Abstract
In male mammals, the X and Y chromosomes are transcriptionally silenced in primary spermatocytes by meiotic sex chromosome inactivation (MSCI) and remain repressed for the duration of spermatogenesis. Here, we test the longstanding hypothesis that disrupted MSCI might contribute to the preferential sterility of heterogametic hybrid males. We studied a cross between wild-derived inbred strains of Mus musculus musculus and M. m. domesticus in which sterility is asymmetric: F1 males with a M. m. musculus mother are sterile or nearly so while F1 males with a M. m. domesticus mother are normal. In previous work, we discovered widespread overexpression of X-linked genes in the testes of sterile but not fertile F1 males. Here, we ask whether this overexpression is specifically a result of disrupted MSCI. To do this, we isolated cells from different stages of spermatogenesis and measured the expression of several genes using quantitative PCR. We found that X overexpression in sterile F1 primary spermatocytes is coincident with the onset of MSCI and persists in postmeiotic spermatids. Using a series of recombinant X genotypes, we then asked whether X overexpression in hybrids is controlled by cis-acting loci across the X chromosome. We found that it is not. Instead, one large interval in the proximal portion of the M. m. musculus X chromosome is associated with both overexpression and the severity of sterility phenotypes in hybrids. These results demonstrate a strong association between X-linked hybrid male sterility and disruption of MSCI and suggest that trans-acting loci on the X are important for the transcriptional regulation of the X chromosome during spermatogenesis.
Keywords: Haldane’s rule, meiosis, postmeiotic sex chromatin, speciation, spermatogenesis
FORTY years ago, Lifschytz and Lindsley (1972) proposed the provocative hypothesis that the X chromosome is inactivated during male meiosis and that disruption of this process could lead to sterility. The idea that failure of meiotic sex chromosome inactivation (MSCI) might cause sterility is significant in speciation genetics since it provides a possible explanation for the widespread observation that in crosses between species, sterility typically appears first in the heterogametic sex (Haldane 1922; Forejt 1996; Presgraves 2008).
Whether MSCI takes place in Drosophila has been debated for several decades (Kremer et al. 1986; McKee and Handel 1993; Hense et al. 2007; Vibranovski et al. 2009). Current evidence suggests that expression on the X chromosome is suppressed relative to the autosomes during spermatogenesis but that this suppression precedes meiosis and thus does not reflect MSCI (Meiklejohn et al. 2011; but see Vibranovski et al. 2012). Likewise, the sex chromosomes are not differentially silenced in chicken oocytes (Guioli et al. 2012). Therefore, failed MSCI cannot explain hybrid sterility in all female-heterogametic taxa. In therian mammals, however, MSCI is well established (Solari 1974; McKee and Handel 1993; Namekawa et al. 2007). Although no MSCI-essential loci have been found on the sex chromosomes, many of the autosomal genes that regulate this process have been identified, and the cellular progression of MSCI is well defined in the lab mouse (e.g., Turner et al. 2004; Ichijima et al. 2011).
In reproductively normal male mice, both sex chromosomes are transcriptionally active during mitosis and early prophase I of meiosis (leptotene–zygotene). At the start of pachytene, the X and Y undergo global chromatin remodeling and transcriptional silencing of all protein-coding genes, and are sequestered from the autosomes in a heterochromatin domain known as the sex body (reviewed in Handel 2004; Turner 2007). The sex body is disassembled at the end of prophase I but the sex chromosomes remain condensed and transcriptionally repressed through the two meiotic divisions (Namekawa et al. 2006; for exceptions on the Y see Vernet et al. 2011, 2012). Importantly, MSCI is essential for male fertility. Cells with defective MSCI are eliminated by late pachytene (Turner et al. 2006; Royo et al. 2010). Thus, global failure of MSCI causes meiotic arrest and complete sterility. Whether failed MSCI is a cause of hybrid male sterility is an open question.
While some sex-linked genes are expressed in haploid spermatids (Hendriksen et al. 1995; Namekawa et al. 2006; Mueller et al. 2008; Reynard et al. 2009), most remain repressed for the duration of spermatogenesis (Namekawa et al. 2006; Turner et al. 2006), a state termed postmeiotic sex chromatin repression (PSCR). Male lab mice with defective PSCR are subfertile or sterile due to reduced sperm count and morphologically abnormal sperm (Roest et al. 1996; Cocquet et al. 2009; Achame et al. 2010). Although different genes are implicated in chromosome-wide repression in primary spermatocytes (e.g., H2ax, Fernandez-Capetillo et al. 2003; Brca1, Turner et al. 2004; MDC1, Ichijima et al. 2011) vs. postmeiotic spermatids (e.g., Hr6b, Achame et al. 2010; Sly, Coquet et al. 2009), it is thought that PSCR is a downstream consequence of MSCI at the epigenetic level (Turner et al. 2006).
We previously studied gene expression in sterile male hybrids between the house mouse subspecies, Mus musculus musculus and M. m. domesticus (Good et al. 2010). In the cross between wild-derived inbred strains musculusPWK and domesticusLEWES, hybrid male sterility is asymmetric and strongly X linked: both F1 and late backcross males with a musculusPWK X chromosome are sterile or nearly so whereas males with a domesticusLEWES X are normal (Good et al. 2008a,b). Strikingly, a large number of genes on the musculusPWK X chromosome are overexpressed in the testes of sterile F1 males (Good et al. 2010). However, because whole testes comprise germ cells from all stages of spermatogenesis, together with somatic cells, we could not determine whether overexpression is specific to germ cells in which the X is normally silenced or repressed (Good et al. 2010).
Here, we test the hypothesis that the timing of X chromosome overexpression in sterile hybrid male house mice is consistent with disrupted MSCI. We used fluorescence-activated cell sorting (FACS) to isolate discrete germ cell populations from reciprocal F1 males and quantified the expression of X-linked genes using real-time reverse-transcription PCR (qRT–PCR). If disrupted MSCI explains overexpression of the musculusPWK X in sterile F1 males then X-linked genes should be overexpressed in meiotic cells subject to MSCI, whereas expression should be normal in cells in which the X is transcriptionally active. We found strong support for this prediction. Therefore, we then sought to define the genetic architecture of disrupted MSCI on the musculusPWK X. We hypothesized that disrupted MSCI in hybrid males results from local mismatch between loci in the domesticusLEWES autosomal genome and cis-acting sequences distributed along the musculusPWK X. To test this, we selected genes along the length of the X that were previously shown to be overexpressed in sterile F1 testes (Good et al. 2010) and measured whole-testis expression in F1 males with recombinant X chromosomes (X introgression F1s). If MSCI is disrupted in cis, only musculusPWK alleles should be overexpressed. Contrary to this prediction, we found that both musculusPWK and domesticusLEWES alleles were overexpressed in some introgression lines, identifying a region of the X chromosome likely to contain trans-acting loci that are important for the regulation of gene expression on the X chromosome during spermatogenesis.
Materials and Methods
Animals
The wild-derived inbred strains used in this study were purchased from the Jackson Laboratory (http://www.jax.org) and were maintained at the University of Arizona Central Animal Facility. Pure F1 males were produced from reciprocal crosses between PWK/PhJ (musculusPWK) and LEWES/EiJ (domesticusLEWES). To generate F1 males with recombinant X chromosome genotypes we crossed homozygous musculusPWK or domesticusLEWES X introgression line females to pure heterosubspecific males. Sex chromosome genotypes for the seven X introgression F1s are shown in Figure 2A; see Good et al. (2008b) and Campbell et al. (2012) for details. Intrasubspecific controls were produced by crossing musculus strains PWK/PhJ and CZECHII/EiJ (musculusCZECHII), and domesticus strains LEWES/EiJ and WSB/EiJ (domesticusWSB). These intrasubspecific crosses were generated to avoid confounding expression differences within and between subspecies with differences between inbred and heterozygous genotypes (i.e., to remove the effects of inbreeding depression).
Figure 2.
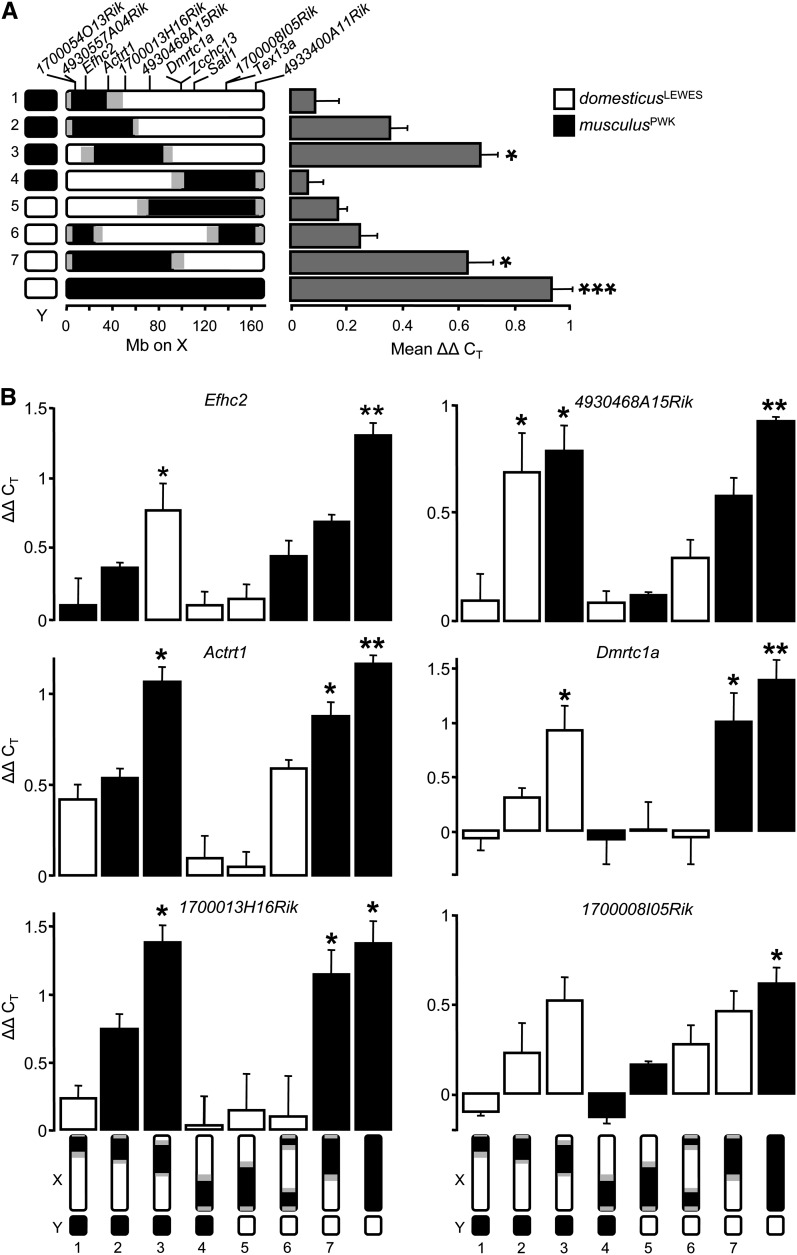
X overexpression in F1 hybrid male testes is associated with a musculusPWK introgression on the proximal X. (A) Sex chromosome genotypes and mean X overexpression in experimental males. X introgression genotypes are numbered 1–7; regions of uncertainty between domesticusLEWES (open) and musculusPWK (solid) recombination break points on the X are shown shaded; all genoytpes share the same F1 autosomal background. Expression was normalized relative to beta-actin, Acrv1, and Protamine 1 (ΔCT); mean ΔΔCT is the difference in expression between experimental genotypes and the fertile F1, averaged across the 12 X-linked genes indicated on the left side. Error bars are +1 SE. Tests for significant differences between experimental genotypes and control were performed on ΔCT values with ANOVA followed by Dunnett post hoc tests, (*) P < 0.01, (***) P < 0.0001. (B) Overexpression of individual X-linked genes does not require a musculusPWK allele. Data are presented as genotype means (±1 SE) for six representative genes. Sex chromosome genotypes are shown on the x-axis. Solid bars denote the musculusPWK allele at a given locus; open bars denote the domesticusLEWES allele. Significance was tested as in 2A, (*) P ≤ 0.05, (**) P ≤ 0.001. See also Figure S2.
All males were maintained in cages containing a maximum of three same-sex sibs until 50 days, after which they were caged singly for at least 20 days and killed by cervical dislocation. The reciprocal F1 males used in the targeted cell-type expression experiment (n = 5/genotype) were 70–73 days old. The reciprocal F1 and X introgression F1 males used in the whole-testis expression experiment were 70 days old (n = 3/genotype). Testes were dissected immediately after euthanasia, cut into two sections, and stored in RNAlater (Ambion) at −20° until processing. Data collection for individual measures of testis mass, sperm count, and sperm abnormality is described in Campbell et al. (2012).
Preparation and staining of testicular single cell suspensions for FACS
Single cell suspensions were prepared according to Getun et al. (2011) with minor modifications. Briefly, interstitial cells were removed by digesting freshly dissected detunicated testes with 1mg/ml collagenase I (Sigma) in Gey’s balanced salt solution (GBSS, Sigma). Tubules were digested with 1 mg/ml trypsin (Worthington) in GBSS with 1 mg/ml collagenase and germ cells were released by gentle pipetting with a wide orifice transfer pipette for 90 sec/testis. Cells were prestained with 0.16 mg/ml Hoechst 33343 (Invitrogen) and 0.8 mg/ml trypsin in GBSS. Trypsin was inactivated with 0.16 mg/ml fetal calf serum, and final staining was performed by bringing Hoechst concentration to 0.36 mg/ml. Cells were filtered twice through 70-μm nylon mesh, stained with propidium iodide (0.002 mg/ml), and stored on ice protected from light until sorting. All digestion and staining steps were carried out at 33° for 15 min with agitation at 120 rpm; DNase (0.004 mg/ml) was added at every step to eliminate cell clumps. Immediately before processing, cells were spun down for 4 min at ∼200 × g in a clinical centrifuge and resuspended in plain GBSS.
FACS and validation
Cell sorting was performed using a BD FACSAria IIu (BDBiosciences, San Jose, CA). Propidium iodide discriminates dead from viable cells whereas Hoechst differentiates viable cells based on DNA content and chromatin structure (see Bastos et al. 2005 for details). Propidium iodide was excited at 488 nm and detected at 610/20 with a 595LP filter. Hoechst was excited at 405 nm, with Hoechst Blue being detected at 450/50 nm and Hoechst Red at 630/30 nm with a 600LP filter. All samples were run with an 85-μm nozzle at <5000 events/sec. Cell populations enriched for spermatogonia and primary spermatocytes from early prophase I (LZ, leptotene/zygotene), mid-pachytene–diplotene primary spermatocytes (PD), secondary spermatocytes (SS), and postmeiotic round spermatids (RS) were collected into RLT buffer (Qiagen) with 1% betamercaptoethanol and stored on ice until processing.
We validated the purity of targeted cell populations in two ways. First, a subset of each cell population was collected into fixative (3:1 methanol:glacial acetic acid), spread on slides, and stained with 5% Giemsa (ENG Scientific). Cells were examined at 100× on a phase-contrast microscope and the number of cells of the targeted type was counted for a minimum of 50 cells/population. Second, five autosomal genes that are enriched in different spermatogenic cell types were assayed in each cell population with qRT–PCR as described below: Igfr1 (spermatogonia/early prophase I), Hormad1 (pachytene), Hist1h1t (mid-late pachytene), and Acrv1 and Protamine 1 (round spermatids).
RNA extraction and cDNA synthesis
RNA from FACS cells was extracted within 24 hr of each sort using the RNeasy Plus kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized the same day using SuperScript III (Invitrogen) with oligo(dT) and ∼200 ng template RNA per 50 µl reaction. RNA extraction and cDNA synthesis from whole testis were as for cells, but with ∼10 µg template RNA per 40 µl cDNA reaction.
Selection of experimental loci and primer design
We assayed seven X-linked genes in FACS cells. To determine whether X overexpression was restricted to genes associated with particular spermatogenic time points we chose three genes that are normally highly expressed prior to MSCI in mitotic spermatogonia and early prophase I spermatocytes (Ammecr1, Rps6ka6, Fmr1), three postmeiotic genes (Actrt1, 4930557A04Rik, 4930468A15Rik), and one gene with both mitotic/early meiotic and postmeiotic enrichment (1700013H16Rik) (Namekawa et al. 2006; Chalmel et al. 2007; Achame et al. 2010). The 12 X-linked genes assayed in whole testis were a subset of the 32 genes with predominantly postmeiotic expression that were significantly overexpressed in the testes of sterile F1 males in an earlier microarray experiment (Good et al. 2010). These genes were chosen to maximize the range of coverage on the X and the representation of both musculusPWK and domesticusLEWES alleles in X introgression genotypes (Figure 2A).
Primers were designed in Primer3 (v. 0.4.0) spanning at least one intron or across exon–UTR boundaries. Whenever possible, primers were placed in exons present in all splice variants listed in the Ensembl database (NCBIM37). All primer sequences were checked for SNPs in musculusPWK and domesticusWSB strains using the Wellcome Trust Sanger Institute database and BLASTed against the mouse G+T database using megablast. Sequences with multiple polymorphic sites or homology to nonspecific targets were discarded. Primer sequences and amplicon sizes are provided in Supporting Information, Table S1.
qRT–PCR
qRT–PCR was performed with PerfeCTa SYBR Green FastMix for iQ (Quanta Biosciences) on a MyiQ2 light cycler (Bio-Rad). PCR efficiency for each primer pair was evaluated with standard curve analysis using an eight point dilution series from whole-testis cDNA. Efficiencies of 95–105% were obtained for all primers. For experimental assays, 16-µl reactions with a final concentration of 300 nM of each primer were run in 96-well plates for 40 cycles with annealing at 60°. We analyzed three to five (FACS cells) or three (testes) biological replicates per genotype with three technical replicates per sample. All standard deviations (SDs) for technical replicates were <0.30. For each locus, all genotypes from a single biological replicate were run on the same plate with appropriate negative controls; duplicate reactions for reference genes (Ubc or beta-actin) were included on each plate. Melt curve analysis was performed at the end of each run to check for multiple peaks indicative of nonspecific amplification. Single peaks were obtained for all loci. Crossing threshold (CT) values were calculated automatically with iQ5 Optical System Software (v. 2.1). CT values were normalized relative to Ubc or beta-actin values for each plate (ΔCT). Because postmeiotic cells are depleted in the testes of sterile males (Good et al. 2008a), whole-testis expression was normalized a second time relative to two postmeiotic autosomal genes, Acrv1 and Protamine 1.
Choice of control genotypes for whole-testis expression assays
We analyzed expression in experimental genotypes relative to both the fertile F1 (domesticusLEWES × musculusPWK) and mean expression in intrasubspecific controls (musculusCZECHII × musculusPWK and domesticusWSB × domesticusLEWES). There were minor differences in patterns of significance for individual genes but the results of analyses based on average relative expression (mean ΔΔCT) were the same (data not shown). We chose to present analyses with the fertile F1 control because this eliminates any potential differences in X expression due to autosomal background.
Results
Validation of FACS purity
Based on Giemsa staining, mean percentage purity in the four targeted cell populations was 89–69% (leptotene/zygotene, n = 6 males, 89% SD 8.0%; pachytene/diplotene, n = 4, 80% SD 6.0%; secondary spermatocytes, n = 4, 69% SD 20%; round spermatids, n = 6, 85% SD 7.0%; Figure S1A). Lower purity in secondary spermatocyte samples was mainly due to contamination with round spermatids. Importantly, the cellular composition of targeted populations did not differ between reciprocal F1 genotypes (ANOVA, P ≥ 0.6). The relative expression of autosomal genes enriched at different spermatogenic time points corresponded to expectations for each cell population (Figure S1B).
X overexpression is consistent with defective MSCI and PSCR
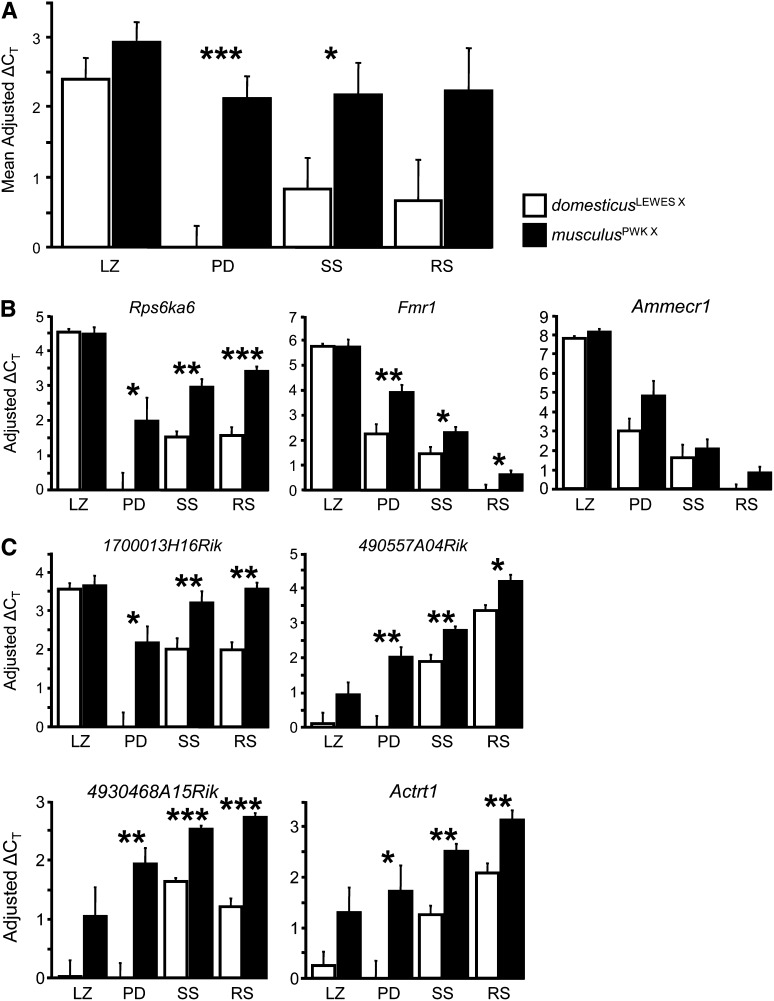
If disrupted MSCI explains overexpression of the musculusPWK X in sterile F1 males, then X-linked genes should be overexpressed only in meiotic cells subject to MSCI (i.e., overexpressed in pachytene/diplotene cells but not in the leptotene/zygotene population). We would also expect to observe overexpression in secondary spermatocytes. Consistent with these predictions, six of the seven X-linked genes assayed (86%) were significantly overexpressed in pachytene/diplotene and secondary spermatocyte cells from sterile F1 males, whereas expression in leptotene/zygotene cells did not differ between sterile and fertile genotypes for any gene (Figure 1). Across all seven genes, the average expression difference between genotypes was highly significant in the pachytene/diplotene population (PD, ANOVA, F(63,1) = 24.2, P < 0.0001), moderately significant in secondary spermatocytes (SS, F(62,1) = 4.46, P = 0.04), and nonsignificant in the leptotene/zygotene population (LZ, F(60,1) = 1.47, P = 0.2) (Figure 1A). The same six X-linked genes were overexpressed in round spermatids (RS, Figure 1, B and C). Although genotype means for this population were not significantly different (RS, F(62,1) = 3.50, P = 0.07), there was a clear trend toward overexpression in the sterile F1 (Figure 1A).
Figure 1.
Overexpression of X-linked genes in sterile F1 hybrid males is coincident with the onset of MSCI in primary spermatocytes and persists in postmeiotic round spermatids. (A–C) Expression in fertile F1 (open bars, domesticusLEWES X chromosome) and sterile F1 (solid bars, musculusPWK X chromosome) hybrid males in four germ cell populations in which the X is normally expressed (LZ, spermatogonia/early prophase I), silenced by MSCI (PD, pachytene/diplotene; SS, secondary spermatocytes), and repressed by PSCR (RS, round spermatids). Expression was normalized relative to Ubc (ΔCT) and adjusted to produce positive values by setting the lowest expression level to zero. Data are presented as genotype means +1 SE; pairwise differences between genotypes were tested with ANOVA, (*) P ≤ 0.05, (**) P ≤ 0.005, (***) P ≤ 0.0001. (A) Mean expression of the seven X-linked genes shown in B and C. (B) Expression of three X-linked genes that are normally highly expressed prior to MSCI in spermatogonia and early prophase I primary spermatocytes and do not escape PSCR. (C) Expression of four X-linked genes that are normally moderately expressed in postmeiotic round spermatids. Note that 1700013H16Rik is also normally enriched in spermatogonia (Namekawa et al. 2006).
We found previously that a large number of X-linked genes are overexpressed in sterile F1 male testes (Good et al. 2010). However, because progressive cell loss throughout spermatogenesis in sterile F1 males could bias the cellular composition of the testis toward early spermatogenic cell types we excluded those genes normally expressed in mitotic/early meiotic cells and focused on postmeiotic genes only (i.e., genes that escape PSCR). Here, using discrete cell populations, we show that both postmeiotic (Figure 1C) and mitotic/early meiotic genes (Figure 1B) are overexpressed in meiotic spermatocytes and in postmeiotic spermatids. Thus, both MSCI and PSCR appear to be disrupted and this likely affects the expression of a broad array of X-linked genes.
Overexpression of X-linked genes is not limited to musculusPWK alleles
We used a series of musculusPWK X introgression lines (Good et al. 2008b; Campbell et al. 2012) to test the hypothesis that disrupted MSCI in hybrid males results from local mismatch between loci in the domesticusLEWES autosomal genome and cis-acting sequences distributed along the musculusPWK X. If MSCI is disrupted in cis, only musculusPWK alleles should be overexpressed. We did not find support for this hypothesis: there was no relationship between whole-testis expression and allelic species identity in X introgression F1 males (ANOVA, F(250,1) = 0.4, P = 0.5). There was also no effect of the origin of the Y chromosome (F(19,1) = 0.3, P = 0.6). Instead, most instances of overexpression were concentrated in two genotypes (3 and 7) with a musculusPWK introgression on the proximal X (Figure 2B and Figure S2). Across all 12 genes assayed in this experiment, the average difference in expression between these two genotypes and controls was highly significant (Dunnett post hoc on ANOVA, genotype 3, P = 0.005, genotype 7, P = 0.003; Figure 2A). Nonetheless, overexpression was consistently most pronounced in the F1 with the complete musculusPWK X (Figure 2B and Figure S2). These results indicate that cis-regulatory differences between the musculusPWK and domesticusLEWES X chromosomes do not fully account for X overexpression in sterile hybrids and suggest that proper regulation of MSCI depends in part on a trans-acting locus or loci on the proximal part of the X.
Strong association between X overexpression and sterility phenotypes
Although all X introgression F1’s used in this study have subfertile reproductive phenotypes, variance in the severity of these deficits is large and is mostly explained by the location of musculusPWK introgressions on the X (Campbell et al. 2012). Notably, the interval on the proximal X associated with overexpression in genotypes 3 and 7 contains QTL for which musculusPWK alleles have negative effects on testis weight and sperm head morphology on an F1 background (Campbell et al. 2012). This prompted us to ask whether there was a correlation between the degree of X overexpression and the severity of reproductive deficits.
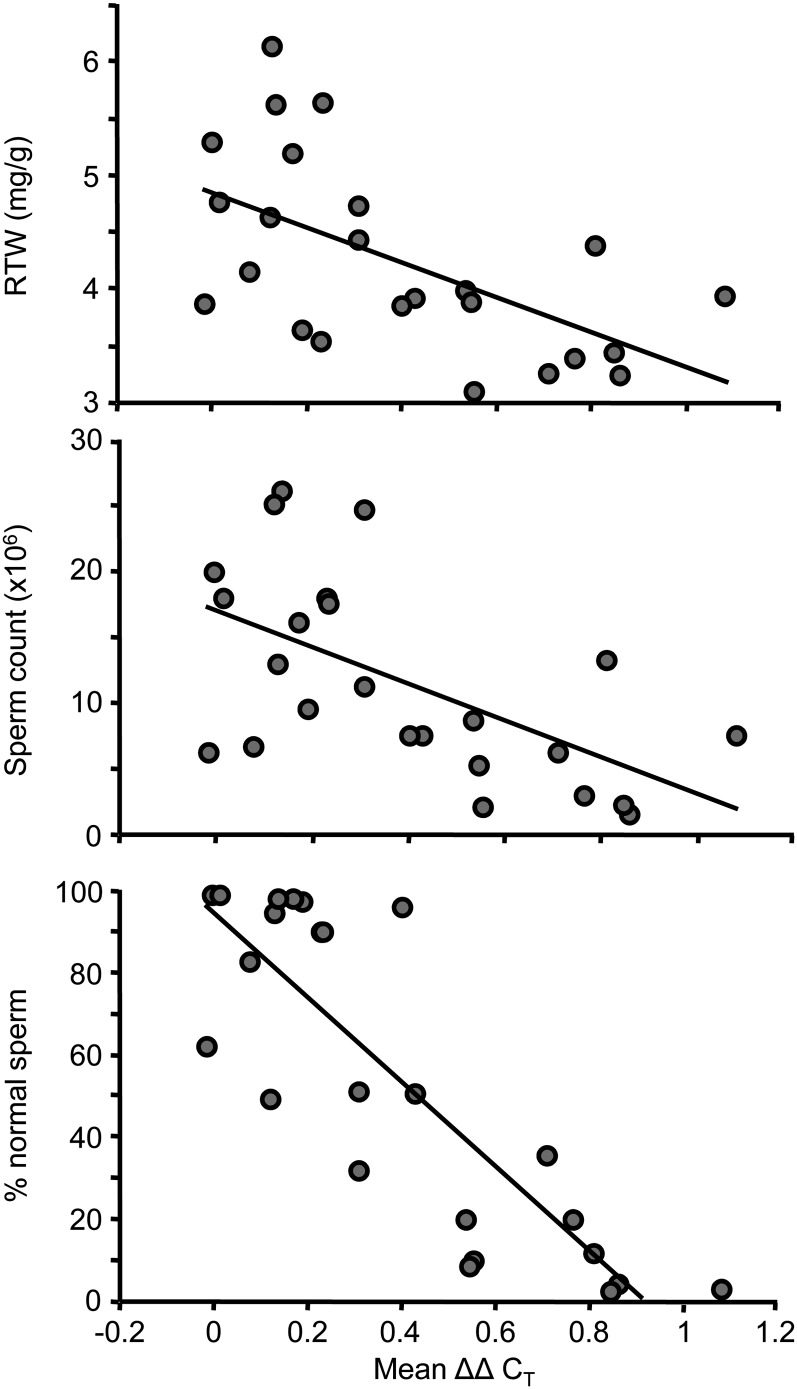
Using phenotypic data for the males assayed in this study, we found highly significant negative relationships between average X overexpression and body mass-corrected testis weight (RTW, R2 = 0.33, P = 0.003), sperm count (R2 = 0.34, P = 0.003), and the percentage of normal sperm (R2 = 0.71, P < 0.0001; Figure 3). This pattern suggests that hybrid male sterility and disrupted MSCI share a common genetic basis on the X.
Figure 3.
Hybrid males with more X overexpression have worse reproductive phenotypes. Significant negative correlations between X overexpression (mean ΔΔCT) and relative testis weight (RTW, R2 = 0.33, P = 0.003), sperm count (R2 = 0.34, P = 0.003), and the percentage of normal sperm (R2 = 0.71, P < 0.0001), measured in the 24 F1 males used in the whole-testis expression study (see Figure 2A for genotypes). Mean ΔΔCT is the difference in expression between experimental genotypes and the fertile F1, averaged across the 12 X-linked genes shown in Figure 2A.
Discussion
We present evidence that MSCI is defective in sterile hybrid house mice with a M. m. musculus X chromosome. Persistent X overexpression in round spermatids indicates that postmeiotic sex chromatin repression is also incomplete. These results, together with a strong association between X overexpression and the severity of sterility phenotypes, provide support for the longstanding hypothesis that failed X inactivation during spermatogenesis could contribute to hybrid male sterility (Lifschytz and Lindsley 1972; Forejt 1996; Presgraves 2008). While most work on the genetic mechanisms of mammalian MSCI has focused on autosomal genes, the finding that MSCI is disrupted in F1 males with a musculusPWK X chromosome but is normal in the reciprocal F1 indicates that MSCI-essential loci may exist on the X chromosome. The significant increase in X expression in males with a large musculusPWK introgression on the proximal X suggests that this interval contains a trans-acting locus or loci important for MSCI. We discuss alternative mechanisms that could cause MSCI and PSCR defects in sterile hybrids, and the relationship between MSCI and the large role of the X chromosome in the origins of reproductive isolation.
Disruption of MSCI
Transcriptional silencing of the sex chromosomes in early pachytene is essential for mammalian male fertility (Royo et al. 2010). In lab mouse mutants, primary spermatocytes with continued expression of the X, or the Y, or even specific Y-linked genes (e.g., Zfy2), are eliminated by apoptosis before the end of pachytene (Turner et al. 2005, 2006; Royo et al. 2010). Thus, global failure of MSCI causes meiotic arrest and complete absence of postmeiotic cells. In sterile F1 hybrid males, X overexpression coincident with the onset of MSCI strongly suggests that this process is disrupted. However, both the reproductive phenotypes and the pattern of X overexpression in these hybrids indicate that failure of MSCI is not complete. Some germ cells progress through meiosis and a smaller number survive spermiogenesis, albeit with severe abnormalities in sperm differentiation (Good et al. 2008a; Campbell et al. 2012). Likewise, while the difference in average X expression between the sterile and fertile F1 is most extreme in the late prophase I (PD) cells in which MSCI is in effect (Figure 1A), for any individual gene, the level of expression in the sterile F1 is lower in PD cells than in the cell population in which that gene is normally expressed (Figure 1, B and C).
There are at least three possible explanations for the pattern of incomplete disruption of MSCI. First, “leaky” MSCI in all or most primary spermatocytes results in moderate levels of X overexpression that are not sufficient to trigger the apoptotic response. The fact that failed silencing of several Y-linked genes does not cause meiotic arrest in transgenic lab mice suggests that misexpression of some genes can be tolerated (Royo et al. 2010). However, a large number of X-linked genes are overexpressed in sterile F1 testes (Good et al. 2010). Given that all protein-coding genes on the X are subject to MSCI, it is likely that even moderate overexpression of many X-linked genes would be toxic to primary spermatocytes. A second alternative is that MSCI fails completely in a subset of cells, and these are eliminated before the first meiotic division. In this case, overexpression in secondary spermatocytes could be a direct consequence of disrupted localization of heterochromatin proteins to the sex chromosomes in primary spermatocytes, or an indirect effect of an unhealthy cellular environment. Finally, because the late prophase I population was enriched for both pachytene and diplotene cells, the data are also consistent with a scenario in which MSCI is initiated normally but is not properly maintained. Quantification of stage-specific meiotic cell loss, together with immunostaining for epigenetic markers of the sex body, heterochromatin, and transcriptional repression, will pinpoint the timing and cellular distribution of MSCI defects in sterile hybrids.
What are the causes and consequences of MSCI disruption in hybrid males?
In the musculusPWK × domesticusLEWES cross, both male sterility and defective MSCI segregate with the musculusPWK X chromosome. Therefore, incompatibilities explaining these phenotypes must involve X-linked loci whose allelic identity, or distribution, differs between musculusPWK and domesticusLEWES. These findings raise two key questions: (1) Is defective MSCI the primary cause of hybrid male sterility; in other words, are the genes underlying defective MSCI and the genes underlying hybrid male sterility the same in this cross? and (2) Is defective MSCI a direct consequence of mismatch between loci that regulate this process, or is it a secondary consequence of earlier-acting incompatibilities? The strong association between X overexpression and male reproductive defects hints at a common genetic basis. However, evaluating this hypothesis awaits a better understanding of the genetic basis of MSCI defects. Therefore, we focus our discussion on the alternatives raised in the second question.
Although several autosomal genes essential for the initiation and progression of MSCI have been identified (e.g., H2ax, Fernandez-Capetillo et al. 2003; Brca1, Turner et al. 2004; MDC1, Ichijima et al. 2011), it is not known whether these loci interact with specific sequences on the X. In fact, evidence that MSCI is a special case of meiotic silencing of unsynapsed chromatin (MSUC), a generalized response to asynapsis at the zygotene/pachytene transition (Baarends et al. 2005; Turner et al. 2005, 2006), argues against the necessity of MSCI-essential loci on the X. However, based on the finding that most micro(mi)RNAs on the mouse X are highly expressed when all X-linked protein coding genes are silenced by MSCI (Song et al. 2009), Yan and McCarrey (2009) proposed that these MSCI-escaping miRNAs play a key role in regulating X inactivation in the male germ line. Preliminary support for this hypothesis is provided by an association between failed MSCI and disproportionate downregulation of miRNAs on the X vs. the autosomes in male lab mice lacking AGO4, a key protein in the miRNA pathway (Modzelewski et al. 2012).
The expression patterns in the seven X introgression F1’s assayed in this study demonstrate a strong association between X overexpression and an interval on the proximal half of the musculusPWK X between ∼37.1 and 68.7 Mb. If hybrid defects in MSCI are caused by incompatibilities between loci that control this process, then this interval contains a locus (or loci) whose trans-acting effects are essential to MSCI. Notably, while this interval contains ∼15% of protein-coding genes on the X, it contains ∼40% of X-linked miRNAs. As such, musculusPWK-derived miRNAs on the proximal X are promising candidates for targeted fine-scale mapping of the genetic basis of disrupted MSCI in hybrid males.
A nonmutually exclusive alternative is that MSCI defects are a consequence of genetic incompatibilities that act earlier in meiosis. In reproductively normal males, the intricate process of recombination immediately precedes MSCI. Recombination is initiated at leptotene by the formation of DNA double-strand breaks (DSBs) required for synapsis between homologous chromosomes, and culminates at the zygotene to pachytene transition when autosomal synapsis and DSB repair is complete. Notably, in lab mouse mutants with extensive autosomal asynapsis at pachytene, the sex chromosomes are not properly condensed or silenced and meiosis arrests midway through pachytene (Mahadevaiah et al. 2008; Burgoyne et al. 2009; Homolka et al. 2012). The link between autosomal asynapsis and failed MSCI may be explained by the integral role of several DNA damage response proteins in both DSB repair and initiation of MSCI (e.g., γH2AX, Fernandez-Capetillo et al. 2003; BRCA1, Turner et al. 2004). Asynapsis delays DSB repair and can promote the formation of additional breaks; persistent unrepaired DSBs on asynapsed autosomes sequester BRCA1 (Mahadevaiah et al. 2008). Burgoyne and colleagues proposed that MSCI fails under these conditions because this key protein is unavailable (Mahadevaiah et al. 2008; Burgoyne et al. 2009). A recent report of excess asynapsis in musculusPWK × C57BL/6J pachytene spermatocytes (Modzelewski et al. 2012) indicates that this phenotype may exist in the musculusPWK × domesticusLEWES cross, a hypothesis that we plan to evaluate by quantifying asynapsis, DSBs, and BRCA1 distribution in pachytene chromosome spreads.
Postmeiotic overexpression
In normal postmeiotic cells, dozens of multicopy genes on both sex chromosomes (Mueller et al. 2008; Reynard et al. 2009), together with ∼13% of single copy genes on the X (Namekawa et al. 2006), are expressed in round spermatids. Nevertheless, chromosome-wide expression on the X is substantially lower than that on the autosomes, and both sex chromosomes remain condensed and are enriched for epigenetic markers of transcriptional repression (Namekawa et al. 2006; Turner et al. 2006). In the sterile F1 males, two of three mitotic/early meiotic X-linked genes (Rps6ka6, Fmr1) were significantly overexpressed in round spermatids, although the signal of overexpression was stronger in postmeiotic genes (4930557A04Rik, 4930468A15Rik, Actrt1, 1700013H16Rik). These data suggest that PSCR is also defective in the sterile F1 males.
PSCR is thought to depend on MSCI at the epigenetic level: repressive histone marks established on the sex chromosomes at pachytene are transmitted through the two meiotic divisions (Namekawa et al. 2006; Turner et al. 2006). Thus, defective PSCR in the sterile F1 may be a secondary consequence of disrupted MSCI. It is possible, however, that X overexpression in meiotic cells vs. postmeiotic cells is caused by distinct genetic incompatibilities between domesticusLEWES-derived loci and loci on the musculusPWK X.
Several lab mouse mutants with normal MSCI exhibit global derepression of the sex chromosomes in postmeiotic spermatids (reviewed in Montellier et al. 2011). In particular, the multicopy Y-linked gene, Sly, is essential for normal sperm differentiation and is a key regulator of PSCR (Reynard et al. 2009; Cocquet et al. 2009). Sly-deficient male lab mice are subfertile, produce female-biased litters, and exhibit disruption of PSCR and widespread overexpression of X-linked postmeiotic genes, including the multicopy Slx and Slxl1 genes that are closely related to Sly (Ellis et al. 2005; Cocquet et al. 2009). In contrast, Slx/Slxl1-deficiency results in reduced male fertility and male-biased litters, but does not disrupt PSCR (Cocquet et al. 2012). Simultaneous ablation of all three genes significantly improves reproductive phenotypes and restores sex ratio parity and PSCR, suggesting that Slx/Slxl1 and Sly are involved in an intragenomic conflict (Cocquet et al. 2012). These antagonistic interactions appear to underlie the dramatic copy number expansion of Slx/Slxl1, Sly, and several other X-linked genes in house mice (Ellis et al. 2011).
The M. m. musculus X and Y carry at least twice as many copies of Slx/Slxl1 and Sly, respectively, as the M. m. domesticus X and Y (Scavetta and Tautz 2010; Ellis et al. 2011). Thus, the ratio of Slx/Slxl1 to Sly gene copies is unbalanced in hybrids, and sterile F1 males with a musculusPWK X and domesticusLEWES Y are potentially Sly deficient. Given phenotypic similarities between sterile F1 hybrids and Sly-deficient lab mice (i.e., X overexpression and sperm abnormalities), Cocquet and colleagues proposed that Slx/Slxl1:Sly copy number imbalance contributes to hybrid male sterility in the musculusPWK × domesticusLEWES cross (Cocquet et al. 2012; see also Ellis et al. 2011). Unfortunately, the distribution of musculus- vs. domesticus-derived Sly, Slx, and Slxl1 genotypes among the X introgression F1’s used in this study precludes a statistical test for the effect of copy number imbalance on X expression: only genotype 7 carries the mismatch between musculus-derived Slx and Slxl1 and domesticus-derived Sly (Figure 2A). However, the observation that genotype 7 and genotype 3, which carries musculus-derived Slxl1 and Sly, exhibit comparable levels of X overexpression (Figure 2A) and reproductive deficits (Campbell et al. 2012), suggests that any negative effect of Slx/Slxl1:Sly copy number imbalance is outweighed by interactions between other loci on the musculusPWK X and loci in the domesticusLEWES autosomal genome.
Conclusions
Intrinsic fitness deficits in hybrids are caused by negative interactions between loci that function normally on the genetic background in which they evolved (Bateson 1909; Dobzhansky 1937; Muller 1942). In species with a heterogametic sex, these incompatibilities often involve the X chromosome; identifying the genetic mechanisms and evolutionary processes that cause this pattern is a major focus in speciation genetics. Here we show that sterility in hybrid males is associated with disruption of MSCI. Thus, X-linked genetic incompatibilities can have epigenetic consequences. Moreover, we find that a proximal portion of the M. m. musculus X chromosome is involved, suggesting that disrupted MSCI in sterile males reflects mismatch between M. m. musculus alleles at X-linked loci essential for MSCI and M. m. domesticus alleles at autosomal loci that regulate this process.
Given that MSCI evolved ∼180 million years ago in the common ancestor of eutherian and metatherian mammals (Kumar and Hedges 1998; Potrzebowski et al. 2008) and is an essential component of male fitness, it might seem surprising that loci involved in this process should vary between house mouse subspecies that diverged less than half a million years ago (Geraldes et al. 2011). However, it has been proposed that MSCI evolved as a genome defense against selfish genetic elements such as transposons or sex ratio distorters (Kelly and Aramayo 2007; Meiklejohn and Tao 2010; Presgraves 2010), both of which accumulate preferentially on the X. Under this scenario, it is plausible that antagonistic coevolution between loci that regulate MSCI and selfish genetic elements, whose transmission depends on MSCI escape, could produce functional divergence over short evolutionary timescales. Thus, identification of X-linked loci required for MSCI promises insight into the proposed relationship between intragenomic conflict and the process of speciation (Presgraves 2010; Cocquet et al. 2012).
Supplementary Material
Acknowledgments
This manuscript was significantly improved by comments from Paul Burgoyne, an anonymous reviewer, and Bret Payseur; and by discussion with Mary Ann Handel. We are very grateful to Paula Campbell (University of Arizona Flow Cytometry Core) for her help optimizing and running the FACS assay. Dan Vanderpool and Bivian Torres also gave key advice on germ cell preps and cell sorting. C. William Birky provided equipment used during the course of the experiment. P.C. is supported by a G. G. Simpson Postdoctoral Fellowship from the University of Arizona. J.M.G. was supported by a National Institutes of Health (NIH) grant (1-R01HD73439-01). This work was funded by National Science Foundation and NIH grants to M.W.N.
Footnotes
Communicating editor: B. A. Payseur
Literature Cited
- Achame E. M., Wassenaar E., Hoogerbrugge J. W., Sleddens-Linkels E., Ooms M., et al. , 2010. The ubiquitin-conjugating enzyme HR6B is required for maintenance of X chromosome silencing in mouse spermatocytes and spermatids. BMC Genomics 11: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarends W. M., Wassenaar E., van der Laan R., Hoogerbrugge J., Sleddens-Linkels E., et al. , 2005. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol. Cell. Biol. 25: 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos H., Lassalle B., Chicheportiche A., Riou L., Testart J., et al. , 2005. Flow cytometric characterization of viable meiotic and postmeiotic cell by Hoechst 33342 in mouse spermatogenesis. Cytometry A 65: 40–49. [DOI] [PubMed] [Google Scholar]
- Bateson W., 1909. Heredity and variation in modern lights, pp. 85–101 in Darwin and Modern Science, edited by Seward A. C. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Burgoyne P. S., Mahadevaiah S. K., Turner J. M. A., 2009. The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet. 10: 207–215. [DOI] [PubMed] [Google Scholar]
- Campbell P., Good J. M., Dean M. D., Tucker P. K., Nachman M. W., 2012. The contribution of the Y chromosome to hybrid male sterility in house mice. Genetics 191: 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel F., Rolland A. D., Niederhauser-Wiederkehr C., Chung S. W., Demougin P., et al. , 2007. The conserved transcriptome in human and rodent spermatogenesis. Proc. Natl. Acad. Sci. USA 104: 8346–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J., Ellis P. J. I., Yamauchi Y., Mahadevaiah S. K., Affara N. A., et al. , 2009. The multicopy gene Sly repressed the sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 7: e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J., Ellis P. J. I., Mahadevaiah S. K., Affara N. A., Vaiman D., et al. , 2012. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 8: e1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1937. Genetics and the Origin of Species. Columbia University Press, New York. [Google Scholar]
- Ellis P. J. I., Clemente E. J., Ball P., Touré A., Ferguson L., et al. , 2005. Deletions on mouse Yq lead to upregulation of multiple X- and Y-linked transcripts in spermatids. Hum. Mol. Genet. 14: 2705–2715. [DOI] [PubMed] [Google Scholar]
- Ellis P. J. I., Bacon J., Affara N. A., 2011. Association of Sly with sex-linked gene amplification during mouse evolution: A side effect of genomic conflict in spermatids? Hum. Mol. Genet. 20: 3010–3021. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O., Mahadevaiah S. K., Celeste A., Romanienko P. J., Camerini-Otero R. D., et al. , 2003. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 4: 497–508. [DOI] [PubMed] [Google Scholar]
- Forejt J., 1996. Hybrid sterility in the mouse. Trends Genet. 12: 412–417. [DOI] [PubMed] [Google Scholar]
- Geraldes A., Basset P., Smith K. L., Nachman M. W., 2011. Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Mol. Ecol. 20: 4722–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getun I. V., Torres B., Bois P. R. J., 2011. Flow cytometry purification of mouse meiotic cells. JoVE 50: . 10.3791/2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guioli S., Lovell-Badge R., Turner J. M. A., 2012. Error-prone ZW pairing and no evidence for meiotic sex chromosome inactivation in the chicken germ line. PLoS Genet. 8: e1002560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Handel M. A., Nachman M. W., 2008a Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution 62: 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Dean M. D., Nachman M. W., 2008b A complex genetic basis to X- linked hybrid male sterility between two species of house mice. Genetics 179: 2213–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Giger T., Dean M. D., Nachman M. W., 2010. Widespread over-expression of the X chromosome in sterile F1 hybrid mice. PLoS Genet. 6: e1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1922. Sex ratio and unisexual sterility in animal hybrids. J. Genet. 12: 101–109. [Google Scholar]
- Handel M. A., 2004. The XY body: a specialized chromatin domain. Exp. Cell Res. 296: 57–63. [DOI] [PubMed] [Google Scholar]
- Hendriksen P. J., Hoogerbrugge J. W., Themmen A. P., Koken M. H., Hoeijmakers J. H., et al. , 1995. Postmeiotic transcription of X and Y chromosomal genes during spermatogenesis in the mouse. Dev. Biol. 170: 730–733. [DOI] [PubMed] [Google Scholar]
- Hense W., Baines J. F., Parsch J., 2007. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 5: e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolka D., Jansa P., Forejt J., 2012. Genetically enhanced asynapsis of autosomal chromatin promotes transcriptional dysregulation and meiotic failure. Chromosoma 121: 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijima Y., Ichijima M., Lou Z., Nussenzweig A., Camerini-Otero R. D., et al. , 2011. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev. 25: 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Aramayo R., 2007. Meiotic silencing and the epigenetics of sex. Chromosome Res. 15: 633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer H., Hennig W., Dijkhof R., 1986. Chromatin organization in the male germ line of Drosophila hydei. Chromosoma 94: 147–161. [Google Scholar]
- Kumar S., Hedges S. B., 1998. A molecular timescale for vertebrate evolution. Nature 392: 917–920. [DOI] [PubMed] [Google Scholar]
- Lifschytz E., Lindsley D. L., 1972. The role of X-chromosome inactivation during spermatogenesis. Proc. Natl. Acad. Sci. USA 69: 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah S. K., Bourc’his D., de Rooij D. G., Bestor T. H., Turner J. M. A., et al. , 2008. Extensive meiotic asynapsis iin mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J. Cell Biol. 182: 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee B. D., Handel M. A., 1993. Sex chromosomes, recombination, and chromatin conformation. Chromosoma 102: 71–80. [DOI] [PubMed] [Google Scholar]
- Meiklejohn C. D., Tao Y., 2010. Genetic conflict and sex chromosome evolution. Trends Ecol. Evol. 25: 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Landeen E. L., Cook J. M., Kingan S. B., Presgraves D. C., 2011. Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol. 9: e1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modzelewski A. J., Holmes R. J., Hilz S., Grimson A., Cohen P. E., 2012. AGO4 regulates entry into meiosis and influences silencing of the sex chromosomes in the male mouse germline. Dev. Cell 23: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montellier E., Rousseaux S., Zhao Y., Khochbin S., 2011. Histone crotonylation specifically marks the haploid male germ cell gene expression program. BioEssays 34: 187–193. [DOI] [PubMed] [Google Scholar]
- Mueller J. L., Mahadevaiah S. K., Park P. J., Warburton P. E., Page D. C., et al. , 2008. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 40: 794–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1942. Isolating mechanisms, evolution and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Namekawa S. H., Park P. J., Zhang L. F., Shima J. E., McCarrey J. R., et al. , 2006. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16: 660–667. [DOI] [PubMed] [Google Scholar]
- Namekawa S. H., VandeBerg J. L., McCarrey J. R., Lee J. T., 2007. Sex chromosome silencing in the marsupial male germ line. Proc. Natl. Acad. Sci. USA 104: 9730–9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski L., Vinckenbosch N., Marques A. C., Chalmel F., Jégou B., et al. , 2008. Chromosomal gene movements reflect the recent origin and biology of the therian sex chromosomes. PLoS Biol. 6: 709–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24: 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180. [DOI] [PubMed] [Google Scholar]
- Reynard L. N., Cocquet J., Burgoyne P. S., 2009. The multi-copy mouse gene Sycp3-like Y-linked (Sly) encodes an abundant spermatid protein that interacts with a histone acetyltransferase and an acrosomal protein. Biol. Reprod. 81: 250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roest H. P., van Klaveren J., de Wit J., van Gurp C. G., Koken M. H. M., et al. , 1996. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 86: 799–810. [DOI] [PubMed] [Google Scholar]
- Royo H., Polikiewicz G., Mahadevaiah S. K., Prosser H., Mitchell M., et al. , 2010. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr. Biol. 20: 2117–2123. [DOI] [PubMed] [Google Scholar]
- Scavetta R. J., Tautz D., 2010. Copy number changes of CNV regions in intersubspecific crosses of the house mouse. Mol. Biol. Evol. 27: 1845–1856. [DOI] [PubMed] [Google Scholar]
- Solari A. J., 1974. The behaviour of the XY pair in mammals. Int. Rev. Cytol. 38: 273–317. [DOI] [PubMed] [Google Scholar]
- Song R., Ro S., Michaels J. D., Park C., McCarrey J. R., et al. , 2009. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat. Genet. 41: 488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. M. A., 2007. Meiotic sex chromosome inactivation. Development 10: 1823–1831. [DOI] [PubMed] [Google Scholar]
- Turner J. M. A., Aprelikova O., Xu X., Wang R., Kim S., et al. , 2004. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr. Biol. 14: 2135–2142. [DOI] [PubMed] [Google Scholar]
- Turner J. M. A., Mahadevaiah S. K., Fernandez-Capetillo O., Nussenzweig A., Xu X., et al. , 2005. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 37: 41–47. [DOI] [PubMed] [Google Scholar]
- Turner J. M. A., Mahadevaiah S. K., Ellis P. J. I., Mitchell M. J., Burgoyne P. S., 2006. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev. Cell 10: 521–529. [DOI] [PubMed] [Google Scholar]
- Vernet N., Mahadevaiah S. K., Ellis P. J. I., Ojarikre O. A., Longepied G., et al. , 2011. The Y-encoded gene Zfy2 acts to remove cells with unpaired chromosomes at the first meiotic metaphase in male mice. Curr. Biol. 21: 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet N., Mahadevaiah S. K., Ellis P. J. I., de Rooij D. G., Burgoyne P. S., 2012. Spermatid development in X0 male mice with varying Y chromosome short- arm gene content: evidence for a Y gene controlling the initiation of sperm morphogenesis. Reproduction 144: 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski M. D., Lopes H. F., Karr T. L., Long M., 2009. Stage-specific expression profiling if Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 5: e1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski M. D., Zhang Y. E., Kemkemer C., Lopes H. F., Karr T. L., et al. , 2012. Re-analysis of the larval testis data on meiotic sex chromosome inactivation revealed evidence for tissue-specific gene expression related to the drosphila X chromosome. BMC Biol. 10: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., McCarrey J. R., 2009. Sex chromosome inactivation in the male. Epigenetics 4: 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.