Abstract
Following the irradiation of nondividing yeast cells with ultraviolet (UV) light, most induced mutations are inherited by both daughter cells, indicating that complementary changes are introduced into both strands of duplex DNA prior to replication. Early analyses demonstrated that such two-strand mutations depend on functional nucleotide excision repair (NER), but the molecular mechanism of this unique type of mutagenesis has not been further explored. In the experiments reported here, an ade2 adeX colony-color system was used to examine the genetic control of UV-induced mutagenesis in nondividing cultures of Saccharomyces cerevisiae. We confirmed a strong suppression of two-strand mutagenesis in NER-deficient backgrounds and demonstrated that neither mismatch repair nor interstrand crosslink repair affects the production of these mutations. By contrast, proteins involved in the error-prone bypass of DNA damage (Rev3, Rev1, PCNA, Rad18, Pol32, and Rad5) and in the early steps of the DNA-damage checkpoint response (Rad17, Mec3, Ddc1, Mec1, and Rad9) were required for the production of two-strand mutations. There was no involvement, however, for the Pol η translesion synthesis DNA polymerase, the Mms2-Ubc13 postreplication repair complex, downstream DNA-damage checkpoint factors (Rad53, Chk1, and Dun1), or the Exo1 exonuclease. Our data support models in which UV-induced mutagenesis in nondividing cells occurs during the Pol ζ-dependent filling of lesion-containing, NER-generated gaps. The requirement for specific DNA-damage checkpoint proteins suggests roles in recruiting and/or activating factors required to fill such gaps.
Keywords: DNA damage, mutagenesis, lesion bypass, checkpoint, nucleotide excision repair
MUTAGENESIS associated with induced DNA damage is generally considered to occur during S phase, when polymerase-blocking lesions are bypassed in an error-prone manner by a translesion synthesis (TLS) DNA polymerase. In the yeast Saccharomyces cerevisiae, ultraviolet (UV)-induced mutagenesis is dependent on Pol ζ (Rev3-Rev7) and Rev1, with rev1, rev3, or rev7 mutants exhibiting a “reversionless” phenotype in response to UV irradiation (Lawrence 2002). Just as induced mutagenesis is considered to be a consequence of error-prone TLS, nucleotide excision repair (NER) is thought to counteract such mutagenesis by removing UV-induced lesions before they can be encountered during DNA replication.
Although NER is generally considered to be an error-free, mutation-avoidance mechanism, data obtained >30 years ago demonstrated a pro-mutation role of NER in the UV-induced mutagenesis that occurs in nondividing yeast cells (Eckardt and Haynes 1977; James and Kilbey 1977; James et al. 1978; Eckardt et al. 1980). Either the induction of recessive lethal mutations in diploid strains (James and Kilbey 1977; James et al. 1978) or the induction of forward mutations in the de novo adenine biosynthetic pathway in haploid strains was monitored (Eckardt and Haynes 1977; Eckardt et al. 1980). Despite the differences in assay systems used, the authors reached the same conclusions. First, UV-induced mutations in nondividing cells affected both strands of the DNA duplex (referred as “two-strand” mutations) and hence must have occurred prior to S phase. Second, a lack of NER suppressed the generation of pre-S, two-strand mutations but did not reduce the frequency of canonical, one-strand mutations that occur during replicative bypass of UV-induced lesions. A similar NER-associated phenomenon has been described in Escherichia coli under conditions where the SOS system is constitutively activated (Cohen-Fix and Livneh 1992). Finally, a requirement of NER for UV-induced adaptive mutagenesis in nongrowing yeast cells was recently reported (Heidenreich et al. 2010).
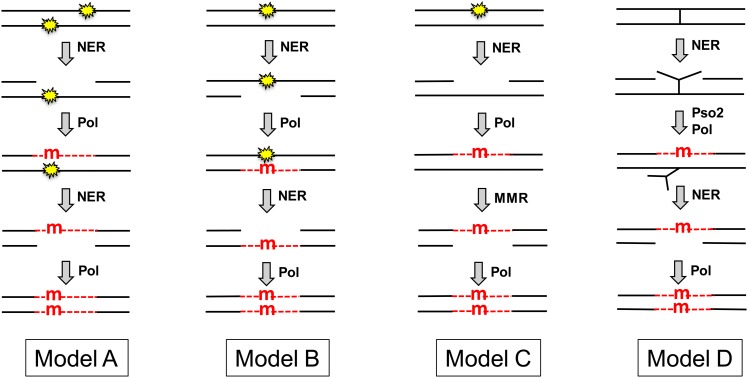
The molecular mechanism of NER-dependent, two-strand mutations remains poorly understood, although several speculative models have been put forth to explain this phenomenon (Figure 1 and see Abdulovic et al. 2006). The first model proposes the occurrence of closely spaced lesions on opposing strands of a duplex DNA molecule (Kilbey et al. 1978). As illustrated in model A, removal of one lesion by NER produces a gap that contains the second lesion. A mutation would then be introduced opposite the second lesion during the gap-filling stage of NER. Following completion of the first round of NER, a second round of repair would be initiated to remove the remaining lesion. Use of the mutation-containing strand as a template to fill the second NER-generated gap would introduce the mutation into the complementary strand of the duplex. In relation to the likelihood of this model, data suggest that closely spaced UV-induced lesions can occur in opposing DNA strands in vivo (Reynolds 1987). A second possible scenario is that, instead of there initially being two closely spaced lesions on opposite DNA strands, the NER machinery incorrectly removes the undamaged strand instead of the lesion-containing strand (model B). This mistake would necessitate an error-prone gap-filling process to bypass the lesion in the gap, which would then be followed by a second round of NER to remove the lesion. As in the first model, a mutation in the complementary DNA strand would be introduced during the second round of NER-associated repair synthesis. The third model (model C) proposes that the NER-dependent mutations originate simply because any type of DNA synthesis has an inherent error frequency (Eckardt et al. 1980). The DNA polymerase that fills NER-generated gaps thus might introduce a mutation during the gap-filling phase of NER, thereby creating a mismatch in the vicinity of the original lesion. Repair of such a mismatch by the mismatch repair (MMR) machinery would then convert the mismatched segment to the mutant homoduplex. That the MMR machinery operates in nondividing cells was recently demonstrated (Rodriguez et al. 2012). The fourth model (model D) suggests that two-strand mutations occur during repair of rare DNA-interstrand crosslinks generated by UV light (see Friedberg et al. 2006). In nongrowing cells, the repair of DNA-interstrand crosslinks is initiated by NER-dependent dual incision of one of the DNA strands and then proceeds mainly via a mutagenic Rev3- and Pso2-dependent repair pathway (Sarkar et al. 2006). The initial mutation would be introduced during the gap-filling process that occurs opposite the crosslinked oligonucleotide, which would then be removed in a second round of NER. As in the first two models, a complementary mutation in the opposing DNA strand would be introduced during the filling of the second, NER-generated gap.
Figure 1.
Models for NER-associated mutagenesis. The NER machinery is recruited either by a UV-induced CPD or (6-4) photoproduct (yellow stars) or by an interstrand crosslink (|). NER excises an oligonucleotide either from the strand containing the lesion (models A, C, and D) or from the undamaged strand (model B). The resulting gap is filled by a DNA polymerase (Pol), which introduces a mutation (red “m”) opposite a lesion within the gap (models A, B, and D) or opposite an undamaged template (model C). Finally, the mutation is introduced into both DNA strands by a second round of NER (models A, B, and D) or by MMR (model C). The Pso2 protein is specifically required for the bypass of an interstrand crosslink. Newly synthesized DNA within NER or MMR-generated gaps is indicated by red dashed lines.
To gain insight into the molecular mechanism that produces two-strand mutations in nondividing cells, we have examined the genetic control of this unique type of NER-dependent, UV-induced mutagenesis. In addition, dose–response curves for the production of one- vs. two-strand mutations were compared in wild-type (WT) and NER-defective backgrounds. When considered together, our data support models in which UV-induced mutagenesis in nondividing cells is initiated during the filling of lesion-containing gaps generated by NER. These data have broad implications for how error-free repair processes can be used to initiate damage-induced mutagenesis, thereby allowing nongrowing cells to acquire genetic changes.
Materials and Methods
Media
Yeast strains were grown in YPDA medium (1% yeast extract, 2% peptone, 2% D-glucose, 250 mg/liter of adenine) or synthetic-complete (SC) medium (Rose et al. 1990). Solid media contained 2% agar. For selection of drug-resistant clones, YPDA medium contained 200 mg/liter geneticin (G418) or 300 mg/liter hygromycin B. Ura− auxotrophs were selected on 5-fluoroorotic acid (FOA) plates prepared as described (Rose et al. 1990). Red/white colony screening was performed on YPD10 plates containing five times the normal amount of glucose and no extra adenine. On this medium, ρ+ ade2 ADEX colonies accumulate red pigment; ρ− ade2 ADEX mutants (which form white colonies on standard 2%-glucose-containing YPD plates) accumulate dark brown pigment; and ρ+ and ρ− ade2 adeX mutants form white colonies (Eckardt and Haynes 1977). Due to poor accumulation of red pigment in the rpb9Δ rad26Δ background on regular YPD10 plates, colony-color screening for this strain was performed on YPD10 plates containing 0.1–0.2% of yeast extract.
Yeast strains and plasmids
All S. cerevisiae strains used in this study were derivatives of strain SJR2308 and are listed in Table 1. SJR2308 is a RAD5 derivative of W303-1A that contains the lys2ΔA746 mutation, which was introduced by two-step allele replacement using the plasmid pSR582 (Harfe and Jinks-Robertson 1999). Complete gene deletions were accomplished by PCR-mediated gene replacement using either the hphMX4 hygromycin-resistance module of plasmid pAG32 (Goldstein and McCusker 1999) or the kanMX6 G418-resistance module of plasmid pFA6a-kanMX6 (Wach et al. 1994). Each deletion was confirmed by PCR amplification and, whenever possible, by phenotype.
Table 1. Yeast strains.
| Strain | Genotype |
|---|---|
| SJR2308 | MATa leu2-3,112 ura3-1 can1-100 his3-11,15 ade2-1 lys2ΔA746 |
| SJR2328 | SJR2308 rad17Δ::hphMX4 |
| SJR2371 | SJR2308 rad1Δ::kanMX4 |
| SJR2975 | SJR2308 rad14Δ::hphMX4 |
| SJR2976 | SJR2308 mlh1Δ::hphMX4 |
| SJR2977 | SJR2308 pol32Δ::hphMX4 |
| SJR2979 | SJR2308 rad30Δ::hphMX4 |
| SJR2980 | SJR2308 rev3Δ::hphMX4 |
| SJR3006 | SJR2308 rad5Δ::hphMX4 |
| SJR3007 | SJR2308 rad18Δ::hphMX4 |
| SJR3059 | SJR2308 pso2Δ::hphMX4 |
| SJR3060 | SJR2308 ddc1Δ::hphMX4 |
| SJR3061 | SJR2308 mec3Δ::hphMX4 |
| SJR3062 | SJR2308 rad9Δ::hphMX4 |
| SJR3063 | SJR2308 mms2Δ::hphMX4 |
| SJR3089 | SJR2308 sml1Δ::hphMX4 |
| SJR3090 | SJR2308 sml1Δ::hphMX4 mec1Δ::kanMX6 |
| SJR3091 | SJR2308 dun1Δ::hphMX4 |
| SJR3092 | SJR2308 ubc13Δ::hphMX4 |
| SJR3162 | SJR2308 rad7Δ::hphMX4 |
| SJR3163 | SJR2308 rad26Δ::hphMX4 |
| SJR3164 | SJR2308 chk1Δ::hphMX4 |
| SJR3200 | SJR2308 rev1Δ::hphMX4 |
| SJR3201 | SJR2308 LEU2:pol30-K164R |
| SJR3202 | SJR2308 rad16Δ::kanMX6 |
| SJR3204 | SJR2308 rad26Δ::hphMX4 rpb9Δ::kanMX6 |
| SJR3205 | SJR2308 sml1Δ::hphMX4 rad53Δ::kanMX6 |
| SJR3255 | SJR2308 exo1Δ::hphMX4 |
| SJR3256 | SJR2308 exo1-D173A |
| SJR3304 | SJR2308 rev1-S31A |
The pol30-K164R allele, which is marked with a nearby LEU2 gene, was introduced by transformation with SacI-digested pSR870. Following selection of Leu+ transformants, presence of the pol30-K164R allele was confirmed by DNA sequencing and by UV sensitivity. To construct pSR870, a POL30-containing SacI/MluI fragment from pBL205 (Bauer and Burgers 1990) and a LEU2-containing SacI/MluI fragment from pRDK925 (Lau et al. 2002) were cloned into SacI-digested pBluescript-SK(+) (Stratagene). Codon 174 of POL30 was then changed from AAA (lysine) to AGG (arginine) by site-directed mutagenesis, yielding pSR870.
The exo1-D173A allele was introduced by two-step allele replacement using SmaI-digested pSR737. pSR737 was constructed by inserting an exo1-D173A-containing XhoI/SacII fragment from pSM638 (Moreau et al. 2001) into XhoI/SacII-digested pRS306 (Sikorski and Hieter 1989). Following selection of Ura+ transformants, plasmid loss events were selected on FOA plates. Presence of the exo1-D173A allele was confirmed by DNA sequencing.
Plasmid pSR1021 was used for introducing the rev1-S31A allele and was created as follows. First, REV1-containing plasmid pFL41 (Larimer et al. 1989) was used as a template to separately amplify the ≈0.5-kb left and right arms that overlap in the region containing serine codon 31 (5′-AGC). The left arm was amplified using primer pairs rev1-Xho (5′-cat act cga gtt ctt tca ttt gaa ttg aat gc-3′) and rev1-S31A-XR (5′-c act ttg ctg GGC aag gca att g-3′); the right arm was amplified using rev1-Spe (5′-tac tac tag t ag cca cta tgt gag taa ccg-3′) and rev1-S31A-SF (5′-c aat tgc ctt GCC cag caa agt g-3′). The rev1-S31A-XR and rev1-S31A-SF primers are complementary to each other and contain mutations that convert 5′-AGC (serine) to 5′-GCC (alanine; uppercase letters in the primers indicate codon 31). In a second round of PCR, the left and right arms were mixed together and amplified in the presence of the two external primers (rev1-Xho and rev1-Spe) to obtain a full-length, 1-kb product. Obtaining a sufficient amount of the full-length product required a third round of PCR, which used the product of round 2 plus the external primers. The resulting fragment was digested with XhoI and SpeI and cloned into XhoI/SpeI-digested pRS306 (Sikorski and Hieter 1989), yielding pSR1021. The rev1-S31A allele was introduced by a two-step allele replacement using ClaI-digested pSR1021, and its presence was confirmed by DNA sequencing.
UV irradiation and mutagenesis
Yeast strains were grown in liquid YPDA media for 7 days at 30° with shaking. Cells were harvested from 20-ml cultures, washed once with 20 ml of distilled water, and resuspended in 20 ml of water. Cell suspensions were sonicated for 2 min at 20 W using a 50VT ultrasonic homogenizer with a stepped titanium microtip of 3.9-mm diameter (BioLogics). The cell suspension was centrifuged for 3 min at 120 × g in an Eppendorf 5810R centrifuge, and the upper 3 ml, which contains only unbudded G0 cells, was removed (Eckardt and Haynes 1977). The absence of budded (non-G0) cells in the upper 3- ml fraction was confirmed microscopically.
To investigate the genetic control of UV-induced mutagenesis, G0 cells were diluted into water containing 1% YPDA to a final cell density of ≈3⋅103 cells/ml. Ten-milliliter aliquots were UV-irradiated in 100- × 15-mm polystyrene petri dishes using either a UVC-515 ultraviolet crosslinker (Ultra-Lum, Inc.) at a dose rate of ≈25 J/m2/sec or a germicidal G8T5 8W lamp at a dose rate of 0.054 J/m2/sec. The UV dose used to irradiate a given strain yielded 35–55% survival. In a typical experiment, 100-μl aliquots of the irradiated sample were plated on each of 200 YPD10 plates; 50-μl aliquots of a non-irradiated control sample were plated on 200 YPD10 plates. Prior to plating, cell suspensions were kept on ice in the dark. Plates were incubated for 5 days in the dark at 30° and then for an additional 1–2 days at 4° before inspecting colony color. The median values of the number of colonies per plate in the irradiated (Mir) and control (Mco) samples were obtained by counting colonies on 21 randomly chosen plates of each sample. The percentage survival was calculated as [Mir/(2⋅Mco)]⋅100. Mutant frequency was calculated as the ratio of pure white or sectored red/white ade2 adeX colonies on the 200 plates to the estimated total number of colonies on the 200 plates (calculated as Mir⋅200 and Mco⋅200). The UV-induced mutant frequency was calculated as a difference between the mutant frequencies in the paired irradiated and non-irradiated (control) samples. Supporting information, Table S1 lists all the raw data obtained in these analyses.
The frequencies of induced mutants in irradiated populations of different tested strains were compared using two-tailed Fisher’s exact test for 2 × 2 contingency tables. In these analyses, Nmi(1) and (NUV(1) − Nmi(1)) vs. Nmi(2) and (NUV(2) − Nmi(2)) were compared, where Nmi(x) is the number of induced mutant colonies and NUV(x) is the total number of colonies counted in irradiated samples of strains 1 and 2. The numbers of induced mutants were calculated as Nmi = [NmUV − Nm0⋅NUV/N0], where NmUV is the actual number of mutant colonies counted in the irradiated sample, and Nm0 and N0 are the numbers of mutant and total colonies in the non-irradiated sample of the same strain. For the strains displaying very low induced mutant frequencies, the significance of mutant induction was examined by Fisher’s exact test for 2 × 2 contingency tables, comparing Nm0 and (N0 − Nm0) vs. NmUV and (NUV − NmUV). In all cases, we assumed that two-tail P-values >0.05 indicated no significant differences between the compared populations.
UV-induced dose–response curves
G0 cells were diluted in water containing 1% YPDA to a final cell density of ≈3⋅105 cells/ml and were then irradiated as described above. At each UV dose tested, appropriately diluted aliquots of the irradiated sample were plated on 50 YPD10 plates; for the non-irradiated control, 50-μl aliquots of a 100-fold dilution were plated on 50 YPD10 plates. Plating and post-plating incubation was performed as above. The total number of colonies on the 50 plates and the number of ade2 adeX mutants, at each UV dose tested, were counted. UV-induced killing and mutagenesis curves were analyzed as described by Haynes and Eckardt (1979). The surviving fraction of cells [S(x)] at each UV dose (x) was calculated as a ratio of the number of viable cells per unit volume in the UV-irradiated sample [Ns(x)] to the number of viable cells per unit volume in the unirradiated sample (N0). Induced mutant yields were calculated as Y(x) = [Nm(x) − Nm0⋅S(x)]/N0, where Nm(x) is the number of mutant colonies counted at dose x and Nm0 is the number of spontaneous mutant colonies counted in the non-irradiated sample. Induced mutant frequencies were calculated as M(x) = Nm(x)/Ns(x) − Nm0/N0. Survival data were fitted with (i) linear S(x) = exp(−k1⋅x); (ii) linear-quadratic S(x) = exp[−(k1⋅x + k2⋅x2)]; and (iii) quadratic S(x) = exp(−k2⋅x2) approximation functions (where k1 and k2 are constant coefficients of killing for one- and two-hit processes, respectively). Induced mutant yields were fitted with (i) linear Y(x) = m1⋅x⋅S(x), (ii) linear-quadratic Y(x) = (m1⋅x + m2⋅x2)⋅S(x), and (iii) quadratic Y(x) = m2⋅x2⋅S(x) approximation functions (where m1 and m2 are constant coefficients of mutability for one- and two-hit processes, respectively). Finally, induced mutant frequencies were fitted with (i) linear M(x) = m1⋅x, (ii) linear-quadratic M(x) = m1⋅x + m2⋅x2, and (iii) quadratic M(x) = m2⋅x2 approximation functions (where m1 and m2 are constant coefficients of mutability for one- and two-hit processes, respectively). Least-square-based curve fitting was performed using GraphPad Prism software (GraphPad Software).
Results
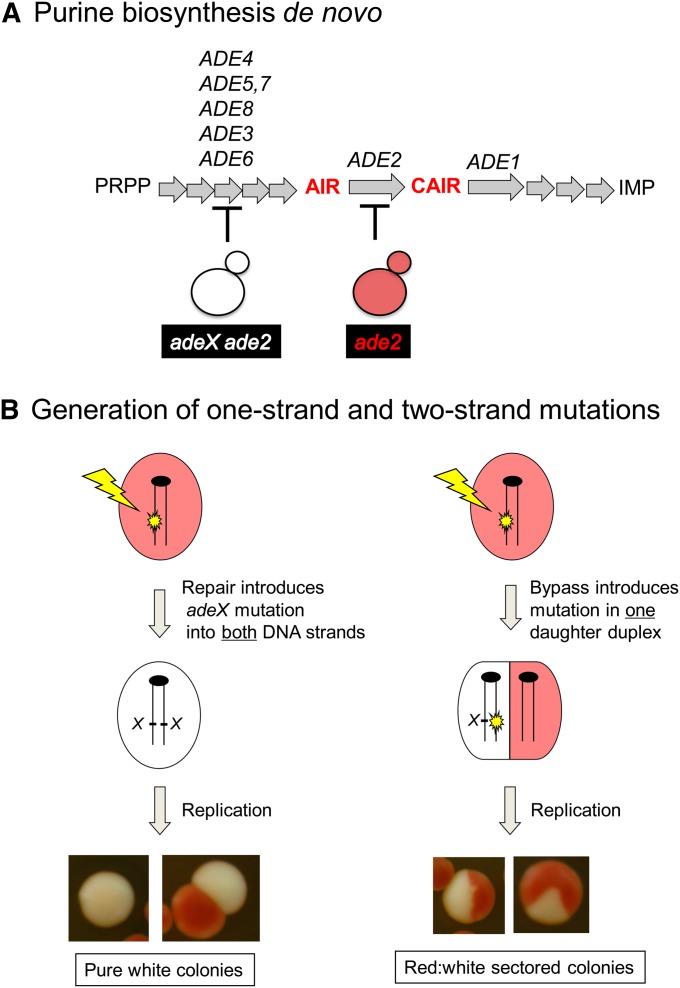
A nonselective ade2 adeX forward mutation system similar to that used previously (Eckardt and Haynes 1977) was used to determine the genetic requirements of UV-induced mutagenesis in nondividing (G0) cells. As illustrated in Figure 2A, the Ade2 protein is required for de novo purine biosynthesis and its absence blocks the pathway at a point where a red precursor accumulates. Inactivation of any one of the five ADE genes whose product functions prior to Ade2 in the biosynthetic pathway prevents the accumulation of the red pigment. An ade2 single mutant thus forms red colonies when adenine is limited in the growth medium, while an ade2 adeX double mutant forms white colonies. Following the isolation of a G0 population of the ade2 strain of interest, cells were irradiated with UV and then immediately plated to single colonies on rich medium. Because the strains used here varied greatly in their UV sensitivity, mutagenesis was assessed in each strain using a UV dose that reduced viability ≈50%. In the wild-type strain, the relevant UV dose was 50 J/m2, whereas only 1.2 J/m2 was sufficient to reduce viability of the NER-deficient rad1Δ and rad14Δ mutants to a comparable level. According to published data, these doses generate ≈14,000 cyclobutane pyrimidine dimers (CPDs) per haploid genome in the wild-type strain and 460 CPDs per genome in NER-deficient mutants (Unrau et al. 1973; Reynolds 1987). Following irradiation with an approximately equitoxic UV dose, we screened for white ade2 adeX double-mutant colonies among the parental background of red ade2 single-mutant colonies. As illustrated in Figure 2B, an adeX mutation that is present in both strands of a duplex prior to DNA replication (a two-strand mutation) give rises to a pure white colony, whereas an adeX mutation generated during DNA replication will affect only one of the resulting daughter duplexes (a “one-strand” mutation) and give rise to a red-white sectored colony.
Figure 2.
The ade2 adeX mutation system. (A) Relevant genes in the adenine biosynthetic pathway are indicated. A red pigment accumulates in the absence of either the ADE1 or the ADE2 gene product; production of the pigment is blocked by inactivation of any gene product that functions prior to Ade1/Ade2 in the pathway. PRPP, phosphoribosyl pyrophosphate; AIR, aminoimidazole ribotide; CAIR, carboxyaminoimidazole ribotide; IMP, inosine monophosphate. (B) Following DNA damage, pure white colonies are produced only if an adeX mutation is present in both strands of duplex DNA. Bypass of damage during DNA replication results in a red-white sectored colony.
NER is required for two-strand mutations in G0 cells
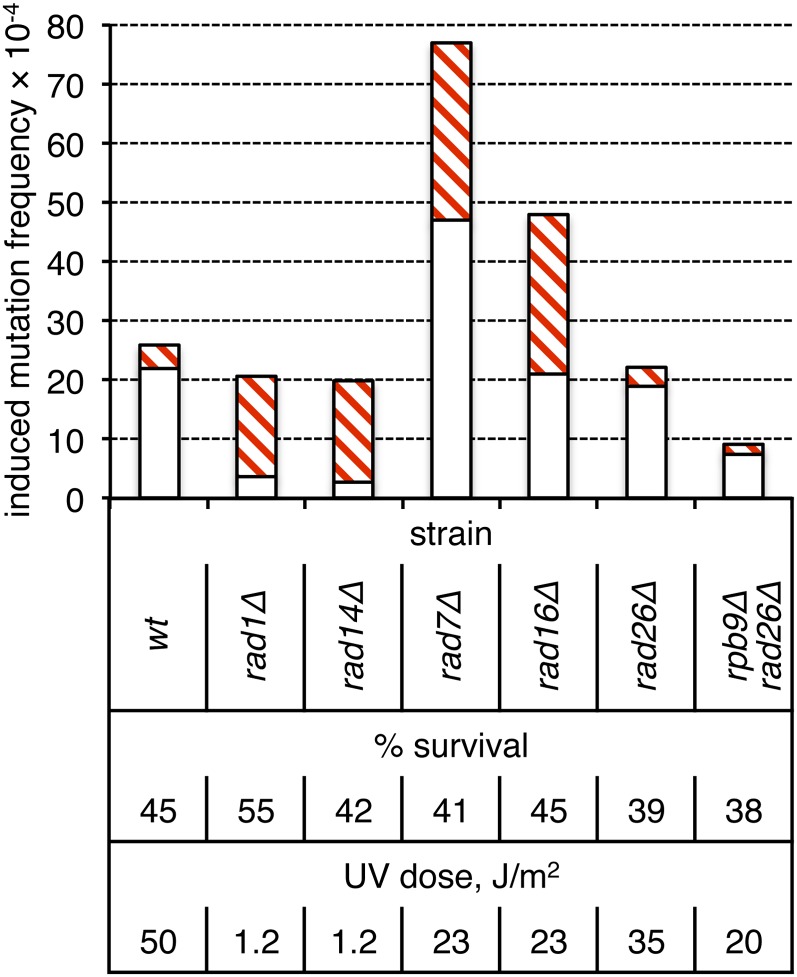
The frequencies and types of ade2 adeX mutants were compared in WT vs. NER-defective rad1Δ or rad14Δ backgrounds. The frequency of UV-induced pure white ade2 adeX colonies (presumably originating from two-strand mutations) was sixfold higher than the frequency of red-white sectored colonies (originating from one-strand mutations) in the WT, NER-proficient background (Figure 3). At an equitoxic UV dose, the total frequency of UV-induced mutants in the rad1Δ and rad14Δ strains was the same as in WT (P ≈ 0.34 and P ≈ 0.22, respectively). There was, however, a reversal in the relative proportions of one- and two-strand mutations upon loss of NER, with five- to six-fold more sectored than pure-white colonies in the rad1Δ and rad14Δ backgrounds. Thus, consistent with the findings of Eckardt and Haynes (1977), most UV-induced mutagenesis in nondividing, G0 cells occurs by an NER-dependent mechanism that presumably introduces mutations into both strands of duplex DNA prior to replication.
Figure 3.
Role of NER in the production of two-strand mutations in nondividing cells. Induction frequencies of white (open bars) and red-white sectored (red cross-hatched bars) ade2 adeX mutants in a G0 population of cells are shown.
In yeast and other organisms, there are two discrete subpathways of NER: global-genome repair (GGR) and transcription-coupled repair (TCR) (reviewed by Friedberg et al. 2006). The difference between these two pathways resides in the initial damage-recognition step; all subsequent steps involve common NER factors such as Rad1 and Rad14. During GGR, recruitment of the NER machinery to the site of a lesion requires the Rad7-Rad16 complex (Verhage et al. 1994). To initiate TCR, the NER machinery can be recruited by two redundant mechanisms that are mediated by the Rpb9 subunit of RNA polymerase II or by Rad26 (Li and Smerdon 2002). Elimination of either Rad7 or Rad16 elevated the frequency of UV-induced sectored ade2 adeX colonies seven- to eight-fold, which was slightly, but significantly greater than the elevation observed in the complete absence of NER (P < 0.001). In contrast to what was observed in the complete absence of NER, there was no compensatory decrease in the frequency of pure-white colonies in either the rad7Δ or the rad16Δ background. The frequency of pure-white colonies in the rad16Δ mutant was similar to that in WT (P ≈ 0.66), but was twofold higher in the rad7Δ mutant than in WT (P < 0.001). The net result was that the overall level of UV-induced mutagenesis relative to WT or NER-defective increased when GGR was eliminated (P < 0.001).
In contrast to what was observed upon elimination of GGR, loss of TCR in a rad26Δ rpb9Δ double mutant was associated with a threefold reduction of the frequency of two-strand UV-induced mutations (P < 0.001); the frequency of one-strand mutations was not affected (P ≈ 0.25). The reduction in frequency of pure-white colonies observed in the absence of TCR was not as extreme as that seen in the complete absence of NER (P ≈ 0.017). In the rad26Δ single mutant, there was no change in UV-associated mutagenesis (P ≈ 0.40), consistent with functional redundancy between the two subpathways of TCR. Together, these data suggest that the TCR subpathway of NER is more error prone than GGR, with the former promoting and the latter limiting NER-associated mutagenesis.
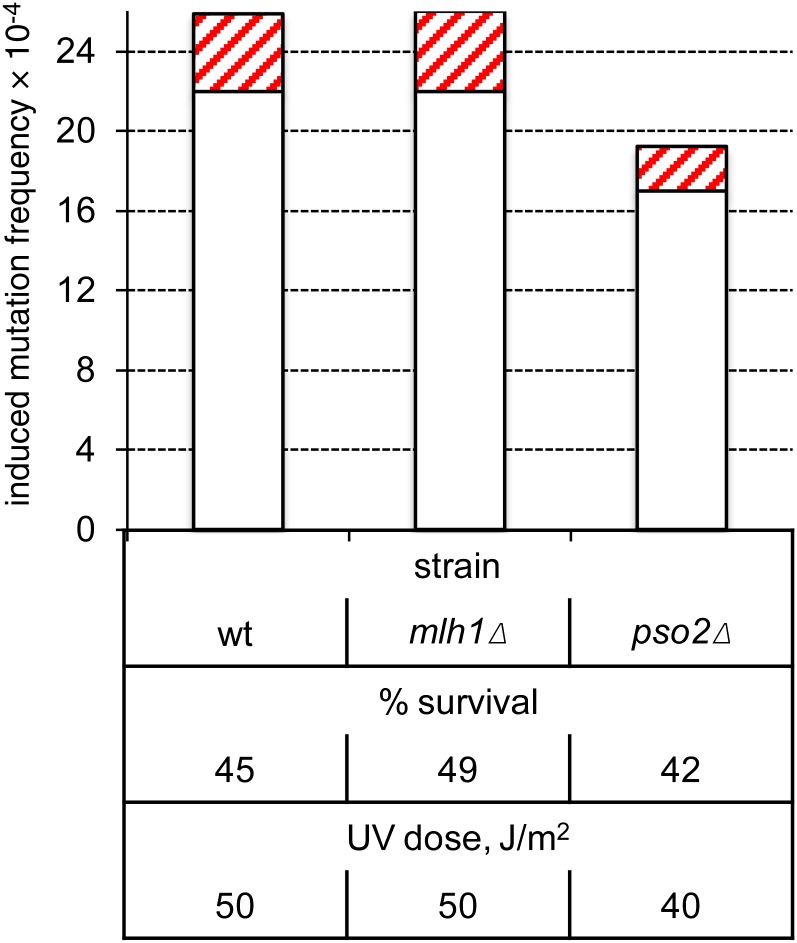
Neither MMR nor interstrand crosslink repair affect mutagenesis in G0
Models C and D in Figure 1 postulate a specific requirement of the MMR and interstrand crosslink repair (ICLR) pathways, respectively, for UV-induced, two-strand mutations. To test these models, we inactivated MMR or ICLR by deleting MLH1 (Prolla et al. 1994) or PSO2 (Sarkar et al. 2006), respectively. The very similar characteristics of UV-induced mutagenesis in the WT and either the mlh1Δ (P = 1) or pso2Δ (P ≈ 0.07) strain indicate that neither pathway is a major contributor to UV-induced mutagenesis in nondividing cells (Figure 4).
Figure 4.
Neither MMR nor ICLR is required for two-strand mutations in nondividing cells. Induction frequencies of pure white (open bars) and red-white sectored (red cross-hatched bars) ade2 adeX mutants in G0 populations of WT, mlh1Δ, and pso2Δ cells are shown.
Role of TLS and postreplication DNA repair
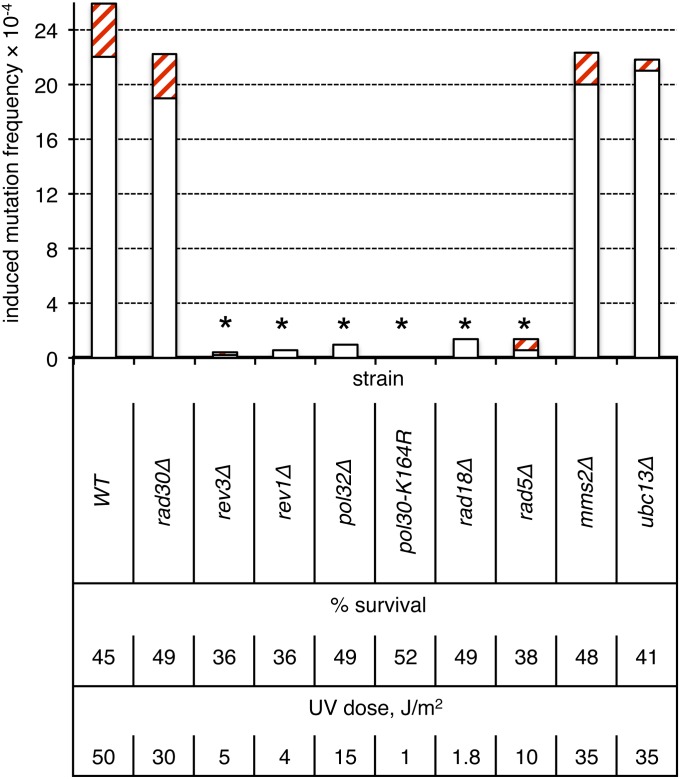
Both models A and B (Figure 1) propose that two-strand UV-induced mutations in nondividing cells are generated by error-prone bypass of photodamage during the filling of NER-generated gaps. Pol η (Rad30) is primarily involved in error-free bypass of photolesions, whereas Pol ζ (Rev3-Rev7), in conjunction with the Rev1 protein, is responsible for error-prone TLS across UV-induced damage and hence is critically important for UV-induced mutagenesis, at least in proliferating cells (reviewed by Waters et al. 2009). In the ade2 adeX system, elimination of Pol η had no effect on UV-induced mutagenesis in G0 cells (Figure 5; P ≈ 0.51). By contrast, there was no detectable UV-induced mutagenesis in a rev3Δ or rev1Δ strain (irradiated to unirradiated sample comparison, P = 1 and P ≈ 0.18, respectively), demonstrating that Pol ζ, together with Rev1, provides the major source of UV-induced mutations in nongrowing cells.
Figure 5.
Requirements of error-prone and error-free bypass pathway components for two-strand mutations in nondividing cells. Induction frequencies of pure white (open bars) and red-white sectored (red cross-hatched bars) ade2 adeX mutants in G0 populations of cells are shown. Asterisks above the bars indicate a lack of UV-induced mutagenesis (irradiated to unirradiated sample comparison, P > 0.1 in all cases).
Monoubiquitination of PCNA (Pol30) at lysine 164 (K164) by the Rad18-Rad6 complex is an essential prerequisite for Pol ζ-dependent lesion bypass of acute UV damage in dividing cells (Hoege et al. 2002). Extension of the ubiquitin at K164 by Rad5 and Mms2-Ubc13 into a regulatory polyubiquitin chain directs error-free bypass using an undamaged DNA template. In nondividing cells, there was no UV-induced mutagenesis in either a rad18Δ or a pol30-K164R strain (irradiated to unirradiated sample comparison, P ≈ 0.36 and P = 1, respectively; Figure 5). Pol32, a nonessential subunit of DNA pol δ, is also required for Pol ζ-dependent UV-induced mutagenesis in dividing cells (Gerik et al. 1998; Gibbs et al. 2005). Recent work suggests that it, together with Pol31, comprises two additional subunits of the Pol ζ holoenzyme (Johnson et al. 2012; Makarova et al. 2012). Accordingly, there was no induction of mutagenesis in G0-irradiated pol32Δ cells. Finally, we investigated the role of Rad5, Mms2, and Ubc13 in UV-induced mutagenesis in G0 cells. As noted above, Rad5 is required for polyubiquitination of PCNA and hence is involved primarily in the error-free, template-switch pathway. Although loss of the error-free bypass pathway in dividing cells typically is associated with a mutator phenotype, Rad5 has been implicated in Pol ζ-dependent bypass of spontaneous lesions (Liefshitz et al. 1998; Cejka et al. 2001; Minesinger and Jinks-Robertson 2005) and of UV-lesion bypass in a gapped-plasmid transformation assay (Gangavarapu et al. 2006; Pagès et al. 2008). We observed no UV-induced mutagenesis in a rad5Δ mutant (irradiated to unirradiated sample comparison, P ≈ 0.50), indicating that Rad5, like Rad18, is required for UV-induced mutagenesis in nondividing cells. By contrast, the level of UV-induced mutagenesis in an mms2Δ or ubc13Δ mutant was the same as in WT (P ≈ 0.62 and P ≈ 0.49, respectively). While the absence of an effect of Mms2 or Ubc13 loss on two-strand mutations, which presumably occur prior to the initiation of replication, is not surprising, the lack of an increase in one-strand mutations is unexpected.
Role of DNA-damage checkpoint proteins
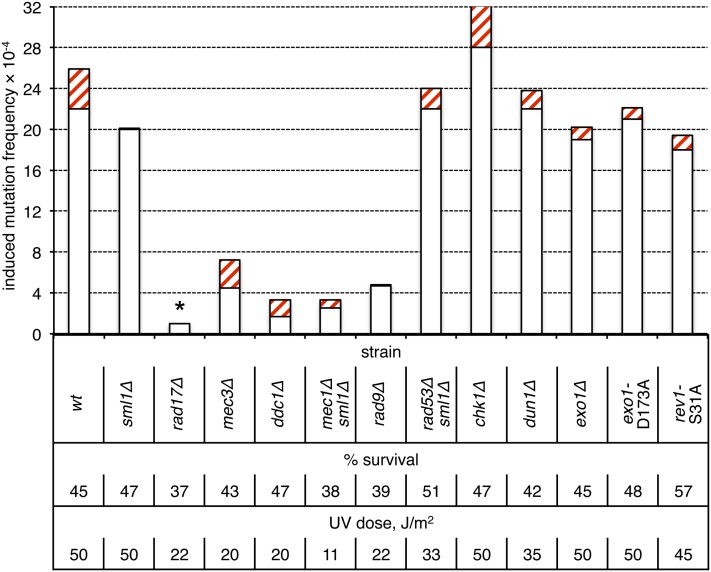
The DNA damage checkpoint response is activated by NER-generated single-strand gaps (Giannattasio et al. 2004, 2010). In brief, RPA-coated single-strand DNA (ssDNA) recruits the Mec1-Ddc2 kinase complex, while the 5′ ssDNA/double-strand DNA junction of gaps recruits the 9-1-1 checkpoint clamp, which is composed of Rad17, Mec3, and Ddc1 (for a review, see Harrison and Haber 2006). The 9-1-1 complex directly activates the Mec1 kinase (Majka et al. 2006), which then phosphorylates multiple targets including Rad9, Rad53, and Chk1. Rad9 serves as a platform for subsequent autophosphorylation of the Rad53- and Chk1-signaling kinases, which phosphorylate downstream effectors leading to G1/S and S/G2 cell-cycle progression delays and G2/M cell-cycle arrest. The Rad53 and Dun1 protein kinases are involved in additional DNA-damage-induced transcriptional responses. We found that inactivation of Mec1, Rad9, or any component of the 9-1-1 checkpoint clamp strongly reduced UV-induced two-strand mutagenesis (Figure 6; P < 0.001 in each case, with no induction of mutagenesis by UV in the rad17Δ strain). By contrast, loss of Rad53, Chk1, or Dun1 had little, if any, effect (P > 0.10 in each case). These data indicate that only the apical DNA damage checkpoint factors (9-1-1, Mec1, and Rad9) are critical for UV-induced mutagenesis in nongrowing cells, whereas the downstream effectors, which regulate cell cycle progression and transcription (Rad53, Chk1, Dun1) are not required.
Figure 6.
Roles of checkpoint proteins in the production of two-strand mutations in nondividing cells. Induction frequencies of pure white (open bars) and red-white sectored (red cross-hatched bars) ade2 adeX mutants in G0 populations of cells are shown. Presence of sml1Δ is required for the viability of mec1Δ and rad53Δ strains and does not affect UV sensitivity. The asterisk above the rad17Δ bar indicates a lack of UV-induced mutagenesis (irradiated to unirradiated sample comparison, P > 0.8).
The 5′ to 3′ Exo1 exonuclease has been identified as an additional factor with an early role in checkpoint activation in UV-irradiated, G1-arrested cells (Giannattasio et al. 2010). Loss of Exo1 leads to a defect in the G1/S transition delay and is associated with reduced phosphorylation of several Mec1 substrates. It was suggested that Exo1-mediated expansion of NER-generated gaps is an important prerequisite for DNA-damage checkpoint activation and might be relevant to the two-strand mutations that occur in nondividing cells. In the case of UV-induced mutagenesis in the ade2 adeX system used here, however, neither deletion of EXO1 nor loss of Exo1 catalytic activity (exo1-D173A allele) had an effect on UV-induced mutagenesis in G0-arrested cells (Figure 6; P ≈ 0.22 and P ≈ 0.43, respectively). We suggest that NER-generated gaps per se are sufficient to activiate a 9-1-1-, Mec1-, and Rad9-dependent pathway that controls UV-induced mutagenesis, whereas full activation of the Rad53 pathway may require Exo1-mediated processing of NER-generated gaps.
Dose–response curves for UV-induced ade2 adeX mutants in WT and NER-deficient strains
The exquisite sensitivity of NER-defective strains to UV necessitated the use of equitoxic rather than equivalent doses of UV when comparing the mutagenesis profile to that of WT. Although a direct involvement of NER in generating two-strand mutations has been inferred, it is formally possible that the specific reduction of two-strand mutations observed upon loss of NER is simply a reflection of the much lower load of UV damage sustained. To address this issue, we examined the dose–response curves for UV-induced pure and sectored ade2 adeX mutants in WT and rad14Δ strains. In the simplest case, linearity of a dose–response curve would indicate that one mutational hit is sufficient for the generation of a mutant clone, whereas a quadratic dose–response curve implies a requirement for two independent mutational hits.
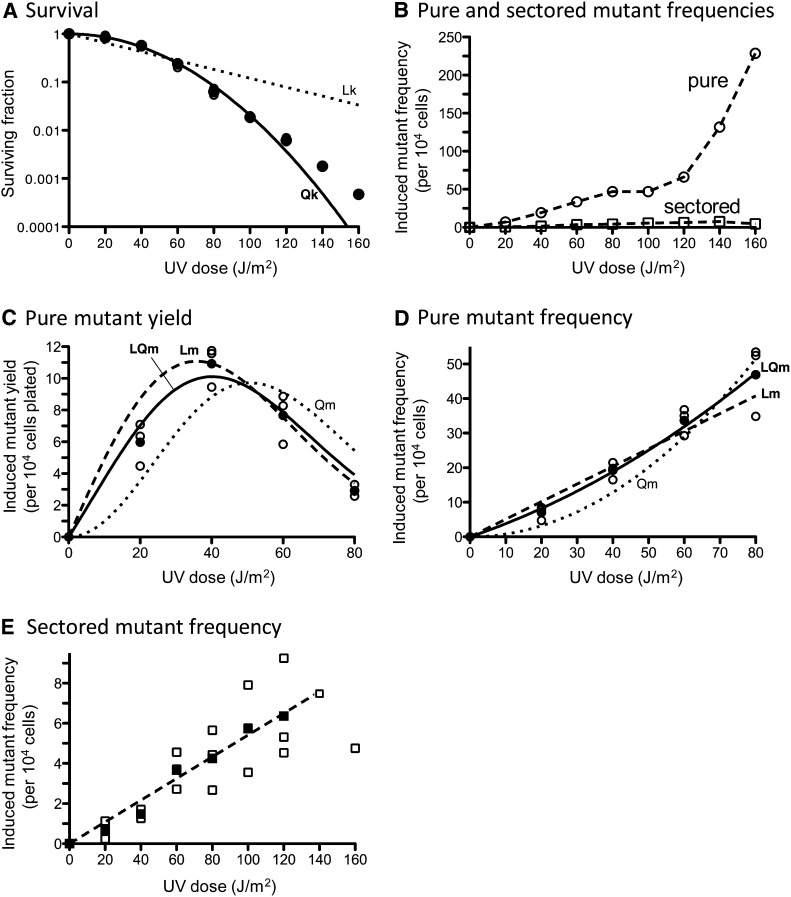
In the WT background, the survival curve obtained over a wide range of UV doses was better fit by a quadratic rather than a linear function (Qk and Lk, respectively, in Figure 7A). Induction of pure white ade2 adeX mutants up to a UV dose of 80 J/m2 was consistent with either linear-quadratic (LQm) or linear (Lm) mutagenesis curves (Figure 7, B–D). However, at UV doses of 100–160 J/m2, the frequencies of pure white ade2 adeX mutants increased more rapidly than the first power of dose (Figure 7B). The relatively weak, UV-induced increase in the frequency of red-white sectored colonies in the WT strain was linear (Figure 7E).
Figure 7.
UV-induced killing and mutagenesis in a WT strain. (A) UV-induced killing. For each UV dose, survival was determined in three independent experiments; these values are plotted and overlap at each dose. The data are best fit (R2 = 0.9986) with a quadratic killing curve (Qk, solid line) with the coefficient of lethality k2 = 0.000389 (J/m2)−2. Approximation of the data with a linear function (Lk) is represented as dotted lines. Approximation with a linear-quadratic function is not represented due to negative k1 value. (B) Average values of UV-induced pure white ade2 adeX mutant frequencies (circles) and red-white sectored ade2 adeX mutant frequencies (squares) obtained in three independent experiments. (C) Yields of UV-induced pure white ade2 adeX mutants. For each UV dose, values obtained in three independent experiments (open circles) as well as average values (solid circles) are plotted. The data are best fit (R2 = 0.9604) with a linear-quadratic mutagenesis curve (LQm, solid line) with the coefficients of mutability m1 = 0.3503 (J/m2)−1 and m2 = 0.003015 (J/m2)−2. Approximation of the data with a linear mutagenesis function (Lm) is represented as a dashed line [R2 = 0.8907; m1 = 0.5095 (J/m2)−1]. Approximation with a quadratic function (Qm) is represented as a dotted line. (D) Frequencies of UV-induced pure white ade2 adeX mutants. For each UV dose, values obtained in three independent experiments (open circles) and average values (solid circles) are plotted. The data are best fit (R2 = 0.9966) with a linear-quadratic mutagenesis curve (LQm, solid line) with the coefficients of mutability m1 = 0.3503 (J/m2)−1 and m2 = 0.003015 (J/m2)−2. Approximation of the data with a linear mutagenesis function (Lm) is represented as a dashed line [R2 = 0.96; m1 = 0.5095 (J/m2)−1]. Approximation with a quadratic function (Qm) isrepresented as a dotted line. (E) Frequencies of UV-induced red-white sectored ade2 adeX mutants. For each UV dose, values obtained in three independent experiments (open squares) and average values (solid squares) are plotted. The data are well fit with a linear mutagenesis curve (R2 = 0.9828).
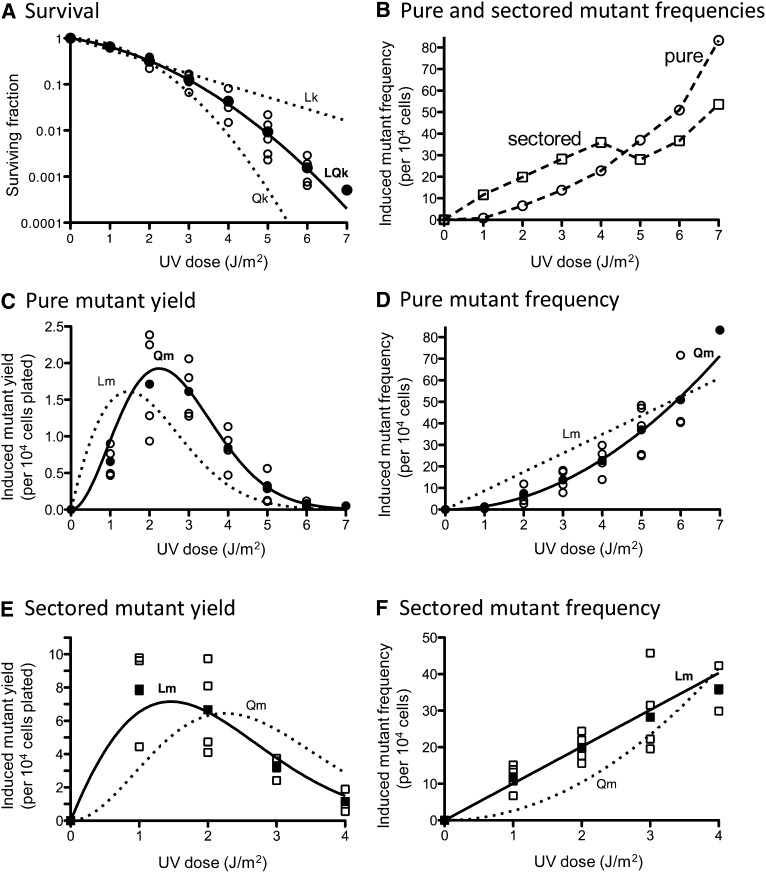
In the rad14Δ background, the UV survival curve followed the linear-quadratic killing model (LQk; Figure 8A). The induction of pure white ade2 adeX mutants at all doses tested was best fit with by a purely quadratic mutagenesis curve (Qm; Figure 8, B–D). Over the dose range from 1 to 4 J/m2, the yield and the frequency of red-white sectored mutants rose linearly with dose (Lm; Figure 8, E and F). Departure from initial dose–response dependence was observed, however, at UV doses >4 J/m2 (Figure 8B).
Figure 8.
UV-induced killing and mutagenesis in an NER-deficient (rad14Δ) strain. (A) UV-induced killing. Survival values obtained in three to five independent experiments (open symbols) and average values (solid symbols) are plotted. The data are best fit (R2 = 0.9999) with a linear-quadratic killing curve (LQk, solid line) with coefficients of lethality k1 = 0.3024 (J/m2)−1 and k2 = 0.1305 (J/m2)−2. Approximation of the data with linear (Lk) or quadratic (Qk) functions is represented as dotted lines. (B) The average values of UV-induced pure white (circles) and red-white sectored ade2 adeX mutant frequencies (squares) obtained in three to five independent experiments are shown. (C) Yields of UV-induced pure white ade2 adeX mutants. For each UV dose, values obtained in three to four independent experiments (open circles) and average values (solid circles) are plotted. The data are best fit (R2 = 0.9657) with a quadratic mutagenesis curve (Qm, solid line) with the coefficient of mutability m2 = 1.455 (J/m2)−2. An approximation of the data with a linear mutagenesis function (Lm) is represented as a dotted line. The approximation with a linear-quadratic function is not represented due to a negative m1 value. (D) Frequencies of UV-induced pure white ade2 adeX mutants. For each UV dose, values obtained in three to four independent experiments (open circles) and average values (solid circles) are plotted. The data are best fit (R2 = 0.9747) with a quadratic mutagenesis curve (Qm, solid line) with the coefficient of mutability m2 = 1.455 (J/m2)−2. Approximation of the data with a linear mutagenesis function (Lm) is represented as dotted line. An approximation with a linear-quadratic function is not shown due to a negative m1 value. (E) Yields of UV-induced red-white sectored ade2 adeX mutants. For each UV dose, the values obtained in three to four independent experiments (open squares) and average values (solid squares) are plotted. The data are best fit (R2 = 0.9503) with a linear mutagenesis curve (Lm, solid line) with the coefficient of mutability m1 = 10.06 (J/m2)−1. Approximation of the data with a quadratic mutagenesis function (Qm) is represented as dotted line. An approximation with a linear-quadratic function is not represented due to a negative m1 value. (F) Frequencies of UV-induced red-white sectored ade2 adeX mutants. For each UV dose, values obtained in three to four independent experiments (open squares) and the average values (solid squares) are plotted. The data are best fit (R2 = 0.9683) with a linear mutagenesis curve (Lm, solid line) with the coefficient of mutability m1 = 10.06 (J/m2)−1. Approximation of the data with a quadratic mutagenesis function (Qm) is represented as dotted line. An approximation with a linear-quadratic function is not shown due to a negative m1 value.
As observed previously (Eckardt and Haynes 1977; Eckardt et al. 1980) and confirmed here (Figure 8), the frequency of pure white ade2 adeX colonies in an NER-defective background increased with UV dose and, as in WT, even outnumbered that of sectored mutants at the highest doses examined. An explanation suggested previously for the dose-dependent phenomenon in an NER-defective background is that pure ade2 adeX colonies reflect lethal sectoring (a mutational hit in one strand and a lethal hit in the other) (Eckardt et al. 1980). If this is the case, then the induction of pure mutant colonies in NER-deficient strains is expected to the follow “two-hit” quadratic function. In our analyses, the induction of pure ade2 adeX mutants in the rad14Δ strain clearly followed the quadratic function (Figure 8D). We note, however, that the induction curve of pure ade2 adeX mutants in early studies with a rad2-20 strain did not reveal a quadratic component (Eckardt and Haynes 1977), although a clear tendency toward a quadratic mechanism was observed in a subsequent study with rad1-1 strains (Eckardt et al. 1980). One possible explanation for the variable induction curves of pure mutant colonies is that residual NER remained in the specific rad point mutants used in early studies. As expected, and in contrast to the quadratic induction curve for pure ade2 adeX mutants, we observed a linear curve (up to a UV dose yielding 4% survival) for the induction of sectored mutants in a rad14Δ background (Figure 8E).
In our study, the induction of pure white ade2 adeX mutants in the WT strain (up to a UV dose yielding 5% survival) fit well with both linear-quadratic (with a large linear component and a small quadratic component) and linear mutagenesis curves (Figure 7D). A linear dose–response pattern in a WT strain (up to a UV dose yielding 4% survival) also was reported in earlier studies (Eckardt and Haynes 1977). By contrast, the induction of Arg+ revertants in the arg4-17 reversion system was purely quadratic (Kilbey et al. 1978). The arg4-17 data led to the proposal of a two-hit mutagenesis mechanism reflecting the induction of two closely spaced lesions on complementary DNA strands. It was subsequently demonstrated, however, that the induction of such closely opposed lesions in vivo (detected as double-strand breaks when DNA was treated with a glycosylase that nicks at sites of UV damage) was linear (Reynolds 1987). To explain the unexpected linearity, it was suggested that the induction of closely opposed lesions at a single site follows a sigmoidal (quadratic response followed by plateau phase) dose–response curve, but that the summation of multiple sigmoidal functions across a genome yields a linear dose–response curve. This interpretation can account for the linearity of dose–response curves in the ade2 adeX mutation system, where there are multiple (Reynolds 1987) potential mutation sites across the genome. By contrast, a quadratic response is expected in the case of arg4-17 reversion because there is only a single possible mutation site.
Discussion
In the studies reported here, a nonselective ade2 adeX forward mutation system was used to investigate the genetic control of UV-induced mutagenesis in nondividing haploid cells. The significance of this system is that it allows the frequencies of one- and two-strand mutations to be monitored via the production of red-white sectored and pure-white colonies, respectively (see Figure 2). Following UV irradiation of G0 cells, 85% of induced mutations in a WT background were two-strand mutations and, therefore, must have arisen prior to DNA replication. In the complete absence of NER (rad1Δ or rad14Δ background), the total frequency of ade2 adeX mutations was unchanged, but 80–85% were one-strand events (Figure 3). We note that this was a more dramatic reduction in two-strand mutation frequency than observed in the earlier study (Eckardt and Haynes 1977) and suggest that this may have reflected use of the potentially non-null rad2-20 allele.
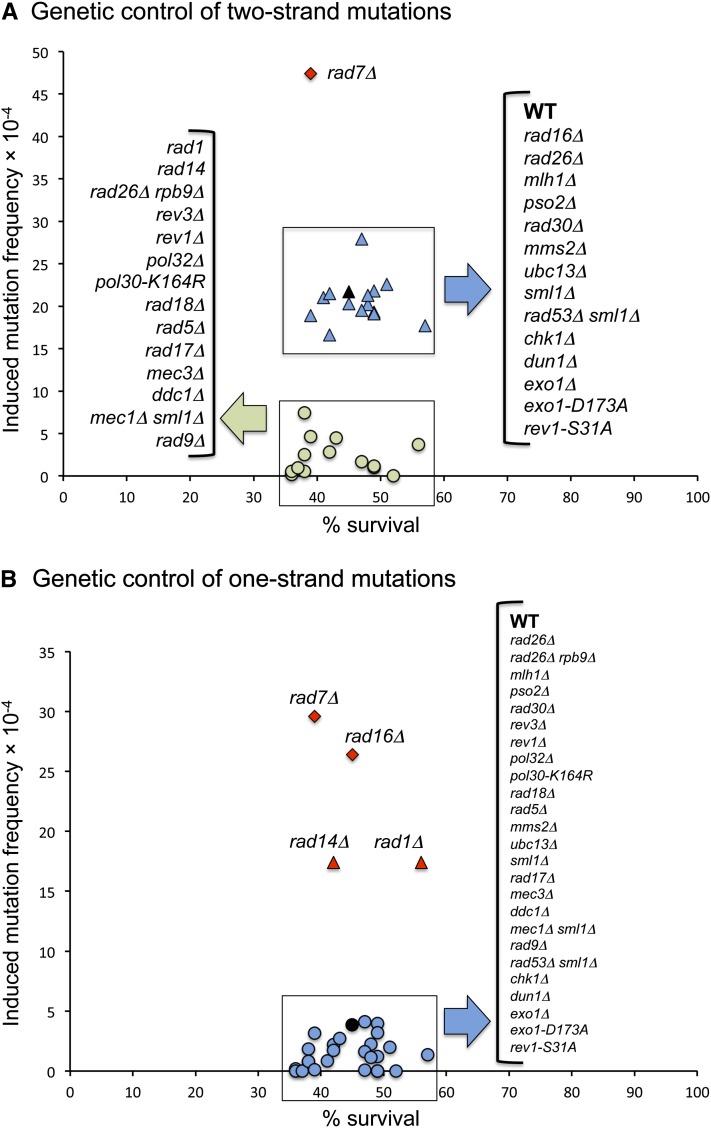
Following confirmation that UV-induced two-strand mutations require a functional NER pathway, we examined the effects of deleting genes involved in NER subpathways, DNA MMR, interstrand crosslink repair, error-free lesion bypass, error-prone lesion bypass, and the DNA damage checkpoint response. Although the primary focus was on two-strand mutations, data were also generated for the one-strand mutations; the two- and one-strand frequency data are summarized in Figure 9, A and B, respectively. In the case of one-strand mutations, their numbers were in a minority in a WT background, making it difficult to detect subtle changes in the corresponding frequencies. It was clear, however, that the production of one-strand mutations required the Pol ζ-dependent TLS pathway, consistent with error-prone bypass of damage during DNA replication. The absence an increase in one-strand mutations upon loss of Ubc13-Mms2 likely reflects compensatory mechanisms of error-free bypass (Choi et al. 2010).
Figure 9.
Summary of one- and two-strand mutation frequencies in defined mutant backgrounds. (A) Genetic control of two-strand mutations. (B) Genetic control of one-strand mutations. Black symbols indicate WT frequencies. Frequencies similar to WT are plotted as blue symbols, those less than WT are in green, and those greater than WT are in red.
Models A and B in Figure 1 posit that two-strand mutagenesis initiates when a lesion is contained within an NER-generated gap and is bypassed during the gap-filling reaction. The mutation-containing strand is then used as a template in a subsequent round of NER that removes the lesion. By contrast, model C proposes that a mutation is introduced during error-prone filling of a lesion-free, NER-generated gap and then is introduced into the complementary strand by DNA MMR. This model does not involve a lesion-bypass step and hence is not expected to be Pol ζ-dependent. Finally, model D suggests that mutations are generated during repair of UV-induced DNA interstrand crosslinks, which is also expected to require lesion bypass and hence Pol ζ activity. We eliminated models C and D as major contributors to two-strand mutations by inactivation of MMR (mlhΔ strain) and ICLR (pso2Δ strain), respectively.
A requirement for the catalytic subunit of Pol ζ (Rev3), as well as an accessory subunit (Pol32), and for Rev1 in the production of two-strand mutations is consistent with bypass of a UV-induced lesion within an NER-generated gap, as proposed in models A and B. NER can be triggered either though direct damage recognition or through blockage of the transcription machinery—the GGR and TCR subpathways, respectively. The elimination of GGR did not affect the frequency of two-strand mutations, but there was a threefold reduction in these events in a TCR-deficient background (rad26Δ rpb9Δ). This suggests that the TCR subpathway is primarily responsible for the production of two-strand mutations. One interesting possibility is that the TCR subpathway is more likely than GGR to aberrantly remove the undamaged strand, thereby forcing bypass of a UV lesion and triggering a subsequent round of NER. Although our data cannot discriminate between models A and B, these models could, in principle, be distinguished by engineering an appropriate lesion into one or both strands of a transforming plasmid. If model A is correct, two-strand mutations should predominate only when closely opposed lesions are present in both strands.
Given the requirement of Pol ζ in the production of two-strand mutations, we examined the relevance of proteins/complexes previously implicated in regulating Pol ζ-dependent mutagenesis either by directly promoting Pol ζ activity or by facilitating an alternative, error-free bypass mechanism. The error-free bypass mechanisms require an undamaged duplex (usually the sister chromatid) as template and hence operate either at or behind the replication fork. If two-strand mutations indeed arise prior to replication, their frequency should not be affected by the presence/absence of error-free alternatives. The Rad6-Rad18 complex monoubiquitinates PCNA at lysine 164 (K164), a modification that precedes polyubiquitination by Ubc13-Mms2 and is required for both error-prone and error-free bypass in dividing cells (Hoege et al. 2002). In G0 cells, either elimination of Rad18 or mutation of K164 of PCNA (pol30-K164R strain) eliminated two-strand mutations, indicating that monoubiquitination of PCNA is required, most likely for Pol ζ recruitment to a lesion-containing gap. As expected for mutations that originate during G0, however, the frequency of two-strand mutations was unchanged in an mms2Δ or ubc13Δ strain. The major role of Rad5 is as the E3 ubiquitin ligase for the Ubc13-Mms2 E2 ubiquitin conjugase, but Rad5 also has been implicated in the Pol ζ bypass of some lesions (Gangavarapu et al. 2006; Pagès et al. 2008). In the ade2 adeX system used here, two-strand mutations were dependent on Rad5 as well as on Rad18. A dual requirement of both Rad5 and Rad18 for two-strand mutations in nondividing cells is unique and is in contrast to spontaneous lesion tolerance, where Rad5 and Rad18 seem to be involved in separate pathways of Pol ζ-dependent bypass (Liefshitz et al. 1998; Cejka et al. 2001; Minesinger and Jinks-Robertson 2005).
The DNA damage response (DDR) is activated by ssDNA produced during repair processes or contained within replication-generated gaps rather than by the primary DNA damage (reviewed by Harrison and Haber 2006; Lazzaro et al. 2009). DDR requires apical sensor proteins (the Mec1-Ddc2 complex and the 9-1-1 checkpoint clamp), mediators that amplify the signal (Rad9), and downstream effectors (Rad53 and Chk1) that phosphorylate target proteins. Complete activation of the pathway delays or arrests the cell cycle and facilitates repair by inducing expression of, or post-translationally modifying, target proteins. In budding yeast, activation of DDR is typically detected via hyper-phosphorylation of Rad53. In a previous study of DDR activation in G1-arrested cells treated with UV, both NER and the 5′ to 3′ exonuclease Exo1 were required (Giannattasio et al. 2010). It was suggested that Exo1 enlarges the ≈30-nt gaps created by NER when the gap-filling process is delayed, which might occur if the gap contains a lesion. Our analyses demonstrated a critical role only for apical DDR factors (Mec1 and 9-1-1) and for the Rad9 sensor in the generation of two-strand mutations in nondividing cells; elimination of the downstream factors Rad53, Chk1, or Dun1 did not alter the frequency of events (Figure 6). In addition, neither deletion of Exo1 nor loss of its exonuclease activity affected two-strand mutations. At least in the system used here, NER-generated gaps appear to be sufficient to activate a 9-1-1/Mec1/Rad9-dependent pathway that promotes two-strand mutations. The observation that the 9-1-1 clamp physically interacts with Pol ζ and stimulates its recruitment to UV-damaged chromosomes (Sabbioneda et al. 2005) may indicate a structural role for 9-1-1 (and presumably Mec1 and Rad9) that is unrelated to the function of these DNA-damage checkpoint proteins in a phosphorylation cascade.
How might the DDR pathway promote UV-induced mutagenesis in nondividing cells? First, a connection between DDR components and the efficiency of NER has been previously reported. In yeast, for example, there is a moderate reduction in the rate of CPD removal in rad9 (Al-Moghrabi et al. 2001) and mec1 mutants, but not in rad53 or chk1 mutants (Taschner et al. 2010). A significant reduction in the rate of removing CPDs and pyrimidine (6-4) pyrimidone photoproducts (6-4PPs) has been reported in ATR (the yeast Mec1 homolog)-deficient human fibroblasts (Auclair et al. 2008), and a moderate reduction in the rate of CPD removal has been observed in human cells carrying the XPA (the yeast Rad14 homolog) S196A allele that lacks an ATR-phosphorylation site (Shell et al. 2009). It is possible that a general impairment of NER in DDR-deficient mutants accounts for the suppression of two-strand mutagenesis; this effect may be limited to TCR, as it is this subpathway of NER that contributes most to two-strand mutations. A second possibility is that the relevant DDR factors are required for activation of the Pol ζ-dependent pathway of mutagenic lesion bypass. Indeed, 9-1-1/Mec1-dependent phosphorylation of Rev1 (Sabbioneda et al. 2007; Pages et al. 2009), as well as a moderate reduction of UV-induced mutagenesis in a rad14Δ rev1-S31A mutant lacking the Mec1-phosphorylation site of Rev1, has been reported in yeast (Pages et al. 2009). In our system, however, it is unlikely that Mec1-dependent phosphorylation of Rev1 is important for the production of two-strand mutations in G0 cells; the rev1-S31A mutant was as UV-mutable as the WT strain (Figure 6). There are, however, potential Mec1-phosphorylation sites (SQ/TQ) present in other proteins involved in mutagenic bypass of UV lesions, including Rev3, Rev7, Rad6, and Rad18 (Pages et al. 2009). Although no damage-induced phosphorylation of these proteins has been detected biochemically in cycling cells (Pages et al. 2009), it may still be relevant to UV-induced mutagenesis in nondividing cells. Potential Mec1-phosphorylation sites also are present in two other proteins required for error-prone lesion bypass: PCNA (one SQ site) and Rad5 (five SQ and two TQ sites) (data not shown).
Together, our data suggest the following model for the UV-induced mutagenesis in nondividing cells. First, mutagenesis is initiated when NER generates a lesion-containing gap. In principle, this could reflect either two closely opposed lesions or rare removal of the undamaged strand (models A and B in Figure 1, respectively). Second, error-prone filling of the lesion-containing gap requires Rev1, Pol ζ, monoubiquitinated PCNA, and Rad5; the presence/absence of Pol η does not affect the frequency of this process. Third, a second round of NER is initiated to remove the remaining UV lesion. Filling of the gap thus created uses the mutation-containing strand synthesized during the first round of NER as a repair template, and this introduces the mutation into both strands of the duplex. Finally, genome duplication yields daughter cells that each contains the mutation at the ADEX locus, and a pure colony is produced. Intriguingly, there is a partial requirement for the 9-1-1 checkpoint clamp, Mec1, Ddc2, and Rad9 for two-strand mutagenesis, but there is no involvement either of Exo1 or of the more downstream components of checkpoint signaling (Rad53, Chk1, and Dun1). We suggest that the requirement for checkpoint proteins, but not a full-blown checkpoint response, reflects a role in recruiting and/or activating functions required to fill the lesion-containing gap created by NER.
The mechanisms and proteins involved in DNA damage repair, bypass, and checkpoint activation are highly conserved, suggesting that the yeast studies reported here will be widely applicable. Importantly, we have confirmed that NER, which normally promotes genetic stability by removing bulky DNA damage, is required for most UV-induced mutagenesis in nondividing cells; we have identified additional proteins relevant to this process; and we have excluded possible models for how a genetic change can be introduced into both strands of duplex DNA prior to the resumption of growth. Whether NER-dependent mutagenesis is a more general response to DNA damage in nondividing cells is not clear and likely will be related to the details of the underlying molecular mechanism. If only a single lesion is required, then the inherent strand-discrimination error rate of NER will initiate a permanent genetic change at any lesion that is a cognate substrate. If closely opposed lesions are a prerequisite, however, then NER-dependent mutagenesis may be more limited. Importantly, because this type of mutagenesis occurs in the absence of cell division, it potentially provides a way for cells to escape either adverse growth conditions or normal growth inhibition. It thus may contribute to responses or be relevant to genetic changes required for tumorigenesis, especially those that occur in post-mitotic tissues.
Supplementary Material
Acknowledgments
We thank Kevin Lehner for his critical reading of the manuscript and Yi Yin for helpful discussions. This research was supported by the National Institutes of Health grants 5R01-GM064769-09 and 3R01-GM-064769-09S1-P3034122 to S.J.R.
Footnotes
Communicating editor: N. M. Hollingsworth
Literature Cited
- Abdulovic A., Kim N., Jinks-Robertson S., 2006. Mutagenesis and the three R’s in yeast. DNA Repair (Amst.) 5: 409–421. [DOI] [PubMed] [Google Scholar]
- Al-Moghrabi N. M., Al-Sharif I. S., Aboussekhra A., 2001. The Saccharomyces cerevisiae RAD9 cell cycle checkpoint gene is required for optimal repair of UV-induced pyrimidine dimers in both G(1) and G(2)/M phases of the cell cycle. Nucleic Acids Res. 29: 2020–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auclair Y., Rouget R., Affar El B., Drobetsky E. A., 2008. ATR kinase is required for global genomic nucleotide excision repair exclusively during S phase in human cells. Proc. Natl. Acad. Sci. USA 105: 17896–17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer G. A., Burgers P. M., 1990. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Res. 18: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Vondrejs V., Storchova Z., 2001. Dissection of the functions of the Saccharomyces cerevisiae RAD6 postreplicative repair group in mutagenesis and UV sensitivity. Genetics 159: 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Szakal B., Chen Y. H., Branzei D., Zhao X., 2010. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol. Biol. Cell 21: 2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O., Livneh Z., 1992. Biochemical analysis of UV mutagenesis in Escherichia coli by using a cell-free reaction coupled to a bioassay: identification of a DNA repair-dependent, replication-independent pathway. Proc. Natl. Acad. Sci. USA 89: 3300–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt F., Haynes R. H., 1977. Induction of pure and sectored mutant clones in excision-proficient and deficient strains of yeast. Mutat. Res. 43: 327–338. [DOI] [PubMed] [Google Scholar]
- Eckardt F., Teh S. J., Haynes R. H., 1980. Heteroduplex repair as an intermediate step of UV mutagenesis in yeast. Genetics 95: 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., et al. , 2006. DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- Gangavarapu V., Haracska L., Unk I., Johnson R. E., Prakash S., et al. , 2006. Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 7783–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik K. J., Li X., Pautz A., Burgers P. M. J., 1998. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 273: 19747–19755. [DOI] [PubMed] [Google Scholar]
- Giannattasio M., Lazzaro F., Longhese M. P., Plevani P., Muzi-Falconi M., 2004. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 23: 429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M., Follonier C., Tourriere H., Puddu F., Lazzaro F., et al. , 2010. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol. Cell 40: 50–62. [DOI] [PubMed] [Google Scholar]
- Gibbs P. E., Mcdonald J., Woodgate R., Lawrence C. W., 2005. The relative roles in vivo of Saccharomyces cerevisiae Polη, Polζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169: 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. L., Mccusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Jinks-Robertson S., 1999. Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4766–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J. C., Haber J. E., 2006. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40: 209–235. [DOI] [PubMed] [Google Scholar]
- Haynes R. H., Eckardt F., 1979. Analysis of dose-response patterns in mutation research. Can. J. Genet. Cytol. 21: 277–302. [DOI] [PubMed] [Google Scholar]
- Heidenreich E., Eisler H., Lengheimer T., Dorninger P., Steinboeck F., 2010. A mutation-promotive role of nucleotide excision repair in cell cycle-arrested cell populations following UV irradiation. DNA Repair (Amst.) 9: 96–100. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S., 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141. [DOI] [PubMed] [Google Scholar]
- James A. P., Kilbey B. J., 1977. The timing of UV mutagenesis in yeast: a pedigree analysis of induced recessive mutation. Genetics 87: 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. P., Kilbey B. J., Prefontaine G. J., 1978. The timing of UV mutagenesis in yeast: continuing mutation in an excision-defective (rad1–1) strain. Mol. Gen. Genet. 165: 207–212. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Prakash L., Prakash S., 2012. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc. Natl. Acad. Sci. USA 109: 12455–12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbey B. J., Brychcy T., Nasim A., 1978. Initiation of UV mutagenesis in Saccharomyces cerevisiae. Nature 274: 889–891. [DOI] [PubMed] [Google Scholar]
- Larimer F. W., Perry J. R., Hardigree A. A., 1989. The REV1 gene of Saccharomyces cerevisiae: isolation, sequence, and functional analysis. J. Bacteriol. 171: 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P. J., Flores-Rozas H., Kolodner R. D., 2002. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol. Cell. Biol. 22: 6669–6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. W., 2002. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair (Amst.) 1: 425–435. [DOI] [PubMed] [Google Scholar]
- Lazzaro F., Giannattasio M., Puddu F., Granata M., Pellicioli A., et al. , 2009. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair (Amst.) 8: 1055–1067. [DOI] [PubMed] [Google Scholar]
- Li S., Smerdon M. J., 2002. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 21: 5921–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefshitz B., Steinlauf R., Friedl A., Eckardt-Schupp F., Kupiec M., 1998. Genetic interactions between mutants of the ‘error-prone’ repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat. Res. 407: 135–145. [DOI] [PubMed] [Google Scholar]
- Majka J., Niedziela-Majka A., Burgers P. M., 2006. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell 24: 891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova A. V., Stodola J. L., Burgers P. M., 2012. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 40: 11618–11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minesinger B. K., Jinks-Robertson S., 2005. Roles of RAD6 epistasis group members in spontaneous Polζ-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics 169: 1939–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S., Morgan E. A., Symington L. S., 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159: 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès V., Bresson A., Acharya N., Prakash S., Fuchs R. P., et al. , 2008. Requirement of Rad5 for DNA polymerase ζ-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics 180: 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès V., Santa Maria S. R., Prakash L., Prakash S., 2009. Role of DNA damage-induced replication checkpoint in promoting lesion bypass by translesion synthesis in yeast. Genes Dev. 23: 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla T. A., Christie D.-M., Liskay R. M., 1994. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol. Cell. Biol. 14: 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R. J., 1987. Induction and repair of closely opposed pyrimidine dimers in Saccharomyces cerevisiae. Mutat. Res. 184: 197–207. [DOI] [PubMed] [Google Scholar]
- Rodriguez G. P., Romanova N. V., Bao G., Rouf N. C., Kow Y. W., et al. , 2012. Mismatch repair-dependent mutagenesis in nondividing cells. Proc. Natl. Acad. Sci. USA 109: 6153–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P., 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sabbioneda S., Minesinger B. K., Giannattasio M., Plevani P., Muzi-Falconi M., et al. , 2005. The 9–1-1 checkpoint clamp physically interacts with polzeta and is partially required for spontaneous polzeta-dependent mutagenesis in Saccharomyces cerevisiae. J. Biol. Chem. 280: 38657–38665. [DOI] [PubMed] [Google Scholar]
- Sabbioneda S., Bortolomai I., Giannattasio M., Plevani P., Muzi-Falconi M., 2007. Yeast Rev1 is cell cycle regulated, phosphorylated in response to DNA damage and its binding to chromosomes is dependent upon MEC1. DNA Repair (Amst.) 6: 121–127. [DOI] [PubMed] [Google Scholar]
- Sarkar S., Davies A. A., Ulrich H. D., McHugh P. J., 2006. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase ζ. EMBO J. 25: 1285–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell S. M., Li Z., Shkriabai N., Kvaratskhelia M., Brosey C., et al. , 2009. Checkpoint kinase ATR promotes nucleotide excision repair of UV-induced DNA damage via physical interaction with xeroderma pigmentosum group A. J. Biol. Chem. 284: 24213–24222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M., Harreman M., Teng Y., Gill H., Anindya R., et al. , 2010. A role for checkpoint kinase-dependent Rad26 phosphorylation in transcription-coupled DNA repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 30: 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unrau P., Wheatcroft R., Cox B., Olive T., 1973. The formation of pyrimidine dimers in the DNA of fungi and bacteria. Biochim. Biophys. Acta 312: 626–632. [DOI] [PubMed] [Google Scholar]
- Verhage R., Zeeman A. M., De Groot N., Gleig F., Bang D. D., et al. , 1994. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 6135–6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P., 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Waters L. S., Minesinger B. K., Wiltrout M. E., D’Souza S., Woodruff R. V., et al. , 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73: 134–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.