to the standard Rsps chromosome (K = 0.995). Thus, it appears that aub and piwi mutants enhance the sensitivity to distortion in a manner analogous to increasing Rsp repeat copy number.
Mutation of the gene cutoff (cuff) also significantly enhances distortion (Figure 3C) (P < 0.0005). The function of this gene is unknown; however, it colocalizes with Aub and Vasa in the nuage of both ovaries and testes (Chen et al. 2007). Mutation of cuff results in the upregulation of Het-A and Tart retrotransposons but does not affect the production of Het-A– or Tart-derived piRNAs, suggesting a role in targeting or silencing of TEs but not in piRNA biogenesis (Chen et al. 2007; Kibanov et al. 2011).
Mutations of zucchini (zuc) and squash (squ) have differential effects on distortion (Figure 3C). Zuc is an endoribonuclease thought to be involved in the processing of primary piRNA precursors in ovarian somatic and germline cells (Pane et al. 2007; Malone et al. 2009; Haase et al. 2010; Nishimasu et al. 2012). The zuc mutant chromosome did not significantly alter distortion in the SD-5* and SD-72 backgrounds. However, an enhancement was observed in the SD-LA background. In the somatic cells of the ovary Zuc functions in production of Piwi-associated primary piRNAs and may serve to promote localization of the piRNA processing machinery to mitochondria (Pane et al. 2007; Malone et al. 2009; Saito et al. 2009; Haase et al. 2010; Watanabe et al. 2011; Nishimasu et al. 2012). The role of Zuc in the testis has not been extensively studied; however, loss of zuc has been shown to have little effect on the abundance of AT-chX-1 and Su(ste)-4, the two most common piRNAs in the Drosophila testes (Nagao et al. 2010).
The allele squpp, on the other hand, exhibited suppression of distortion with both SD-5* and SD-72 chromosomes (Figure 3C). Interestingly, Squ is a component of a testis-specific, electron-dense, perinuclear structure known as the piRNA Nuage Giant Body (piNG-body), thought to be a major site of piRNA processing and/or targeting in spermatocytes (Kibanov et al. 2011). Squ mutant males exhibit a small reduction in levels of At-chX-1 and Su(ste)-4 piRNAs (Nagao et al. 2010). In squ mutant females overall piRNA levels remain largely the same, with a slight decrease detected in some studies (Pane et al. 2007; Malone et al. 2009; Haase et al. 2010). However, expression of some transposons is increased in these mutants, suggesting that, at least in females, Squ acts downstream of piRNA biogenesis (Pane et al. 2007; Haase et al. 2010). Unexpectedly, in this study, introduction of a mutant squ allele led to a significant suppression of drive compared to the Rsps control in two of three genetic backgrounds (Figure 3C) (P < 0.0005). However, since only single mutations of piwi, zuc, and squ were tested, we cannot completely rule out the possibility of confounding background influences in our analysis of their effects on distortion.
The contributions of aub and Rsp to SD sensitivity are genetically separable
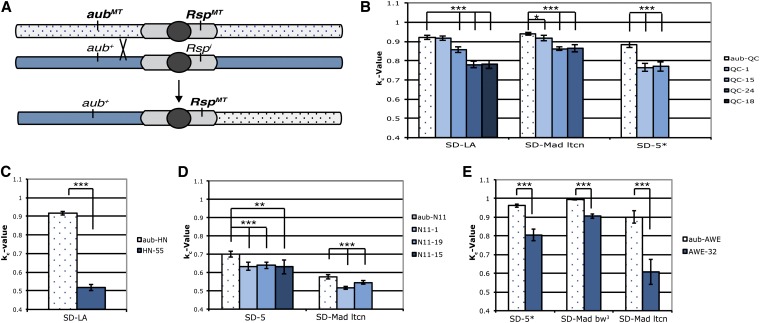
D. melanogaster second chromosomes are known to harbor several unidentified enhancers and suppressors of SD (Figure 1A). Therefore we wanted to determine whether the enhancement in drive seen in the aub mutant chromosomes was specifically due to the introduced mutations vs. other properties of the chromosome (Sandler and Hiraizumi 1959; Hiraizumi et al. 1980; Brittnacher and Ganetzky 1984; Hiraizumi and Thomas 1984). We sought to genetically separate the contributions of the aub mutation from the cis-associated Rsp repeats. This parsing was achieved by crossing flies with aubmt chromosomes containing an associated Rsp locus (Rspmt) with an aub+ Rspi stock. Crossing to a CyO/Sconoc balancer stock isolated individual chromosomes and these isolates were screened by Southern blot for Rsp status and by sequencing for the presence or absence of the associated aub mutation. Recombinant chromosomes that contained the original Rspmt repeat region, but lacked the associated aubergine mutation (aub+ Rspmt) were then crossed to SD lines and tested for kc value according to the scheme in Figure 3B.
Four alleles of aubergine (aubHN, aubQC, aubN11, and aubAWE) were tested. Most recombinant (aub+ Rspmt) chromosomes show a significant decrease in drive relative to the parental chromosome once the aubergine mutation is removed. Tests with several SD chromosomes and the awe-32 recombinant chromosome, an aub+ RspAWE derivative of the parental aubAWE chromosome, gave kc values that are between 9.0% and 32.5% lower than the parental chromosome kc value (P < 0.00003) (Figure 4E). Due to its relatively low repeat copy number, the aubN11 parental chromosome exhibits only low levels of sensitivity to Sd (Figure 3A). However, aub+ RspN11 recombinants show a further reduction in drive when tested with SD-5 or SD-Mad lt cn chromosomes (Figure 4D). The aub+ RspHN recombinant chromosome shows a reduction in drive of >50% compared to the aubHN RspHN chromosome (Figure 4C). For aubQC, three of four recombinants show a decrease in k value with all SD lines tested. Line QC-1 did not show a significant change when tested with SD-Los Arrenos and showed only a 2.3% decrease with SD-Mad lt cn. When tested against SD 5*, however, line QC-1 shows a 13% decrease in kc. (Figure 4B). The cause of this discrepancy is not entirely clear because the break points in these recombination events are unknown. It is possible, however, that the recombination event altered the status of another unknown factor that modifies SD in some circumstances.
Figure 4.
The contributions of aub mutants and associated repeats are genetically separable. (A) Recombinant chromosomes of the type aub+ RspMT were generated by allowing recombination between the aubMT RspMT and aub+ Rspi chromosomes. (B) kc values for recombinant chromosomes generated from aubQC. QC-1 showed no significant reduction in k when crossed with SD-Los Arrenos (SD-LA), but showed a significant reduction with both SD-Mad lt cn and SD-5*. QC-15, QC-18, and QC-24 showed significant reductions in drive with all SD chromosomes tested. (C) After testing >80 individual chromosomes, only one aub+ RspHN allele was recovered. This allele showed a significant reduction in drive when tested with SD-LA. (D) AubN11 chromosomes contain only an intermediate level of repeats. However, when the aub mutation was removed, the amount of drive experienced by these chromosomes was further reduced. (E) AWE-32 showed a reduction in kc compared with the parental chromosome with all three SD lines tested. All tests represent the total kc value for the total progeny of 20–30 individual males. All recombinants, with the exception of QC-1 crossed with SD-LA, showed a significant reduction in kc value, P < 0.0001 (two-tailed Z-test).
Addition of an aub mutation to the canonical Rsps chromosome enhances distortion in trans
The observation that the aubN11 chromosome has many fewer Rsp repeats than the standard Rsps chromosome provided an opportunity to test whether the addition of the aubN11 allele to the standard Rsps repeat is sufficient to increase drive. This experiment was carried out by allowing recombination between the aubN11 RspN11 chromosome and the aub+Rsps chromosome followed by screening for aubN11 Rsps recombinants (Figure 5A).
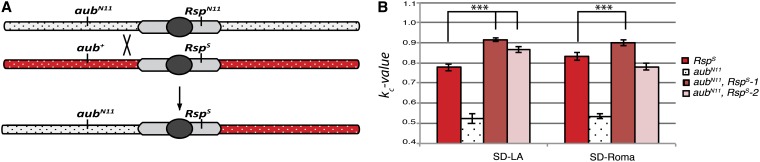
Figure 5.
Addition of an aub mutation to a Rsps chromosome enhances distortion. (A) Recombinant chromosomes of the type aubN11 Rsps were generated by allowing recombination between aubN11 RspN11 and aub+ Rsps chromosomes. Potential recombinant chromosomes were tested for Rsp status by Southern blot and for the presence of the 110-bp deletion in aubN11 by PCR assay. (B) Both recombinants show a significant increase in distortion over that of a standard Rsps chromosome when tested against SD-Los Arrenos (SD-LA). Only aubN11 Rsps-2 showed a significant reduction when crossed to SD-Roma. All tests represent the total kc for 20–30 males (P < 0.00001, two-tailed Z-test).
If the aub mutation contributes to the degree of drive observed for this chromosome, we would expect to see an increase in kc over that of the Rsps control chromosome. This enhancement was observed in two recovered recombinant lines (Figure 5B). Both aubN11 Rsps-1 and aubN11 Rsps-2 show a statistically significant increase in k value compared to the Rsps control in the SD-Los Arrenos background; however, only aubN11 Rsps-1 showed a significant enhancement in the SD-Roma background.
Taken together these data strongly argue that the contributions of the specific aubergine mutation and its associated repeat array to the overall drive phenotype are genetically separable. Replacement of an aub mutation with a wild-type allele leads to a reduction in kc for that chromosome without alteration of its Rsp repeat array. Further, placing the Rsps array in the context of a RspN11 allele leads to a significant increase in drive over that of Rsps alone in three of four independent tests (Figure 5B).
However, given that multiple known modifiers of SD, including M(SD), E(SD), and St(SD), have been mapped to the second chromosome, it is difficult to completely rule out the contributions of these unidentified loci using standard genetic recombination (Sandler and Hiraizumi 1959; Hiraizumi et al. 1980; Brittnacher and Ganetzky 1984; Hiraizumi and Thomas 1984).
An aub mutation placed on an SD chromosome by homologous recombination enhances distortion in cis
To remove the confounding effects of unknown modifiers and enhancers from interpretation of genetic experiments, we specifically targeted aub mutations to an SD-Los Arrenos chromosome, using ends-out homologous recombination (HR) (Figure 2) (Rong and Golic 2000; Staber et al. 2011). This technique allows the precise replacement of an endogenous locus with an engineered construct. The p[w25-AUB] construct contains two 2.5-kb “homology arms” cloned from the aub locus of Canton-S flies. Arm1 contains mostly intronic sequence from the gene CG16833, as well as a small portion of the 5′-UTR. The mini-white selectable marker was placed between the 3′ end of aub and the 5′-UTR of CG16833 to reduce the possibility of interference from this insert (Figure 2B).
In addition to a wild-type construct, two mutations were engineered: an aubHR-HN mutation, which changes Q622 to an amber stop codon (CAG → TAG), and the mutation aubHR-E721A, which changes the catalytic glutamic acid required for RNA slicer activity to an alanine (GAG → GCG) (Figure 2C) (Harris and MacDonald 2001; Liu et al. 2004). We selected SD-Los Arrenos as the target chromosome because its relatively low k value would allow for a sensitized background in which to detect enhancement of distortion.
Following targeting, integration events were verified by PCR followed by sequencing across the length of the arms to ensure no unintended mutations were induced. Additionally chromosomes were tested for the presence of Sd-RanGAP by genomic PCR across the junction of wild-type RanGAP and the Sd-RanGAP duplication (Merrill et al. 1999; Robinson et al. 2008). Chromosomes identified as carrying both Sd and the HR insert were crossed into a w+ background and then used for subsequent k tests.
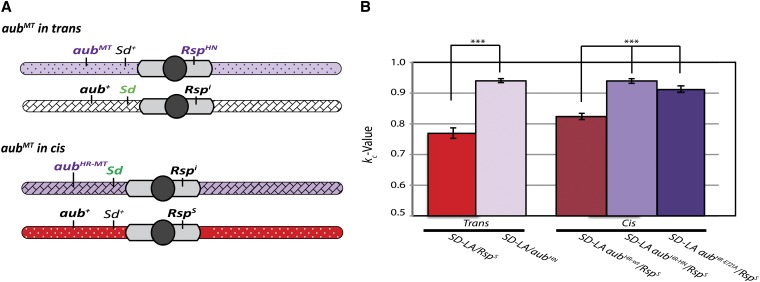
When placed in cis with Sd by HR, the aubHR-HN mutation recapitulated the effect seen in trans with males carrying both an aubHN mutant second chromosome and an SD- Los Arrenos chromosome. The average kc for two independent mutations tested against a Rsps chromosome is 0.940, identical to the kc for the same mutation in trans (Figure 6B).
Figure 6.
Engineered aub mutations on an SD chromosome enhance distortion in cis. (A) Schematic of chromosomes used to assay the effect of aub mutations placed on an SD chromosome. (B) The aubHN mutation targeted to an SD-Los Arrenos (SD-LA) chromosome by HR (aubHR-HN) enhances distortion of a Rsps compared to a targeted SD-LA aubHR-wt mutation. The aubHR-E721A allele, in which a catalytic glutamic acid residue is replaced with alanine, also shows a significant enhancement of drive when compared to a wild-type targeted chromosome (SD-LA aubHN-wt), although the increase is smaller than that seen with the SD-LA aubHR-HN targeted allele. The SD-LA aubHR-HN and SD-LA aubHN-wt kc values each represent the pooled progeny counts for two independently derived alleles, ∼60–70 males total. The SD-LA aubE721A data represent a single allele with n = 27 males (***P < 0.00005, two-tailed Z-test).
Interestingly the catalytic mutant aubHR-E721A also showed an enhancement of k value (kc = 0.913) although this effect was not as strong as that from the premature stop codon in the aubHR-HN mutant. The average kc of the wild-type construct (kc = 0.824) showed a small, but significant enhancement of drive over that of the standard Rsps chromosome (kc = 0.769, P < 0.0005). This enhancement was observed in two independent lines. The aub locus of the parental SD-Los Arrenos chromosome shows several polymorphisms not found in Canton-S or in the GenBank reference sequence (BDGP R5.12) (C. Staber, personal communication). Although two of these polymorphisms are intronic and one is a silent mutation, it is possible that replacement of this locus with a wild-type aub sequence results in this enhancement of drive.
It is also possible that the presence of mini-white in the intergenic region 3′ to aub affects drive. However, removal of the mini-white with Cre-recombinase causes a slight increase, not a decrease, in drive (our unpublished observation). While the cause of the small alteration in kc value for SD-Los Arrenos aubHR-WT chromosomes remains unclear, the change in kc for the wild-type construct is small when compared to the increase caused by the presence of an aub mutation.
To further examine the role of the endoribonuclease Zuc and the putative ribonuclease Squ, mutant alleles of each gene were tested in the SD-LA aubHR-WT and SD-LA aubHR-HN backgrounds. SD-LA males carrying either the wild-type or the aubHR-HN alleles showed significant suppression of distortion in the presence of zuc or squ mutations (Figure S1).
Discussion
In this study, we have shown that mutations to aubergine act as genetic enhancers of Segregation Distorter. Chromosomes carrying both a mutation in the aub locus and a Rsps repeat array in cis are more strongly distorted than chromosomes with Rsps alone. Removal of the aub mutation by recombination reverses this effect. Further, addition of a Rsps repeat array to an aubN11 chromosome, which naturally contains significantly fewer repeats than Rsps, results in a marked enhancement of drive compared with that of the Rsps repeats alone. These independent lines of evidence demonstrate that both mutations to the aub locus and the repeat array contribute to the severity of drive and that these contributions are genetically separable.
Additionally, mutations specifically targeted to the aubergine locus of an SD chromosome show that the effect of this mutation can be recapitulated in cis and is therefore not specific to the Rsp-bearing chromosome. A mutation to one of the three catalytic residues of aub also produces a significant enhancement in drive, although this effect is not as large as that of the aubHR-HN allele that contains a premature stop codon, truncating the protein upstream of the catalytic residues. Although we have not completely eliminated the possibility that the effects of aub mutations on SD are indirect, evidence that disruption of aub catalytic activity alters distortion suggests that aub’s function as a ribonuclease is required for suppression of SD.
Silencing of repetitive transposon targets in the germline of both testes and ovaries utilizes a mechanism known as “ping-pong” piRNA production because it requires reciprocal cleavage of sense and antisense RNAs by piRNA-guided AGO3 and Aub proteins, respectively (Brennecke et al. 2007; Gunawardane et al. 2007; Nagao et al. 2010). In the testes, these two proteins associate with other piRNA pathway components including Vasa (Vas), Armitage (Armi), and Tudor (Tud) in large electron-dense perinuclear bodies, dubbed piNG-bodies, which are thought to regulate the processing and/or targeting of piRNAs to transposable elements via sequence homology (Lim and Kai 2007; Lim et al. 2009; Nishida et al. 2009; Kibanov et al. 2011). The localization of both Aub and AGO3 to this perinuclear region is codependent; loss of one protein through mutation results in mislocalization of the other (Cox et al. 2000; Brennecke et al. 2007; Nishida et al. 2007; Li et al. 2009; Nagao et al. 2010).
Interestingly, Rsps piRNAs have mostly been detected in association with AGO3 in wild-type testes. These piRNAs are derived from a single strand of the Rsp repeat sequence (Nagao et al. 2010). The genetic data presented here suggest that wild-type function of the piRNA processing machinery suppresses distortion. Hypomorphic mutations, such as aubHN, may more profoundly disrupt processing in this pathway than the catalytic mutation aubE721A, which could potentially permit proper complex formation, but disrupt piRNA processing or targeting.
This interpretation explains the differential effects of the aubHN and the aubE721A mutations on drive; the catalytically inactive mutant may allow AGO3-dependent processing at the piNG-body. In contrast, the hypomorph, because of the reduced amount of Aub protein or the lack of the C-terminal region, may have a more significant effect on complex formation and therefore compromise both Aub- and AGO3-dependent processing.
Work in both the Stellate and the Winters sex ratio drive systems has implicated RNAi in mediating suppression of drive (Aravin et al. 2001, 2004; Tao et al. 2007a,b). In the Stellate sex ratio system antisense transcripts of the Y chromosome Suppressor of Stellate [Su(Ste)] locus are processed by Aub and AGO3 in the testis of D. melanogaster males to generate piRNAs that silence the X-linked Stellate (Ste) locus by homology-mediated cleavage of sense Ste transcripts (Aravin et al. 2001, 2004; Nagao et al. 2010).
In the Winters sex ratio system of D. simulans, the Distorter on X (Dox) is suppressed by Not much yang (Nmy) on chromosome 3R. Suppression requires the presence of two inverted repeats (IRs) in Nmy. Loss of a single copy of this 360-bp IR element results in activation of Dox and the loss of Y-bearing sperm (Tao et al. 2007a,b). The two IR copies may be required to permit folding of Nmy RNA into a dsRNA stem-loop structure that could serve as a substrate for generation of endo-siRNAs or piRNAs (Tao et al. 2007b). Dox itself carries a single copy of this repeat, suggesting a target for homology-mediated post-transcriptional gene silencing (Tao et al. 2007a). Both Winters and Stellate provide salient examples of systems where RNAi activity is thought to suppress meiotic drive. In line with the observations reported here, the piRNA pathway or other germline RNAi mechanisms appear to act protectively to suppress the potentially deleterious effects of these drivers.
In a recent publication, Tao et al. (2007a) propose a model in which RanGAP mislocalization prevents nuclear import of Rsp-primed piRNA-induced silencing complexes (piRISC) needed to silence the Rsp locus (Ferree and Barbash 2007; Tao et al. 2007a; Larracuente and Presgraves 2012). This failure to silence the Rsp locus could lead to a generalized defect in chromatin condensation originating from the second chromosome. While the data presented here certainly support the supposition that the piRNA pathway works to silence Rsp transcription during spermatogenesis, we suggest the following modification to this model.
The defects caused by Sd-RanGAP are primarily associated with nuclear accumulation and not with exclusion of RanGTP-mediated transport cargo from the nucleus (Kusano et al. 2001, 2003). piRNA biogenesis is thought to require export of a piRNA precursor molecule to the cytoplasm where it is processed into mature piRNAs in the nuage of the ovary or the piNG-body of the testis (Figure 7A) (Lim and Kai 2007; Klattenhoff and Theurkauf 2008; Klattenhoff et al. 2009; Khurana and Theurkauf 2010; Saito et al. 2010; Kibanov et al. 2011; Watanabe et al. 2011). Nuclear export of RNA–protein complexes (RNPs) requires a class of proteins known as exportins that facilitate directional transit through the nuclear pore complex (NPC) (Gorlich and Kutay 1999a; Kohler and Hurt 2007). To export miRNA precursor RNPs from the nucleus, the exportin must also bind RanGTP (Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004; Kohler and Hurt 2007). In SD, accumulation of nuclear Sd-RanGAP protein depletes nuclear RanGTP through increased GTP hydrolysis, reducing the effective concentration of RanGTP available to bind the exportin–precursor-piRNA complex. Without RanGTP to facilitate exit through the nuclear pore, more RNP complexes are retained in the nucleus, reducing the availability of precursor-piRNA substrates for processing in the cytoplasm (Figure 7B). Interestingly, wild-type RanGAP has been observed to colocalize with piNG-bodies in spermatocytes (Figure S2), suggesting piRNA processing may be tightly coupled with nuclear transport.
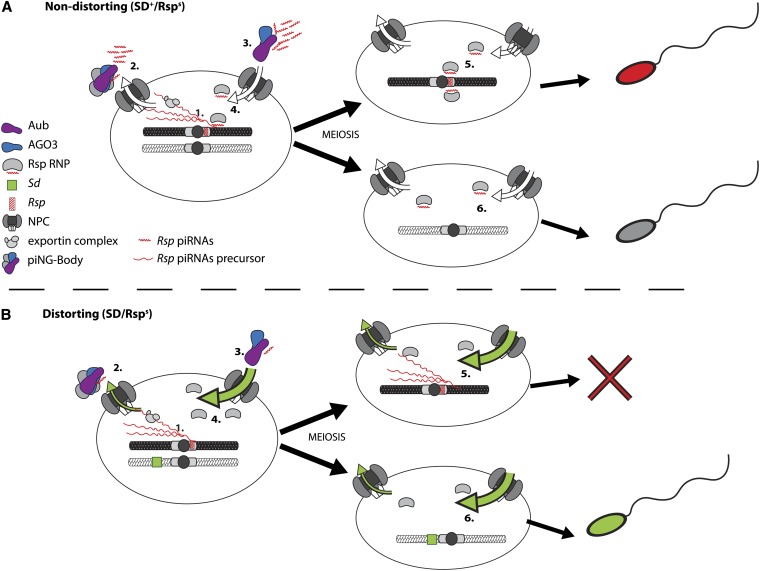
Figure 7.
A model for the role of the piRNA pathway in segregation distortion. (A) 1, in SD+ Rsps nuclei precursor Rsp piRNA transcripts are made in spermatocytes; 2, following export through the nuclear pore complex (NPC), these precursor Rsp piRNAs are processed into mature piRNAs by Aub and AGO3 in the piNG-body; 3, mature Rsp piRNAs reenter the nucleus as part of a RNA–protein silencing complex (Rsp RNP); 4, inside the nucleus Rsp piRNAs are then used by the Rsp RNP to target the Rsps locus for silencing; 5, this silencing is maintained through meiosis and is required for proper condensation of Rsps-bearing spermatids; 6, in Rspi spermatids, Rsp piRNA complexes have no target. (B) 1, under distorting conditions, nuclear transport is perturbed (green arrows), causing nuclear retention of Rsp precursor piRNAs; 2, this retention prevents cytoplasmic processing of precursors into mature piRNAs; 3, thus there is no Rsp-piRNA complex primed and ready to enter the nucleus; 4, therefore the Rsps locus is not properly silenced in these cells; 5, as a result, Rsps-bearing spermatids fail to properly condense following meiosis and are eliminated; 6, lacking a Rsps locus, the SD-bearing spermatids condense normally and are individualized into sperm.
In the germline cells of the ovary, Piwi enters the nucleus where it is thought to mediate epigenetic silencing through direct interaction with HP1 and homology-guided targeting of transposon transcripts (Pal-Bhadra et al. 2004; Brower-Toland et al. 2007; Klattenhoff and Theurkauf 2008). Further, the role of the interplay between RNAi and maintenance of epigenetic states has been well characterized in Schizosaccharomyces pombe (Volpe et al. 2002; Motamedi et al. 2004; Verdel et al. 2004; Sugiyama et al. 2005; Buhler et al. 2006; Verdel et al. 2009). It is unknown how the generation of piRNAs in the cytoplasm of spermatocytes might contribute to epigenetic silencing. Yet, it seems likely given the known role of RNAi in general, and piRNAs specifically in chromatin regulation, that mature Rsp piRNAs contribute to the epigenetic silencing of the Rsp locus through alteration of chromatin states.
Proper silencing of Rsp may be required to allow chromatin compaction as well as the transition from histones to protamines, a sperm-specific histone variant, necessary for sperm maturation (Ferree and Barbash 2007; Tao et al. 2007a; Larracuente and Presgraves 2012). In spermatids bearing chromosomes containing large numbers of Rsp repeats, silencing may require a significant contribution from the piRNA pathway. The defect in nuclear export caused by Sd-RanGAP may ultimately result in the import of too few mature Rsp-piRNA–primed RNP complexes to facilitate effective silencing, leading to defective chromatin compaction and the specific destruction of Rsp-bearing spermatids (Figure 7B). Mutation of piRNA pathway components would exacerbate this defect, resulting in a further reduction of the functional pool of piRNAs available for Rsp targeting. The data presented here show that as predicted by our proposed model, disruption of piRNA processing through mutation of aubergine leads to an enhancement of distortion. Still, not all components that influence piRNA biogenesis may have comparable effects on distortion, as our results on the effects of mutations in zuc and squ demonstrate. However, given the complexities of processing and transport of piRNAs on precursors and final small RNA products, we envision that different mutations could have opposing effects on distortion.
The piRNA pathway is a sophisticated adaptive defense against mobilization of selfish genetic elements in the germline. Thus far, identified targets of this pathway have mostly been restricted to transposable elements that reproduce through retrotransposition into novel locations in the genome. The data presented here strongly suggest that the piRNA pathway is also able to protect against other types of selfish elements, such as meiotic drivers, which propagate selfishly by destroying gametes of the alternative genotype. Understanding the molecular interaction between piRNA biogenesis and segregation distortion could provide significant insight into the biology of gametogenesis and the etiology of meiotic drive, as well as the evolution of mechanisms to defend against invasion of the genome by this type of ultraselfish genetic element.
Supplementary Material
Acknowledgments
We thank Cynthia Staber for her technical expertise, helpful discussions, and thoughtful commentary on this article. Additionally we thank Arthur Sugden and Lauren Alpert for their help with software and statistical analysis, respectively; Kasia Sierzputowska for the counting of many fruit flies; and Yiannis Savva, Leila Rieder, and the two anonymous reviewers for their constructive commentary and help in the editing of this work.
Footnotes
Communicating editor: C.-ting Wu
Literature Cited
- Aravin A. A., Naumova N. M., Tulin A. V., Vagin V. V., Rozovsky Y. M., et al. , 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11: 1017–1027. [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Klenov M. S., Vagin V. V., Bantignies F., Cavalli G., et al. , 2004. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 24: 6742–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A. A., Hannon G. J., Brennecke J., 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764. [DOI] [PubMed] [Google Scholar]
- Bohnsack M. T., Czaplinski K., Gorlich D., 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., et al. , 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Brittnacher J. G., Ganetzky B., 1984. On the components of segregation distortion in Drosophila melanogaster. III. Nature of enhancer of SD. Genetics 107: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B., Findley S. D., Jiang L., Liu L., Yin H., et al. , 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 21: 2300–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M., Verdel A., Moazed D., 2006. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125: 873–886. [DOI] [PubMed] [Google Scholar]
- Burt A., Trivers R., 2008. Genes in Conflict: The Biology of Selfish Genetic Elements. Belknap Press and; Harvard University Press, Cambridge, MA. [Google Scholar]
- Cabot E. L., Doshi P., Wu M. L., Wu C. I., 1993. Population-genetics of tandem repeats in centromeric heterochromatin - unequal crossing-over and chromosomal divergence at the responder locus of Drosophila-melanogaster. Genetics 135: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M. A., Xuan Z., Zhang M. Q., Hannon G. J., 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16: 2733–2742. [DOI] [PubMed] [Google Scholar]
- Chen Y., Pane A., Schupbach T., 2007. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr. Biol. 17: 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. N., Chao A., Lin H., 2000. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514. [DOI] [PubMed] [Google Scholar]
- Dunn L. C., 1953. Variations in the segregation ratio as causes of variations in gene frequency. Acta Genet. Stat. Med. 4: 139–147. [DOI] [PubMed] [Google Scholar]
- Ferree P. M., Barbash D. A., 2007. Distorted sex ratios: a window into RNAi-mediated silencing. PLoS Biol. 5: e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky B., 1977. On the components of segregation distortion in Drosophila melanogaster. Genetics 86: 321–355. [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Kutay U., 1999a Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15: 607–660. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Kutay U., 1999b Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15: 607–660. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Mattaj I. W., 1996. Nucleocytoplasmic transport. Science 271: 1513–1518. [DOI] [PubMed] [Google Scholar]
- Gunawardane L. S., Saito K., Nishida K. M., Miyoshi K., Kawamura Y., et al. , 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. [DOI] [PubMed] [Google Scholar]
- Haase A. D., Fenoglio S., Muerdter F., Guzzardo P. M., Czech B., et al. , 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 24: 2499–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A. N., Macdonald P. M., 2001. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128: 2823–2832. [DOI] [PubMed] [Google Scholar]
- Hartl D. L., 1973. Complementation analysis of male fertility among the segregation distorter chromosomes of Drosophila melanogaster. Genetics 73: 613–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D. L., 1974. Genetic dissection of segregation distortion. I. Suicide combinations of SD genes. Genetics 76: 477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D. L., Hiraizumi Y., Crow J. F., 1967. Evidence for sperm dysfunction as the mechanism of segregation distortion in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 58: 2240–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraizumi Y., Thomas A. M., 1984. Suppressor systems of Segregation Distorter (SD) chromosomes in natural populations of Drosophila melanogaster. Genetics 106: 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraizumi Y., Martin D. W., Eckstrand I. A., 1980. A modified model of segregation distortion in Drosophila melanogaster. Genetics 95: 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtchens K., Lyttle T. W., 2003. Responder (Rsp) alleles in the segregation distorter (SD) system of meiotic drive in Drosophila may represent a complex family of satellite repeat sequences. Genetica 117: 291–302. [DOI] [PubMed] [Google Scholar]
- Kalab P., Weis K., Heald R., 2002. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295: 2452–2456. [DOI] [PubMed] [Google Scholar]
- Khurana J. S., Theurkauf W., 2010. piRNAs, transposon silencing, and Drosophila germline development. J. Cell Biol. 191: 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibanov M. V., Egorova K. S., Ryazansky S. S., Sokolova O. A., Kotov A. A., et al. , 2011. A novel organelle, the piNG-body, in the nuage of Drosophila male germ cells, is associated with piRNA-mediated gene silencing. Mol. Biol. Cell 22: 3410–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C., Theurkauf W., 2008. Biogenesis and germline functions of piRNAs. Development 135: 3–9. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C., Xi H., Li C., Lee S., Xu J., et al. , 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A., Hurt E., 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8: 761–773. [DOI] [PubMed] [Google Scholar]
- Kusano A., Staber C., Ganetzky B., 2001. Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Dev. Cell 1: 351–361. [DOI] [PubMed] [Google Scholar]
- Kusano A., Staber C., Ganetzky B., 2002. Segregation distortion induced by wild-type RanGAP in Drosophila. Proc. Natl. Acad. Sci. USA 99: 6866–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano A., Staber C., Chan H. Y., Ganetzky B., 2003. Closing the (Ran)GAP on segregation distortion in Drosophila. Bioessays 25: 108–115. [DOI] [PubMed] [Google Scholar]
- Larracuente A. M., Presgraves D. C., 2012. The selfish Segregation Distorter gene complex of Drosophila melanogaster. Genetics 192: 33–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Vagin V. V., Lee S., Xu J., Ma S., et al. , 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A. K., Kai T., 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104: 6714–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A. K., Tao L., Kai T., 2009. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 186: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Carmell M. A., Rivas F. V., Marsden C. G., Thomson J. M., et al. , 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- Lund E., Guttinger S., Calado A., Dahlberg J. E., Kutay U., 2004. Nuclear export of microRNA precursors. Science 303: 95–98. [DOI] [PubMed] [Google Scholar]
- Lyttle T. W., 1991. Segregation distorters. Annu. Rev. Genet. 25: 511–557. [DOI] [PubMed] [Google Scholar]
- Lyttle T. W., Brittnacher J. G., Ganetzky B., 1986. Detection of Rsp and modifier variation in the meiotic drive system Segregation distorter (SD) of Drosophila melanogaster. Genetics 114: 183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C. D., Brennecke J., Dus M., Stark A., McCombie W. R., et al. , 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean J. R., Merrill C. J., Powers P. A., Ganetzky B., 1994. Functional identification of the Segregation distorter locus of Drosophila melanogaster by germline transformation. Genetics 137: 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill C., Bayraktaroglu L., Kusano A., Ganetzky B., 1999. Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila. Science 283: 1742–1745. [DOI] [PubMed] [Google Scholar]
- Motamedi M. R., Verdel A., Colmenares S. U., Gerber S. A., Gygi S. P., et al. , 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802. [DOI] [PubMed] [Google Scholar]
- Nagao A., Mituyama T., Huang H., Chen D., Siomi M. C., et al. , 2010. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 16: 2503–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K. M., Saito K., Mori T., Kawamura Y., Nagami-Okada T., et al. , 2007. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13: 1911–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K. M., Okada T. N., Kawamura T., Mituyama T., Kawamura Y., et al. , 2009. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 28: 3820–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H., Ishizu H., Saito K., Fukuhara S., Kamatani M. K., et al. , 2012. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491: 284–287. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M., Leibovitch B. A., Gandhi S. G., Rao M., Bhadra U., et al. , 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672. [DOI] [PubMed] [Google Scholar]
- Pane A., Wehr K., Schupbach T., 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12: 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli S., Dimitri P., 1989. Cytogenetic analysis of segregation distortion in Drosophila melanogaster: the cytological organization of the Responder (Rsp) locus. Genetics 121: 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson B., Wiseman N., Zhang M., Staber C. J., Woodruff R. C., 2008. Identification of selfish genetic elements in natural populations of Drosophila melanogaster. II. Segregation Distorter (SD). Dros. Inf. Serv. 91: 183–185. [Google Scholar]
- Rong Y. S., Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Saito K., Inagaki S., Mituyama T., Kawamura Y., Ono Y., et al. , 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299. [DOI] [PubMed] [Google Scholar]
- Saito K., Ishizu H., Komai M., Kotani H., Kawamura Y., et al. , 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 24: 2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sandler L., Hiraizumi Y., 1959. Meiotic drive in natural populations of Drosophila melanogaster. Ii. Genetic variation at the Segregation-Distorter locus. Proc. Natl. Acad. Sci. USA 45: 1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L., Hiraizumi Y., 1960a Meiotic drive in natural populations of Drosophila melanogaster. IV. Instability at the Segregation-Distorter locus. Genetics 45: 1269–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L., Hiraizumi Y., 1960b Meiotic drive in natural populations of Drosophila melanogaster. V. On the nature of the Sd region. Genetics 45: 1671–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L., Novitski E., 1957. Meiotic drive as an evolutionary force. Am. Nat. 91: 105–110. [Google Scholar]
- Sandler L., Hiraizumi Y., Sandler I., 1959. Meiotic drive in natural populations of Drosophila melanogaster. I. The cytogenetic basis of Segregation-Distortion. Genetics 44: 233–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T., Wieschaus E., 1991. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129: 1119–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti K. A., Brennecke J., 2010. The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet. 26: 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staber C. J., Gell S., Jepson J. E., Reenan R. A., 2011. Perturbing A-to-I RNA editing using genetics and homologous recombination. Methods Mol. Biol. 718: 41–73. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Cam H., Verdel A., Moazed D., Grewal S. I., 2005. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl. Acad. Sci. USA 102: 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Araripe L., Kingan S. B., Ke Y., Xiao H., et al. , 2007a A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. PLoS Biol. 5: e293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Masly J. P., Araripe L., Ke Y., Hartl D. L., 2007b A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. PLoS Biol. 5: e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin R. G., 1991. The independent distorting ability of the Enhancer of Segregation Distortion, E(SD), in Drosophila melanogaster. Genetics 128: 339–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin R. G., Marthas M., 1984. Factors influencing the effect of Segregation Distortion in natural populations of Drosophila melanogaster. Genetics 107: 375–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin R. G., Ganetzky B., Powers P. A., Lyttle T. W., Pimpinelli S., et al. , 1991. Segregation distortion in Drosophila-melanogaster - genetic and molecular analyses. Am. Nat. 137: 287–331. [Google Scholar]
- Tokuyasu K. T., Peacock W. J., Hardy R. W., 1977. Dynamics of spermiogenesis in Drosophila melanogaster. VII. Effects of segregation distorter (SD) chromosome. J. Ultrastruct. Res. 58: 96–107. [DOI] [PubMed] [Google Scholar]
- Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., et al. , 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37: D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., et al. , 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A., Vavasseur A., Le Gorrec M., Touat-Todeschini L., 2009. Common themes in siRNA-mediated epigenetic silencing pathways. Int. J. Dev. Biol. 53: 245–257. [DOI] [PubMed] [Google Scholar]
- Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I., et al. , 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Chuma S., Yamamoto Y., Kuramochi-Miyagawa S., Totoki Y., et al. , 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev. Cell 20: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. I., Lyttle T. W., Wu M. L., Lin G. F., 1988. Association between a satellite DNA sequence and the Responder of Segregation Distorter in D. melanogaster. Cell 54: 179–189. [DOI] [PubMed] [Google Scholar]
- Yi R., Qin Y., Macara I. G., Cullen B. R., 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.