Abstract
The increasing ability to sequence and compare multiple individual genomes within a species has highlighted the fact that copy-number variation (CNV) is a substantial and underappreciated source of genetic diversity. Chromosome-scale mutations occur at rates orders of magnitude higher than base substitutions, yet our understanding of the mechanisms leading to CNVs has been lagging. We examined CNV in a region of chromosome 5 (chr5) in haploid and diploid strains of Saccharomyces cerevisiae. We optimized a CNV detection assay based on a reporter cassette containing the SFA1 and CUP1 genes that confer gene dosage-dependent tolerance to formaldehyde and copper, respectively. This optimized reporter allowed the selection of low-order gene amplification events, going from one copy to two copies in haploids and from two to three copies in diploids. In haploid strains, most events involved tandem segmental duplications mediated by nonallelic homologous recombination between flanking direct repeats, primarily Ty1 elements. In diploids, most events involved the formation of a recurrent nonreciprocal translocation between a chr5 Ty1 element and another Ty1 repeat on chr13. In addition to amplification events, a subset of clones displaying elevated resistance to formaldehyde had point mutations within the SFA1 coding sequence. These mutations were all dominant and are proposed to result in hyperactive forms of the formaldehyde dehydrogenase enzyme.
Keywords: copy-number variation, nonallelic homologous recombination, gene amplification, chromosomal rearrangements
AS a consequence of studies that utilize genomic microarrays and next-generation DNA sequencing, it has become clear that much of the natural genetic variation that exists between individuals is due to alterations in the number of copies of genes rather than differences in the nucleotide sequence (Girirajan et al. 2011; Veltman and Brunner 2012). It has been calculated that 8–25 kb of DNA are deleted or duplicated per generation in humans compared to about 100 bp of point mutations (Itsara et al. 2010). Although deletions and duplications vary in size from a few base pairs (for example, changes in the lengths of microsatellites) to many megabases (for example, changes in ploidy), Girirajan et al. (2011) define copy-number variants (CNVs) in humans as changes that vary in size between 50 bp and 1 Mb. While most CNV events have no obvious effect, a significant number are associated with human diseases including Charcot–Marie–Tooth syndrome, autosomal dominant leukodystrophy, Williams syndrome, several cancer predispositions, and autism-related and other neurodevelopmental disorders (Abrahams and Geschwind 2008; Girirajan et al. 2011; Krepischi et al. 2012; Malhotra and Sebat 2012; Sullivan et al. 2012). There is also strong evidence suggesting that CNVs played a significant role in human evolution (Iskow et al. 2012). Finally, multiple somatic CNV events are frequently observed in the altered karyotypes of cancer cells (Stratton et al. 2009).
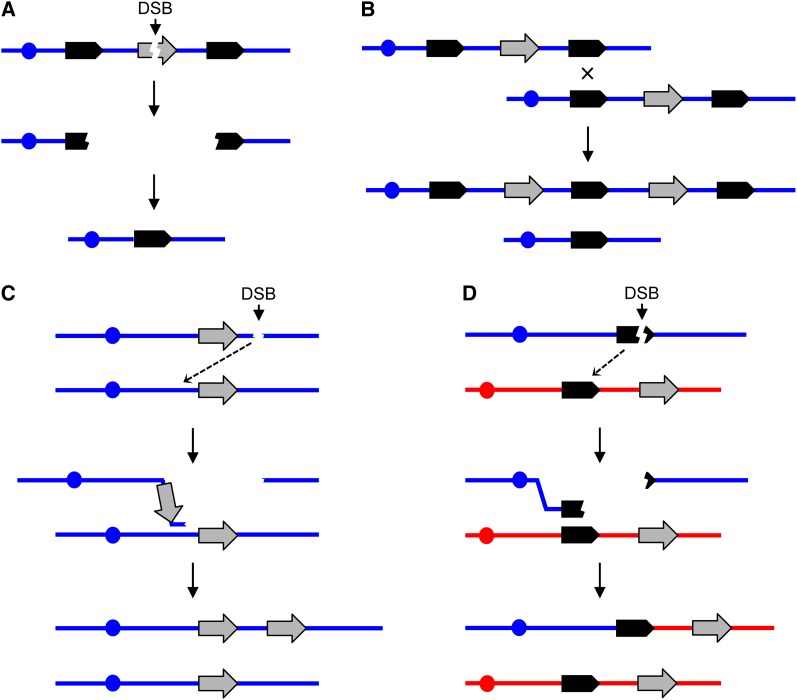
CNVs have also been widely observed in natural populations and have been studied in detail in model organisms. In the discussion below, we analyze genomic deletions and duplications in the yeast Saccharomyces cerevisiae. We define a CNV as a deletion or duplication that includes at least one gene (average of about 2 kb) and no more than one chromosome arm (average of about 400 kb). The most frequently observed mechanisms for the generation of deletions and duplications are illustrated in Figure 1.
Figure 1.
General mechanisms for the generation of deletions and duplications. In this figure, chromosomes are shown as horizontal blue or red lines, DNA repeats as solid arrows, reporter genes as shaded arrows, and centromeres as circles. Single DNA strands are not represented. Variations of these models are also possible. (A) Deletion formation by the single-strand annealing (SSA) pathway. A double-strand DNA break (DSB) between the flanking repeats results in two broken ends that are resected 5′ to 3′. When complementary single-strand regions of the flanking repeats are exposed, reannealing occurs, resulting in loss of the sequences between the repeats (Paques and Haber 1999). (B) Unequal crossovers resulting in deletions and duplications. This homologous recombination event could involve either sister chromatids in haploids or diploids or homologous chromosomes in diploids. (C) Microhomology-mediated replication. A free 3′ end associated with centromere-containing DNA fragment invades a sister chromatid or a homologous chromosome using a nonallelic microhomology sequence. The subsequent break-induced replication (BIR) event generates a duplication in which the breakpoints share little sequence homology. It is assumed that the DNA fragment without a centromere is lost. (D) Coupled terminal duplication and deletion resulting in a nonreciprocal translocation. The different homologs are shown in blue and red. A DSB in or distal to a repeat in the blue chromosome is repaired by a BIR event using a repeat on a nonhomologous chromosome (red). This event results in a duplication of sequences from the red chromosome and a deletion of sequences from the blue chromosome.
Perhaps the most notable early example of CNV in yeast was the discovery of a large deletion on the right arm of chromosome 3 (chr3) that spanned the mating-type locus and distal genes linked to it (Hawthorne 1963). Later studies demonstrated that this ∼100-kb deletion was mediated by homologous recombination between two directly oriented nontandem repeats at the MAT and HMR loci (Herskowitz 1988). The Hawthorne deletion was unusual in that it was originally isolated in a diploid strain. In contrast, most early deletion studies were performed in haploids. As a result, the identified deletions typically involved small segments spanning only a few loci, since larger deletions often include essential genes. For the well-characterized HIS4 and CYC1 genes, only a small fraction (<1%) of the mutations were deletions (Fink and Styles 1974; Sherman et al. 1974), which occurred at these loci at a frequency <10−8/cell division.
Strain-specific hotspots for deletions were identified at two genetic locations. Although deletions of CYC1 are observed at very low frequency in most wild-type strains, Liebman and colleagues (Liebman et al. 1979) found a strain in which deletions of three linked genes (one of which was CYC1) occurred at a relatively high rate (10−5–10−6 events/division). Subsequently, it was shown that in strains that had a high deletion rate, the CYC1 gene was flanked by two 6-kb Ty retrotransposons in direct orientation and that the deletion events were a consequence of homologous recombination between the Ty elements (Liebman et al. 1981). Similarly, recombination between flanking delta LTR elements (the long-terminal-repeat sequences associated with Ty1 and Ty2 elements) was implicated in the high frequency of deletions at the SUP4 locus (Rothstein et al. 1987). These deletions could reflect single-strand annealing between the direct-flanking repeats (Figure 1A) or represent one of the products of unequal crossing over (Figure 1B).
Assay systems have been developed to study the environmental and genetic control of deletions in yeast. One such assay was developed to study deletions mediated by homologous recombination between direct flanking repeats (DEL assay) (Schiestl 1989). In this system, recombination between two directly repeated mutant his3 genes, one with a deletion at the 5′ end and one with a deletion at the 3′ end, produced a functional copy of HIS3 at a very high frequency (10−4/division). In contrast, a different deletion selection system that relied on recombination between micro- or nonhomologous sequences to reactivate a mutant ura2 gene yielded recombinants at a much lower frequency (10−10/division) (Tourrette et al. 2007).
Chen and Kolodner (1999) developed an assay for detecting large deletions involving simultaneous loss of two counterselectable markers (URA3 and CAN1) located ∼10 kb apart (gross chromosomal rearrangements, GCR, assay). These markers were located near the left end of chr5, about 10 kb distal to PCM1, the first essential gene on the chromosome. The types of genetic events detected by this assay include interstitial deletions, terminal deletions followed by telomere addition, and translocations between chr5 and other chromosomes (Chen and Kolodner 1999). For both the translocations and interstitial deletions, the rate of events in a wild-type strain is very low (<10−9/division); the breakpoints of the events occur between CAN1 and PCM1 and typically involved micro- or no sequence homology. Most of these rearrangements reflect nonhomologous end-joining (NHEJ) events, although homologous-recombination (HR) events involving very small homologies are also observed (Putnam et al. 2005). More recent versions of the GCR assay showed that the introduction of homology either from a dispersed repeated gene family member (HXT13) or from a Ty element inserted between CAN1 and PCM1 elevated the rate of URA3–CAN1 codeletions by two to three orders of magnitude. Most of these deletions were a consequence of nonreciprocal translocations between the repeat on chr5 and a homologous sequence elsewhere in the genome and were associated with amplification of the terminal segment of the donor chromosome (Figure 1D) (Putnam et al. 2009; Chan and Kolodner 2011, 2012). In summary, the frequency of deletions in yeast is highly dependent on the context of the reporter gene. In regions in which the reporter is flanked by directly repeated sequences, deletions are frequent and occur through homologous recombination; in regions without repeats, deletions are rare and often involve NHEJ or other microhomology pathways.
Although duplication events in yeast have been less investigated than deletions, there appear to be more diverse types of mechanisms involved. One distinction between duplication events is whether they involve an increase in copy number of a preexisting duplication or duplication of a sequence that is initially present in single copy. Alterations in copy number of the tandemly repeated ribosomal RNA (rRNA) genes or CUP1 genes represent the first class of events. For these types of genes, alterations in copy number occur as a consequence of HR of a variety of types including unequal crossovers (Petes 1980; Szostak and Wu 1980; Welch et al. 1990), gene conversion (Welch et al. 1990; Gangloff et al. 1996), and single-strand annealing (SSA)(Ozenberger and Roeder 1991). Some of these events are remarkably frequent. For example, unequal crossovers within the rRNA gene array occur at a frequency of at least 10−2/mitotic division (Szostak and Wu 1980) and a frequency of ∼10−1/meiotic division (Petes 1980).
In contrast, the frequency of duplicating sequences that are initially present in single copy is much lower, although very dependent on chromosome context and ploidy. A number of reporter systems have been used to detect duplications; differences in the types of events recovered are likely to reflect the chromosome context of the reporter gene rather than the reporter gene per se. Duplications of the ADH2 or ADH4 genes can be selected on medium containing antimycin A (Dorsey et al. 1992). Duplications of RPL20B can be selected as normal-growing derivatives that display a rescue of the slow-growth phenotype of their rpl20AΔ progenitor strain (Koszul et al. 2004; Payen et al. 2008), and duplications of a segment of the ura2-15-30-72 allele can be recovered as Ura+ revertants (Schacherer et al. 2005, 2007). Narayanan et al. (2006), through an approach similar to the one used in the present study, identified cells with amplification of a cassette containing CUP1 and SFA1, using sequential selection on medium that contained high levels of copper and formaldehyde. Finally, high-order amplification (>10 extra copies) of the partly defective leu2-d allele can be selected as Leu+ prototrophs (Erhart and Hollenberg 1983; Watanabe and Horiuchi 2005). A duplication screening system has also been developed based on the color displayed by cells carrying one (pink) or two or more copies (red) of the ade3-2p reporter gene (Koshland et al. 1985; Green et al. 2010).
Less specific selection regimens also have been used to obtain duplications. For example, growth of yeast cells in medium with low levels of phosphate result in duplication of the linked PHO3 and PHO5 genes (Hansche et al. 1978), cells grown in limited glucose have duplications of the hexose transporter encoded by HXT6 (Brown et al. 1998; Dunham et al. 2002), and cells selected in medium with low sulfur concentrations amplified the SUL1 gene (Gresham et al. 2008). Amplification of the HTA2–HTB2 histone genes has also been identified as a dosage compensation mechanism for the deletion of HTA1–HTB1 (Libuda and Winston 2006). Finally, duplications (and deletions) in yeast have been detected without any selection in irradiated diploids (Argueso et al. 2008), in strains with an HO-induced DNA break (Hoang et al. 2010), and in genetically unstable tel1mec1 strains (Vernon et al. 2008; McCulley and Petes 2010).
The types of duplications commonly observed can be classified into six groups: (1) interstitial segmental duplications in which the breakpoints are in repeated sequences in direct orientation (Koszul et al. 2004; Libuda and Winston 2006; Argueso et al. 2008; Payen et al. 2008; McCulley and Petes 2010); (2) segmental duplications unassociated with repeats at the duplication breakpoints (Koszul et al. 2004; Schacherer et al. 2005, 2007; Payen et al. 2008); (3) interstitial duplications in which the duplicated segments are in an inverted orientation (Moore et al. 2000; Rattray et al. 2005; Watanabe and Horiuchi 2005; Narayanan et al. 2006); (4) terminal duplications of part of one chromosome arm associated with a terminal deletion of another chromosome (Dunham et al. 2002; Umezu et al. 2002; Argueso et al. 2008; Kim et al. 2008; Vernon et al. 2008; Hoang et al. 2010; McCulley and Petes 2010; Chan and Kolodner 2011, 2012); (5) linear extrachromosomal plasmids with the amplified copies in inverted orientation (Dorsey et al. 1992; Narayanan et al. 2006); and, (6) interstitial triplications of a genomic segment containing an origin of replication flanked by short inverted repeats (Brewer et al. 2011).
Class 1 duplications can be generated by unequal crossovers (Figure 1B) or by break-induced replication (BIR) in which a double-strand break (DSB) in one repeat is repaired using a nonallelic repeat on a sister chromatid or a homolog. In one system (Payen et al. 2008), duplications of this type were dependent on Pol32p, arguing that these events occurred by BIR. Class 2 duplications are likely to reflect BIR events in which the invading end utilizes very small (<10 bp) regions of homology (Figure 1C) (Schacherer et al. 2005, 2007; Payen et al. 2008); such events have been termed “microhomology-induced replication” (Hastings et al. 2009). Class 3 duplications are associated with the processing or replication of inverted repeats located near the reporter gene. In one study, the initiating event is likely to be processing of a cruciform (Narayanan et al. 2006). Most class 4 events are a consequence of a DSB within or near a repeat that is repaired by a BIR event involving a repeat located elsewhere in the genome (resulting in a translocation as in Figure 1D). Since the net result of the translocation event is a large terminal deletion of one chromosome arm and a large duplication of another chromosome arm, class 4 events are more often observed in diploid strains (Dunham et al. 2002; Umezu et al. 2002; Argueso et al. 2008; Kim et al. 2008; Vernon et al. 2008; Hoang et al. 2010; McCulley and Petes 2010). In haploids, only repeats very close to the end of chromosomes can support this type of event (Putnam et al. 2009; Chan and Kolodner 2011, 2012). In the two class 5 events analyzed, the reporter genes were located near the telomeres (Dorsey et al. 1992; Narayanan et al. 2006). The formation of the palindromic plasmid in one system reflected the processing of a small palindromic repeat located centromere proximal to the reporter gene (Narayanan et al. 2006). Finally, class 6 inverted triplication events observed at the SUL1 locus have been proposed to occur through the extrusion of a circular DNA intermediate formed during replication, followed by reintegration into the genome via homologous recombination (Brewer et al. 2011).
In summary, there are a variety of mechanisms that produce and select for CNVs in S. cerevisiae. The relative importance of these mechanisms is dependent on the chromosome context (in particular, whether the reporter gene is flanked by repeats) and ploidy. In our study, we optimized a system to select for low-order amplification events (one extra copy) that more closely resemble the duplications associated with human disease. We used this system to examine gene duplications in both haploids and diploids and in wild-type and mismatch-repair defective strains. In the chromosome context that we examined (reporter gene in the middle of the right arm of chr5), most duplications reflected homologous recombination between flanking Ty repeats in haploids, whereas diploids displayed mostly nonreciprocal translocations between Ty elements on chr5 and Ty elements on other chromosomes, particularly chr13.
Materials and Methods
Yeast strains and plasmids
All strains used in the CNV assay were isogenic with strain MS71, except for noted locus-specific changes introduced by transformation (Lemoine et al. 2005). Allele combinations were obtained by mating isogenic haploid strains of opposite mating types and selecting segregant spores with the desired genotypes. MS71 is essentially isogenic to the CG379 strain background (Morrison et al. 1991; Argueso et al. 2008). The specific strains and oligonucleotides used in this study are listed in supporting information, Table S1 and Table S2, respectively. A detailed description of the constructions of specific strains and plasmids is presented in File S1.
Culture media and CNV selection conditions
Unless otherwise noted, yeast cells were grown in YPD-rich media or in SC drop-out media (Rose et al. 1990). The selection of clones carrying amplification events was performed by first streaking cells to single colonies in YPD plates (2 days, 30°), and then inoculating these colonies in 5 ml liquid YPD (18 hr, 30°). Dilutions containing ∼2 × 106 cells from these cultures were plated on SC supplemented with a complete drop-out mix, 150 μM CuSO4, and 1 mM formadehyde. Formadehyde is not stable in aqueous solution; therefore, 1 M dilutions [813 μl of 37% methanol stabilized stock (Fischer Scientific BP-531) in 9.187 ml of sterile water] were prepared fresh for each batch of media and mixed in immediately before pouring the plates. Plates were always used within 24 hr of pouring. Cells were incubated in this media at 30° and FA/Cu-resistant colonies were observed after 3–5 days. Only one colony per culture was examined in downstream experiments to ensure independence. Candidate CNV clones (CFRs) were streaked to single colonies nonselectively in YPD without FA or Cu for 2 days, and then the purified clones were patched in YPD, grown overnight, and then frozen. The FA/Cu-resistance phenotype of each clone was retested by growing overnight cultures in 5 ml liquid YPD, and then serial dilutions from the cultures were spotted on to SC-complete Cu 150 μM/FA 1 mM alongside their corresponding parental strain. Plates were incubated at 30° for 4 days before the differential growth phenotype was assessed.
The nature of the chromosomal rearrangements present in the selected CFR clones was determined using a molecular karyotype analysis, including PFGE and array CGH, as described previously (Argueso et al. 2008). The detailed procedures are presented in File S1. The full set of copy-number variation events detected by array CGH in all clones examined is shown in Figure S1.
Analysis of loss-of-heterozygosity
Independent clones derived from MS71xYJM789 hybrid background diploids carrying allelic mitotic recombination products resulting in loss-of-heterozygosity (LOH) on the right arm of chr13 were selected in SC-complete plates with 5-FOA. The LOH breakpoints were analyzed from two sets of clones. The first set of 5-FOA-resistant clones was derived from strain JAY408 and was analyzed with custom SNP genotyping microarrays and data analysis methods described previously (St Charles et al. 2012). We also isolated a second set of LOH clones derived from strain JAY800. This strain was the one used to measure the rate of LOH on the right arm of chr13, by counting colonies growing in 5-FOA (nonpermissive) and in SC complete (permissive) and by using the values in calculations through the method of the median (Lea and Coulson 1949). LOH breakpoints from this set were analyzed by PCR and restriction fragment length polymorphisms at EcoRI (primers JAO1031/JAO1032) and BglII (primers JAO1029/JAO1030) as described in Results.
Results
A new selection system for CNV
De novo CNV is a pervasive source of genetic diversity that has been increasingly associated with phenotypic consequences in humans, including disease. We set out to develop a new assay system to detect chromosomal rearrangements involving de novo gene amplification in yeast, especially those events resulting in only one additional copy of the affected genomic segments. To do this, we took advantage of a previous system that used the SFA1 and CUP1 genes to detect amplification through their gene dosage-dependent phenotypes. SFA1 encodes formaldehyde dehydrogenase, which detoxifies formaldehyde (FA), and CUP1 encodes methallothionein, which sequesters copper ions (Cu) from solution. Cells harboring amplification of these genes display increased tolerance to formaldehyde and copper, respectively, and these phenotypes have been used to select for gene amplification (Narayanan et al. 2006). A limitation of this earlier system, however, was that it primarily identified clones carrying multiple copies of the reporter genes (>10-fold increase in some cases) because the difference in phenotype between amplification levels was subtle. We therefore decided to optimize the selection conditions with the goal of identifying clones with genome rearrangements that more closely resembled those seen in human de novo CNV (i.e., only one extra copy of the amplified genes).
We first deleted the endogenous copy of SFA1 and its regulatory regions, and we also removed the entire cluster of tandem CUP1 repeats. Next, we constructed a CNV reporter cassette containing one copy of the SFA1 and CUP1 genes, as well as a selective marker for integration (Hph, resulting in hygromycin B resistance). This cassette was integrated on chr5 (Figure 2A) between the DDI1 and UBP5 genes. This region is also flanked by the YERCTy1-1 and YERCTy1-2 repetitive elements that we had previously shown to be the most active for the formation of CNV-associated nonallelic homologous recombination (NAHR) chromosomal rearrangements following induction of DNA DSBs by ionizing radiation (Argueso et al. 2008). This region, and the YERCTy1-1 element in particular, had also been shown by other studies to frequently participate in NAHR under various conditions, including spontaneous rearrangements (Narayanan et al. 2006; McCulley and Petes 2010; Chan and Kolodner 2011, 2012; Cheng et al. 2012).
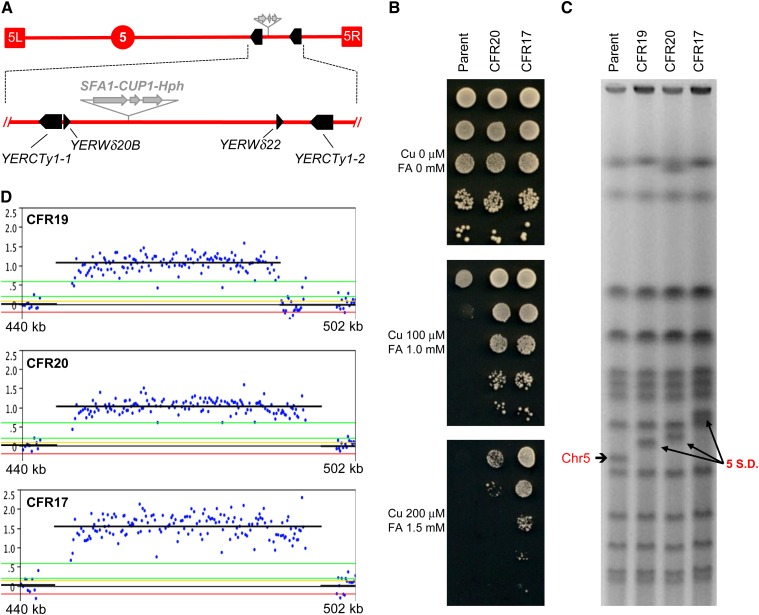
Figure 2.
Identification of chr5 amplifications in haploids: Segmental duplications. (A) Schematic representation of the region of chr5 where the SFA1–CUP1 CNV reporter was inserted (shaded arrows). The reporter also contained a drug-resistance marker, either Hph (hygromycin B), as shown, or Kan (G418–geneticin). Terminal boxes correspond to the left and right chr5 telomeres (5L and 5R, respectively), and the circle represents CEN5. Bottom, expanded view of the region involved in segmental amplifications (440–502 kb), showing full-length Ty1 elements as solid arrows and solo LTRs as arrowheads according to their orientation. (B) FA/Cu-resistance phenotypic differential between the parental haploid strain (JAY372) containing one copy of the reporter and CFR clones carrying two (CFR20) or three (CFR17) copies. Serial dilutions of these strains were spotted on plates containing Cu and FA at the concentrations indicated to the left. (C) PFGE showing the karyotypes of the parental haploid and three CFR clones carrying different segmental amplifications on chr5. The parental size chr5 is indicated to the left and the segmental amplifications are indicated to the right (5 SD). (D) Detailed view of chr5 array-CGH plots showing the Log2 Cy5/Cy3 signal intensity (copy number) and the rearrangement breakpoints for the three CFRs from C. The plots are aligned directly below to the expanded section of A such that the breakpoints correspond to the repeat sequences. Each blue dot corresponds to the signal of a specific probe in the region. The black horizontal lines correspond to the average signal for probes in a region of amplification. Neutral signal corresponds to no copy-number change (1× in haploids); positive signals corresponding to two copies (∼1.0) or three copies (∼1.5) are shown.
Previous studies that used SFA1 and/or CUP1 as amplification reporters used either single or sequential selection for resistance to FA or Cu. Given that these genetic resistance mechanisms are completely independent of each other, we reasoned that simultaneous selection for FA and Cu might create a synergistic effect that would provide a more discrete phenotypic differential between strains carrying one, two, or three copies of the reporter. To test this hypothesis, we built both haploid and diploid strains carrying known numbers of reporter insertions either at the chr5 site described above or on chr4, at the native SFA1 locus. These strains were then tested in media containing incremental concentrations of FA and Cu at various combinations to identify optimal conditions that would inhibit the growth of haploid cells carrying one copy of the reporter, but allow normal growth of cells carrying two or more copies. Likewise, we looked for a combination of FA and Cu that inhibited the growth of diploid cells carrying two homozygous copies of the reporter, but supported growth of cells carrying three or more copies. Representative FA/Cu concentration optimization assays showing the gene dosage-dependent resistance phenotype are presented in Figure S2 and Figure S3. We found that haploid cells with one copy of the reporter had a similar level of FA/Cu resistance as diploids carrying two homozygous copies and that minimal media containing 1.0 mM FA and 150 μM CuSO4 prevented colony formation by these strains. However, this inhibition could be reliably maintained only below a critical plating density of ∼200 cells/mm2, or ∼106 cells per standard Petri dish. Above this cell concentration, we observed sporadic growth of small background colonies, possibly due to a filtering effect by the excess stagnant cells.
Once the optimal conditions for simultaneous FA/Cu selection had been established, we set out to isolate clones carrying spontaneous chr5 amplification events. We grew cultures of the haploid and diploid test strains, plated appropriate dilutions on FA/Cu, and incubated them for 4 days at 30° to select for spontaneous copy-number variants. To ensure independence, we selected only one FA/Cu resistant colony from each culture, and isolated single-colony derivatives of these strains using rich media without FA/Cu. These purified candidate clones were named CFR (for copper formaldehyde resistant) and were retested in media containing FA/Cu comparing their phenotype to that of the respective parent strain (Figure 2B and Figure 3A). The majority of the clones retested positive (102 resistant of 115 CFRs in the full strain set; Table 1), confirming that the selection conditions worked as intended. Even though the sporadic growth of background colonies on densely plated cultures prevented us from calculating high confidence CNV mutation rates, we estimated that gene amplification on chr5 occurred spontaneously at a rate ranging from 10−6 to 10−7 events/cell division. We focused the remainder of our study on the characterization of the spectra of chromosomal rearrangements associated with the various experimental conditions described below.
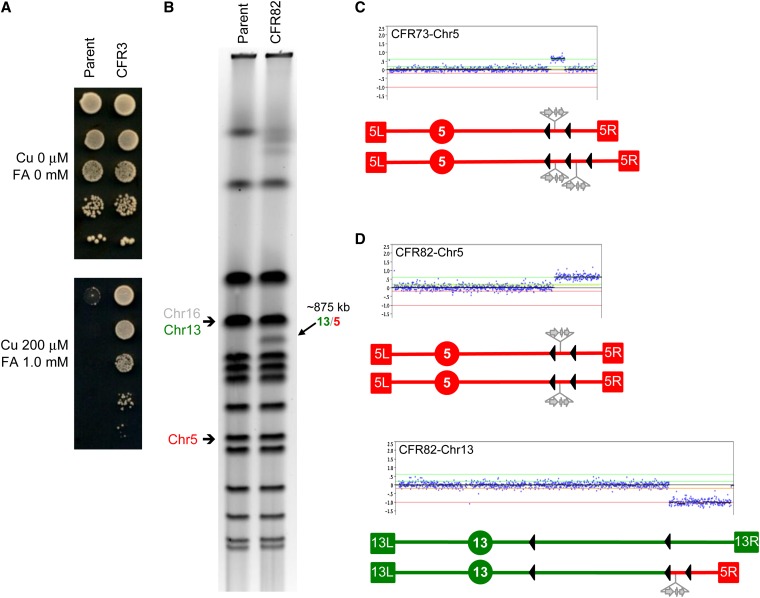
Figure 3.
Identification of chr5 amplifications in diploids: chr13/chr5 translocation. Schematic representations are as in Figure 2. Chr5 is shown in red and chr13 is shown in green. (A) FA/Cu-resistance phenotype in the parental diploid strain JAY350 (two reporter copies) compared to a derivative resistant clone CFR3 (three copies). (B) PFGE showing the karyotype of the parental diploid strain JAY350 and of the derivative resistant clone CFR82 carrying the indicated recurrent 875 kb chr13/chr5 translocation. An identical translocation was also present in clone CFR3 shown in A. The parental size chr5 and chr13 are indicated to the left; chr16 comigrates with chr13 and is also indicated. (C) chr5 array-CGH plot (top) and schematic representation (bottom) of the karyotype in the CFR75 clone showing one parental size homolog and one homolog containing a segmental duplication between the YERCTy1-1 and YERCTy1-2 elements (solid arrowheads). (D) Array-CGH plots and schematic representation of chr5 and chr13 in the CFR82 clone. The breakpoints correspond to YERCTy1-1 in chr5 and YMRCTy1-5 in chr13. For position reference, the chr13 centromere-proximal (left) element YMRCTy1-4 is also represented (see also Figures 4C and 5B).
Table 1. Summary of the karyotype changes associated with the CNV reporter in the CFR clones.
| Haploids |
Diploids |
||||
|---|---|---|---|---|---|
| WT | msh2Δ | WT | msh2Δ | ymrcty1-5Δ | |
| CFR clones isolated | 26 | 22 | 29 | 22 | 16 |
| Positive CFR upon retest | 24 | 21 | 26 | 21 | 10 |
| Karyotype changes | |||||
| Chr5 segmental duplicationa | 22 | 5 | 5 | 2 | 0 |
| Chr13/chr5 translocationb | 0 | 0 | 20 | 8 | 3 |
| Chr7/chr5 translocation | 0 | 0 | 0 | 0 | 5 |
| Chr5 isochromosomec | 0 | 0 | 0 | 0 | 1 |
| Chr5 aneuploidyd | 0 | 0 | 0 | 0 | 1 |
| No karyotype change | 2 | 16 | 1 | 11 | 0 |
| CFR clones with CGH data | 9 | 5 | 9 | 10 | 10 |
Longer than normal chr5 molecules harboring tandem duplications or triplications of a segment of chr5 containing the CNV reporter flanked by direct repetitive full-length Ty1 or solo δ LTR elements.
CFR511, a clone derived from the ymrcty1-5Δ homozygous diploid (JAY510), had a complex rearrangement consisting of a deletion of the right arm of chr13 from YMRCTy1-4 to the right telomere, joined by a segment of chr5 containing a quadruplication of the segment containing the CNV reporter between YERCTy1-1 and YERCδ26, in addition to one copy of the terminal segment between YERCδ26 and TEL05R.
CFR510, a clone derived from the ymrcty1-5Δ homozygous diploid (JAY510), had an isochromosome 5 formed by loss of the left arm from TEL05L to the Watson Ty1 element inserted at the ura3-52 allele, joined by the segment from right arm from YERCTy1-1 to TEL05R. This isochromosome was present in addition to the two parental size copies of chr5, both of which were retained in CFR510.
CFR512, a clone derived from the ymrcty1-5Δ homozygous diploid (JAY510), had three copies of the parental size chr5, a simple aneuploidy of chr5.
Characterization of gene amplification chromosomal rearrangements
After confirming the resistance phenotype, we analyzed all CFR clones for possible chromosome size changes using pulsed-field gel electrophoresis (PFGE). Among the haploid CFRs, we observed loss of the parental-sized chr5 coupled with the appearance of larger new chromosomal bands (Figure 2C). In addition, we also analyzed several CFRs by array CGH to directly test whether the FA/Cu-resistance phenotype was due to CNV (Figure 2D). As expected, most CFRs had additional copies of the chr5 region where the CNV reporter had been inserted. The parallel analysis of the PFGE and array-CGH data showed that in haploids the CNV events were all segmental duplications and that the rearrangement breakpoints were two flanking Ty1 elements or flanking delta elements (the long-terminal repeats associated with Ty1) (Figure 2, A and D and Figure S1). For example, clone CFR19 had breakpoints at the Watson-oriented LTR elements YERWdelta20B and YERWdelta22, while CFR20 had breakpoints at the Crick-oriented full-length Ty1 elements YERCTy1-1 and YERCTy1-2. The size increases in the chromosomal molecules carrying these two segmental duplications (∼40 and ∼50 kb, for CFR19 and CFR20, respectively), and the log2 Cy5/Cy3 amplification signals (+1.07 and +1.09, for CFR19 and CFR20, respectively) were consistent with a doubling of the amplified chr5 region mediated by NAHR between the direct repeats.
Another class of rearrangements observed was exemplified by the pattern seen in CFR17. In this case, the breakpoints were exactly the same as in CFR20, but the rearranged chromosome was ∼100 kb longer than the parental chr5, and the array-CGH amplification signal was also higher (Log2 ratio of +1.57). This result confirmed that CFR17 contained a segmental triplication, mediated by the two Ty1 elements in the region. Accordingly, as shown in Figure 2B, the FA/Cu-resistance phenotype of CFR17, with its three copies of the CNV reporter, was noticeably stronger than that of CFR20 with two copies. It is unclear whether the triplication events occurred prior to the initial FA/Cu selection plating, or if they arose initially in cells carrying a duplication that then underwent a secondary NAHR event during the growth of the colony to expand to the 3× level. Since 3× cells are hyperresistant to FA/Cu, they would be expected to rapidly outnumber the 2× parent cells in the colony. It is also possible that such secondary rearrangements could be further stimulated by the growth in media containing FA, a known genotoxic agent (Kumari et al. 2012).
The phenotypic difference between the 2× and 3× reporter dosage levels in haploid strains, as well as our optimization trials, encouraged us to attempt the selection of spontaneous amplification rearrangements in diploid strains homozygous for the chr5 reporter insertion. We isolated several independent WT diploid CFR clones in FA/Cu media, repurified them under nonselective conditions, and found that most retested positive for FA/Cu resistance (Figure 3A and Table 1). In analyzing the karyotype changes in these diploid clones, we found that 5 of 25 CFRs had one chr5 homolog containing Ty1-mediated segmental duplications that were essentially identical to those seen in haploids, in addition to one parental copy of chr5 (e.g., CFR73 in Figure 3C and CFR85 in Figure S6).
The majority of the diploid CFR clones, however, had a class of rearrangements not observed in haploids: nonreciprocal translocations, exemplified by CFR82 (Figure 3, B and D). In this case, the two parental copies of chr5 were unchanged, but one of the copies of chr13 was rearranged by the loss of a 176-kb segment between YMRCTy1-5 and the right telomere (TEL13R) and the addition of a 128 kb chr5 segment from YERCTy1-1 to TEL05R, which included the CNV reporter insertion. YMRCTy1-5 is a Crick-oriented full Ty1 retrotransposon insertion that is present in our strain background near the tR(UCU)M1 tRNA gene (at SGD coordinate 748,219), but that is absent in the S288c S. cerevisiae reference genomic sequence. This chr13/chr5 translocation results in an ∼875-kb chromosome visible in PFGE, with an overall reduction in size of ∼48 kb relative to the parental chr13. The presence of one remaining parental copy of chr13 made the loss of the large chr13 segment viable in the diploids, while the amplified segment of chr5 was responsible for the third copy of the CNV reporter that conferred the FA/Cu-resistance phenotype. This type of event is also shown schematically in Figure 1D.
Surprisingly, 20 of 25 independent WT diploid CFRs had the same 875-kb translocation band as CFR82. We used array CGH to analyze genomic DNA from three other of these clones and found the same chr13 deletion and chr5 amplification pattern in all three. These results indicated that the 875-kb chr13/chr5 translocation was a recurrent NAHR event. The fact that no other reporter-associated translocations were observed in the WT diploids suggested that 875-kb event must occur spontaneously at a rate substantially higher than that of other possible ectopic recombination interactions.
In addition to the chromosomal rearrangements that were associated with amplification of the CNV reporter, in a few cases, we also observed unselected karyotype changes in other regions of the genome. These unselected events were observed only in diploids and are shown in Figure S1. They included three trisomies (chr1, chr3, chr16) and five chromosomal rearrangements, four segmental duplications (chr3, chr4, chr12, chr13), and one nonreciprocal translocation between the right arm of chr3 and the left arm of chr5 in clone CFR100. The breakpoints in all five unselected rearrangements occurred at full-length Ty or LTR repeats; therefore, they were similar in nature to the selected FA/Cu-resistant chr5 amplifications.
One or more extra copies of the SFA1–CUP1 gene dosage reporter were detected in all but two haploid and one diploid CFR clones (CFR16, 42, and 77, respectively). These three exceptions retested positive in the selective plates, but did not show any detectable karyotype changes in PFGE. In addition, we examined CFR16 by array CGH and also did not detect any gene dosage changes. These results suggested the existence of a secondary mechanism of FA/Cu resistance. We later found that these clones had acquired dominant point mutations in the SFA1 gene. These mutations are presented at the end of the Results section.
Investigation of mechanisms for the recurrent chr13/chr5 translocation
We considered two hypotheses to explain the recurrence of the 875-kb translocation. The first was that YMRCTy1-5 might be more similar in sequence to YERCTy1-1 than any other Ty element in the genome. Since the YMRCTy1-5 sequence was not available in the reference genome, we PCR amplified and fully sequenced this element from our strain. We then constructed a sequence alignment containing YERCTy1-1, YMRCTy1-5, and the other 30 existing full-length Ty1 element sequences in the S288c reference genome (Figure S4). The alignments showed that YMRCTy1-5 was indeed among the most similar Ty1 elements to YERCTy1-1 (99.6% identity; with a 2981-bp segment of perfect homology). Nonetheless, we did find three other Ty1 elements (YMLWTy1-2, YLRCTy1-1, and YPLWTy1-1) that had even higher identity and had the same orientation with respect to the centromere as YERCTy1-1. These elements should in principle be competent to participate in NAHR with YERCTy1-1 to generate other stable chromosomal translocations.
One function of the DNA mismatch repair system is to reduce the frequency of recombination between repeated genes that have sequence differences (Harfe and Jinks-Robertson 2000; George and Alani 2012). If near perfect sequence identity was important in affecting the recombination partners of YERCTy1-1, then by removing the DNA mismatch repair system we might be able to relax this constraint and observe translocations involving diverged Ty1 sequences from other chromosomes. We therefore tested haploid and diploid derivatives with a deletion of the MSH2 gene, which is required for all functions of the mismatch repair system, including antirecombination activity (Surtees et al. 2004). We recovered 44 msh2Δ and msh2Δ/msh2Δ CFRs and examined their karyotypes by PFGE and array CGH. Only two classes of rearrangements were found in these mutants, and they were the same seen in the WT strains: chr5 segmental duplications and chr13/chr5 translocations. This result, coupled with the existence of other suitable Ty1 elements in the genome (>99% identity to YERCTy1-1), ruled out the recurrence of the chr13/chr5 translocation due to a sequence identity mechanism. However, one of the msh2Δ/msh2Δ clones (CFR99) had a chr13/chr5 translocation chromosome that was slightly larger than the others in its class (∼895 kb vs. 875 kb), and had a chr13 array-CGH deletion breakpoint at YMRWdelta19, a solo LTR element positioned 20.6 kb distal to YMRCTy1-5 (File S1 and Figure S5). The amplified region from chr5 was the same as in the other translocations, but the recombination event likely involved YERWdelta20B, which is in the compatible orientation to generate a stable monosomic rearrangement.
Even though there were no major qualitative differences in the chromosomal rearrangements found in the mismatch repair-deficient CFRs, we did find a substantial shift in their abundance relative to the number of clones without any detectable PFGE or array-CGH changes (e.g., CFR93, Figure S6). Clones with normal karyotypes accounted for ∼65% of all msh2Δ and msh2Δ/msh2Δ CFRs, suggesting the prevalence of a nucleotide mutation FA/Cu-resistance mechanism, rather than CNV (Table 1 and discussed below).
The second possibility we considered to explain the recurrence of chr13/chr5 875-kb translocation was the presence of a putative fragile site on the right arm chr13 located near the right telomere. By this explanation, YMRCTy1-5 would not be actively responsible for this translocation, but instead, it would simply be the predominant site of repair of the precursor DSB associated with this fragile site. This scenario is plausible because YMRCTy1-5 is the most distal full-length Ty1 element on the right arm of chr13 and, therefore, the first large NAHR substrate available to repair a DSB being resected from the right end of the chromosome. This explanation was also consistent with the observation of the CFR99 clone (Figure S5), which, possibly due to a relaxed sequence identity requirement in msh2Δ/msh2Δ, was repaired at an LTR repeat located distal (SGD coordinate 768,548) to the larger homology at YMRCTy1-5.
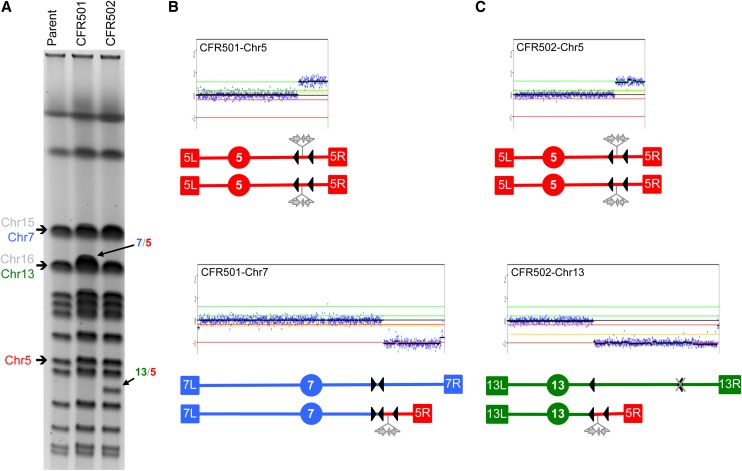
To test the fragile site hypothesis, we generated isogenic diploids with homozygous deletions of YMRCTy1-5. In this strain, DNA lesions formed at distal positions on the right arm and resected toward the centromere would no longer find a suitable recombination substrate, preventing the formation of the 875-kb chr13/chr5 translocation seen in WT cells. We analyzed the karyotypes of 10 independent CFRs derived from this strain and observed that the pattern of genome rearrangements had changed (Table 1). Remarkably, three of these CFRs still displayed deletions on the right arm of chr13, but the breakpoint in all three was at YMRCTy1-4 (e.g., CFR502, Figure 4C) located near SGD coordinate 375,000. In the absence of YMRCTy1-5, YMRCTy1-4 became the first available full-length Ty1 NAHR substrate for repair of a lesion being resected from the right. Assuming that such lesion originated at a putative fragile site distal to YMRCTy1-5, this resection would have to span at least an additional 370 kb to reach YMRCTy1-4. Chromosomal rearrangements that involve long-range processing of a distally located DSB up to a Ty element have also been observed previously (Hoang et al. 2010).
Figure 4.
Alternative translocations in diploids lacking the YMRCTy1-5 repeat element. Schematic representations are as in Figure 2. Chr5 is shown in red, chr13 is shown in green, and chr7 is shown in blue. (A) PFGE showing the karyotypes of parent diploid strain JAY510 and of derivative clones CFR501 (containing a chr7/chr5 translocation) and CFR502 (containing a chr13/chr5 translocation). The parental bands for chr5, chr7, and chr13 involved in the rearrangements are indicated to the left; also indicated are chr16 and chr15, which comigrate with chr13 and chr7, respectively. (B) Array-CGH plots and schematic representation of chr5 and chr7 in the CFR501 clone. The breakpoints correspond to YERCTy1-1 in chr5 and YGRWTy2-2/YGRCTy1-3 in chr7. (C) Array-CGH plots and schematic representation of chr5 and chr13 in the CFR502 clone. The breakpoints correspond to YERCTy1-1 in chr5 and YMRCTy1-4 in chr13. Note the deletion of the YMRCTy1-5 element in this strain, represented by the gray X over the normal position of this element (distal/right arrowhead).
Another interesting observation in the strain with the deletion of YMRCTy1-5 was the appearance of a different recurrent rearrangement. Five of 10 CFRs had the same 925-kb chr7/chr5 translocation that comigrated with chr16 and chr13 in PFGE (e.g., CFR501; Figure 4A). This translocation was composed of a deletion of chr7 right arm sequences distal to YGRCTy1-3 and amplification of chr5 sequences from YERCTy1-1 to the right telomere (Figure 4B). In addition, we observed one strain that was trisomic for chr5 and one strain with a chr5/chr5 translocation (isochromosome) involving YERCTy1-1 and the Watson-oriented Ty1 element inserted at the ura3-52 allele on the left arm.
One explanation for the changes observed in the CNV spectrum in the ymrcty1-5Δ/ymrcty1-5Δ diploids was that the extensive resection required for ectopic repair of the distal DNA lesion lowered the recovery of CFRs with chr13 deletions. This lengthier and more time-consuming resection would provide additional opportunities for allelic repair of the DSB using the intact sister chromatid or homolog, an outcome that is not detectable in the CNV assay. As a consequence, other less-abundant NAHR events became more prevalent among the FA/Cu-resistant clones. The results obtained with the ymrcty1-5Δ/ymrcty1-5Δ diploids also uncovered the existence of a second recurrent translocation event, involving the right arm of chr7.
Supporting evidence and initial mapping of a candidate chr13 right arm fragile site
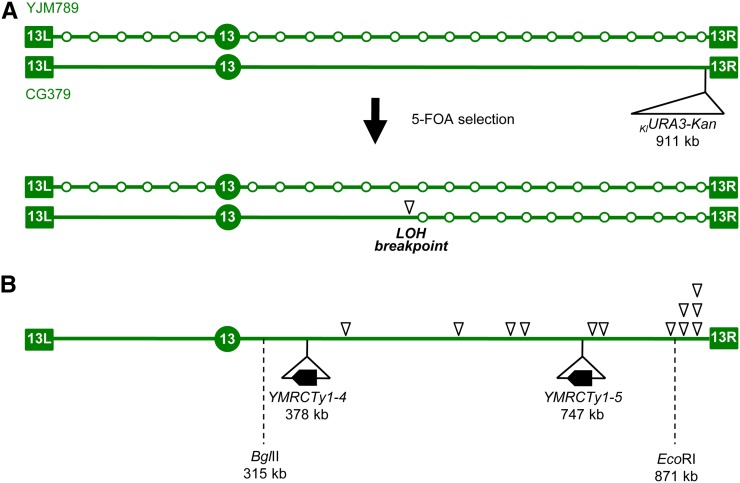
The results of the CNV assays described above suggested the existence of a frequent DNA lesion on the right end of chr13, responsible for triggering the recurrent translocations observed in the diploid CFRs. To obtain independent support for this possibility, we generated a hybrid diploid strain marked to allow the detection of LOH events on chr13. Allelic recombination repair is a much more frequent event than NAHR; however, because our diploid CNV strain is isogenic, it was not suitable to detect LOH. We therefore crossed our haploid strain to a haploid isogenic with YJM789, a highly diverged strain background (Wei et al. 2007). The resulting hybrid diploid has a large number of heterozygous single-nucleotide polymorphisms (SNPs) that can be followed to analyze recombination on a genome-wide basis (Lee et al. 2009). In addition to the SNPs, we inserted a URA3–Kan cassette on the CG379-derived homolog of chr13, downstream of the ADH6 gene about 10 kb from the right telomere (Figure 5A). A DSB formed in the CG379 homolog and repaired through allelic recombination with the YJM789 homolog may result in LOH of the markers between the site of recombination repair and the right telomere, including the loss of the URA3 marker. The presumption in this experiment was that a discrete fragile site in this region would produce a bias in the distribution of sites of LOH recombination, causing an excess of breakpoints to be detected in its vicinity (Tang et al. 2011).
Figure 5.
Initial mapping of a candidate fragile site on the right arm of chr13. (A) Experimental rationale. Schematic representation of chr13 in the hybrid diploid strain (JAY800 and JAY801), formed by mating haploids of the diverged strain backgrounds YJM789 and CG379. The open circles in the YJM789-derived chromosome represent SNP positions. The KlURA3-Kan marker was inserted in the CG379-derived chromosome, downstream of the ADH6 gene, near the right telomere. Mitotic crossovers initiated by DNA breaks in the CG379-derived chromosome and repaired using the YJM789 homolog as template result in clones that are resistant to 5-FOA and homozygous for the right end of the YJM789-derived chr13. Genotyping with SNP microarrays can determine the precise site of allelic mitotic recombination (LOH breakpoint, open arrowhead). This site should occur in close proximity to the precursor DNA lesion. (B) LOH breakpoint positions for 12 independent 5-FOA-resistant clones determined by SNP microarray genotyping showing a clustering of breakpoints near the right end of chr13. Also indicated are the relative positions of the two Ty1 elements involved in the chr13/chr5 translocations described in Figures 3 and 4 and the position of the SNP–RFLP markers BglII and EcoRI. The EcoRI marker is described in the Results. The genotype at BglII marker site was also determined and found to be heterozygous in most 5-FOAR clones (117/131; 89.3%).
Independent spontaneous LOH events were selected in plates containing 5-FOA, and the rate of these events was determined to be 2.4 × 10−5 LOH events/cell division (95% confidence interval: 1.7–3.9 × 10−5). We then analyzed 12 FOAR clones using custom allele-specific microarrays to determine their genome-wide SNP genotype (CG379, YJM789, or heterozygous) (St Charles et al. 2012). As expected from the 5-FOA selection, all 12 clones analyzed displayed terminal LOH on the right arm of chr13. Fifty percent of the detected LOH breakpoints (inverted triangles in Figure 5B) were found in the 41-kb region immediately proximal to the URA3–Kan insertion site, which corresponded to only 6.3% of the possible recombination window (distance CEN13 to URA3-Kan = 643 kb). The other half of the 5-FOAR clones had breakpoints scattered through the rest of the right arm.
Since the pattern observed in the SNP microarray experiment was suggestive of a bias in the distribution of LOH breakpoints, we examined a larger number of FOAR clones to assess the significance of this find. We genotyped these additional clones for a SNP marker 40 kb proximal to URA3–Kan. This SNP was located at the approximate boundary of the breakpoint cluster detected by microarrays and corresponded to an EcoRI restriction site (SGD chr13 coordinate 870,864) in the CG379 homolog that was absent in the YJM789 homolog. Using primers that flanked the polymorphism, we generated a PCR fragment for 130 independent 5-FOAR diploids, treated the fragments with EcoRI, and examined the products by gel electrophoresis. Of the 130 new LOH clones, 109 were homozygous for the YJM789 SNP, 20 were heterozygous, and 1 was homozygous for the CG379 SNP (possibly within a mitotic gene conversion tract associated with LOH). Therefore, 21 of 130 clones had an LOH breakpoint distal to the EcoRI site. Based on the size of this interval relative to the distance between CEN13 and the URA3–Kan insertion, we expect only eight events. By chi-square analysis, this difference is very significant (P = 2.8 × 10−6). This deviation was even more pronounced when we considered the PCR–RFLP and microarray data together (P = 2.5 × 10−9). We interpret these results as evidence for the presence of a frequent recombination-initiating lesion near the right end of chr13, perhaps a DSB formed at a chromosomal fragile site. Alternatively, it is possible that the right telomere of chr13 is poorly maintained and frequently becomes “uncapped,” allowing resection beginning at the telomere itself and extending into telomere-proximal chromosome sequences.
Characterization of new SFA1 dominant alleles
As discussed above, we detected CFR clones that retested positive for the FA/Cu-resistance phenotype but did not have any detectable chromosomal rearrangements. This class was particularly abundant in the mismatch-repair deficient strains suggesting a nucleotide mutation-based mechanism of resistance. In addition, because we identified several such isolates from diploid strains, these point mutations would have to be dominant/gain-of-function alterations to display the resistance phenotype in the presence of the second wild-type allele.
To characterize this possible mutation, we first crossed two msh2Δ haploid CFR isolates from this class (CFR49 and CFR50, MATα) to a wild-type strain (JAY377, MATa). All three haploid strains had only one copy of the CNV reporter, but CFR49 and CFR50 had the Kan G418 resistance marker linked to the reporter, whereas JAY377 had the Hph hygromycin B resistance marker at the same position (Figure 2A). We tested the corresponding diploids for FA/Cu resistance. Both diploids derived were FA/Cu resistant, confirming that the mutations in the two CFR clones were dominant (Figure 6A). Next, we sporulated and dissected tetrads from these diploids to examine the segregation pattern of the resistance phenotype. We observed two resistant and two sensitive spores, indicating that the mutant phenotype was controlled by only one locus. In addition, we also scored the spores for the Kan and Hph markers associated with CNV reporter. As expected, both markers segregated 2:2, and spores were either G418R HygS or G418S HygR. However, we noted that FA/Cu resistance always cosegregated with G418 resistance. Since both CFR49 and CFR50 had CNV reporters marked with Kan, this result indicated that their FA/Cu-resistance mutations were tightly linked to the CNV reporter, presumably in the SFA1 or CUP1 genes.
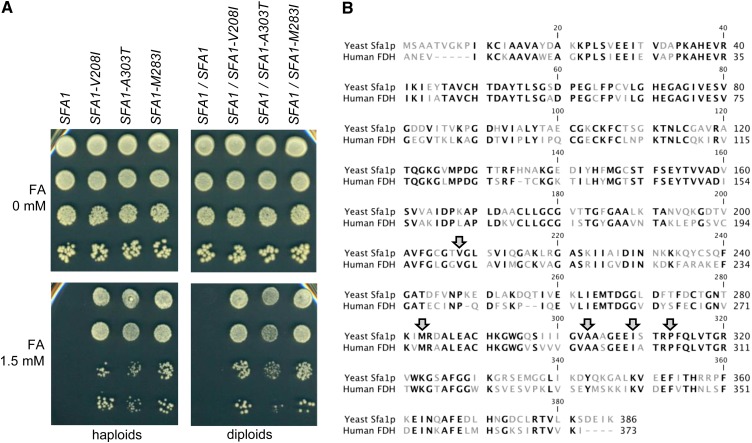
Figure 6.
Characterization of dominant mutant alleles of the SFA1 gene. (A) Formaldehyde (FA)-resistance phenotype in haploid (left) and heterozygous diploid (right) strains carrying three different mutant alleles of SFA1. The genotypes are indicated at the top. Serial dilutions were plated in media containing 0 or 1.5 mM FA as indicated to the left. No CuSO4 was added. (B) Amino acid residue sequence alignment between the S. cerevisiae Sfa1p and the human FDH enzymes. Conserved residues (60.9% identity) are shown in black letters, and diverged residues are shown in gray. Shaded arrows point to the five residues that were substituted in the dominant SFA1 mutations.
We then PCR amplified and sequenced the CNV reporter from 13 independent CNV-less CFRs to find the nucleotide mutations. This analysis showed that all sequenced clones carried mutations in the SFA1 gene. Since the mutations were in SFA1, we repeated the resistance assays in plates containing FA or Cu separately. This analysis showed that the SFA1 mutations did not alter resistance to Cu (data not shown) and that their effect on resistance to FA was quite pronounced (Figure 6A). In fact, a single copy of the mutant SFA1 alleles conferred a level of FA resistance comparable to that seen in a strain carrying three copies of the wild-type SFA1 gene (data not shown).
A total of five different dominant SFA1 alleles were identified. Two of the alleles arose independently multiple times: SFA1–Val208Ile (nucleotide mutation G622A) occurred in nine cases, and SFA1–Ala303Thr (nucleotide mutation G907A) occurred three times. Alleles SFA1–Met283Ile, SFA1–Ile309Met, and SFA1–Pro313Ala occurred once each. All alleles sequenced were isolated from msh2Δ haploids, except for SFA1–Met283Ile, which was isolated in a MSH2 haploid (CFR16).
To gain insight into the potential effect of these mutations on the enzymatic activity of Sfa1p, we analyzed them in the context of the human glutathione-dependent formaldehyde dehydrogenase enzyme (hFDH) for which a crystal structure has been solved (Sanghani et al. 2002a,b). All five dominant alleles corresponded to residues that are conserved between the human and yeast enzymes (Figure 6B). Interestingly, the two alleles most frequently isolated (SFA1–Val208Ile and SFA1–Ala303Thr) are located in the active site of hFDH, in direct contact with the NAD(P)+ coenzyme. This structural arrangement raises the possibility that mutation at these specific residues may destabilize the binding of the coenzyme, facilitating its release from the active site after the reaction with the S-hydromethylglutathione. This would accelerate the rate at which FA is detoxified, therefore enhancing FA resistance without increasing the number of copies of the SFA1 gene.
Discussion
A new assay for copy-number variation
In this study, we took advantage of the gene dosage-dependent phenotypes of the SFA1 and CUP1 genes to develop an optimized assay for the detection of gene amplifications. While several analogous systems have been previously reported in yeast, few of them have the sensitivity necessary to select low-order amplification events. In contrast, our system is capable of identifying spontaneous chromosomal rearrangements, resulting in a simple doubling of the region of interest, going from one to two copies, and also subtler (50%) increases in gene dosage, going from two to three copies. This sensitivity makes the system equally suited for the analysis of amplifications in haploid and diploid cells, whereas most previous systems function exclusively in haploids. We believe our system is an attractive and more germane model for the somatic CNVs commonly observed in humans (Frohling and Dohner 2008; Girirajan et al. 2011) and represents a valuable new tool for the investigation of structural genomic variation in a biomedically relevant context.
Another distinctive quality of this assay is that it allows the detection of chromosomal rearrangements initiated by rare spontaneous DNA lesions, regardless of where in the genome they occur. These lesions may include not only DSBs, but also uncapped telomeres, collapsed replication intermediates that switch DNA template, or any other recombination-initiating event. For example, in the case of the translocations detected in diploids, the precursor lesions likely occurred on chr13 or on chr7, but the CNV reporter was inserted on chr5. Conventional assays that are based on deletions such as DEL or GCR (Schiestl 1989; Chen and Kolodner 1999), or amplification assays using haploids (Payen et al. 2008), are thought to detect rearrangements initiated predominantly by lesions in the immediate vicinity of their respective reporter sequences. The same issue is even more pronounced in assays that rely on the induction of site-specific DSBs (e.g., HO, I-SceI endonucleases) to raise the frequency of recombination up to a high level required for detection. The spontaneous and genome-wide nature of the FA/Cu-resistance assay is a highly desirable feature because, in principle, it should provide experimental results that more broadly represent global genomic instability processes, rather than the local properties of specific loci that may not necessarily apply to other regions.
The versatility of this detection system allowed us to conduct parallel genome stability experiments in the same chromosomal context in haploids and diploids. We observed that the classes of chromosomal rearrangements associated with each ploidy state were quite specific and distinct. In haploids we identified only segmental duplications and, as expected, no associated large deletions. Segmental duplications were generally also observed in most other previously reported haploid genome rearrangement assays, with the notable exception of the translocations detected in the recent iterations of the GCR assay, in which a deletion of the relatively small and nonessential terminal region of chr5 left arm is associated with the duplication of a terminal segment from a different chromosome (Putnam et al. 2009; Chan and Kolodner 2011, 2012). The diploid CFR clones displayed two main classes of rearrangements: ∼15% were segmental duplications similar to those found in the haploids, but most others were nonreciprocal translocations resulting in amplification of the right arm of chr5 (including the CNV reporter) coupled with a deletion of a terminal segment of a different chromosome. We also detected a clone (CFR512) containing a trisomy of chr5, and incidentally, this same clone also carried two unselected trisomies of chr3 and chr16. These and other unselected karyotype changes were observed only in diploid clones. In summary, these data confirmed that diploid cells can tolerate a more diverse repertory of rearrangements than haploids and therefore provide a richer platform to study genome stability.
Another key observation was that all chromosomal rearrangement breakpoints detected in this study contained dispersed repetitive DNA sequences, either full-length Ty or solo LTR elements. No breakpoints were detected at single-copy or microhomology sequences. This pattern was consistent with several other studies that implicated Ty sequences as hotspots for chromosomal rearrangements (Mieczkowski et al. 2006; Scheifele et al. 2009) and confirmed that NAHR is the primary DSB repair pathway responsible for such events in yeast. Interestingly, the spontaneous rearrangements selected in the CFR diploid clones were reminiscent of those we found in isogenic, G2-synchronized diploids that survived exposure to 800 Gy of ionizing radiation resulting in ∼250 DSBs per cell (Argueso et al. 2008). The NAHR-mediated segmental duplications and translocations identified in that high-dose γ-ray experiment were qualitatively indistinguishable from the ones selected spontaneously in this study. The only meaningful difference was the massive drop in the overall frequency of occurrence: two to three rearrangements per 800 Gy surviving cell, compared to one rearrangement per approximately 106 viable cells at the 0-Gy dose. This suggests that the FA/Cu-resistance assay may be successfully used to study CNV formation following low doses of radiation, as well as other clastogenic environmental exposures.
Dominant formaldehyde hyperresistant alleles of SFA1
During the course of our study, we unexpectedly identified dominant mutations in the SFA1 gene that increased the resistance of cells to formaldehyde, presumably through increased activity of the formaldehyde dehydrogenase (FDH) enzyme. This was intriguing and unusual, since mutations that result in the substitution of conserved amino acid residues at the core of an enzyme’s active site are often associated with reduction or complete loss of activity and are typically recessive. This result also suggests that the natural selection forces that have acted on the sequence of SFA1 stopped short of the maximum possible biochemical activity, therefore implying that a super-active FDH may somehow be associated with negative fitness consequences. Regardless of the their role in the evolution of SFA1, the fact that the mutations have significantly increased activity, and consequently a larger FA-resistance phenotypic differential between one and two mutant copies, provided an opportunity for the improvement of this gene as a reporter for gene dosage. Recent work in our laboratories using the SFA1 mutant alleles in the CNV reporter showed a pronounced reduction in background colony growth when cultures were plated at high cell densities (to be presented elsewhere), in effect enabling future versions of the CNV assay that will be fully quantitative.
Detection of recurrent chromosomal rearrangements
Our analysis of spontaneous amplifications selected from diploid clones revealed a remarkable recurrence of a specific chromosomal rearrangement between the right arms of chr13 and chr5. We took two experimental approaches to address this result. First, we created a derivative strain deleted for a central component of the DNA mismatch repair system (msh2Δ/msh2Δ) that should relax sequence identity constraints imposed on recombination partner choice (George and Alani 2012). This strain produced the very same rearrangements as the wild type, indicating that sequence similarity is not an important factor driving the recurrence of this translocation.
The second factor we investigated was the possible presence of a chromosomal fragile site on the right arm of chr13. Previous studies from our group and others have shown that frequent breaks are associated with certain Ty sequences under conditions of replication stress and that these breaks can be potent inducers of chromosomal rearrangements (Cha and Kleckner 2002; Lemoine et al. 2005). Replication stress is a well-characterized activator of fragile site expression in humans (Arlt et al. 2012). However, it is important to note that the fragile nature may not be entirely dependent on replication stress. For example, the same yeast fragile sites have also been detected as hotspots for NAHR even under normal growth conditions (Hoang et al. 2010; Chan and Kolodner 2012).
We tested the candidate chr13R fragile site in two ways. We deleted the YMRCTy1-5 element and observed that in some cases the rearrangements recovered involved a proximal Ty element on the right arm of chr13. We then examined spontaneous allelic recombination clones containing LOH on the right arm of chr13 and found a statistically significant clustering of breakpoints consistent with the presence of a candidate fragile site in the region. Taken together, these results suggested the possibility of a frequent DSB distal to the YMRCTy1-5 element, which could be the trigger for the recurrent chr13/chr5 translocations. Alternatively, a telomere with a partial defect in capping might also account for the high incidence of chr13R rearrangements.
In either the fragile site or weak telomere scenarios, a region of chr13 at least ∼150 kb would have to be resected to expose the homology at YMRCTy1-5 and initiate NAHR. Chromosomal rearrangements involving similarly long resection tracts (break-distal recombination, BDR) have been recently reported (Hoang et al. 2010; Tan et al. 2012). While the DSB at chr13R explanation is attractive, it is unlikely to be the only factor involved. Several other Ty repeats exist in the genome that should in principle be competent to participate in translocations, yet none of those other sites were detected with the wild-type diploid strain. This suggests that there may be other significant contributing cellular mechanisms.
We considered three other factors that may be favoring the detection of this specific NAHR interaction in the FA/Cu-resistance assay. The first is not related to a recombination mechanism, but rather may be associated with the viability of clones. This would be possible if cells carrying the chr13/chr5 translocation somehow had higher FA/Cu resistance and would therefore form colonies more often than cells with other rearrangements. We do not think this is the case for two reasons. We compared the relative viability and growth of clones carrying the chr13/chr5 rearrangements with clones carrying the chr7/chr5 translocation or the chr5 SDs in media containing FA/Cu (data not shown). These three classes of clones appeared to be just as viable between them. In addition, even if chr13/chr5 translocation had a more robust growth, this would not prevent the recovery of other, less robust rearrangements that should also be viable in diploid cells.
Another factor that could create the observed detection bias is the relative proximity between potential recombination partner sequences in the tridimensional structure of the genome. This mechanism has been invoked as a contributor to the formation of recurrent chromosomal rearrangements in cancer cells (Wijchers and de Laat 2011). Thus, we analyzed the available structure of the S. cerevisiae genome (Duan et al. 2010) and asked whether proximity between the right arms of chr5 and chr13 might explain the observed rearrangements. This analysis showed that the reverse is true: these two chromosomes are relatively farther apart from each other than from other chromosomes. This observation suggests that the relative position of a sequence in the static model of the genome may not be as important for recombination as its ability to move around the nucleus during DSB repair. Two recent studies have elegantly demonstrated that yeast DNA sequences undergo a rapid transition from a relatively constrained nuclear localization to an increased mobility state immediately after a nearby DSB is induced (Dion et al. 2012; Mine-Hattab and Rothstein 2012). According to this model, a lesion on chr13 would engage the chr5 donor sequence not because of static proximity, but rather through a higher-than-normal degree of freedom to explore the nuclear space in search for homology. It would be interesting to investigate if the newly discovered transition from static to mobile status varies between different regions of the genome and whether some sequences are able to explore larger nuclear volumes than others following breakage.
Finally, we note that a recent haploid yeast study also identified recurrent nonreciprocal translocations involving dispersed Ty1 elements (Chan and Kolodner 2012). In that study, six Ty1 elements (including YERCTy1-1) were used as donor sequences repeatedly (∼70% of cases) at frequencies much higher than those expected from random choice. As was the case in our analysis, the authors did not find a significant correlation between the relative spatial proximity in nucleus of the recombining elements or their degree of nucleotide sequence similarity. However, their results and our own clearly showed that strong, yet unknown, mechanisms exist in the yeast nucleus that drive specific Ty repeats toward preferential NAHR with others. Understanding these processes could shed some much needed light into the origins of recurrent chromosomal rearrangements in cancer cells.
Supplementary Material
Acknowledgments
We thank Thomas Hurley for helpful discussions on the modeling on the SFA1 dominant mutations onto the crystal structure of the hFDH enzyme. The CNV research in the Petes, Argueso, and Mieczkowski laboratories was supported by an American Recovery and Reinvestment Act National Institutes of Health (NIH)–National Institute of Environmental Health Sciences Challenge Grant (5RC1ES018091-02). In addition, T.D.P. was supported by NIH grants GM24110 and GM52319, and J.L.A. was supported by American Cancer Society grant ACS IRG no. 57-001-53.
Footnotes
Communicating editor: C.-ting Wu
Literature Cited
- Abrahams B. S., Geschwind D. H., 2008. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 9: 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso J. L., Westmoreland J., Mieczkowski P. A., Gawel M., Petes T. D., et al. , 2008. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. USA 105: 11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt M. F., Wilson T. E., Glover T. W., 2012. Replication stress and mechanisms of CNV formation. Curr. Opin. Genet. Dev. 22: 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Payen C., Raghuraman M. K., Dunham M. J., 2011. Origin-dependent inverted-repeat amplification: a replication-based model for generating palindromic amplicons. PLoS Genet. 7: e1002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. J., Todd K. M., Rosenzweig R. F., 1998. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol. Biol. Evol. 15: 931–942. [DOI] [PubMed] [Google Scholar]
- Cha R. S., Kleckner N., 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606. [DOI] [PubMed] [Google Scholar]
- Chan J. E., Kolodner R. D., 2011. A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet. 7: e1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. E., Kolodner R. D., 2012. Rapid analysis of Saccharomyces cerevisiae genome rearrangements by multiplex ligation-dependent probe amplification. PLoS Genet. 8: e1002539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Kolodner R. D., 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23: 81–85. [DOI] [PubMed] [Google Scholar]
- Cheng E., Vaisica J. A., Ou J., Baryshnikova A., Lu Y., et al. , 2012. Genome rearrangements caused by depletion of essential DNA replication proteins in Saccharomyces cerevisiae. Genetics 192: 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V., Kalck V., Horigome C., Towbin B. D., Gasser S. M., 2012. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 14: 502–509. [DOI] [PubMed] [Google Scholar]
- Dorsey M., Peterson C., Bray K., Paquin C. E., 1992. Spontaneous amplification of the ADH4 gene in Saccharomyces cerevisiae. Genetics 132: 943–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y. J., et al. , 2010. A three-dimensional model of the yeast genome. Nature 465: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham M. J., Badrane H., Ferea T., Adams J., Brown P. O., et al. , 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 16144–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhart E., Hollenberg C. P., 1983. The presence of a defective LEU2 gene on 2 mu DNA recombinant plasmids of Saccharomyces cerevisiae is responsible for curing and high copy number. J. Bacteriol. 156: 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G. R., Styles C. A., 1974. Gene conversion of deletions in the his4 region of yeast. Genetics 77: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohling S., Dohner H., 2008. Chromosomal abnormalities in cancer. N. Engl. J. Med. 359: 722–734. [DOI] [PubMed] [Google Scholar]
- Gangloff S., Zou H., Rothstein R., 1996. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 15: 1715–1725. [PMC free article] [PubMed] [Google Scholar]
- George C. M., Alani E., 2012. Multiple cellular mechanisms prevent chromosomal rearrangements involving repetitive DNA. Crit. Rev. Biochem. Mol. Biol. 47: 297–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S., Campbell C. D., Eichler E. E., 2011. Human copy number variation and complex genetic disease. Annu. Rev. Genet. 45: 203–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B. M., Finn K. J., Li J. J., 2010. Loss of DNA replication control is a potent inducer of gene amplification. Science 329: 943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D., Desai M. M., Tucker C. M., Jenq H. T., Pai D. A., et al. , 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansche P. E., Beres V., Lange P., 1978. Gene duplication in Saccharomyces cerevisiae. Genetics 88: 673–687. [PMC free article] [PubMed] [Google Scholar]
- Harfe B. D., Jinks-Robertson S., 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34: 359–399. [DOI] [PubMed] [Google Scholar]
- Hastings P. J., Ira G., Lupski J. R., 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne D. C., 1963. A deletion in yeast and its bearing on the structure of the mating type locus. Genetics 48: 1727–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I., 1988. The Hawthorne deletion twenty-five years later. Genetics 120: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang M. L., Tan F. J., Lai D. C., Celniker S. E., Hoskins R. A., et al. , 2010. Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS Genet. 6: e1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow R. C., Gokcumen O., Lee C., 2012. Exploring the role of copy number variants in human adaptation. Trends Genet. 28: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A., Wu H., Smith J. D., Nickerson D. A., Romieu I., et al. , 2010. De novo rates and selection of large copy number variation. Genome Res. 20: 1469–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Narayanan V., Mieczkowski P. A., Petes T. D., Krasilnikova M. M., et al. , 2008. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 27: 2896–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Kent J. C., Hartwell L. H., 1985. Genetic analysis of the mitotic transmission of minichromosomes. Cell 40: 393–403. [DOI] [PubMed] [Google Scholar]
- Koszul R., Caburet S., Dujon B., Fischer G., 2004. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 23: 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepischi A. C., Pearson P. L., Rosenberg C., 2012. Germline copy number variations and cancer predisposition. Future Oncol. 8: 441–450. [DOI] [PubMed] [Google Scholar]
- Kumari A., Lim Y. X., Newell A. H., Olson S. B., McCullough A. K., 2012. Formaldehyde-induced genome instability is suppressed by an XPF-dependent pathway. DNA Repair 11: 236–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D. E., Coulson C. A., 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 28: 491–511. [DOI] [PubMed] [Google Scholar]
- Lee P. S., Greenwell P. W., Dominska M., Gawel M., Hamilton M., et al. , 2009. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 5: e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F. J., Degtyareva N. P., Lobachev K., Petes T. D., 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587–598. [DOI] [PubMed] [Google Scholar]
- Libuda D. E., Winston F., 2006. Amplification of histone genes by circular chromosome formation in Saccharomyces cerevisiae. Nature 443: 1003–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman S., Shalit P., Picologlou S., 1981. Ty elements are involved in the formation of deletions in DEL1 strains of Saccharomyces cerevisiae. Cell 26: 401–409. [DOI] [PubMed] [Google Scholar]
- Liebman S. W., Singh A., Sherman F., 1979. A mutator affecting the region of the iso-1-cytochrome c gene in yeast. Genetics 92: 783–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D., Sebat J., 2012. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 148: 1223–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley J. L., Petes T. D., 2010. Chromosome rearrangements and aneuploidy in yeast strains lacking both Tel1p and Mec1p reflect deficiencies in two different mechanisms. Proc. Natl. Acad. Sci. USA 107: 11465–11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski P. A., Lemoine F. J., Petes T. D., 2006. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair 5: 1010–1020. [DOI] [PubMed] [Google Scholar]
- Mine-Hattab J., Rothstein R., 2012. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 14: 510–517. [DOI] [PubMed] [Google Scholar]
- Moore I. K., Martin M. P., Paquin C. E., 2000. Telomere sequences at the novel joints of four independent amplifications in Saccharomyces cerevisiae. Environ. Mol. Mutagen. 36: 105–112. [PubMed] [Google Scholar]
- Morrison A., Bell J. B., Kunkel T. A., Sugino A., 1991. Eukaryotic DNA polymerase amino acid sequence required for 3′-5′ exonuclease activity. Proc. Natl. Acad. Sci. USA 88: 9473–9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan V., Mieczkowski P. A., Kim H. M., Petes T. D., Lobachev K. S., 2006. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125: 1283–1296. [DOI] [PubMed] [Google Scholar]
- Ozenberger B. A., Roeder G. S., 1991. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol. Cell. Biol. 11: 1222–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Miol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C., Koszul R., Dujon B., Fischer G., 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 4: e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., 1980. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell 19: 765–774. [DOI] [PubMed] [Google Scholar]
- Putnam C. D., Pennaneach V., Kolodner R. D., 2005. Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype. Mol. Cell. Biol. 25: 7226–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Hayes T. K., Kolodner R. D., 2009. Specific pathways prevent duplication-mediated genome rearrangements. Nature 460: 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A. J., Shafer B. K., Neelam B., Strathern J. N., 2005. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 19: 1390–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P., 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Rothstein R., Helms C., Rosenberg N., 1987. Concerted deletions and inversions are caused by mitotic recombination between delta sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghani P. C., Bosron W. F., Hurley T. D., 2002a Human glutathione-dependent formaldehyde dehydrogenase: structural changes associated with ternary complex formation. Biochemistry 41: 15189–15194. [DOI] [PubMed] [Google Scholar]
- Sanghani P. C., Robinson H., Bosron W. F., Hurley T. D., 2002b Human glutathione-dependent formaldehyde dehydrogenase: structures of apo, binary, and inhibitory ternary complexes. Biochemistry 41: 10778–10786. [DOI] [PubMed] [Google Scholar]
- Schacherer J., de Montigny J., Welcker A., Souciet J. L., Potier S., 2005. Duplication processes in Saccharomyces cerevisiae haploid strains. Nucleic Acids Res. 33: 6319–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J., Tourrette Y., Potier S., Souciet J. L., de Montigny J., 2007. Spontaneous duplications in diploid Saccharomyces cerevisiae cells. DNA Repair 6: 1441–1452. [DOI] [PubMed] [Google Scholar]
- Scheifele L. Z., Cost G. J., Zupancic M. L., Caputo E. M., Boeke J. D., 2009. Retrotransposon overdose and genome integrity. Proc. Natl. Acad. Sci. USA 106: 13927–13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., 1989. Nonmutagenic carcinogens induce intrachromosomal recombination in yeast. Nature 337: 285–288. [DOI] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Jackson M., Gilmore R. A., Parker J. H., 1974. Mutants of yeast defective in iso-1-cytochrome c. Genetics 77: 255–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Charles J., Hazkani-Covo E., Yin Y., Andersen S. L., Dietrich F. S., et al. , 2012. High-resolution genome-wide analysis of irradiated (UV and gamma rays) diploid yeast cells reveals a high frequency of genomic loss of heterozygosity (LOH) events. Genetics 190: 1267–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton M. R., Campbell P. J., Futreal P. A., 2009. The cancer genome. Nature 458: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan P. F., Daly M. J., O’Donovan M., 2012. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet. 13: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surtees J. A., Argueso J. L., Alani E., 2004. Mismatch repair proteins: key regulators of genetic recombination. Cytogenet. Genome Res. 107: 146–159. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Wu R., 1980. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature 284: 426–430. [DOI] [PubMed] [Google Scholar]
- Tan F. J., Hoang M. L., Koshland D., 2012. DNA resection at chromosome breaks promotes genome stability by constraining non-allelic homologous recombination. PLoS Genet. 8: e1002633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Dominska M., Greenwell P. W., Harvanek Z., Lobachev K. S., et al. , 2011. Friedreich’s ataxia (GAA)n*(TTC)n repeats strongly stimulate mitotic crossovers in Saccharomyces cerevisae. PLoS Genet. 7: e1001270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrette Y., Schacherer J., Fritsch E., Potier S., Souciet J. L., et al. , 2007. Spontaneous deletions and reciprocal translocations in Saccharomyces cerevisiae: influence of ploidy. Mol. Microbiol. 64: 382–395. [DOI] [PubMed] [Google Scholar]
- Umezu K., Hiraoka M., Mori M., Maki H., 2002. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160: 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman J. A., Brunner H. G., 2012. De novo mutations in human genetic disease. Nat. Rev. Genet. 13: 565–575. [DOI] [PubMed] [Google Scholar]
- Vernon M., Lobachev K., Petes T. D., 2008. High rates of “unselected” aneuploidy and chromosome rearrangements in tel1 mec1 haploid yeast strains. Genetics 179: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Horiuchi T., 2005. A novel gene amplification system in yeast based on double rolling-circle replication. EMBO J. 24: 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., McCusker J. H., Hyman R. W., Jones T., Ning Y., et al. , 2007. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. USA 104: 12825–12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J. W., Maloney D. H., Fogel S., 1990. Unequal crossing-over and gene conversion at the amplified CUP1 locus of yeast. Mol. Gen. Genet. 222: 304–310. [DOI] [PubMed] [Google Scholar]
- Wijchers P. J., de Laat W., 2011. Genome organization influences partner selection for chromosomal rearrangements. Trends Genet. 27: 63–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.