Abstract
The switch from an outcrossing mode of mating enforced by self-incompatibility to self-fertility in the Arabidopsis thaliana lineage was associated with mutations that inactivated one or both of the two genes that comprise the self-incompatibility (SI) specificity-determining S-locus haplotype, the S-locus receptor kinase (SRK) and the S-locus cysteine-rich (SCR) genes, as well as unlinked modifier loci required for SI. All analyzed A. thaliana S-locus haplotypes belong to the SA, SB, or SC haplotypic groups. Of these three, the SC haplotype is the least well characterized. Its SRKC gene can encode a complete open-reading frame, although no functional data are available, while its SCRC sequences have not been isolated. As a result, it is not known what mutations were associated with inactivation of this haplotype. Here, we report on our analysis of the Lz-0 accession and the characterization of its highly rearranged SC haplotype. We describe the isolation of its SCRC gene as well as the subsequent isolation of SCRC sequences from other SC-containing accessions and from the A. lyrata S36 haplotype, which is the functional equivalent of the A. thaliana SC haplotype. By performing transformation experiments using chimeric SRK and SCR genes constructed with SC- and S36-derived sequences, we show that the SRKC and SCRC genes of Lz-0 and at least a few other SC-containing accessions are nonfunctional, despite SCRC encoding a functional full-length protein. We identify the probable mutations that caused the inactivation of these genes and discuss our results in the context of mechanisms of S-locus inactivation in A. thaliana.
Keywords: S locus, self-incompatibility, switch to self-fertility, S-locus receptor kinase, S-locus cysteine-rich protein
THE switch from an outcrossing mode of mating to self-fertility was a major transition in the evolutionary history of Arabidopsis thaliana. Recent studies have shown that this switch was accompanied by multiple independent losses of self-incompatibility (SI), the major mechanism that promotes outcrossing in the Brassicaceae (Sherman-Broyles et al. 2007; Shimizu et al. 2008; Boggs et al. 2009a). In this family, SI is controlled by numerous haplotypes of the S locus. Within each S-locus haplotype (hereafter S haplotype), are two genes that determine specificity in the SI response: one gene encodes the stigma-expressed S-locus receptor kinase (SRK) and the other encodes the pollen coat-localized ligand for SRK, the S-locus cysteine-rich (SCR) protein. The SRK and SCR proteins are highly polymorphic and co-evolving proteins (Sato et al. 2002) and their haplotype-specific interaction is responsible for the specific recognition and inhibition by the stigma epidermis of self-related pollen (i.e., pollen derived from the same flower, other flowers on the same plant, or plants expressing the same S haplotype) (reviewed in Rea and Nasrallah 2008). Consequently, an understanding of the genetic events associated with the switch to self-fertility in the A. thaliana lineage was sought through analysis of SRK and SCR sequences harbored by various A. thaliana geographical accessions (Kusaba et al. 2001; Shimizu et al. 2004, 2008; Sherman-Broyles et al. 2007; Tang et al. 2007; Boggs et al. 2009a,b; Tsuchimatsu et al. 2010) and comparisons to orthologous sequences from A. thaliana’s close self-incompatible relatives A. lyrata and A. halleri (Bechsgaard et al. 2006).
These comparisons have suggested that A. thaliana has retained only three of the many S haplotypes that must have existed in A. thaliana’s self-incompatible ancestor and that still exist in A. lyrata and A. halleri. These three S haplotypes are designated SA, SB, and SC (Shimizu et al. 2004), and correspond, respectively, to the A. lyrata S37, S16, and S36 haplotypes (Bechsgaard et al. 2006). All A. thaliana accessions analyzed to date contain nonfunctional versions of these three S haplotypes (Shimizu et al. 2008; Boggs et al. 2009a) or hybrid haplotypes derived by recombination between the SA and SC haplotypes (Sherman-Broyles et al. 2007; Boggs et al. 2009a). Additionally, A. thaliana harbors disruptive mutations at other loci required for SI that apparently arose stochastically in different populations of the species. Indeed, transformation with functional SRK-SCR gene pairs isolated from self-incompatible A. lyrata demonstrated that, while some A. thaliana accessions express robust and developmentally stable SI similar to A. lyrata, other accessions express transient SI or weak SI or fail to express SI (Nasrallah et al. 2002, 2004; Liu et al. 2007; Boggs et al. 2009b). In view of this complex genetic architecture of self-fertility, it has been difficult to determine if loss of SI in the A. thaliana lineage was caused by inactivation of the S locus or by a mutation in another locus that spread through the species, causing relaxation of selective constraints on the S locus and allowing its degradation. Whatever the nature of the initial event(s) that caused loss of SI, the data are consistent with multiple independent events that inactivated the SA, SB, and SC haplotypes. Thus, the path to self-fertility in A. thaliana was very different from that described for Capsella rubella (Foxe et al. 2009; Guo et al. 2009), which has retained only one S haplotype and may have been founded by a single self-fertile individual (Guo et al. 2009).
Analysis of the SA and SB haplotypes and their A. lyrata counterparts, for which both SRK and SCR sequences have been identified (Kusaba et al. 2001; Shimizu et al. 2004, 2008; Boggs et al. 2009a), has demonstrated the presence of disruptive mutations or rearrangements in one or both of these genes (Sherman-Broyles et al. 2007; Shimizu et al. 2008; Boggs et al. 2009a; Tsuchimatsu et al. 2010). By contrast, for the SC haplotype, only SRK sequences, some of them encoding full-length open reading frames, are available, and neither SCRC sequences nor the corresponding A. lyrata SCR36 sequences have been isolated to date. Thus, although there is clear evidence that the SC haplotype is nonfunctional, at least in some accessions (Boggs et al. 2009a), it is not known what mutations were associated with inactivation of this locus in the 11 A. thaliana accessions known to harbor the SC haplotype (Shimizu et al. 2004; Sherman-Broyles et al. 2007), how these mutations compare with those that inactivated the SA and SB haplotypes, and if SA-SC recombinant haplotypes have retained SCRC sequences. Thus, our view of the events that remodeled the A. thaliana S locus is incomplete.
We set out to address this issue by isolating SCRC sequences and assessing the functionality of both the SRKC and SCRC genes. Here we report on our analysis of the SC haplotype of the Lz-0 accession and the isolation of its SCRC sequence. We also describe the use of this sequence to isolate the A. lyrata SCR36 allele and to characterize the SCRC alleles of other SC-containing A. thaliana accessions. Moreover, we present transformation experiments aimed at determining if the Lz-0 SRKC and SCRC sequences, which encode apparently functional proteins, have retained the ability to confer SI.
Materials and Methods
Plant materials
Seeds for the A. thaliana SC-containing accessions Lz-0 (CS1354), Br-0 (CS22628), Bur-0 (CS22656), Ita-0 (CS1244), Kas-2 (CS1264), Mr-0 (CS1372), Pro-0 (CS22649), Ra-0 (CS22649), RRS-10 (CS22565), and Wt-5 (CS22637) were obtained from the Arabidopsis Biological Resource Center (ABRC, Columbus, Ohio). DNA from A. lyrata S36 plants was kindly provided by J. Bechsgaard and M. Schierup, Bioinformatics Research Center, University of Aarhus, DK-8000 Aarhus, Denmark.
Construction of a genomic library and isolation of S-locus sequences from Lz-0
A genomic library was constructed in λDASH II (Stratagene, La Jolla, CA) using DNA isolated from Lz-0 plants. The library, in which inserts averaged 15 kb in size, was screened with DNA probes that were generated by polymerase chain reaction (PCR) amplification using the primer pairs shown in Supporting Information, Table S1 and labeled with 32P using the Random Primed DNA Labeling kit (Roche Diagnostics, Indianapolis). Screening of the genomic library was performed as described in File S1). The sequences of the full-length SRKC-Lz and SCRC-Lz genes (GenBank accession nos. KC207414 and KC207415) and their flanking DNA were derived by sequencing of clone inserts at the Cornell University Life Sciences Core Laboratories Center (Ithaca, NY).
RT–PCR of SRK and SCR transcripts
The SRK gene is expressed in pistils, most intensely in the stigma epidermis and to a lesser extent in the style, and the SI response is evident in stigmas of buds at stage 13 of flower development (staging according to Smyth et al. 1990), which corresponds to one day prior to flower opening (hereafter referred to as the −1 bud stage). The SCR gene is expressed in the anther tapetum and, for some alleles, also in the developing microspores. Therefore, RNA was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA) from 25 pistils dissected from floral buds at the −1 stage of development for analysis of SRKC transcripts, or from 25 young floral buds at a stage in which the tapetum is still intact for analysis of SCRC transcripts. After treatment with amplification grade DNaseI (Invitrogen), RT–PCR reactions were performed with the Superscript III One Step RT PCR system (Invitrogen) using the following intron-flanking primers (Table S1): for SRKC, the SRK-e1fp forward primer specific for exon 1 and one of three reverse primers SRK-e3rp, SRK-e5rp, or SRK-e7rp, which are specific for exon 3, exon 5, or exon7/3′-UTR, respectively; for SCRC, the forward SCRLzF4 and reverse SCR-LzR1 primers, which flank the two exons of the gene.
Amplification of SCRC genes from various A. thaliana accessions and the SCR36 and SRK36 alleles from A. lyrata S36 DNA
SCRC sequences were amplified from genomic DNA isolated from various Sc-containing A. thaliana accessions using the SCRLzF4 and SCRLzR1 primers (Table S1), while SCR36 was amplified from DNA of A. lyrata S36-containing plants using the SCRLzF1 and SCRLzRP primers (Table S1). SRK36 sequences were amplified from A. lyrata S36-containing plants using two PCR primer pairs (Table S1): (1) AtSRKC36fp (located at the beginning of exon 1) and AtSRKC36rp2 (within exon 2); and AlSRK36fp1 (specific to intron 1) and AtSRKCKrp1 (located at the end of the coding region in exon 7). PCR products were cloned in the pGEMT-Easy or pCR2.1 plasmids (Invitrogen) and multiple samples of each clone were analyzed to build a consensus sequence for each allele. The resulting SRK36 and SCR36 sequences have been deposited in GenBank (accession nos. KC207416 and KC207417).
DNA and protein gel blot analysis
Genomic DNA from each A. thaliana accession was digested with EcoRI, run on a 1% (w/v) agarose gel, and transferred to Hybond N+ membranes (GE Healthcare Life Sciences, Piscataway, NJ) using an alkaline transfer method. The blot was hybridized with a 32P-labeled Lz-0 SCRC probe, exposed to phosphor screens, and developed using a STORM 860 PhosphorImager (GE Healthcare Life Sciences).
For protein immunoblot analysis, the AtS1pr::YFP-SRKb/SCRb transformation plasmid, designated p594, and immunoblot analysis of the YFP-SRKb protein, were described previously (Kitashiba et al. 2011).
Transgenes and plant transformation
Plant transformation constructs were generated as described in File S1. The constructs were introduced into Agrobacterium strain GV3101 (Koncz and Schell 1986) and subsequently used to transform A. thaliana plants by the floral dip method (Zhang et al. 2006). Hygromycin-resistant transformants were analyzed by reciprocal pollination assays as previously described (Boggs et al. 2009b,c).
Results and Discussion
Structure of the Lz-0 SC haplotype
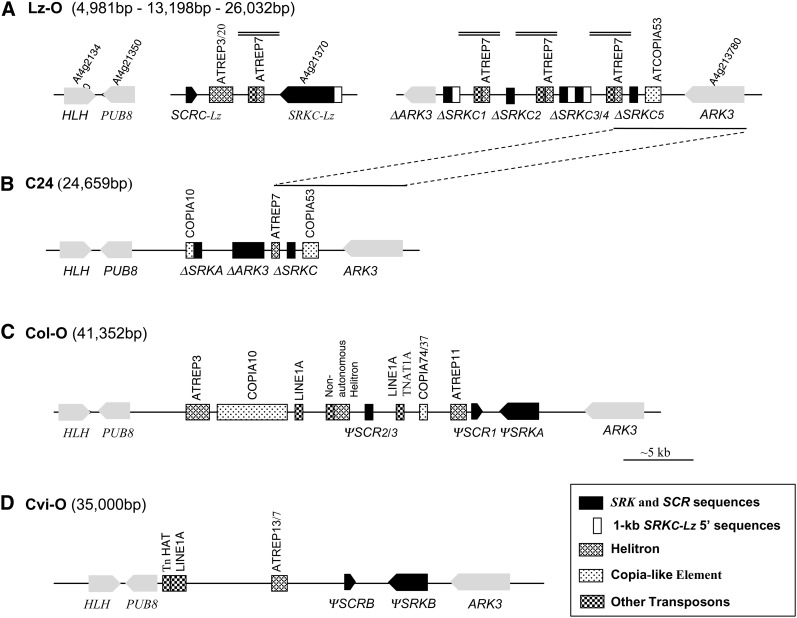
SRKC sequences have been detected in the Br-0, Bur-0, Ita-0, Kas-2, Kr-0, Lz-0, Mr-0, Pro-0, Ra-0, RRS-10, and Wt-5 accessions of A. thaliana (Shimizu et al. 2004; Sherman-Broyles et al. 2007). Truncated SRKC sequences were also detected in several accessions in which the S haplotype was derived by recombination between SA and SC haplotypes (Sherman-Broyles et al. 2007; Boggs et al. 2009a). In an attempt to identify SCRC sequences and to compare the organization of an SC haplotype to that of previously characterized S haplotypes, we focused on the Lz-0 accession. We constructed a bacteriophage library of genomic DNA derived from this accession and isolated the S-locus region, which is flanked on one side by PUB8 (Plant U Box 8; At4g21350) and on the other side by ARK3 (Arabidopsis Receptor Kinase 3; At4g21380) (Kusaba et al. 2001; Goubet et al. 2012). The library screens (described in File S1) resulted in the cloning of three nonoverlapping segments of the Lz-0 SC haplotype (Figure 1A): (1) segment 1, consisting of ∼12 kb of DNA ending with the PUB8 gene at one boundary of the S haplotype; (2) segment 2, consisting of ∼13 kb of DNA containing full-length SRKC and SCRC sequences; and (3) segment 3, consisting of ∼26 kb of DNA ending with the ARK3 gene at the other boundary of the S haplotype.
Figure 1.
Comparison of S-haplotype structure in Lz-0 (A), C24 (B), Col-0 (C), and Cvi-0 (D). Numbers in parentheses to the right of the accession name denote the lengths of each of the three cloned segments of the Lz-0 pseudo-S-haplotype, and the PUB8-ARK3 regions of C24, Col-0, and Cvi-0. The gray bars above the map in A indicate the locations of the 2.6-kb 3′ SRKC sequence repeats (see text for details). The dashed lines between A and B link regions of high sequence similarity found in the Lz-0 S haplotype and the recombinant SA-SC haplotype of C24. Transposon sequences were identified using RepBase (Kohany et al. 2006).
The structure of the Lz-0 SC haplotype was compared to that of the previously characterized C24 SA-SC recombinant haplotype (Figure 1B; Sherman-Broyles et al. 2007), the Col-0 SA haplotype (Figure 1C; Kusaba et al. 2001), and the Cvi-0 SB haplotype (Figure 1D; Tang et al. 2007). Among these S haplotypes, the Lz-0 S haplotype has the longest PUB8-ARK3 region (>44 kb). As is typical for A. thaliana S haplotypes, it is rich in helitron sequences, including ATREP3, ATREP20, and especially ATREP7 (Sherman-Broyles et al. 2007). Also, as found in the Col-0 SA and Cvi-0 SB haplotypes, it contains a full-length SRK sequence (designated AtSRKC-Lz) oriented in a head-to-head arrangement with an SCR sequence (designated AtSCRC-Lz). However, the Lz-0 S haplotype stands alone among these haplotypes in containing five truncated SRKC sequences (labeled ΔSRKC1–ΔSRKC5) located in a highly rearranged region near the ARK3 boundary of the S locus (Figure 1A). In each of these repeats, the ΔSRKC’s are associated with ATREP7 sequences that are preceded by an identical mix of CRI-35, ATTIRTA1, Polinton3 SM, BOM2H2, and Gypsy9-SM1 sequences. The ΔSRKC1 and ΔSRKC4 sequences are identical to the full-length SRKC-Lz gene over 1309 bp, including the first 309 bp at the start of the coding region (Figure S1) and 1 kb of upstream DNA, the latter being also repeated in the intergenic DNA between ΔSRKC3 and ΔSRKC4 (Figure 1A). In contrast, the ΔSRKC2, ΔSRKC3, and ΔSRKC5 sequences are identical to the SRKC-Lz over a segment that encompasses 160 bp at the 3′ end of the coding region (Figure S1) and 2.6 kb of 3′ flanking DNA (Figure 1A).
Thus, despite containing two gaps, the Lz-0 S haplotype differs in overall organization, sequence content, and placement of transposon sequences from the other A. thaliana S haplotypes analyzed (Figure 1). In particular, it differs from the C24 S haplotype, as well as the Kas-2 S haplotype (not shown), both of which contain SC- and SA-derived sequences and were clearly produced by recombination between SA and SC haplotypes (Sherman-Broyles et al. 2007; Boggs et al. 2009a). We found no evidence that the Lz-0 SC haplotype was the product of such interhaplotypic recombination events. Rather, its multiple truncated SRK sequences and repeats in the ARK3-proximal region suggest that several intrahaplotypic unequal recombination events occurred during the genesis of this haplotype.
Despite these differences, the ARK3-proximal regions of the Lz-0 and C24 S haplotypes are highly similar. In fact, the two S haplotypes are >99% identical over a span of ∼10 kb (see dashed lines indicating the location of homologous regions in Figure 1, A and B), starting at ∼1 kb past ΔSRKC4 through ΔSRKC5, interrupted by a 212-bp insertion in the Lz-0 sequence, and then continuing throughout the 3′ flanking region and the entire ARK3 gene. Indeed, the predicted C24 and Lz-0 ARK3 proteins differ by only one amino-acid substitution (phenylalanine-590 to tyrosine) (Figure S2). This low divergence supports the previous conclusion that the ARK3 gene in the C24 SA-SC recombinant haplotype is likely derived from an SC haplotype (Sherman-Broyles et al. 2007).
Another similarity between the C24 and Lz-0 S haplotypes is that they both contain an internal truncated ARK3 sequence, designated ΔARK3, in addition to the full-length ARK3 gene that flanks the S haplotype. ARK3 encodes a serine/threonine receptor protein kinase belonging to the same S Domain Receptor-Like Kinase (SD-RLK) family as SRK (Dwyer et al. 1994; Shiu and Bleecker 2001, 2003) but it does not function in SI. Like SRK and several other members of the SD-RLK gene family, the ARK3 gene is composed of seven exons and six introns, and its predicted protein product consists of an extracellular S domain encoded by exon 1, a transmembrane domain encoded by exon 2, and a cytoplasmic domain encoded by exons 3 through 7. The ΔARK3 sequences of the Lz-0 and C24 S haplotypes appear to have arisen independently because they have drastically different structures. In Lz-0, ΔARK3 lacks exon 1 entirely but it retains most of intron 1 and exons 2–7, each of which contains uninterrupted open reading frames exhibiting >97% nucleotide sequence identity with the corresponding regions of the full-length Lz-0 ARK3 gene (Figure S2). In contrast, the C24 ΔARK3 is a hybrid sequence apparently generated by the SA-SC recombination event that created the C24 S haplotype (Sherman-Broyles et al. 2007), which contains the last 206 bp of exon 1 and all of intron 1, single base pair deletions in each of exons 2 and 3, and a deletion of all of exon 6. It should be noted that the ARK3 gene seems to be prone to duplication because some functional S haplotypes of A. lyrata were found to contain both an ARK3 gene and an ARK3 pseudogene (Charlesworth et al. 2003; Hagenblad et al. 2006; Goubet et al. 2012).
The AtSRKC-Lz gene
The full-length AtSRKC-Lz gene contains an apparently intact open reading frame. Like functional SRK genes, its 3932-bp transcriptional unit is predicted to produce a full-length protein of 854 amino acids that lacks obvious null mutations (Figure S1 and Figure S3). Furthermore, RT–PCR of pistil RNA using primers pairs that spanned all predicted exon–intron junctions of the gene (see Materials and Methods) demonstrated that the AtSRKC-Lz gene is transcribed. Importantly, sequence analysis of the RT–PCR products confirmed that the predicted splice junctions are indeed used in the plant. Thus, the AtSRKC-Lz gene, like the SRKC genes derived from the Ita-0, Kas-1, and Kr-0 accessions (Shimizu et al. 2004), is an expressed gene that potentially produces a functional SRK protein.
Amino-acid sequence alignments (Figure S3) demonstrated that this predicted AtSRKC-Lz protein is very similar (>99%) to previously reported A. thaliana SRKC variants (Figure S4). Furthermore, comparison of AtSRKC-Lz and AlSRK36, the A. lyrata SRK allele that is most similar to A. thaliana SRKC (Bechsgaard et al. 2006), and was cloned here in its entirety using PCR primers derived from the AtSRKC-Lz gene sequence (Table S1), revealed that their S domains are 90.6% identical. As expected, the sequence similarity among the SRKC variants is much higher than that shared by these variants with the SRKA variant of Col-0/Wei-0 (60%) or with the SRKB variant of Cvi-0 (61.9%). As is typically the case for SRKs, this intraspecific sequence divergence is as great as that observed in interspecific comparisons of SRKs having different SI specificities (Figure S4).
The SCRC gene in Lz-0 and other A. thaliana accessions
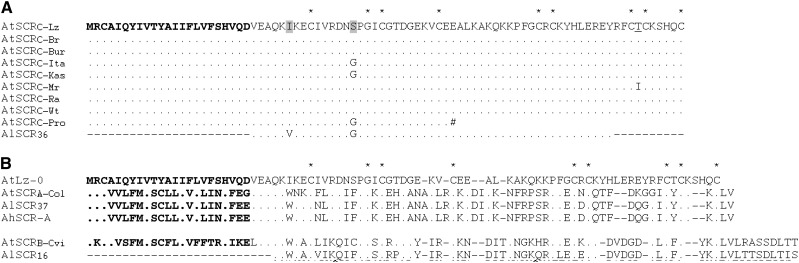
Similar to other SCR genes, the AtSCRC-Lz gene (432 bp in length) is composed of two exons (67 bp and 188 bp) separated by a 177-bp intron. The gene encodes a full-length open reading frame that is predicted to produce a secreted protein of 61 amino acids after removal of the signal sequence (Figure 2), and contains the eight cysteines and one glycine that are conserved in all known SCR proteins. To determine if other SC-containing accessions of A. thaliana harbor SCRC sequences, genomic DNA isolated from the Br-0, Bur-0, Ita-0, Kas-0, Mr-0, Pro-0, Ra-0, RRS-10, and Wt-5 accessions were subjected to PCR using primers designed from the AtSCRC-Lz sequence (Table S1). PCR products were obtained from all accessions, except for RRS-10, and the predicted amino-acid sequences demonstrated high sequence similarity among the amplified SCRC variants. Three categories of SCRC variants were recovered (Figure 2): (1) SCRCs identical to AtSCRC-Lz were found in Br-0, Bur-0, Ra-0, and Wt-5; (2) SCRCs with a full-length open reading frame containing a single amino-acid substitution relative to AtSCRC-Lz were found in Ita-0, Kas-2, and Mr-0; and (3) the SCRC of Pro-0, which contains both an amino-acid substitution and a nonsense mutation that would truncate the SCRC protein. Additionally, the amplified SCRC genes of Ita-0 and Mr-0 included indels relative to AtSCRC-Lz within their introns (not shown). The SCRC variants are highly diverged from the two other A. thaliana SCR variants, SCRA and SCRB (Figure 2; nucleotide sequence identity averages only 35.2 and 32.9%, respectively). Also, unlike SCRA and SCRB, the predicted mature SCRC protein is short and lacks the C-terminal extension after the eighth conserved cysteine.
Figure 2.
Amino-acid sequences of various SCR variants. (A) SCRC variants isolated from various SC-containing A. thaliana accessions are aligned with their functional A. lyrata equivalent, AlSCR36. (B) The Lz-0 SCRC variant is aligned with the SCRA variant of Col-0 along with the corresponding A. lyrata and A. halleri sequences (AlSCR37 and AhSCR-A), and the SCRB variant of Cvi-0 along with the corresponding A. lyrata AlSCR16 sequence. In each alignment, the eight invariant cysteines and one invariant glycine characteristic of SCR proteins are indicated by asterisks above the AtSCRC-Lz sequence, and dots indicate identical amino acids. Signal sequences are shown in boldface type and primer-encoded sequences are represented by dashes at the N- and C termini. Amino-acid positions that differ in the AtSCRC variants relative to AlSCR36 are shaded in the AtSCRC-Lz sequence. The threonine residue in AtSCRC-Lz that is substituted for an isoleucine in AtSCRC-Mr is underlined. The “#” sign in the SCRC variant of the Pro-0 accession indicates the position of a premature stop codon.
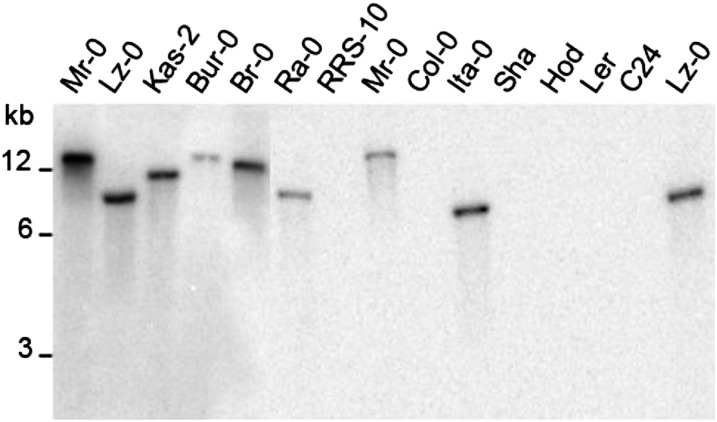
The distribution of SCRC sequences in A. thaliana populations was also assessed by DNA gel blot analysis using a probe corresponding to the AtSCRC-Lz gene. This probe is expected to be specific for SCRC sequences because its high divergence from SCRA and SCRB precludes cross-hybridization. Figure 3 shows that the AtSCRC-Lz probe hybridized with DNA from all tested SC-containing accessions with the exception of RRS-10 (consistent with the PCR results), but not with DNA from SA accessions (Col-0, Ler-0, Sha-0, and Hodja), nor with DNA from C24, indicating that the C24 SA–SC recombinant haplotype has not retained any SCRC sequences.
Figure 3.
DNA gel blot analysis of EcoRI-digested genomic DNA isolated from various A. thaliana accessions. The blot was hybridized with a probe corresponding to Lz-0 SCRC sequences. Molecular length markers are indicated on the left.
To determine how intact the SCRC sequences of A. thaliana are, they must be compared to their A. lyrata SCR36 (AlSCR36) orthologs. However, the AlSCR36 transcriptional unit could not be amplified using primers flanking the coding region (i.e., the primers used for amplification of SCRC variants from A. thaliana accessions) likely due to the divergence of these sequences in the two species. Therefore, a partial AlSCR36 sequence was amplified using primers complementary to the SCRC-Lz coding region (Table S1). As shown in Figure 2, and excluding sequences complementary to the primers, the A. thaliana SCRC amino-acid sequences differ at only two positions from AlSCR36: a conservative valine-to-isoleucine substitution at position 29 of AtSCRC-Lz and a glycine-to-serine substitution at position 38 in the Lz-0, Ita-0, Kas-2, and Pro-0 SCRC sequences. In addition, the AtSCRC-Lz intron sequence contains 18 nucleotide changes relative to AlSCR36.
Functional analysis of SRKC and SCRC sequences and the basis of self-fertility in the Lz-0 and Kas-2 accessions
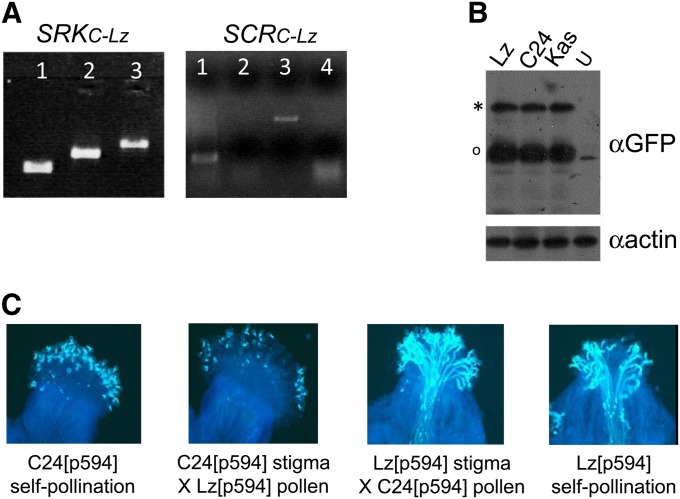
The sequence analyses described above indicate that both AtSRKC-Lz and AtSCRC-Lz encode apparently intact open reading frames. The two genes are also expressed, as determined by RT–PCR (Figure 4A). However, these features are not sufficient indicators of gene functionality, because of the extensive polymorphism characteristic of SRK and SCR variants and because the amino-acid residues that determine specificity in the SRK–SCR interaction are not known. Consequently, the functionality of SRKC and SCRC alleles must be determined in planta by performing pollination assays with tester lines expressing the A. lyrata S36 specificity in stigma and/or pollen.
Figure 4.
Expression of endogenous and transgenic SRK and SCR genes and presence of an SI modifier in Lz-0. (A) RT–PCR of AtSRKC-Lz transcripts in Lz-0 stigmas and of AtSCRC-Lz transcripts in young floral buds. The SRKC-Lz panel shows RT–PCR of AtSRKC-Lz transcripts from floral bud RNA using a forward primer derived from exon 1 in combination with a reverse primer derived from exon 3 (1), or exon 5 (2), or exon 7 (3): the resulting amplicons had the expected sizes for intronless fragments (579 bp, 1028 bp, and 1600 bp, respectively). The SCRC-Lz panel shows RT–PCR of AtSCRC-Lz transcripts from floral bud RNA (1), a no-reverse transcriptase control (2), PCR of genomic DNA (3), and RT–PCR of floral bud RNA from untransformed Lz-0 plants (4). (B) Immunoblot analysis of SRK protein in stigma extracts from a self-fertile Lz-0[AtS1pr::YFP:SRKb-SCRb] transformant and from self-incompatible C24[AtS1pr::YFP:SRKb-SCRb] and Kas[AtS1pr::YFP:SRKb-SCRb] transformants. The “U” lane contains proteins from the stigmas of untransformed Lz-0 plants. The SRKb protein was tagged with yellow fluorescent protein and visualized with anti-GFP antibodies. Antiactin antibodies were used to detect actin as a loading control. Note the equivalent amounts of SRKb in self-compatible and self-incompatible plants. The asterisk indicates the full-length SRK protein; the circle indicates the eSRK, a soluble form of the SRK ectodomain produced from an alternative SRK transcript corresponding to exon 1 and terminating within intron 1. (C) Pollination phenotype of Lz-0[AtS1pr::YFP:SRKb-SCRb] (designated p594) transformants. The absence of pollen tubes indicates an incompatible pollination while the growth of many pollen tubes indicates a compatible pollination.
Generation of tester lines that express S36 specificity
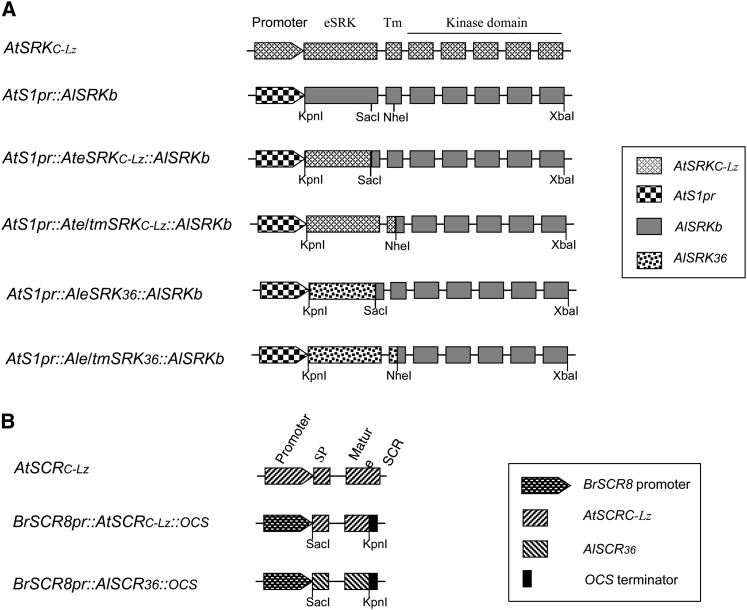
In the absence of genomic libraries for isolation of intact AlSRK36 and AlSCR36 genes, we constructed chimeric versions of these genes (Figure 5) using a strategy we previously described for expression of various SI specificities from A. lyrata or Capsella grandiflora in A. thaliana (Boggs et al. 2009b; File S1). In the case of AlSRK36, two chimeric genes were constructed, in each of which the stigma epidermal cell-specific AtS1 promoter was used to drive expression of an AlSRK36:AlSRKb fusion protein (Figure 5). One construct, AtS1pr::AleSRK36:AlSRKb, was designed to express a protein consisting of most of the extracellular domain of AlSRK36 (AleSRK36) excluding the C-terminal 23 amino acids, with the remainder of the protein derived from AlSRKb. In the second construct, AtS1pr::Ale/tmSRK36:AlSRKb, the predicted protein product consisted of the entire extracellular domain and 15 amino acids of the transmembrane domain of AlSRK36 (Ale/tmSRK36), with the remainder of the protein derived from AlSRKb (Figure 5). In the case of AlSCR36, we constructed BrSCR8pr::AlSCR36::ocs, a chimeric gene in which the promoter of the Brassica rapa SCR8 (BrSCR8) gene is used to drive expression of a mature AlSCR36 protein having 10 AtSCRC-derived amino acids at the C terminus due to incorporation of these sequences in the reverse primer used for amplification of AlSCR36 (Figure 2).
Figure 5.
Diagrams of the SRK (A) and SCR (B) transgenes used in this study.
Pollination assays of C24 plants transformed with these chimeric genes demonstrated that these genes are functional. In 12 of 13 AtS1pr::Ale/tmSRK36:AlSRKb and 4 of 6 AtS1pr::AleSRK36:AlSRKb transformants, the stigmas inhibited the germination of pollen from 8 of 11 BrSCR8pr::AlSCR36::ocs transformants analyzed. In all cases, control pollinations with pollen from untransformed C24 plants or C24[AtS1pr::YFP:SRKb-SCRb] plants produced large numbers of pollen tubes, indicating that the stigmas and pollen of AtS1pr::AleSRK36:AlSRKb, AtS1pr::Ale/tmSRK36:AlSRKb, and BrSCR8pr::AlSCR36::ocs transformants were normal and that these plants exhibited a bona fide SI response. Therefore, we established homozygous AtS1pr::AleSRK36:AlSRKb, AtS1pr::Ale/tmSRK36:AlSRKb, and BrSCR8pr::AlSCR36::ocs lines for use as sources of stigma and pollen in functional analyses of AtSRKC-Lz and AtSCRC-Lz sequences.
A modifier of SI in the Lz-0 genetic background
In principle, testing for the activity of the SRKC-Lz and SCRC-Lz genes would entail performing reciprocal pollinations between Lz-0 and the AlSRK36 and AlSCR36 tester lines. However, this approach is not feasible in the case of Lz-0 plants because they do not become self-incompatible even when transformed with SRK-SCR gene pairs that are known to be functional based on their ability to confer a robust and developmentally stable SI response in plants of the C24 accession (Nasrallah et al. 2004). For example, the previously described AtS1pr::YFP-SRKb/SCRb plasmid (Kitashiba et al. 2011), which contains the A. lyrata SCRb gene and a chimeric SRKb gene designed for stigma-specific expression of a yellow fluorescent protein (YFP)-tagged version of the A. lyrata SRKb protein, is expressed in Lz-0[AtS1pr::YFP:SRKb/SCRb] transformants at levels equivalent to those observed in the stigmas of C24[AtS1pr::YFP:SRKb/SCRb] and Kas[AtS1pr::YFP:SRKb/SCRb] plants (Figure 4B). Yet, this construct confers SI in C24 but not in Lz-0 transformants: in reciprocal pollinations, the pollen of Lz-0[AtS1pr::YFP:SRKb/SCRb] plants was inhibited on the stigmas of C24[AtS1pr::YFP:SRKb/SCRb] plants, but the stigmas of these plants failed to inhibit C24[AtS1pr::YFP-SRKb/SCRb] pollen (Figure 4C). Thus, the Lz-0 accession harbors a stigma-specific modifier of SI that disrupts the SI response without affecting the levels of SRK protein. In view of these results, we assayed for SRKC-Lz and SCRC-Lz function in the C24 genetic background as described below.
Analysis of transgenic plants expressing AtSRKC-Lz and AtSCRC-Lz sequences
C24 plants were transformed with the native AtSRKC-Lz and AtSCRC-Lz genes containing their 5′ and 3′ regulatory sequences (File S1 and Figure 5). However, pollinations of stigmas from 40 AtSRKC-Lz transformants with BrSCR8pr::AlSCR36::ocs pollen and pollinations using pollen from 19 AtSCRC-Lz transformants on AtS1pr::AleSRK36:AlSRKb or AtS1pr::Ale/tmSRK36:AlSRKb stigmas all produced large numbers of pollen tubes (Figure 6). Thus, the native AtSRKC-Lz and AtSCRC-Lz transgenes failed to confer SC specificity in stigma and pollen, respectively. Because this result might be due to mutations in the transcriptional units or regulatory elements of these genes, we transformed C24 plants with the AtS1pr::AteSRKC-Lz:AlSRKb, AtS1pr::Ate/tmSRKC-Lz:AlSRKb, and BrSCR8pr::AtSCRC-Lz::ocs chimeric genes (File S1), which are designed to assay directly for the function of the ligand-binding eSRKC domain of AtSRKC-Lz and of the AtSCRC-Lz transcriptional unit (Figure 5).
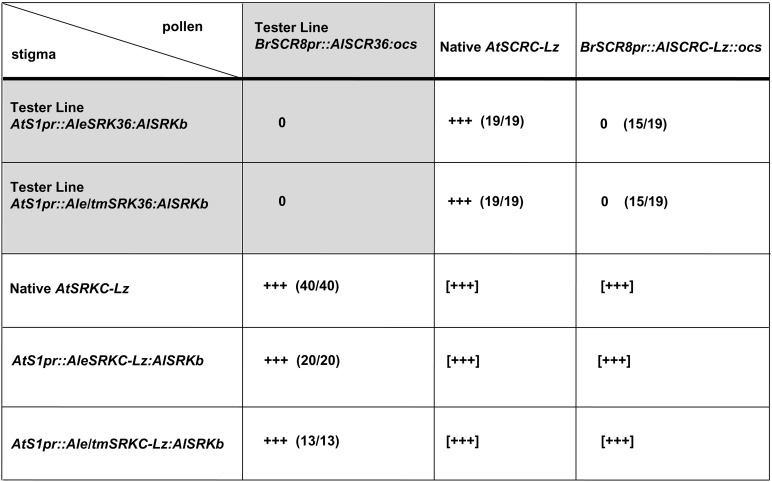
Figure 6.
Pollination analysis of C24 plants transformed with various SRKC-Lz and SCRC-Lz transgenes. Assays were performed by pollinating stigmas from the transformants shown in the first column with pollen from transformants shown in the top row. Shaded blocks show control pollinations of stigmas from the AtS1pr::AleSRK36::AlSRKb and AtS1pr::Ale/tmSRK36::AlSRKb tester lines with pollen from the BrSCR8pr::AlSCR36::ocs tester line. +++, compatible pollination (typically >50 pollen tubes per pollinated stigma); 0, incompatible pollination (typically <5 pollen tubes per pollinated stigma). The numbers in parentheses indicate the number of independent transformants exhibiting the indicated pollination phenotype of the total number of transformants analyzed. [+++] indicates compatible pollinations obtained by performing pollinations between six randomly chosen plants of each genotype.
The BrSCR8pr::AtSCRC-Lz::ocs proved to be functional, as the pollen of 15 of 19 BrSCR8pr::AtSCRC-Lz::ocs transformants analyzed was inhibited by the stigmas of the AtS1pr::AleSRK36:AlSRKb or AtS1pr::Ale/tmSRK36:AlSRKb tester lines (Figure 6). This result confirms that the AtSCRC gene is indeed functionally equivalent to the AlSCR36 gene and consequently, that the A. thaliana SC haplotype and the A. lyrata S36 haplotype encode the same SI specificity. It also demonstrates that the AtSCRC-Lz coding sequence has remained functional and that the two conservative amino-acid substitutions that differentiate AtSCRC-Lz from AlSCR36 do not disrupt SCR function. Further, it may be inferred that the coding sequence is also functional in the SCRC alleles of the Br-0, Bur-0, Ra-0, and Wt-5 accessions, all of which have the same SCR sequence as Lz-0, and likely also in the Kas-2 and Ita-0 accessions, in which the SCR sequences are even more closely related to AlSCR36.
Because replacement of the regulatory sequences of the AtSCRC-Lz 5′ and 3′ sequences with the BrSCR8 promoter and ocs terminator resulted in a functional transgene, we conclude that the nonfunctionality of the AtSCRC-Lz gene, both in its native Lz-0 background and as a transgene in C24 plants, is due to the presence in this gene of one or more mutation in cis-regulatory elements, most likely within the promoter region of the gene, causing suboptimal expression. This conclusion is supported by quantitation of AtSCRC-Lz transcripts by real-time PCR analysis (File S1). AtSCRC-Lz transcript levels were two orders of magnitude lower in Lz-0 and C24[AtSCRC-Lz] plants than the levels that proved effective in conferring SC specificity in the pollen of C24[BrSCR8pr::AtSCRC-Lz::ocs] transformants and also two orders of magnitude lower than the levels of endogenous SCR transcripts observed in the A. lyrata strains analyzed (Figure S5, A and B).
As for AtSRKC-Lz, neither the AtS1pr::AteSRKC-Lz:AlSRKb nor the AtS1pr::Ate/tmSRKC-Lz:AlSRKb transgenes proved to be functional in C24 plants. Indeed, the stigmas of these transformants failed to inhibit the pollen of BrSCR8pr::AtSCRC-Lz::ocs and BrSCR8pr::AlSCR36::ocs transformants (Figure 6), despite being expressed at levels comparable to those of the functional AtS1pr::AleSRK36:AlSRKb and AtS1pr::Ale/tmSRK36:AlSRKb genes and at much higher levels than the nonfunctional native AtSRKC-Lz gene (Figure S5C). Because the AteSRKC-Lz sequence failed to function even when linked to the functional kinase domain of SRKb, we conclude that the extracellular SCR-binding domain of AtSRKC-Lz has accumulated mutations that disrupt receptor function. These mutations might preclude the formation of productive SRKC-Lz/SCRC-Lz complexes either by disrupting the SCR-binding pocket or by changing the eSRK conformation. Comparison of the predicted eSRK amino-acid sequences of AlSRK36 and AtSRKC-Lz (Figure S3) reveals 40 differences, only 18 of which are conservative. At present, it is impossible to determine which of these amino-acid substitutions are disruptive because the residues required for SCR binding have not been identified, apart from the conclusion that three hypervariable regions are essential for receptor function (Boggs et al. 2009c). Only five substitutions, three of which are nonconservative, are found in these regions when AtSRKC-Lz and AlSRK36 are compared (Figure S3): two in hypervariable region II (isoleucine instead of methionine at position 281, and tyrosine instead of aspartic acid at position 301), and three in hypervariable region III (lysine instead of glutamic acid at position 327, valine instead of isoleucine at position 335, and serine instead of aspartic acid at position 341). Only the substitutions at residues 327 and 341 occur in all four sequenced AtSRKC variants, making these changes possible candidates for mutations that caused loss of SCRC recognition by SRKC.
It should be noted, however, that one or more of the other 19 nonconservative amino-acid changes found in the eSRK outside the hypervariable regions might also contribute to the nonfunctionality of the AtSRKC-Lz extracellular domain, especially if they alter folding of the eSRK. Similarly, although mutations in the eSRK that cause loss of SCR recognition are sufficient to abolish receptor function, we cannot exclude the possibility that additional mutations within other regions of the protein sequence in AtSRKC-Lz might also be functionally disruptive. For example, although all available AtSRKC sequences contain the invariant amino acids found in functional kinases, including those specific to serine/threonine kinases (Figure S3), the nonconservative substitutions that differentiate the AtSRKC sequences from AlSRK36 at residues adjoining these conserved regions might disrupt kinase activity. Furthermore, it is likely that mutations have occurred in regulatory sequences of the gene, in view of the very low levels of SRKC transcripts detected in Lz-0 stigmas by quantitative real-time RT–PCR (Figure S5C).
Functional analysis of the Sc haplotype of the Kas-2 accession
Unlike Lz-0, the Kas-2 accession exhibits SI upon transformation with functional SRK–SCR gene pairs (Boggs et al. 2009a). Thus, similar to the C24 accession, Kas-2 harbors a nonfunctional S haplotype and it lacks modifier loci that disrupt expression of the SI trait. Consequently, it is possible to determine if the nonfunctionality of the SC-Kas haplotype is due to disruption of the AtSRKC-Kas gene, the AtSCRC-Kas gene, or both by reciprocal pollination assays of untransformed Kas-2 plants with the C24[BrSCR8pr::AlSCR36::ocs] and C24[AtS1pr::AleSRK36:AlSRKb] tester lines. These pollination assays showed that Kas-2 stigmas failed to inhibit C24[BrSCR8pr::AlSCR36::ocs] pollen and Kas-2 pollen was not inhibited on C24[AtS1pr::AleSRK36:AlSRKb] stigmas. Moreover, transformation of Kas-2 plants with AtS1pr::AleSRK36:AlSRKb or with BrSCR8pr::AlSCR36::ocs did not cause these plants to become self-incompatible as might be expected if the endogenous SRKC-Kas or SCRC-Kas genes were functional. Thus, like their Lz-0 counterparts, the SRKC-Kas and SCRC-Kas alleles are nonfunctional pseudogenes.
Conclusions
The characteristics reported here for the A. thaliana SC haplotype and its SRKC and SCRC genes, together with previously described features of other A. thaliana S haplotypes, underscore the various ways in which the S locus and its genes were inactivated concomitant with, or subsequent to, the switch to self-fertility that occurred in the A. thaliana lineage. Previous studies had shown that in some accessions, such as C24, the SRK and SCR sequences are highly decayed or deleted, and only partial remnants of these sequences have been retained (Sherman-Broyles et al. 2007). In other accessions, SRK sequences are relatively well preserved while SCR sequences are very much degraded or entirely absent (Nasrallah et al. 2004; Sherman-Broyles et al. 2007). For example, in SA-containing accessions, SCRA sequences are nonfunctional and may be repeated, truncated, or rearranged (Nasrallah et al. 2004; Sherman-Broyles et al. 2007; Boggs et al. 2009a), while the SRKA gene has remained much more intact, being transcribed but encoding a truncated open reading frame due to splice site usage as in the Col-0 accession (Kusaba et al. 2001; Shimizu et al. 2004; Tang et al. 2007), or even remaining functional as in the Wei-0 accession (Tsuchimatsu et al. 2010). The situation is reversed in other accessions: for example, in Cvi-0, SRKB encodes a truncated open reading frame (Shimizu et al. 2004) and is thus more decayed than SCRB, which encodes a full-length open reading frame (Tang et al. 2007; Shimizu et al. 2008) but is nonfunctional due to inactivating amino-acid substitutions within the predicted mature SCR protein (Boggs et al. 2009b). The SC haplotype of Lz-0 and of several other SC-containing accessions, presents yet another situation whereby both SRKC and SCRC sequences contain full-length open reading frames. While the predicted SRKC-Lz protein contains disruptive mutations within its extracellular SCR-binding domain and possibly also within other regions of the protein, the SCRC-Lz protein has remained functional, but exhibits suboptimal expression. Although the data do not allow us to infer the sequence of events leading to inactivation of the SC haplotype, they do provide further support for the conclusion that inactivation of the S locus occurred relatively recently in A. thaliana. They also provide the first example of an A. thaliana SCR gene that, instead of being severely decayed, encodes a functional protein but is unable to confer Sc specificity on pollen due to mutations in its regulatory elements.
Acknowledgments
We thank Tiffany Crispell for technical assistance, Jesper Bechsgaard and Mikkel Schierup for A. lyrata DNA, the Arabidopsis Biological Resource Center for seed from A. thaliana accessions, and two anonymous reviewers whose comments improved the manuscript. This article is based upon work supported by National Science Foundation grants IOS-0744579 and IOS-1146725. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Communicating editor: D. Charlesworth
Literature Cited
- Bechsgaard J. S., Castric V., Charlesworth D., Vekemans X., Schierup M. H., 2006. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 23: 1741–1750. [DOI] [PubMed] [Google Scholar]
- Boggs N. A., Nasrallah J. B., Nasrallah M. E., 2009a Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 5(3): e1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs N. A., Dwyer K. G., Shah P., McCulloch A. A., Bechsgaard J., et al. , 2009b Expression of distinct self-incompatibility specificities in Arabidopsis thaliana. Genetics 182: 1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs N. A., Dwyer K. G., Nasrallah M. E., Nasrallah J. B., 2009c In vivo detection of residues required for ligand-selective activation of the S-locus receptor in Arabidopsis. Curr. Biol. 19: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Mable B. K., Schierup M. H., Bartolome C., Awadalla P., 2003. Diversity and linkage of genes in the self-incompatibility family in Arabidopsis lyrata. Genetics 164: 1519–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer K. G., Kandasamy M. K., Mahosky D. I., Acciai J., Kudish B. I., et al. , 1994. A superfamily of S locus-related sequences in Arabidopsis: diverse structures and expression patterns. Plant Cell 6: 1829–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxe J., Slotte T., Stahl E. A., Neuffer B., Hurka H., et al. , 2009. Recent speciation associated with the evolution of selfing in Capsella. Proc. Natl. Acad. Sci. USA 106: 5241–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet P., Berges H., Bellec A., Prat E., Helmstetter N., et al. , 2012. Contrasted patterns of molecular evolution in dominant and recessive self-incompatibility haplotypes in Arabidopsis. PLoS Genet. 8(3): e1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Bechsgaard J. S., Slotte T., Neuffer B., Lascoux M., et al. , 2009. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl. Acad. Sci. USA 106: 5246–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad J., Bechsgaard J., Charlesworth D., 2006. Linkage disequilibrium between incompatibilty locus region genes in the plant Arabidopsis lyrata. Genetics 173: 1057–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitashiba H., Lui P., Nishio T., Nasrallah J. B., Nasrallah M. E., 2011. Functional test of Brassica self-incompatibility modifiers in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 18173–18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O., Gentles A. J., Hankus L., Jurka J., 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Schell J., 1986. The promoter of TL-DNA gene 5 controls the tissue–specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396. [Google Scholar]
- Kusaba M., Dwyer K. G., Hendershot J., Vrebalov J., Nasrallah J. B., et al. , 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13: 627–643. [PMC free article] [PubMed] [Google Scholar]
- Liu P., Sherman-Broyles S., Nasrallah M. E., Nasrallah J. B., 2007. A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr. Biol. 17: 734–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah M. E., Liu P., Nasrallah J. B., 2002. Generation of self- incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297: 247–249. [DOI] [PubMed] [Google Scholar]
- Nasrallah M. E., Liu P., Sherman-Boyles S., Boggs N. A., Nasrallah J. B., 2004. Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc. Natl. Acad. Sci. USA 101: 16070–16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea A. C., Nasrallah J. B., 2008. Self-incompatibility systems: barriers to self-fertilization in flowering plants. Int. J. Dev. Biol. 52: 627–636. [DOI] [PubMed] [Google Scholar]
- Sato K., Nishio T., Kimura R., Kusaba M., Suzuki T., et al. , 2002. Coevolution of the S-locus genes SRK, SLG, SP11/SCR in Brassica oleracea and B. rapa. Genetics 162: 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman-Broyles S., Boggs N., Farkas A., Liu P., Vrebalov J., et al. , 2007. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell 19: 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K. K., Cork J. M., Caicedo A. L., May C. A., Moore K. M., et al. , 2004. Darwinian selection on a selfing locus. Science 306: 2081–2083. [DOI] [PubMed] [Google Scholar]
- Shimizu K. K., Shimizu-Inatsugi R., Tsuchimatsu T., Purugganan M. D., 2008. Independent origins of self-compatibility in Arabidopsis thaliana. Mol. Ecol. 17: 704–714. [DOI] [PubMed] [Google Scholar]
- Shiu S.-H., Bleecker A. B., 2001. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.-H., Bleecker A. B., 2003. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132: 530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. R., Bowman J. L., Meyerowitz E. M., 1990. Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Toomajian C., Sherman-Broyles S., Plagnol V., Guo Y. L., et al. , 2007. The evolution of selfing in Arabidopsis thaliana. Science 317: 1070–1072. [DOI] [PubMed] [Google Scholar]
- Tsuchimatsu T., Suwabe K., Shimizu-Inatsugi R., Isokawa S., Pavlidis P., et al. , 2010. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464: 1342–1346. [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S. S., Niu Q. W., Chua N. H., 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]