Abstract
Genetic diversity is essential for population survival and adaptation to changing environments. Demographic processes (e.g., bottleneck and expansion) and spatial structure (e.g., migration, number, and size of populations) are known to shape the patterns of the genetic diversity of populations. However, the impact of temporal changes in migration on genetic diversity has seldom been considered, although such events might be the norm. Indeed, during the millions of years of a species’ lifetime, repeated isolation and reconnection of populations occur. Geological and climatic events alternately isolate and reconnect habitats. We analytically document the dynamics of genetic diversity after an abrupt change in migration given the mutation rate and the number and sizes of the populations. We demonstrate that during transient dynamics, genetic diversity can reach unexpectedly high values that can be maintained over thousands of generations. We discuss the consequences of such processes for the evolution of species based on standing genetic variation and how they can affect the reconstruction of a population’s demographic and evolutionary history from genetic data. Our results also provide guidelines for the use of genetic data for the conservation of natural populations.
Keywords: genetic diversity, migration, population subdivision, transient dynamics, standing genetic variation
GENETIC diversity in a population of constant size results from the balance between the occurrence of new mutations and the loss of alleles by genetic drift (Fisher 1922; Wright 1931; Kimura and Crow 1964). The expected population genetic diversity can thus be estimated from the effective population size and the mutation rate in the population. In subdivided populations this estimate should further account for the strength of migration (Maruyama 1970; Smith 1970; Nei 1973): limited migration allows for strong differentiation between populations, while strong migration tends to homogenize genetic diversity between populations. Genetic diversity is also known to be affected by population demographic changes; following bottlenecks and founder events, a loss of genetic diversity is expected to occur (Nei et al. 1975). Recently, spatial population expansions were shown to lead to increased differentiation between populations and to generate a low level of genetic diversity at the front of the expansion (Excoffier et al. 2009).
Although theoretical studies on the dynamics of genetic diversity in subdivided populations started appearing in the 1970s (Nei and Feldman 1972; Latter 1973; Nei 1973; Nagylaki 1974, 1977), the transient dynamics and nonequilibrium states of genetic diversity still do not have a good theoretical basis. Early authors characterized the ultimate rate of change of genetic diversity after a perturbation (either a change in population size or gene flow; Nei and Feldman 1972; Latter 1973; Nei 1973; Nagylaki 1974, 1977). They found that changes in genetic diversity are related to the total effective population size, which results in a slow dynamics of genetic diversity change. They thus first highlighted that nonequilibrium states and transient dynamics are expected to act on very large temporal scales. In particular, they showed that decreases in migration rates (population fragmentation or isolation) have long-term effects on genetic diversity: they reduce the amount of genetic diversity within populations and allow for population differentiation (Latter 1973; Takahata and Nei 1985). Additionally, it has been shown that short timescale random fluctuations in migration increase population differentiation (Nagylaki 1979; Whitlock 1992; Rice and Papadopoulos 2009) while cyclic fluctuations of gene flow (such as seasonal fluctuations) mainly affect genetic diversity within populations (Karlin 1982; Shpak et al. 2010). Although the genetic consequences of migration events (admixture) have recently received much attention (e.g., Pritchard et al. 2000; Falush et al. 2003; Price et al. 2009; Gravel 2012), their impact on genetic diversity and more particularly the expected induced transient dynamics have not received much attention.
Genetic diversity has a crucial importance in estimating populations at risk of extinction and species’ adaptive potential. Current genetic diversity characterizes species at risk of extinction through inbreeding depression, loss of genetic diversity, and accumulation of deleterious mutations (Gilpin and Soule 1986; Jimenez et al. 1994; Frankham 1995; Hedrick and Kalinowski 2000). The current level of genetic diversity (or standing genetic variation) is now widely recognized as a determinant for the adaptation of a population to a novel environment (Turner et al. 1993; Feder et al. 2003; Pelz et al. 2005; Colosimo et al. 2005; Hermisson and Pennings 2005; Myles et al. 2005; Hernandez et al. 2011; Jones et al. 2012). First, under new selective pressures, the adaptive value of a preexisting allele can switch from neutral or deleterious to beneficial (Gibson and Dworkin 2004; Hermisson and Pennings 2005). Second, alleles from the standing genetic variation are present at higher frequencies in the population than any newly arisen (de novo) mutation are; thus, they have higher fixation probabilities and lower times to fixation (Barrett and Schluter 2008). Finally, these alleles have already passed successive selective filters and are consequently more likely to be compatible with the background genome (Orr and Betancourt 2001; Schluter et al. 2004; Barrett and Schluter 2008).
Measures of genetic diversity are widely used to understand and infer the demographic and evolutionary history of populations. Indeed, statistical tests using polymorphism data can detect departure from neutrality and infer demographic or selective processes (e.g., Ewens 1972; Watterson 1978; Tajima 1983; Fu and Li 1993; Fay and Wu 2000; see review in Kreitman 2000). Furthermore, due to recent modeling advances in coalescent theory and increased genomic data and computational power, it is now possible to distinguish different demographic scenarios (e.g., population bottleneck and subdivision; Peter et al. 2010) and estimate demographic and selective parameters (e.g., populations size and growth rate, proportion of admixture, selection coefficient) using polymorphism data (Beaumont et al. 2002; Kim and Stephan 2002; Kim and Nielsen 2004; Nielsen et al. 2005; Price et al. 2009). Nevertheless, it is often difficult to distinguish between the transient effects of demographic changes and the effects of selection on polymorphism data (Jensen et al. 2005; Nielsen 2005; Li and Stephan 2006; Kim and Gulisija 2010; Pavlidis et al. 2010). It is also difficult to distinguish between the signatures of different demographic changes such as changes in population size, number, or migration rate (Wakeley 1999). A better understanding of the impact on genetic data of transient dynamics during demographic changes is necessary to disentangle these processes.
Interestingly, although the impact of population subdivision and short timescale population demographic changes on genetic diversity have received a lot of attention, other processes, such as long-term isolation and subsequent population reconnection, have received little attention. Such events have, without a doubt, occurred several times in the past, at long and short timescales. Repeated environmental changes have modified habitats and species distribution and created isolation and reconnection of populations. For example, during the climatic oscillations of the Quaternary period, temperate and tropical species were successively isolated into refugia and experienced habitat and population expansion, allowing for population reconnection (Hewitt 2000, 2004; Zhang et al. 2008; Young et al. 2009). At the same time, the reduction of sea levels (120 m lower than present; Lambeck et al. 2004) allowed the formation of land bridges that connected isolated lands in several parts of the world (Hewitt 2000). Repeated changes in water level resulted in fragmentation and fusion of basins within continents (as in the Great African Lakes; Galis and Metz 1998; Sturmbauer et al. 2001). Similarly, geological events such as volcanic eruptions induced periodic isolation and reconnection of islands (Cook 2008), while tectonic processes such as the formation of mountains isolated populations and reconnected others (Hughes and Eastwood 2006; Antonelli et al. 2009; Antonelli and Sanmartín 2011). More recently, climatic, environmental, and anthropogenic changes (e.g., global warming, urbanization, and agriculture) have also played important roles in modifying the connectivity pattern between populations (Miller and Hobbs 2002; Delaney et al. 2010). Consequently, some species are currently subdivided into poorly connected or completely isolated populations; for examples, ground beetles (Keller et al. 2004), salamanders (Noel et al. 2007), and crickets (Vandergast et al. 2009). In the meantime, other species experience habitat and population expansion (e.g., sparrows, white-tailed deer, zebra mussels; Waples 2010). Isolation and reconnection of populations not only reflect abiotic processes, but they can also represent spatial and temporal interactions of populations (e.g., secondary contacts; Green et al. 2010; Domingues et al. 2012). Consequently, transient states of genetic diversity are expected to be the norm and deserve much more attention.
In this study, we analytically characterized the dynamics of genetic diversity following a change in migration rate between populations, given any migration rate, mutation rate, population size, and degree of fragmentation. We first analyzed how genetic diversity is affected by an event of isolation of populations and by an event of reconnection of populations. We then generalized our results for situations where the migration rate between populations displays strong variation. We demonstrate that temporal changes of migration generate periods where genetic diversity reaches unexpectedly high values that can be maintained over thousands of generations. We also show that migration changes can produce a signature on summary statistics such as Tajima’s D and Ewens–Watterson’s statistics that cannot be differentiated from a signature of population size change or from the signature of selection. Finally, we discuss how such processes can affect observed macroevolutionary patterns of species diversity and how they can affect the reconstruction of populations’ demographic and evolutionary history from genetic data.
Genetic Diversity of Populations
To study the dynamics of genetic diversity after connectivity changes, we consider diploid individuals in a finite island model composed of n random mating populations of size N, so that the total population size is nN. The populations exchange migrants at a rate m. The mutations follow the infinite allele model (each mutation produces a new allele; Kimura and Crow 1964) and occur at a rate μ. The generations are nonoverlapping (Wright–Fisher model; Fisher 1930; Wright 1931).
Genetic diversity, H, is estimated using the identity-by-descent F between pairs of alleles, through the relationship provided by Nei and Feldman (1972):
| (1) |
Further, we characterize within-population genetic diversity Hs and between-population genetic diversity Hb, using within- and between-population genetic identities, respectively. Within-population genetic identity, Fs, corresponds to the probability that two genes randomly chosen from the same population are identical by descent. Between-population genetic identity, Fb, corresponds to the probability that two genes randomly chosen from different populations are identical by descent. Considering that within- and between-population genetic identities Fs and Fb at a given time t are, respectively, Fs,t and Fb,t, their values at the next generation (forward in time), respectively, Fs,t+1 and Fb,t+1, will follow (Smith 1970; Maruyama 1970; Latter 1973)
| (2a) |
where the parameters are
| (2b) |
| (2c) |
| (2d) |
System of Equations 2 can be expressed under matrix notation as
| (3a) |
where
| (3b) |
| (3c) |
| (3d) |
Parameters have the following interpretation: a is the probability that two genes at the same location before migration are still at the same location after migration (either both migrate to the same location or both do not migrate); b is the probability that two genes that were at the same location before migration migrated to different locations; c is the probability that two genes within a population are copies of the same gene; and (1 − μ)2 is the probability that neither of the two randomly chosen genes mutated.
Predicting the Dynamics of Genetic Diversity
To characterize the impact of connectivity changes on genetic diversity, we analyzed the trajectories of within- and between-population genetic identities from any initial genetic identity state. Using Equation 2, Smith (1970) and Maruyama (1970) showed that genetic identities converge toward an equilibrium value Feq (value given in Supporting Information, File S1). Extending the results obtained by Nei and Feldman (1972) for n = 2 populations, we show that the temporal dynamics of genetic identities follow (see Appendix A for more details)
| (4a) |
where
| (4b) |
λ1 and λ2 are respectively the largest and smallest eigenvalues of matrix A, and they follow
| (5) |
C1 and C2 are column vectors of dimension 2 composed of constant values, which depend on the parameters of the model (m, μ, n, and N) and on the initial genetic identity F0 (Appendix A).
In the next section, we provide from Equations 4a and 5 the temporal change of genetic diversity and derive the corresponding time to reach genetic diversity equilibrium after a connectivity change.
Time to Reach Genetic Diversity Equilibrium
The change of genetic diversity can be decomposed in two main temporal dynamics: a long-term and a short-term dynamics. Indeed, the temporal change of genetic diversity depends on two components: and (Equation 4a). They both follow an exponential decay and their rate of change depends on r1 = ln(λ1) and r2 = ln(λ2), respectively (Appendix A). As 1 > λ1 > λ2 > 0, decays more rapidly than (see Appendix A). Thus r1 determines the ultimate (or long-term) change of genetic diversity and r2 determines the transient (or short-term) change of genetic diversity.
When migration and mutation rates are small (i.e., m ≪ 1 and μ ≪ 1) and local population sizes are large (i.e., N ≫ 1), the decay constants r1 and r2 follow
| (6a) |
with
| (6b) |
where M = 4Nm is the scaled migration rate and Ne is the effective population size of the total population (inbreeding, eigenvalue, variance, and mutation effective size are equivalent in the finite island model; Whitlock and Barton 1997). As expected from theory (Whitlock and Barton 1997; Wakeley 1999), in the strong migration limit (M ≫ 1), the effective size is equal to the total population size nN, while in the weak migration limit (M ≪ 1), the effective size is higher than the total population size.
We can estimate the durations of the ultimate and transient changes of genetic diversity, denoted t1 and t2, respectively. Formally, we define t1 and t2 as the times (in number of generations) needed for and to be reduced to a number α, where α ε ]0; 1]:
| (7) |
Assuming that migration and mutation rates are small and population sizes are large, t1 and t2 simplify to (Appendix A):
| (8) |
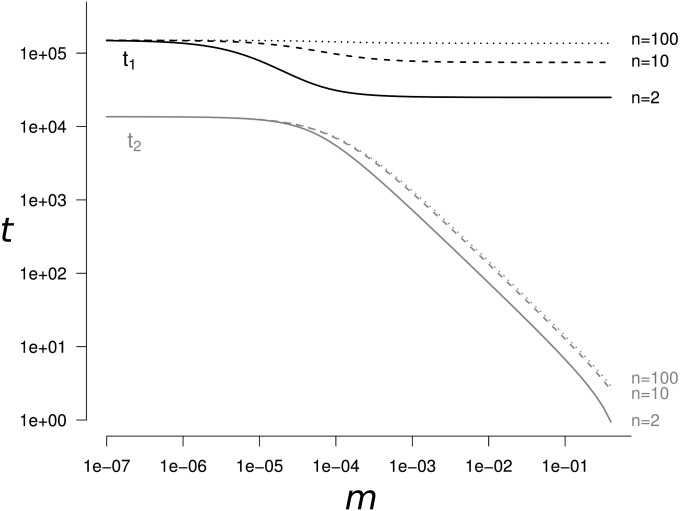
The genetic diversity changes as follows (Figure 1): (i) a convergence of duration t2 from the initial genetic diversity value to a transient genetic diversity value and then (ii) a convergence of duration t1 to the genetic diversity equilibrium Heq. The time to reach genetic diversity equilibrium, t1, depends only on two terms: the mutation rate (term 2μ) and the genetic drift at the total population level (term ). The duration of the transient dynamics, t2, depends on four terms: the mutation rate (term 2μ), the migration rate (term 2m), the genetic drift in each population (term ), and the genetic drift at the total population level (term ). The convergence to the transient and equilibrium values of genetic diversity can occur on separated timescales (i.e., t1 ≫ t2) depending on the parameter values. The timescales t1 and t2 can differ from several orders of magnitude. When n > 14, differences are the highest (t1 ≫ t2), in the domain where and also when and m > μ. When n ≤ 14, the same conditions apply for t1 ≫ t2 except in a restricted domain where (see Appendix A). For example, the duration of the transient dynamics is t2 ≃ 134 and the time to reach equilibrium is t1 ≃ 1.5 × 105 generations (with α = 5%), when 10 populations of size 2500 with a mutation rate of 10−6 are connected with a migration rate of 0.01.
Figure 1.
The time (in number of generations) t1 to reach genetic diversity equilibrium and the length of the transient dynamics period t2 as a function of the migration rate m. The solid line corresponds to n = 2 populations, the dashed line to n = 10, and the dotted line to n = 100. t1 is always at least one order of magnitude higher than t2. This separation of the two periods becomes even greater when . Parameter values are N = 2500, μ = 10−5, α = 5%.
Dynamics of Genetic Diversity After an Isolation Event
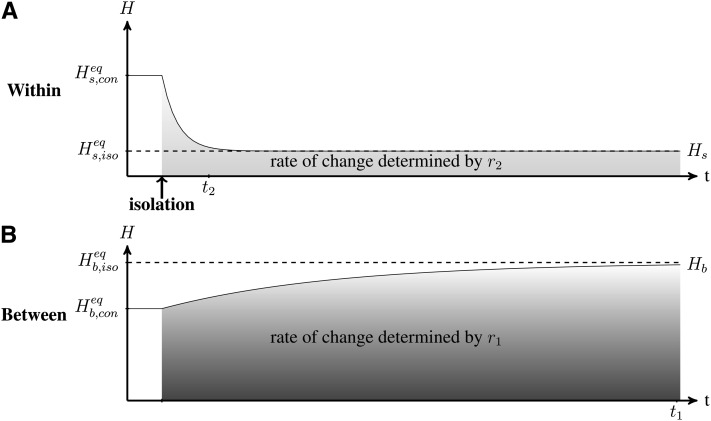
We analyzed the dynamics of genetic diversity after an isolation event, starting with a situation in which populations are connected and at their equilibrium value; i.e., within- and between-population genetic diversity Hs and Hb are at the expected connection equilibrium values and (see Maruyama 1970; Smith 1970; and File S1).
We observe (Figure 2) that immediately after an isolation event, within-population genetic diversity decreases due to genetic drift to the point where it reaches the mutation-drift equilibrium of an isolated population (see File S1 and Kimura and Crow 1964), at a rate determined by r2 (from Equation 6). Meanwhile, between-population genetic diversity slowly increases due to the differentiation of populations induced by mutations (at a rate determined by r1 from Equation 6). Populations ultimately reach complete differentiation (equilibrium value of ). We can show from Equation 2 that following an isolation event, within- and between-population diversities change independently and reach their equilibrium value in t2 and t1 generations, respectively (File S2).
Figure 2.
Dynamics of (A) within-population Hs and (B) between-population Hb genetic diversity after an isolation event. Within- and between-population diversity (solid lines) were previously at their respective connection equilibrium and . After the isolation event, within- and between-population diversities reach their isolation equilibrium and (dashed lines) at rates determined by r2 and r1 (Equation 6). t2 and t1 estimate the time to reach the within- and between-population genetic diversity equilibrium, respectively (Equation 8). Under the effect of genetic drift, within-population diversity reaches its equilibrium value faster than between-population genetic diversity. Parameters are n = 10, N = 2500, m = 10−4 before isolation and m = 0 afterward, and μ = 10−5. t1 ≃ 149,000 generations and t2 ≃ 13,600 generations (for α = 5%).
The decrease of population genetic diversity (within) can occur quickly relative to population differentiation (between-population genetic diversity; see Figure 1). After an isolation event, within-population genetic diversity (Hs) remains above its expected equilibrium , while between-population genetic diversity (Hb) remains below its expected equilibrium . However, the timescales of these nonequilibrium periods differ. When populations are isolated (m = 0), Hs reaches a value close to its equilibrium value in generations, while Hb reaches a value close to its equilibrium value in generations (Equation 8). Therefore, when , Hs converges much more quickly than Hb, and when , both converge in approximately the same amount of time. For example, assuming μ = 10−5 and N = 1,000, Hb is significantly lower than the equilibrium value for approximately t1 ≃ 150,000 generations while Hs is significantly higher than the equilibrium value for approximately t2 ≃ 6000 generations (given α = 5%).
Dynamics of Genetic Diversity After a Connection Event
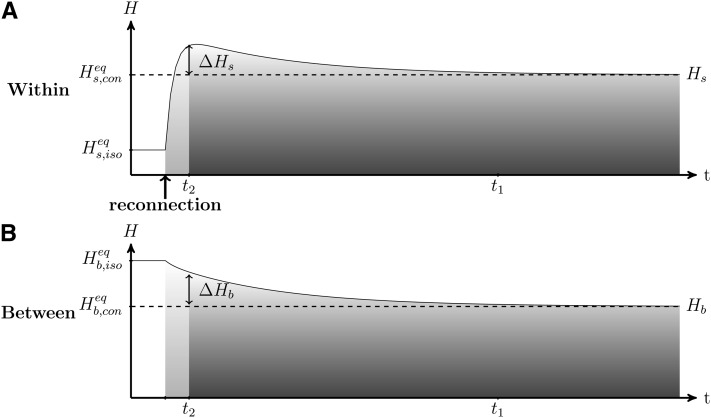
We analyzed the dynamics of genetic diversity after a connection event, starting with a situation in which populations are isolated and at their equilibrium value and (see Smith 1970; Maruyama 1970 and File S1). After a connection event (Figure 3), the genetic diversity accumulated in each population during the isolation period is quickly spread to all populations (Figure 3, fast dynamics in light shading, at a rate determined by r2 from Equation 6). Consequently, within-population genetic diversity quickly increases and reaches a high value that is above its expected connected equilibrium value (, Figure 3; see File S1). This process creates a peak of within-population genetic diversity ΔHs and a transient excess of genetic diversity between populations ΔHb (see Figure 3). Then, due to genetic drift, both the within- and the between-population diversities decrease (slow dynamics in dark shading in Figure 3, at a rate determined by r1 from Equation 6), to the point where the diversities reach the expected value of mutation–migration–drift equilibrium ( and from Equation 4b; Kimura and Crow 1964).
Figure 3.
Dynamics of (A) within-population genetic diversity Hs and (B) between-population genetic diversity Hb after a reconnection event. Within- and between-population diversities were previously at their respective isolation equilibriums and (Equation 4b). After the reconnection event, within- and between-population diversities reach their respective connection equilibriums and (dashed lines). As shown in Equation 8, the time to reach genetic diversity equilibrium t1 and the length of the transient period t2 are well separated. The two periods are: (1) fast convergence at a rate determined by r2 (Equation 6) that is driven by the spread of diversity that had accumulated within populations during isolation, which creates the peak of within-population diversity (ΔHs) and the excess of between-population diversity (ΔHb) and (2) slow dynamics at a rate determined by r1 (Equation 6) that is caused by the gradual loss of genetic diversity. A large number of generations is needed to reach equilibrium. When n = 10, N = 2500, m = 10−4 after reconnection, and μ = 10−5, t1 ≃ 97,000 generations, t2 ≃ 6,900 generations (for α = 5%) and ΔHs ≃ ΔHb ≃ 0.11.
Within- and between-population diversities change successively according to two timescales: first, a fast transient dynamics, followed by a slow asymptotic dynamics (separation of timescales is derived in Appendix A and illustrated in Figure 3). Because the transient dynamics can be shorter than the asymptotic dynamics, the excess of genetic diversity (ΔHs and ΔHb) can be maintained for a very long period (from Figure 1, t1 is longer than 10,000 generations).
Peak of Genetic Diversity Generated by a Connection Event
In this section, we characterize the peak of within-population genetic diversity, ΔHs, and the excess of between-population genetic diversity, ΔHb, observed after a connection event as a function of the mutation rate, the genetic drift, the number of populations, and the migration rate after connection. The exact value of the within-population genetic diversity peak is represented in Figure 4. Assuming that migration and mutation rates are small, we can show that good approximations of the values of ΔHs and ΔHb are (see derivations in Appendix B)
| (9a) |
where
| (9b) |
is the expected equilibrium identity within an isolated population (Kimura and Crow 1964), and
| (9c) |
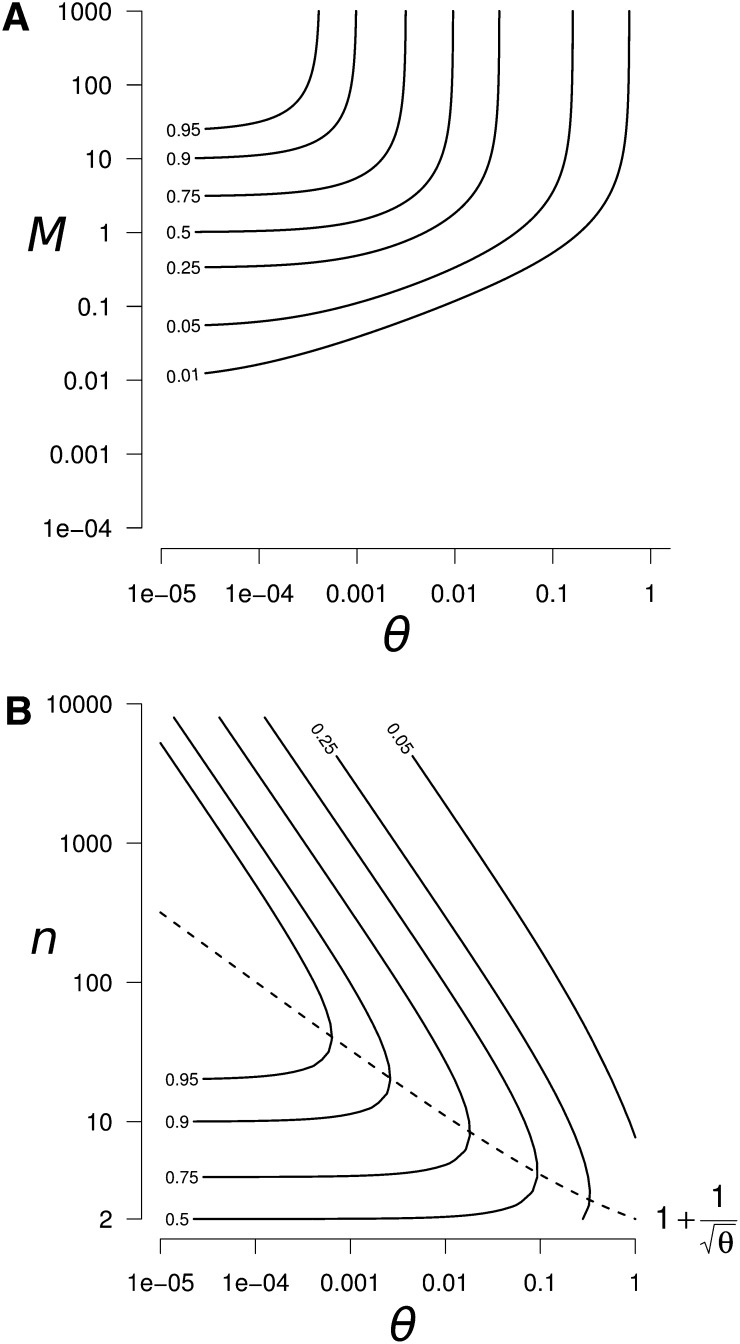
is the expected equilibrium identity between connected populations with the scaled migration rate M, the number of populations n, and the scaled mutation rate θ = 4Nμ (Maruyama 1970; Smith 1970). These approximations lead to the largest absolute error when a small number of populations (n = 2) is combined with weak mutation (θ < 1) and intermediate migration (M ≃ 5). Nevertheless, this error is small (error <0.025 for ΔHs and <0.08 for ΔHb), so Equation 9 provides a good approximation of ΔHs and ΔHb for all n, M, and θ values (see Appendix B for more details about the validity of approximation 9).
Figure 4.
Peak of within-population genetic diversity ΔHs generated by a reconnection event. (A) Contour plot of ΔHs as a function of θ and M, for n = 100. We can clearly see the highest peak of diversity in the high M and low θ region. (B) Contour plot of the peak of genetic diversity after a reconnection event as a function of θ and n, for M ≫ 1 (high M region identified in A). In the high M region, the within-population diversity peak ΔHs and the between-population diversity excess ΔHb are equal. The dashed line represents the number of populations which maximizes the peak of diversity .
The peak of genetic diversity increases with the difference between the two timescales (ΔHs and ΔHb increase with ) as genetic diversity increases during the transient phase and decreases during the asymptotic phase. Indeed, when those two phases are separated there is no loss of genetic diversity caused by the asymptotic decay during the transient phase (terms in Equation 9).
In the domain where the peak is the largest (M ≫ 1 and θ ≪ 1), ΔHs and ΔHb reach the same value:
| (10) |
In this domain, ΔHmax is maximized when the number of populations is (dashed line in Figure 4B):
| (11) |
The corresponding peak of genetic diversity, reached at n*, is
Interestingly, the number of populations and the peak of diversity have a nonmonotonous relationship. The peak of genetic diversity decreases when the number of populations approaches 2 and when it tends to infinity, while an intermediate number of populations n* maximizes the peak of genetic diversity. This can be easily explained by the following processes. During isolation, a small number of populations accumulates less between-population genetic diversity; thus, once reconnected, they share a smaller amount of diversity. In contrast, a large number of populations accumulates a higher level of genetic diversity but also has a higher connection equilibrium value; thus, once reconnected, diversity reaches its expected equilibrium and no peak of diversity is observed.
In summary, high peaks of genetic diversity (ΔHs > 0.25 in Figure 4) can occur for a large range of the parameter space: when mutation is weak (θ < 0.05) and migration is moderate to strong (M > 0.5). Under these conditions, drastic genetic diversity changes can be observed (ΔHs values >0.95; Figure 4B for M ≥ 50 and θ < 5 × 10−4). The number of populations that maximize the peak of diversity, n*, ranges from a few populations when θ ≃ 1, up to a few hundred populations when θ = 10−6 (values of θ < 10−6 are expected to be very rare, and they would require a mutation rate lower than 2.5 × 10−12/bp for a 1-kb gene and a population size of 100). Interestingly, a significant peak of genetic diversity is also observed when only two populations reconnect (ΔHmax|n=2 = 0.5; Figure 4B).
Peak of Genetic Diversity Resulting from a Migration Rate Increase
Complete isolation of populations is not required to generate peaks of genetic diversity. Indeed, an abrupt increase of migration can generate the peak of genetic diversity characterized in the previous sections. In File S3, we determined that if migration crosses a threshold value MT, peaks of genetic diversity can occur. The value of the threshold MT, assuming that m ≪ 1 and μ ≪ 1, is
| (12) |
Consequently, an increase of migration from M0 to M crossing the threshold value MT (i.e., M0 ≪ MT) generates a peak of genetic diversity that can be approximated by Equation 9 (see File S3). For example, in a subdivided population of n = 10 and θ = 0.1, an increase in migration from M0 = 0.01 to M = 10 (which crosses the migration threshold MT = 0.495; Equation 12), generates a peak of within-population diversity of 0.350, while a reconnection event in a similar situation would generate a peak of similar intensity (0.358).
Implications for the Inference of Demography and Selection
To describe the impact of migration changes on the inference of demography and selection from genetic data, we described the dynamics of two broadly used summary statistics: the Ewens–Watterson statistics (Watterson 1978) and Tajima’s D (Tajima 1989). Both the Ewens–Watterson statistics and Tajima’s D are known to detect an excess (resp. deficit) of rare alleles, which induces negative (resp. positive) values of the statistics, compared with the expected neutral equilibrium (constant size population without selection). Usually, an excess of rare alleles is interpreted either as the signature of balancing selection or population expansion, and a deficit of rare alleles is interpreted as the signature of directional selection or as a population bottleneck. We used the Ewens–Watterson statistics, which we denote HEW and follows (Watterson 1978)
| (13) |
where Hs is the genetic diversity, and HA is the expected genetic diversity given the observed number of alleles K. We also used Tajima’s D, which we denote DT and follows (Tajima 1989)
| (14) |
where , l is the sample size, π is the average number of pairwise nucleotide differences, and S is the number of segregating sites.
We simulated samples of 50 sequences of 1 kb, with a per-nucleotide mutation rate of 2 × 10−8, in four populations of size 2500, and ran 5000 replicate simulations. We simulated an isolation event, where the migration rate changed from 0.002 to 0 and a reconnection event in which the migration rate changed from 0 to 0.002. The simulations were performed with the software fastsimcoal (Excoffier and Foll 2011), and the data analysis was performed with Arlequin (Excoffier and Lischer 2010). We simulated samples from the same population (with the parameter values that were used, the sampling scheme had a very weak impact; see Chikhi et al. 2010 for a discussion of how the sampling scheme affects the values of Tajima’s D). To allow for convergence of the coalescent algorithm, we always assumed that the populations were connected prior to the isolation phase. In the reconnection event simulations, we set the duration of the isolated phase to 10N, which allowed the genetic diversity values to reach their equilibrium value.
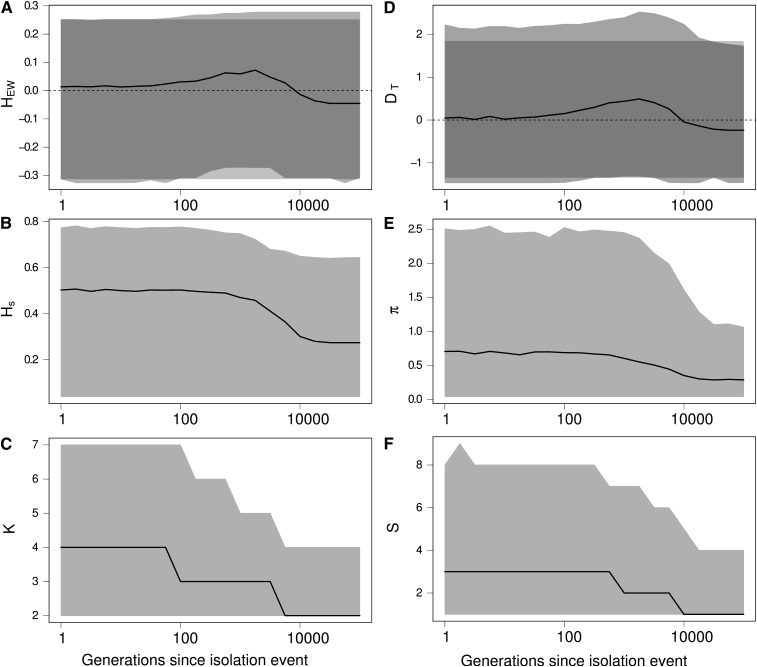
We followed the dynamics of the statistics and estimated their distribution as a function of time. The results in Figure 5 show that an isolation event produces the same signature on the Ewens–Watterson statistics (Figure 5A) and Tajima’s D (Figure 5D), as expected from a bottleneck event and from directional selection. Indeed, following an isolation event, genetic drift first causes the elimination of rare alleles and then eliminates more common alleles. Consequently, the number of alleles K decreases more quickly than genetic diversity Hs (Figure 5, B and C). Similarly, the number of segregating sites S decreases faster than the number of pairwise differencies π (Figure 5, E and F). Therefore, DT and HEW are skewed toward positive values, as expected after a bottleneck or under the effect of directional selection. Moreover, the statistics remain skewed for a long period of time (<10,000 generations in our simulations, see Figure 5).
Figure 5.
Effect of an isolation event on Ewens–Watterson and Tajima’s D neutrality tests and on related summary statistics. (A) Ewens–Watterson statistics (HEW), (B) genetic diversity (Hs), (C) number of alleles (K), (D) Tajima’s D (DT), (E) number of pairwise differences (π), and (F) number of segregating sites (S). For each statistics, the solid line represents the median of the distribution and the light shading represents the 97.5 and 2.5% quantiles of the distribution as a function of the number of generations t after the isolation event. Dark shading in A and D represent the expected distribution of the statistics in an isolated population at equilibrium. Values of HEW and D after an isolation event are skewed toward positive values (signature of a bottleneck or directional selection), while there was no change in the size of the population. K and S decrease more quickly than Hs and π, because rare alleles are eliminated by genetic drift more quickly than common alleles. Coalescence simulations of a 1-kb locus with a mutation rate of 2 × 10−8/bp, where four populations of size 2500 are isolated; 5000 replicates.
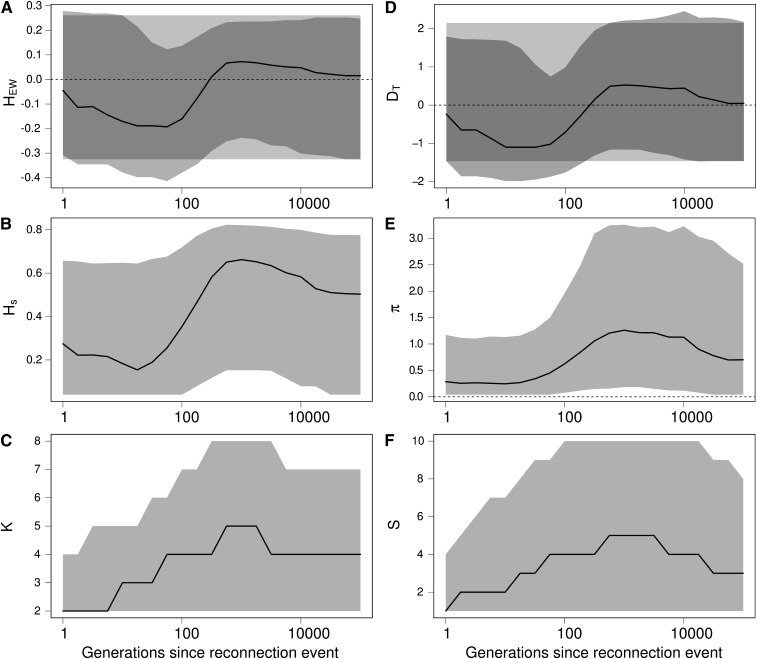
Results in Figure 6 show that a reconnection event can successively produce the same signature as expected from a population expansion or from a bottleneck event on HEW (Figure 6A) and DT (Figure 6, B and C). Indeed, following a reconnection event, migrants first create an excess of rare variants. The number of alleles K increases more quickly than the genetic diversity Hs (Figure 6, B and C), and the number of segregating sites S increases faster than the number of pairwise differences π (Figure 6, E and F), which skews HEW and DT toward negative values. Second, new alleles brought by migrants increase in frequency, creating an excess of common variants. Consequently Hs increases more than K, and π increases more than S, which skews DT and HEW toward positive values.
Figure 6.
Effect of a reconnection event on Ewens–Watterson and Tajima’s D neutrality tests and on related summary statistics. (A) Ewens–Watterson statistics (HEW), (B) genetic diversity (Hs), (C) number of alleles (K), (D) Tajima’s D (DT), (E) number of pairwise differences (π), and (F) number of segregating sites (S). For each statistics, the solid line represents the median of the distribution, and the light shading represents the 97.5% and 2.5% quantiles of the distribution, as a function of the number of generations t after the isolation event. Dark shading in A and D represent the expected distribution of the statistics in an isolated equilibrium population. Values of HEW and D after a reconnection event are first skewed toward negative values (signature of a population expansion or balancing selection) and then toward positive values (signature of a bottleneck or directional selection), while there was no change in the size of the population. K and S first increase more quickly than Hs and π because immigrants bring rare alleles, and then Hs and π reach a higher value because immigrant alleles increase in frequency. Finally, alleles are eliminated by genetic drift until the statistics reach their expected equilibrium value when populations are connected. Coalescence simulations of a 1 kb locus with a mutation rate of 2 × 10−8 per bp, where 4 populations of size 2500 isolated during 25,000 generations are reconnected with a migration rate m = 0.002; 5000 replicates.
Interestingly, the observed duration of the periods in which both statistics are skewed are similar to the expected duration of the dynamics of genetic diversity (from Equation 8). After an isolation event, we observe, in Figure 5, that all statistics reach their equilibrium value within ∼10,000 generations (4N generations). This duration corresponds to the value of the time required to reach within-population genetic diversity equilibrium after an isolation event, t2 ≃ 12,000 generations (4.8N generations, estimated from Equation 8 with α = 5%). In this example, genetic drift is stronger than mutation (2μ ≪ 1/2N) and thus t2 ∼ 2N. t2 corresponds to the duration of the period in which the deficit of rare alleles skews the distribution of HEW and DT. After a reconnection event, we observe (Figure 6) that HEW and DT reach a “peak” within ∼600 generations (0.24N generations). This duration corresponds to the value of the duration of the transient dynamics following a reconnection event, t2 ≃ 540 generations (0.216N generations, estimated from Equation 8 with α = 5%). In this example, migration is stronger than genetic drift and mutation (m ≫ 1/2N and m ≫ μ), and thus, t2 ∼1/2m. t2 corresponds to the period during which the distribution of HEW and DT is skewed. Subsequently, HEW and DT reach their equilibrium value in ∼80,000 generations (32N generations); this duration corresponds to the time required to reach the genetic diversity equilibrium value after a reconnection event, t1 ≃ 75,000 generations (30N generations, estimated from Equation 8 with α = 5%). t1 corresponds to the period during which the deficit of rare alleles is eliminated.
In conclusion, both an isolation and a reconnection event induce changes in the proportion of rare alleles, which skews the values of HEW and DT, thus producing a signature that cannot be differentiated from the signature of past demographic events or of selection.
Discussion
We documented a simple neutral mechanism, which creates long-term peaks of genetic diversity. This peak of genetic diversity appears shortly after an abrupt increase in migration and is conserved for a long time. We also demonstrated that such genetic diversity peaks can occur for a large and plausible range of population sizes, migration rates, mutation rates, and numbers of populations. Subsequent to the genetic diversity peak, the rate of decay of genetic diversity was slow. Consequently, the mechanisms described here leave a strong and long-term footprint on genetic diversity that affects the Ewens–Watterson statistics and Tajima’s D that are commonly used to infer the history of populations from genetic data.
The peak of genetic diversity is due to the spread of the genetic diversity accumulated during (partial) isolation. Therefore, the migration model that is assumed (island model of migration) is a leading factor in determining the strength of the observed genetic diversity peak. Assuming isolation by distance, the within-population genetic diversity is expected to have locally lower peaks. At the same time, under this assumption, the between-population genetic diversity is expected to be higher. Indeed, once populations are connected, each population shares with its neighboring populations alleles accumulated during isolation; thus, differentiation between distant populations will be maintained. Additionally, the amount of genetic diversity accumulated during isolation determines the size of the peak of genetic diversity. The maximum value is reached when populations are completely differentiated, i.e., when no alleles are shared between populations. Our results are robust to the relaxation of the complete isolation and complete differentiation assumptions: when isolation is not complete (because of small migration or nonequilibrium genetic diversity), we show that a genetic diversity peak is still observed (see File S3).
A connection event that occurs after an isolation period might play an important role on species diversification. Indeed, we have demonstrated that such events create an excess of genetic diversity. A high level of genetic diversity has often been hypothesized as being a key factor for species diversification. First, evolution from standing genetic variation might be stronger than from de novo mutation (Gibson and Dworkin 2004; Hermisson and Pennings 2005; Myles et al. 2005; Barrett and Schluter 2008). Second, both theoretical (Gavrilets 2003; Gavrilets and Losos 2009) and empirical work suggest that a high level of preexisting genetic diversity in a population increases its rate of diversification (Harmon et al. 2003; Seehausen 2004; Barrett and Schluter 2008). Interestingly, in several cases of adaptive radiation, a high genetic diversity of founder populations has been documented (e.g., Barrier et al. 1999; Bezault et al. 2011). Several authors argued that the connection of populations after a period of isolation might have played an important role in many adaptive radiations (Hughes and Eastwood 2006; Antonelli and Sanmartín 2011; Bezault et al. 2011; Joyce et al. 2011). Therefore, species that experienced population isolation followed by reconnection events could have benefited from a temporary genetic diversity peak, which has promoted the diversification of that species. Numerous species are known to have experienced such connectivity changes in the past and show remarkable levels of genetic and species diversity (Arnegard et al. 1999). For example, cichlid fishes in the great African lakes experienced periods of habitat fragmentation and reconnection due to lake water level fluctuations (Arnegard et al. 1999); there is some evidence that these processes might have played a role in the explosive radiation of the species (Owen et al. 1990; Young et al. 2009). Additionally, a high rate of speciation is correlated with the timeframe surrounding the uplift of the Northern Andes (Sedano and Burns 2010). The mechanisms described here are thus expected to considerably affect the ability of species to adapt to novel environmental conditions and to diversify over a very long period of time.
Statistics on allelic frequencies such as the Ewens–Watterson statistics (Watterson 1978) and Tajima’s D (Tajima 1989) allow the inference of either selection or population demographic changes. Here, we demonstrate that migration changes can lead to signatures that cannot be differentiated from a selection process or a population size change when using the Ewens–Watterson test and Tajima’s D. Therefore, past migration changes must be considered more carefully and should be viewed as an alternative explanation of bias in neutrality tests and bottleneck or expansion signals. Recently, authors have shown that population structure can bias neutrality tests and produce false bottleneck signals (Leblois et al. 2006; Städler et al. 2009; Chikhi et al. 2010) and that shortly after an isolation event departure from the neutrality can be incorrectly inferred (as shown with simulations by Broquet et al. 2010 and discussed in Waples 2010). The proper interpretation of genetic signatures is crucial for the understanding of the evolutionary history of populations. An interesting extension of this work would be to analyze in more detail the molecular signature of the mechanisms described here and to provide methods that allow the differentiation of such events from selection or demographic changes. Moreover, our results are also relevant for the study of genealogies. Indeed, genetic identities as considered here are commonly used to describe coalescence time distributions (Slatkin 1991; Rousset 1996; Wakeley 1999). Future investigations should also investigate the consequences of isolation and connection events on phylogenetic tree reconstruction. Statistical tools that are available to estimate demographic parameters classically focus on a priori specific scenarios (e.g., population bottleneck, expansion, population with constant migration, population split with subsequent migration; see review in Kuhner 2009). Given the strong impact of migration changes on genetic diversity, accounting for such scenarios is necessary. Recent methods allowing a larger range of population demographic scenarios, such as approximate Bayesian computation (Beaumont et al. 2002; Beaumont 2010), may be powerful tools with which to disentangle the signature of demographic processes from the observed genetic diversity.
One of the major goals of conservation genetics is to maintain genetic diversity, decrease extinction risks, avoid inbreeding depression, maintain species evolutionary potential, and decrease species vulnerability to environmental change (Gilpin and Soule 1986; Newman and Pilson 1997; Jump et al. 2009). In this context, conservationists need to estimate the genetic diversity of a population and its effective size. Such measures are commonly obtained from genetic data and are estimated with standard statistics (Wright 1950; Jorde and Ryman 2007). Although new approaches that consider populations at a nonequilibrium state are emerging, to estimate population size changes and instantaneous migration rates (e.g., Hey and Nielsen 2004), the expected level of genetic diversity is still commonly estimated assuming that populations are at an equilibrium. As shown here, genetic diversity is more likely to be in a transient state. We have demonstrated that reconnecting isolated populations increases genetic diversity above the expected equilibrium value, while isolating populations induces a slow decrease of genetic diversity. Consequently, any estimate inferred from data collected from a population that underwent strong migration changes will not reflect the demographic situation of the population (e.g., census size, genetic diversity). This can have drastic consequences on the selection of conservation strategies and for the management of species (Pearse and Crandall 2004; Caballero et al. 2010).
Acknowledgments
The authors thank two anonymous reviewers and Noah Rosenberg for their valuable comments and suggestions, which increased the clarity and the scope of the article. This project was funded by the Swiss National Research Foundation (SNRF) grant nos. PZ00P3–121702, PZ00P3–139421/1, and 31003A–130065.
Appendix A: Dynamics of Genetic Diversity
In this appendix, we describe the temporal change of genetic diversity (derivation of Equations 4a and 8 and the separation of the dynamics of genetic diversity into two timescales).
Temporal change of genetic diversity
The solution to Equation 3 is
| (A1) |
Denoting P the transformation matrix with eigenvector U1 (associated with λ1) as first column and eigenvector U2 (associated with λ2) as second column,
| (A2) |
and denoting
| (A3) |
Equation A1 becomes
| (A4) |
Further, denoting C1 = y10U1 and C2 = y20U2 leads to Equation 4a.
Ft changes according to two exponential decay functions, and . Their rates of change are and , respectively. Thus r1 = ln(λ1) and r2 = ln(λ2) are the decay constants that determine the rate of change of functions and .
Therefore, the eigenvalues of matrix A can be used to compute the rates of change of genetic diversity. As λ1 > λ2 and both eigenvalues are <1, we have |r2| > |r1|, and thus tends to 0 faster than . r2 determines the transient rate of change of genetic diversity, while r1 determines the asymptotic rate of change of genetic diversity.
We now want to simplify the expression of the rates of change of genetic diversity. To do so, we can rewrite Equation 5 as
With 4bc ≪ (1 − a(1 − c) + b)2 (as m ≪ 1 and N ≫ 1), and given that , Equation 5 further simplifies to
Considering that migration rates and mutation rates are small, we can neglect terms in m2, , μ2, m/N, mμ, and , which leads to
| (A5) |
and thus
| (A6) |
Assuming that migration and mutation rates are small, and that local population sizes are large (N ≫ 1), Equation A6 simplifies to Equation 6 as ln(1 − x) = −x + o(x).
Respective length of the asymptotic and transient dynamics periods
We denote t1 and t2 as the times needed for and to be reduced to a value α, where α ε ]0; 1],
| (A7) |
which leads to
| (A8) |
Assuming small migration and mutation rates and large local population sizes, we can replace the expressions of r1 and r2 from Equation 6 into Equation A8, and we obtain Equation 8. Ft approximately follows
| (A9) |
Timescales separation
This section presents the conditions for t1 ≫ t2. When t1 ≫ t2, and ; thus, Equation A9 simplifies to
| (A10) |
Equation A10 decomposes the dynamics of Ft into two timescales: a transient period of length t2 and an asymptotic period of length t1 − t2 ≃ t1. For t > t1, the genetic identity is close to its equilibrium value Feq, so t1 can be interpreted as the duration of the disequilibrium period.
Equation A10 is true if a t exists such that and . For simplicity, we consider that if and that if . Thus, Equation A10 is a good approximation if a t exists such that
| (A11) |
Thus, showing that
| (A12) |
demonstrates that for , we have and . This provides a sufficient proof of proposition (A11).
We can demonstrate that proposition (A12) depends only on the ratio (see definitions of t1 and t2 Equation 7). Proposition (A12) leads to
Therefore, the condition is necessary and sufficient to prove propositions (A12) and (A11) and the validity of Equation A10. As , Equation A10 is valid when . Considering that m and μ are small, and that N is large, this ratio is approximately equal to (from Equation 8)
| (A13) |
From Equation A13 we can derive the conditions of the timescales separation of the dynamics of genetic diversity (i.e., the parameter values for which t1 ≫ t2). When n > 14, differences are the highest (t1 ≫ t2), in the domain where and also when and m > μ. When n ≤ 14, the same conditions apply for t1 ≫ t2 except in a restricted domain where ( or are required for t1 ≫ t2; see Figure A1). Indeed, denoting , implies that
| (A14a) |
where
| (A14b) |
For θ ≫ 1, we can neglect terms that do not contain θ, and conditions (A14) simplify to
| (A15) |
which simplifies to M ≫ θ.
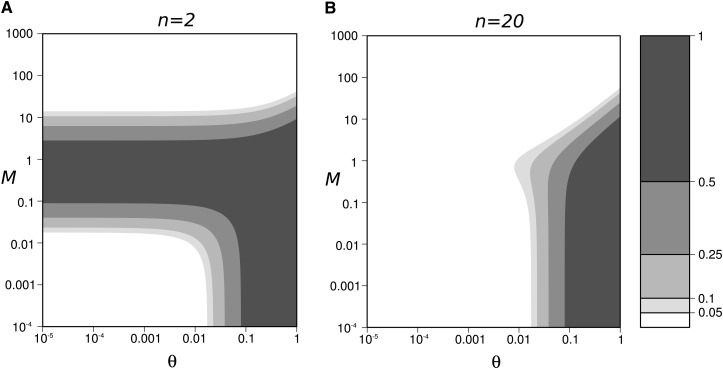
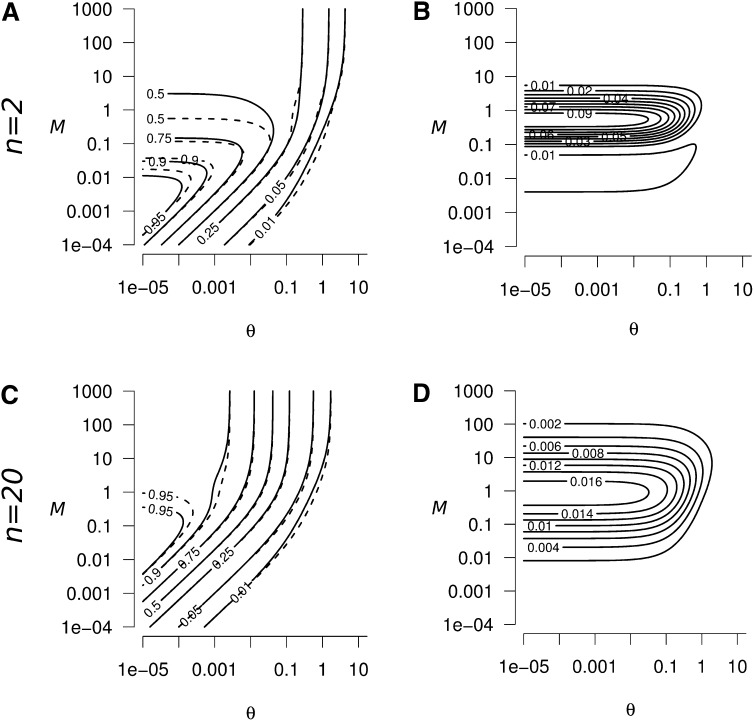
Figure A1.
Domain of validity of Equation A10 (white contours), as a function of the strength of migration (M) and mutation (θ), (A) n = 2, (B) n = 20. The dark shading represent the domains where the validity of Equation A10 is poor (i.e., the exact value of ; from Equation 5).
For θ ≪ 1, terms that contain θ can be neglected in Equation A14, which yields the following conditions:
| (A16) |
Therefore, Equation A10 is not valid when conditions of Equation A16 are not met. This domain is centered around , as
equalize to , for A = 4n − 1. The size of this domain decreases when n increases (Figure A1), and Equation A10 is valid for any value of M in the domain θ ≪ 1 when n > 14 (as conditions (A16) are relaxed when 4n > A + 1, with .
Appendix B: Peak of Genetic Diversity Generated by a Reconnection Event
We derive first the value of the peak of within-population genetic diversity, ΔHs, and the transient excess of between-population genetic diversity, ΔHb, generated by a reconnection event. Second, we characterize their dependency on the migration rate, mutation rate, size, and number of populations.
Derivation of ΔH
We denote the vector of genetic diversity excess at the end of the transient dynamics phase as .
The genetic diversity at the end of the transient dynamics phase is approximately (from Equation A9 when t ≃ t2). Thus ΔH is approximately
| (B1) |
The peak of genetic diversity depends on C1 = y10U1 (from Equations A2 and A3). To derive the value of C1, we compute U1 and y10. The eigenvector is associated with λ1 and follows
| (B2) |
Equation B2 leads to the condition that , where Δred = (1 − a(1 − c) + b)2 − 4bc. We set u11 = 2(1 − a), leading to
Similarly, we can determine the eigenvector associated with λ2:
Denoting the initial value of genetic identity and replacing the expression of U1 and U2 in Equation A3 leads to
Assuming isolation equilibrium for the initial identity leads to and ; thus, ΔH simplifies to
| (B3) |
With 4bc ≪ (1 − a(1 − c) + b)2 (as m ≪ 1 and N ≫ 1), and given that , ΔH further becomes
| (B4) |
Assuming that migration and mutation rates are always small, we can neglect terms in m2, μ2, , mμ, , and , and Equation B4 simplifies to
| (B5) |
By replacing the term (Equation 8 with α = 0.05) in Equation B5, we obtain Equation 9.
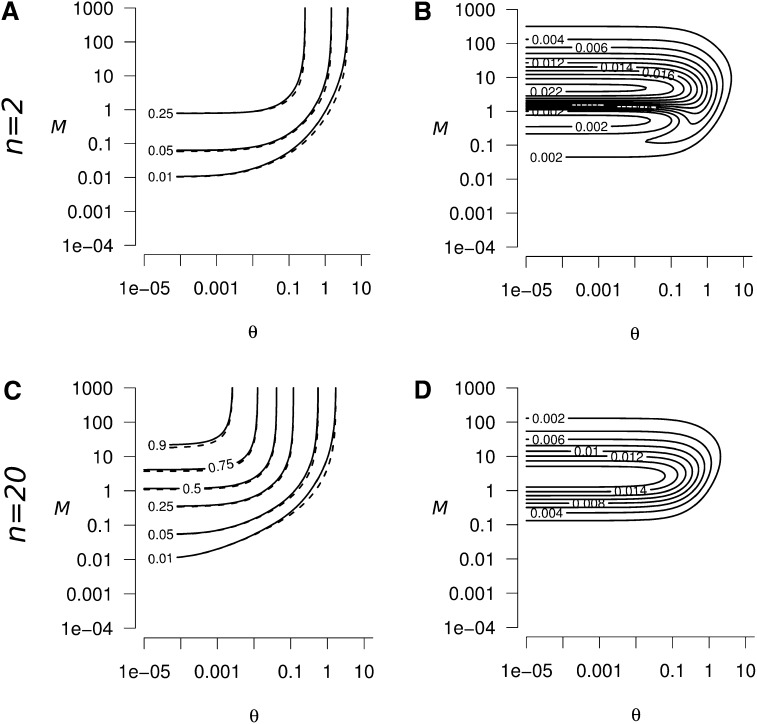
Equation 9 provides a good approximation of the size of the peak of within-population genetic diversity ΔHs and of the transient excess of between-population genetic diversity ΔHb in the entire parameter domain. Figure B1, A and C, and Figure B2, A and C, represent the exact (solid line) and approximate (dashed line; from Equation 9) values of ΔHs and ΔHb, respectively, as a function of θ and M, for n = 2 and n = 20; we can see that the true and approximate values are very close. Figure B1, B and D, and Figure B2, B and D, represent the absolute error resulting from the use of Equation 9 as an approximation of ΔHs and ΔHb, respectively, instead of its exact value, for n = 2 and n = 20. Discrepancies between Equation 9 and the true values of ΔHs and ΔHb can first come from the assumption that m ≪ 1, μ ≪ 1, and N ≫ 1 and second from the assumption of the existence of t2 such that .
Figure B1.
(A and C) Exact (solid lines) and approximate (dashed lines, from Equation 9) values of the peak of genetic diversity ΔHs, as a function of the strength of migration (M) and mutation (θ). In both A and C, the exact and approximate values of ΔHs are very close. (B and D) Absolute error when using Equation 9 to approximate ΔHs, as a function of M and θ. The maximum absolute error is reached when M ≃ 5 and θ < 1 in both B and D. The error decreases when M ≫ 5 or M ≪ 5. The absolute error increases when n decreases, but remains weak: (B) the maximum absolute error is 0.025 for n = 2, and (D) 0.018 for n = 20. Consequently, Equation 9 is a good approximation for the peak of genetic diversity whatever the parameter values of θ, M, and n considered.
Figure B2.
(A and C) Exact (solid lines) and approximate (dashed lines, from Equation 9) values of the transient excess of between-population genetic diversity ΔHb, as a function of the strength of migration (M) and mutation (θ). In both A and C, the exact and approximate values of ΔHb are very close. (B and D) Absolute error when using Equation A9 to approximate ΔHb, as a function of M and θ. The maximum absolute error is reached when M ≃ 1 and θ < 1, the absolute error is 0.09 for n = 2 (B), and 0.016 for n = 20 (D). Equation 9 is a good approximation for ΔHb whatever the parameter values of θ, M, and n considered.
Maximum peak of diversity after a connection event
The peak of genetic diversity increases monotonously with M ( for any value of m, μ, n, and N) and decreases monotonously with θ ( for any value of m, μ, n, and N). The peak of diversity is maximized for intermediate values of n and reached when
| (B6) |
If we neglect terms in μ2, m2, , μm, , and , and assuming small θ and high M, solving Equation B6 using Equation 10 for the peak of genetic diversity leads to Equation 11. When n = n*, Equation 10 leads to a peak of genetic diversity of
Footnotes
Communicating editor: N. A. Rosenberg
Literature Cited
- Antonelli A., Sanmartín I., 2011. Why are there so many plant species in the neotropics? Taxon 60: 403–414. [Google Scholar]
- Antonelli A., Nylander J. A. A., Persson C., Sanmartín I., 2009. Tracing the impact of the andean uplift on neotropical plant evolution. Proc. Natl. Acad. Sci. USA 106: 9749–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnegard M. E., Markert J. A., Danley P. D., Stauffer J. R., Ambali A. J., et al. , 1999. Population structure and colour variation of the cichlid fishes labeotropheus fuelleborni ahl along a recently formed archipelago of rocky habitat patches in southern Lake Malawi. Proc. R. Soc. Lond. B Biol. Sci. 266: 119–130. [Google Scholar]
- Barrett R. D. H., Schluter D., 2008. Adaptation from standing genetic variation. Trends Ecol. Evol. 23: 38–44. [DOI] [PubMed] [Google Scholar]
- Barrier M., Baldwin B. G., Robichaux R. H., Purugganan M. D., 1999. Interspecific hybrid ancestry of a plant adaptive radiation: allopolyploidy of the Hawaiian silversword alliance (Asteraceae) inferred from floral homeotic gene duplications. Mol. Biol. Evol. 16: 1105–1113. [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., 2010. Approximate Bayesian computation in evolution and ecology. Annu. Rev. Ecol. Evol. Syst. 41: 379–406. [Google Scholar]
- Beaumont M. A., Zhang W., Balding D. J., 2002. Approximate Bayesian computation in population genetics. Genetics 162: 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezault E., Mwaiko S., Seehausen O., 2011. Population genomic tests of models of adaptive radiation in Lake Victoria region Cichlid fish. Evolution 65: 3381–3397. [DOI] [PubMed] [Google Scholar]
- Broquet T., Angelone S., Jaquiery J., Joly P., Lena J.-P., et al. , 2010. Genetic bottlenecks driven by population disconnection. Conserv. Biol. 24: 1596–1605. [DOI] [PubMed] [Google Scholar]
- Caballero A., Rodriguez-Ramilo S. T., Avila V., Fernandez J., 2010. Management of genetic diversity of subdivided populations in conservation programmes. Conserv. Genet. 11: 409–419. [Google Scholar]
- Chikhi L., Sousa V. C., Luisi P., Goossens B., Beaumont M. A., 2010. The confounding effects of population structure, genetic diversity and the sampling scheme on the detection and quantification of population size changes. Genetics 186: 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo P. F., Hosemann K. E., Balabhadra S., Villarreal G., Dickson M., et al. , 2005. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Cook L. M., 2008. Species richness in Madeiran land snails, and its causes. J. Biogeogr. 35: 647–653. [Google Scholar]
- Delaney K. S., Riley S. P. D., Fisher R. N., 2010. A rapid, strong, and convergent genetic response to urban habitat fragmentation in four divergent and widespread vertebrates. PLoS ONE 5: e12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues V. S., Poh Y.-P., Peterson B. K., Pennings P. S., Jensen J. D., et al. 2012. Evidence of adaptation from ancestral variation in young populations of beach mice. Evolution 66: 3209–3223. [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 1972. The sampling theory of selectively neutral alleles. Theor. Popul. Biol. 3: 87–112. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Foll M., 2011. fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics 27: 1332–1334. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Lischer H. E. L., 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under linux and windows. Mol. Ecol. Res. 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Foll M., Petit R. J., 2009. Genetic consequences of range expansions. Annu. Rev. Ecol. Evol. Syst. 40: 481–501. [Google Scholar]
- Falush D., Stephens M., Pritchard J. K., 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Wu C. I., 2000. Hitchhiking under positive darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder J. L., Berlocher S. H., Roethele J. B., Dambroski H., Smith J. J., et al. , 2003. Allopatric genetic origins for sympatric host-plant shifts and race formation in rhagoletis. Proc. Natl. Acad. Sci. USA 100: 10314–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1922. Darwinian evolution of mutations. Eugen. Rev. 14: 31–34. [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A., 1930 The Genetical Theory of Natural Selection. Clarendon Press, Oxford. [Google Scholar]
- Frankham R., 1995. Conservation genetics. Annu. Rev. Genet. 29: 305–327. [DOI] [PubMed] [Google Scholar]
- Fu Y. X., Li W. H., 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galis F., Metz J. A., 1998. Why are there so many cichlid species? Trends Ecol. Evol. 13: 1–2. [DOI] [PubMed] [Google Scholar]
- Gavrilets S., 2003. Perspective: models of speciation: What have we learned in 40 years? Evolution 57: 2197–2215. [DOI] [PubMed] [Google Scholar]
- Gavrilets S., Losos J. B., 2009. Adaptive radiation: contrasting theory with data. Science 323: 732–737. [DOI] [PubMed] [Google Scholar]
- Gibson G., Dworkin I., 2004. Uncovering cryptic genetic variation. Nat. Rev. Genet. 5: 681–690. [DOI] [PubMed] [Google Scholar]
- Gilpin M., Soule M., 1986. Minimum viable populations: processes of species extinction, pp. 19–34 in Conservation Biology: The Science of Scarcity and Diversity. Sinauer, Sunderland, MA. [Google Scholar]
- Gravel S., 2012. Population genetics models of local ancestry. Genetics 191: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. E., Krause J., Briggs A. W., Maricic T., Stenzel U., et al. , 2010. A draft sequence of the neandertal genome. Science 328: 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon L. J., Schulte J. A., Larson A., Losos J. B., 2003. Tempo and mode of evolutionary radiation in iguanian lizards. Science 301: 961–964. [DOI] [PubMed] [Google Scholar]
- Hedrick P. W., Kalinowski S. T., 2000. Inbreeding depression in conservation biology. Annu. Rev. Ecol. Syst. 31: 139–162. [Google Scholar]
- Hermisson J., Pennings P. S., 2005. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169: 2335–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez R. D., Kelley J. L., Elyashiv E., Melton S. C., Auton A., et al. , 2011. Classic selective sweeps were rare in recent human evolution. Science 331: 920–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G., 2000. The genetic legacy of the quaternary ice ages. Nature 405: 907–913. [DOI] [PubMed] [Google Scholar]
- Hewitt G. M., 2004. Genetic consequences of climatic oscillations in the quaternary. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359: 183–195, discussion 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J., Nielsen R., 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C., Eastwood R., 2006. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the andes. Proc. Natl. Acad. Sci. USA 103: 10334–10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J. D., Kim Y., DuMont V. B., Aquadro C. F., Bustamante C. D., 2005. Distinguishing between selective sweeps and demography using dna polymorphism data. Genetics 170: 1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J. A., Hughes K. A., Alaks G., Graham L., Lacy R. C., 1994. An experimental study of inbreeding depression in a natural habitat. Science 266: 271–273. [DOI] [PubMed] [Google Scholar]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E., et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde P. E., Ryman N., 2007. Unbiased estimator for genetic drift and effective population size. Genetics 177: 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce D. A., Lunt D. H., Genner M. J., Turner G. F., Bills R., et al. , 2011. Repeated colonization and hybridization in Lake Malawi cichlids. Curr. Biol. 21: R108–R109. [DOI] [PubMed] [Google Scholar]
- Jump A. S., Marchant R., Peñuelas J., 2009. Environmental change and the option value of genetic diversity. Trends Plant Sci. 14: 51–58. [DOI] [PubMed] [Google Scholar]
- Karlin S., 1982. Classifications of selection migration structures and conditions for a protected polymorphism. Evol. Biol. 14: 61–204. [Google Scholar]
- Keller I., Nentwig W., Largiader C. R., 2004. Recent habitat fragmentation due to roads can lead to significant genetic differentiation in an abundant flightless ground beetle. Mol. Ecol. 13: 2983–2994. [DOI] [PubMed] [Google Scholar]
- Kim Y., Gulisija D., 2010. Signatures of recent directional selection under different models of population expansion during colonization of new selective environments. Genetics 184: 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Nielsen R., 2004. Linkage disequilibrium as a signature of selective sweeps. Genetics 167: 1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Crow J. F., 1964. The number of alleles that can be maintained in a finite population. Genetics 49: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M., 2000. Methods to detect selection in populations with applications to the human. Annu. Rev. Genomics Hum. Genet. 1: 539–559. [DOI] [PubMed] [Google Scholar]
- Kuhner M. K., 2009. Coalescent genealogy samplers: windows into population history. Trends Ecol. Evol. 24: 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeck K., Antonioli F., Purcell A., Silenzi S., 2004. Sea-level change along the Italian coast for the past 10,000 yr. Quat. Sci. Rev. 23: 1567–1598. [Google Scholar]
- Latter B. D., 1973. The island model of population differentiation: a general solution. Genetics 73: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblois R., Estoup A., Streiff R., 2006. Genetics of recent habitat contraction and reduction in population size: Does isolation by distance matter? Mol. Ecol. 15: 3601–3615. [DOI] [PubMed] [Google Scholar]
- Li H., Stephan W., 2006. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2: e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., 1970. Effective number of alleles in a subdivided population. Theor. Popul. Biol. 1: 273–306. [DOI] [PubMed] [Google Scholar]
- Miller J., Hobbs R., 2002. Conservation where people live and work. Conserv. Biol. 16: 330–337. [Google Scholar]
- Myles S., Bouzekri N., Haverfield E., Cherkaoui M., Dugoujon J.-M., et al. , 2005. Genetic evidence in support of a shared Eurasian–North African dairying origin. Hum. Genet. 117: 34–42. [DOI] [PubMed] [Google Scholar]
- Nagylaki T., 1974. The decay of genetic variability in geographically structured populations. Proc. Natl. Acad. Sci. USA 71: 2932–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki T., 1977. Decay of genetic variability in geographically structured populations. Proc. Natl. Acad. Sci. USA 74: 2523–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki T., 1979. The island model with stochastic migration. Genetics 91: 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1973. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Feldman M. W., 1972. Identity of genes by descent within and between populations under mutation and migration pressures. Theor. Popul. Biol. 3: 460–465. [DOI] [PubMed] [Google Scholar]
- Nei M., Maruyama T., Chakraborty R., 1975. The bottleneck effect and genetic variability in populations. Evolution 29: 1–10. [DOI] [PubMed] [Google Scholar]
- Newman D., Pilson D., 1997. Increased probability of extinction due to decreased genetic effective population size: experimental populations of Clarkia pulchella. Evolution 51: 354–362. [DOI] [PubMed] [Google Scholar]
- Nielsen R., 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39: 197–218. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Bustamante C., Clark A. G., Glanowski S., Sackton T. B., et al. , 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel S., Ouellet M., Galois P., Lapointe F.-J., 2007. Impact of urban fragmentation on the genetic structure of the eastern red-backed salamander. Conserv. Genet. 8: 599–606. [Google Scholar]
- Orr H. A., Betancourt A. J., 2001. Haldane’s sieve and adaptation from the standing genetic variation. Genetics 157: 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen R. B., Crossley R., Johnson T. C., Tweddle D., Kornfield I., et al. , 1990. Major low levels of Lake Malawi and their implications for speciation rates in cichlid fishes. Proc. R. Soc. Lond. B Biol. Sci. 240: 519–553. [Google Scholar]
- Pavlidis P., Jensen J. D., Stephan W., 2010. Searching for footprints of positive selection in whole-genome snp data from nonequilibrium populations. Genetics 185: 907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse D. E., Crandall K. A., 2004. Beyond fst: analysis of population genetic data for conservation. Conserv. Genet. 5: 585–602 . 10.1007/s10592-004-1863-z. [DOI] [Google Scholar]
- Pelz H.-J., Rost S., Hünerberg M., Fregin A., Heiberg A.-C., et al. , 2005. The genetic basis of resistance to anticoagulants in rodents. Genetics 170: 1839–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter B. M., Wegmann D., Excoffier L., 2010. Distinguishing between population bottleneck and population subdivision by a Bayesian model choice procedure. Mol. Ecol. 19: 4648–4660. [DOI] [PubMed] [Google Scholar]
- Price A. L., Tandon A., Patterson N., Barnes K. C., Rafaels N., et al. , 2009. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 5: e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S. H., Papadopoulos A., 2009. Evolution with stochastic fitness and stochastic migration. PLoS ONE 4: e7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F., 1996. Equilibrium values of measures of population subdivision for stepwise mutation processes. Genetics 142: 1357–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D., Clifford E. A., Nemethy M., McKinnon J. S., 2004. Parallel evolution and inheritance of quantitative traits. Am. Nat. 163: 809–822. [DOI] [PubMed] [Google Scholar]
- Sedano R. E., Burns K. J., 2010. Are the Northern Andes a species pump for neotropical birds?: Phylogenetics and biogeography of a clade of neotropical tanagers (aves: Thraupini). J. Biogeogr. 37: 325–343. [Google Scholar]
- Seehausen O., 2004. Hybridization and adaptive radiation. Trends Ecol. Evol. 19: 198–207. [DOI] [PubMed] [Google Scholar]
- Shpak M., Wakeley J., Garrigan D., Lewontin R. C., 2010. A structured coalescent process for seasonally fluctuating populations. Evolution 64: 1395–1409. [DOI] [PubMed] [Google Scholar]
- Slatkin M., 1991. Inbreeding coefficients and coalescence times. Genet. Res. 58: 167–175. [DOI] [PubMed] [Google Scholar]
- Smith J. M., 1970. Population size, polymorphism, and the rate of non-Darwinian evolution. Am. Nat. 104: 231–237. [Google Scholar]
- Städler T., Haubold B., Merino C., Stephan W., Pfaffelhuber P., 2009. The impact of sampling schemes on the site frequency spectrum in nonequilibrium subdivided populations. Genetics 182: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturmbauer C., Baric S., Salzburger W., Rüber L., Verheyen E., 2001. Lake level fluctuations synchronize genetic divergences of cichlid fishes in African lakes. Mol. Biol. Evol. 18: 144–154. [DOI] [PubMed] [Google Scholar]
- Tajima F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Nei M., 1985. Gene genealogy and variance of interpopulational nucleotide differences. Genetics 110: 325–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R. C., Levy J. C., Clark A., 1993. Complex genetics of type 2 diabetes: thrifty genes and previously neutral polymorphisms. Q. J. Med. 86: 413–417. [PubMed] [Google Scholar]
- Vandergast A. G., Lewallen E. A., Deas J., Bohonak A. J., Weissman D. B., et al. , 2009. Loss of genetic connectivity and diversity in urban microreserves in a southern California endemic Jerusalem cricket (Orthoptera: Stenopelmatidae: Stenopelmatus n. sp ‘‘santa monica’’). J. Insect Conserv. 13: 329–345. [Google Scholar]
- Wakeley J., 1999. Nonequilibrium migration in human history. Genetics 153: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R. S., 2010. Spatial-temporal stratifications in natural populations and how they affect understanding and estimation of effective population size. Mol Ecol Resour 10: 785–796. [DOI] [PubMed] [Google Scholar]
- Watterson G. A., 1978. The homozygosity test of neutrality. Genetics 88: 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock M. C., 1992. Temporal fluctuations in demographic parameters and the genetic variance among populations. Evolution 46: 608–615. [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., Barton N. H., 1997. The effective size of a subdivided population. Genetics 146: 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1950. Genetical structure of populations. Nature 166: 247–249. [DOI] [PubMed] [Google Scholar]
- Young K. A., Whitman J. M., Turner G. F., 2009. Secondary contact during adaptive radiation: a community matrix for Lake Malawi cichlids. J. Evol. Biol. 22: 882–889. [DOI] [PubMed] [Google Scholar]
- Zhang H., Yan J., Zhang G., Zhou K., 2008. Phylogeography and demographic history of Chinese black-spotted frog populations (Pelophylax nigromaculata): evidence for independent refugia expansion and secondary contact. BMC Evol. Biol. 8: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]