Abstract
Bacteria of the genus Arthrobacter are common inhabitants of the soil environment, but can also be recovered from leaf surfaces (the phyllosphere). Using enrichment cultures on 4-chlorophenol, we succeeded in specifically isolating Arthrobacter bacteria from ground cover vegetation in an apple orchard. Based on 16S rRNA gene sequencing, the isolates were found to belong to at least three different species of Arthrobacter. Compared to the model bacterial epiphyte Pantoea agglomerans, the Arthrobacter isolates performed as well or even better in a standardized laboratory test of phyllosphere fitness. A similar performance was observed with the well-characterized soil isolate Arthrobacter chlorophenolicus A6. These findings suggest that the frequently reported presence of Arthrobacter strains on plant foliage can be explained by the capacity to multiply and persist in the phyllosphere environment. As bacteria from the genus Arthrobacter are known for their ability to degrade a wide variety of organic pollutants, their high phyllosphere competency marks them as a promising group for future studies on phyllosphere-based bioremediation, for example, as foliar bioaugmentation on ground cover or buffer-zone vegetation to prevent pesticides from reaching soil, surface-, or groundwater.
Keywords: Arthrobacter, biodegradation, phylloplane, phylloremediation, soil, triadimenol
Introduction
The phyllosphere (Ruinen 1961) is an open habitat that harbors large and diverse communities of bacteria, fungi, and other microorganisms (Leveau 2006). One of the bacterial genera that show up frequently in culture-independent surveys of leaf surface microbiota is Arthrobacter (high %GC Gram-positive, family Micrococcaceae, order Actinomycetales, phylum Actinobacteria). For example, on the leaves of harvest-ready lettuce plants, Arthrobacter sequences were found consistently across samples (Rastogi et al. 2012). Arthrobacter strains have also been isolated from leaves of strawberry (Krimm et al. 2005), sugar beet (Thompson et al. 1995), potato (Heuer and Smalla 1999), the resurrection fern Polypodium polypodioides (Jackson et al. 2006), and olive trees (Ercolani 1991).
Thus, the presence of Arthrobacter on leaf surfaces is an established aspect of phyllosphere microbiology. We are interested in the drivers that underlie this presence, which evokes the basic question whether bacteria from the genus Arthrobacter constitute so-called residual or transient epiphytes (Whipps et al. 2008). Classical examples of residuals are representatives of the genera Pseudomonas, Pantoea, and Erwinia: they are defined (Whipps et al. 2008) by the capacity to multiply in the phyllosphere (Manulis et al. 1998; Mercier and Lindow 2000; Sabaratnam and Beattie 2003). By contrast, transients lack this capacity. For example, Bacillus species have been shown to be poor leaf colonizers even under conducive laboratory conditions (Maduell et al. 2008).
Given their ability to multiply, one expects residual epiphytes to be more abundantly represented than transients in the bacterial communities on plant foliage. Indeed, for genera such as Pseudomonas, Pantoea, and Erwinia, this tends to hold true (Leveau and Tech 2011; Yashiro et al. 2011; Rastogi et al. 2012). Lacking the ability to produce offspring, transients are more likely to be part of the “rare biosphere” component (Kunin et al. 2010) of bacterial communities on plant leaves. This, however, is not a general rule. For example, bacteria of the genus Bacillus can constitute a significant portion of the leaf microbiota (Leveau and Tech 2011). This can be explained by assuming high immigration rates of these bacteria to the leaf surface from other sources, rather than multiplication on the leaf surface (Maduell et al. 2008).
Immigration from soil represents one likely mechanism to explain the presence of Arthrobacter on leaf surfaces of plants. Arthrobacter species are abundant in soil (Mongodin et al. 2006) and soil particles are common on foliage of plants that are grown outdoors (Monier and Lindow 2004). Wind and rain splatter may deliver soil particles to leaf surfaces, especially if the leaves are close to the soil line. In a study that compared bacterial diversity of the lettuce phyllosphere to that of the soil in which these plants were grown, it was revealed that many bacterial species were common between the two compartments (Zwielehner et al. 2008). This was taken as indirect evidence for the movement of soil bacteria to the lettuce canopy. The transport of bacteria by soil particles across larger spatial scales has also been documented (Hua et al. 2007; Polymenakou et al. 2008).
A second contributing factor to the foliar presence of Arthrobacter would be the capacity of Arthrobacter to multiply in the phyllosphere. To the best of our knowledge, a test of such capacity, that is a test of Arthrobacter's residual nature, has not yet been reported. Demonstration of high epiphytic fitness for Arthrobacter would constitute an important finding toward the broader and longer term goal of elucidating the assembly rules that shape phyllosphere communities (Meyer and Leveau 2012).
A particularly interesting property of Arthrobacter species is that they can degrade a wide variety of organic pollutants. These include aromatic hydrocarbons, such as phenols, chlorophenols, BTEX compounds, and phenanthrene (Alvarez and Vogel 1991; Keuth and Rehm 1991; Westerberg et al. 2000; Kotouckova et al. 2004), s-triazines such as atrazine and cyanazine, phenylurea herbicides, glyphosate, and malathion (Kertesz et al. 1994; Strong et al. 2002; Tixier et al. 2002). Nicotine-degrading Arthrobacter strains have been isolated from the tobacco phyllosphere (Sguros 1955) and oil-utilizing Arthrobacter bacteria were isolated from the phyllosphere of crops grown on oil-contaminated soil (Al-Awadhi et al. 2009).
We report here the targeted isolation of Arthrobacter strains from leaf surfaces by exploitation of the fact that Arthrobacter species can grow at the expense of aromatic pollutants including 4-chlorophenol (4-CP). We used 4-CP enrichment cultures to isolate Arthrobacter strains from plant leaves in an apple orchard and we confirmed their epiphytic fitness in laboratory tests. We discuss our findings in the context of exploiting culturable Arthrobacter strains for phylloremediation (Sandhu et al. 2007), that is the removal of foliage-associated organic pollutants by members of the phyllosphere community.
Materials and Methods
Sampling
Epiphytic bacteria were recovered from foliage at an experimental apple orchard (Applied Plant Research or PPO, Randwijk, The Netherlands), which had received weekly treatments with the foliar fungicide triadimenol (Exact®; Bayer CropScience B.V., Monheim, Germany). One of the main photodegradation products of triadimenol is 4-CP (Wang and Lemley 2003; Da Silva and Vieira Ferreira 2004). From each one of six plots (A–F), a composite sample consisting of 16 apple leaves and a composite sample consisting of ground cover (i.e., grass and herb vegetation dominated by Poa pratensis, common meadow grass; Poa annua, annual meadow grass; Stellaria media, common chickweed; Senecio vulgaris, common groundsel) was weighed at 10.9 ± 1.0 and 7.3 ± 1.3 g (average ± standard deviation), respectively, and washed in 100 mL phosphate-buffered saline (PBS) by vortexing (5 sec), sonication (7 min), and vortexing again (5 sec). Leaf washes were concentrated 17-fold by centrifugation and resuspended in PBS, and 0.75-mL aliquots were used to inoculate 15 mL Brunner mineral medium (MM; DSMZ medium no. 457, Braunschweig, Germany) containing 1 mmol/L 4-CP or 0.3 mmol/L triadimenol (Sigma-Aldrich, Zwijndrecht, The Netherlands). Media to which no bacteria were added served as controls. Bottles were incubated at 25°C while shaking at 150 rpm for 4 weeks. Every week, 1 mL of culture was collected and frozen at −20°C for high-performance liquid chromatography (HPLC) analysis of 4-CP and triadimenol.
HPLC measurements
Frozen samples were thawed, filtered over a 0.2-μm filter, and analyzed by HPLC. We used an ASI-100/ASI-100T Autosampler, STH 585 Column Thermostat, UVD 170U/340U UV/VIS Detector, and P680 LPG pump (Dionex, München, Germany). The UV detector was set at 227 nm for 4-CP and at 224 nm for triadimenol. Runs were performed on a reverse phase C-18 column, 3 μm, 150 × 4.6 mm (Grace Davison Discovery Science, Deerfield, IL) at a column temperature of 25°C and a flow rate of 1 mL per min with 50% acetonitrile as the eluent. The injection volume was 25 μL for the 4-CP and 50 μL for the triadimenol samples.
Isolation of bacteria
After 2 weeks of enrichment, serial dilutions of the 4-CP cultures that were inoculated with bacteria from the grass–herb mixture were spread on 1/10 Tryptone Soy Agar (TSA; Oxoid, Cambridge, UK) with 15 g agar per liter. For each one of the six cultures, 12–16 single colonies were transferred to fresh TSA plates and restreaked twice for purity. Care was taken to include a representative from each morphologically distinct colony type. Each isolated strain was checked for its ability to grow in MM with 1 mmol/L 4-CP. Sixteen of those that did were selected for characterization by 16S rRNA gene amplicon sequencing using primers 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′; Lane 1991), and for which primer 1492r was used as the sequencing primer. DNA sequences were aligned with those from closely related type strains of Arthrobacter (Genbank accession numbers AB279889, AB279890, AF102267, AJ512504, X83405, X83406, X80741, X80743, and X83408) over a length of 602 nt in MegAlign (Lasergene; DNAstar, Madison, WI) using the Clustal W algorithm. Rhodococcus pyridinivorans (AF173005) was used as an outlier. TreeView (Page 1996) was used to display the phylogram. Unique sequences were deposited in GenBank under accession numbers JN944570–JN944572.
Growth in liquid medium
Three selected phyllosphere isolates, cp10, cp12, and cp15, were compared with Arthrobacter chlorophenolicus A6 (DSMZ culture collection, Braunschweig, Germany; Westerberg et al. 2000) for growth in MM supplemented with 1 mmol/L 4-CP as the sole source of carbon and energy. Growth was followed by measuring the optical density at 600 nm (OD600) as a function of time. The experiment was performed in triplicate.
Phyllosphere performance test
Spontaneous rifampicin-resistant mutants of cp10, cp12, cp15, and A6 were selected by plating on Luria broth (LB) medium with 15 g agar and 20 mg rifampicin per liter. These derivatives and the Rif-resistant model phyllosphere colonizer Pantoea agglomerans (synonym: Erwinia herbicola) 299R (Brandl et al. 1996) were grown to mid-exponential phase in LB with 20 mg rifampicin per liter at 28°C and 250 rpm and diluted in sterile demineralized water to obtain bacterial suspensions of approximately 1.7 × 104 colony forming units (CFUs)/mL. Two-week-old bean plants (Phaseolus vulgaris, green snap bean, variety Blue Lake Bush 274) with the first two leaves fully expanded were dipped into the bacterial suspension. The plants were then incubated for 1 day at 97% air humidity in a closed box in a growth chamber, followed by 1 day at 50% air humidity in an open box and one more day back at 97% air humidity. The growth chamber was set to maintain a day–night cycle of 16 and 8 h at 21 and 16°C, respectively. Growth and survival of bacteria on the foliage was monitored by sacrificing four leaves for analysis at each time point. Bacteria were recovered from individual leaves in 20 mL PBS by 5-sec vortexing, 7-min sonication, and 5-sec vortexing. Dilutions were spread on LB plates containing rifampicin, and CFUs were counted and normalized per gram of leaf tissue.
In a second test, phyllosphere performance of strain cp15 was compared to that of P. agglomerans 299R, either inoculated separately or mixed in a 1:1 ratio. Bacterial suspensions used for dipping the bean leaves contained approximately 3.3 × 106 CFU/mL of each strain. The inoculation densities were 100-fold higher than in previous experiment in order to ensure interaction between the two strains. On LB agar plates, both strains could easily be distinguished by morphology.
4-Chlorophenol-degradation genes
Primer sets were designed to target homologues of three genes in the A. chlorophenolicus A6 4-CP-degradation cluster, namely Achl_4569 (cphA-I), Achl_4573 (cphC-I), and Achl_4564 (cphC-II). These genes encode for one hydroxyquinol 1,2-dioxygenase and two monooxygenase enzymes (Nordin et al. 2005; Unell et al. 2009). Homologous sequences were obtained from GenBank, aligned using MegAlign (Lasergene; DNAstar) and used to design degenerate primers in conserved regions (Table 1). Genomic DNA from strains cp10, cp12, and cp15 was isolated using the ZR Fungal/Bacterial DNA MiniPrep (Zymo Research, Irvine, CA) after prior incubation for 30 min in TE buffer (30 mmol/L Tris-Cl, 1 mmol/L EDTA, pH 8.0) containing 15 mg of lysozyme and 2 mg of proteinase K per mL. PCR mixtures contained 1 U FastStart Taq DNA polymerase, 1× buffer (Roche Diagnostics, Mannheim, Germany), 0.2 mmol/L of each deoxynucleoside triphosphate, 3 μmol/L of each primer, and 5 ng of genomic DNA in a total volume of 25 μL. The PCR cycling regime was (1) one cycle of 2 min at 95°C, (2) 35 cycles of 30 sec at 95°C, 30 sec at 50°C, and 60 sec at 72°C, and (3) one final extension cycle of 10 min at 72°C. PCR products were verified by agarose gel electrophoresis and purified using a Qiaquick PCR purification kit (Qiagen, Venlo, The Netherlands). The fragments were sequenced (Macrogen, Seoul, Korea) from both directions with the same primers used for PCR amplification. Sequences were deposited in GenBank under accession numbers JN944561–JN944563 and JN944566–JN944569.
Table 1.
Degenerate primers used to amplify Arthrobacter genes involved in 4-chlorophenol degradation
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Size in A6 (bp) | NCBI entries used for alignment |
|---|---|---|---|---|
| cphA-I | CARYTNATGCARGCNYTNAC | CRTCYTCRTCNGCYTCCCA | 385 | YP_002478496; ABL75139; ACX85436; BAI53128 |
| cphC-I | ATGAAYGTNGTNATGTTYAC | GRTACCAYTTNGCRTGNTC | 538 | YP_002478500; ABL75143; ACX85440; BAI53124; BAI53132 |
| cphC-II | GCNCAYATHACNAAYCARMG | GCYTTRAANGTDATRTTCAT | 485 | YP_002478491; YP_831411 |
Results and Discussion
Leaf surface washes from trees and ground vegetation at six plots in an experimental apple orchard were used to seed two sets of enrichment cultures, one with 4-CP and one with triadimenol. Turbidity representing bacterial growth was observed only in the six 4-CP enrichment cultures that were inoculated with bacteria from ground vegetation. Analysis of the supernatants of these cultures by HPLC showed that 4-CP concentrations had fallen below 10 μmol/L within 2 weeks. At this time, enrichments were spread on 1/10 TSA plates and for each one of the six cultures 12–16 bacterial colonies were selected. For more than half of the isolates, we confirmed the ability to grow on MM containing 1 mmol/L 4-CP as the sole source of carbon and energy (Table 2). Of these 4-CP degraders (“cp isolates”), approximately 25% featured a yellow colony type, while the others were white. Sixteen of the cp isolates were selected for characterization by 16S rRNA gene amplicon sequencing and all were identified as Arthrobacter species, belonging to one of three groups (A, B, or C), based on alignment to the 16S rRNA gene sequences of known type strains of Arthrobacter species (Fig. 1). Members of group A showed 100% sequence similarity to 4-nitroguaiacol degrader Arthrobacter nitroguajacolicusT (Kotouckova et al. 2004) and atrazine-degrader Arthrobacter aurescens TC1 (Strong et al. 2002; Mongodin et al. 2006), both soil isolates. All strains with the yellow colony phenotype belonged to this group A. Sequences in group B were identical to those of Arthrobacter polychromogenesT (Schippers-Lammertse et al. 1963) and Arthrobacter oxydans, both of which were isolated from air, whereas sequences in group C were identical to that of Arthrobacter humicolaT, which was recovered from paddy soil (Kageyama et al. 2008).
Table 2.
Bacterial isolates obtained from leaf wash enrichment cultures on 4-chlorophenol (4-CP)
| Growth on 4-CP | No growth on 4-CP | ||
|---|---|---|---|
| Yellow colonies | White colonies | Various morphologies | |
| Plot A | 2 (cp15,18)1 | 8 (cp10,12) | 2 |
| Plot B | 1 (cp27) | 6 (cp23,32) | 5 |
| Plot C | 5 (cp34,49) | 4 (cp41) | 7 |
| Plot D | 2 (cp50) | 7 (cp61) | 3 |
| Plot E | 0 | 3 (cp64,65) | 9 |
| Plot F | 0 | 6 (cp74,76) | 7 |
In parentheses are shown the isolates that were selected for characterization by sequencing of the 16S rRNA gene (Fig. 1).
Figure 1.
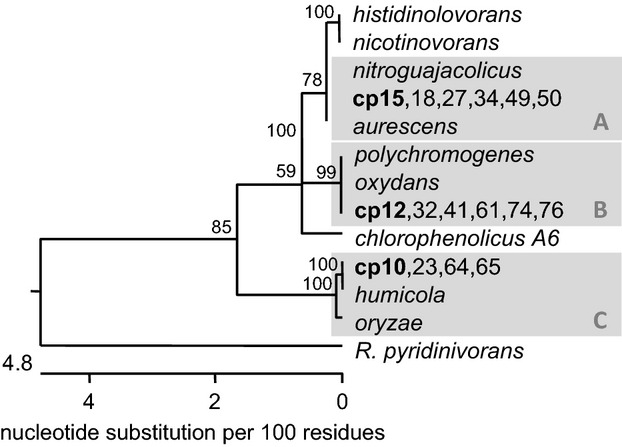
Phylogram based on the alignment of partial 16S rRNA gene sequences of cp isolates and closely related type strains of Arthrobacter. The sequence of Rhodococcus pyridinivorans was used as outlier. Highlighted boxes indicate three groups (A, B, C) of identical cp sequences. Numbers at nodes indicate % bootstrap values (n = 1000 trials).
From each group, one representative was selected for further characterization: cp15 representing group A, cp12 from B, and cp10 from group C. Figure 2 shows the growth of these isolates on MM with 1 mmol/L 4-CP, compared to that of 4-CP model degrader A. chlorophenolicus A6 (DSMZ culture collection, Braunschweig, Germany; Westerberg et al. 2000). Arthrobacter chlorophenolicus A6 was the fastest growing strain in this medium and cp12 the slowest. The maximum specific growth rates (μmax) for A6, cp10, cp12, and cp15 were 0.118, 0.099, 0.062, and 0.089/h, respectively. In addition, we confirmed by PCR analysis that cp10, cp12, and cp15 carried orthologs of cphA-I and cphC-I. These genes encode an intradiol dioxygenase and a monooxygenase, respectively, alleged to be involved in 4-CP degradation by A. chlorophenolicus A6 (Nordin et al. 2005). PCR for the cphC-II gene, encoding another monooxygenase, was positive only for cp12 (Table 3).
Figure 2.
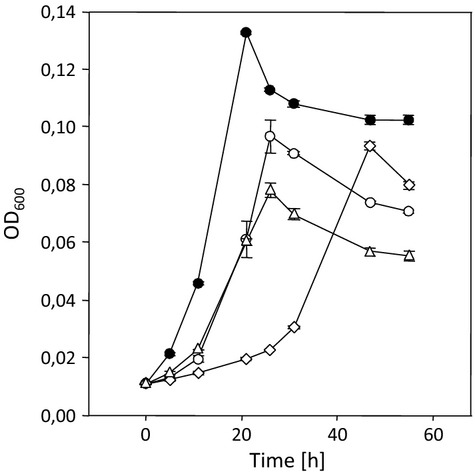
Growth of phyllosphere isolates cp10 (open circles), cp12 (open diamonds), and cp15 (open triangles), compared to that of the 4-chlorophenol (4-CP) model degrader Arthrobacter chlorophenolicus A6 (closed circles) on mineral medium supplemented with 1 mmol/L 4-CP as the sole source of carbon and energy. Error bars represent standard deviations, n = 3.
Table 3.
Partial gene fragments of cphA-I, cphC-I, and cphC-II orthologs amplified by PCR from cp isolates
| Isolate | Gene | Closest match in GenBank | Species | % Identity |
|---|---|---|---|---|
| cp10 | cphA-I | AB530681 (2769..2424) | Arthrobacter sp. IF1 | 100 |
| cp10 | cphC-I | AB530680 (1555..2053) | Arthrobacter sp. IF1 | 100 |
| cp12 | cphA-I | CP001343 (81639..81294) | Arthrobacter chlorophenolicus A6 | 80 |
| cp12 | cphC-I | CP001343 (86948..87446) | A. chlorophenolicus A6 | 85 |
| cp12 | cphC-II | CP001343 (75047..75491) | A. chlorophenolicus A6 | 76 |
| cp15 | cphA-I | AB530681 (2769..2424) | Arthrobacter sp. IF1 | 100 |
| cp15 | cphC-I | AB530681 (8077..8575) | Arthrobacter sp. IF1 | 100 |
These results confirmed that enrichment on 4-CP was very effective in selectively recovering Arthrobacter species from leaf surfaces, at least from ground vegetation in this apple orchard. Prior to the collection of leaf material, the orchard had received weekly applications of the fungicide Exact®, which has triadimenol as an active ingredient. We never found bacterial degraders of triadimenol in enrichment cultures supplemented with triadimenol as sole source of carbon and energy. Under the influence of sunlight, triadimenol can be degraded to 1,2,4-triazole with the release of 4-CP (Iesce et al. 2003; Da Silva and Vieira Ferreira 2004). We do not know to what extent the treatment with triadimenol allowed for the recovery of 4-CP degraders in our study. However, we were never able to recover such degraders from enrichment cultures that were seeded with apple leaves from the same plots. We suspect that the recovery of 4-CP-degrading Arthrobacter strains from ground vegetation but not apple leaves was due to the proximity of the former to soil.
To determine whether our cp isolates represented mere “soil contaminants,” that is transients, or whether they were actually capable of growing epiphytically, we compared isolates cp10, cp12, cp15, and A. chlorophenolicus A6 to a model bacterium for phyllosphere colonization, that is P. agglomerans 299R, in a standard “wet-dry-wet” phyllosphere competency test (Lindow 1993) on bean plants. The results are shown in Figure 3a. Overall, the phyllosphere performance of the four Arthrobacter strains resembled that of P. agglomerans 299R. In all cases, population sizes had increased at least one order of magnitude after 24 h under conditions of high relative humidity, suggesting that these strains were able to access and utilize nutrients on the leaf surface for growth. All strains showed the expected reduction in population size upon exposure to reduced humidity, while P. agglomerans 299R and two of the Arthrobacter isolates (cp 15 and A6) recovered from this stress by increasing population sizes in the subsequent 24-h wet period. Strain cp15 appeared to outperform all other strains, including P. agglomerans 299R, especially in the first 24-h period. In an additional experiment, phyllosphere performance of cp15 was compared to that of P. agglomerans 299R, alone or in competition at high densities (Fig. 3b). Again, the cp15 strain reached higher numbers than 299R, even when they were inoculated together on the same leaf. For both strains, inoculation together with the other strain did not impact the bacterial growth pattern compared to inoculation alone (Fig. 3b).
Figure 3.
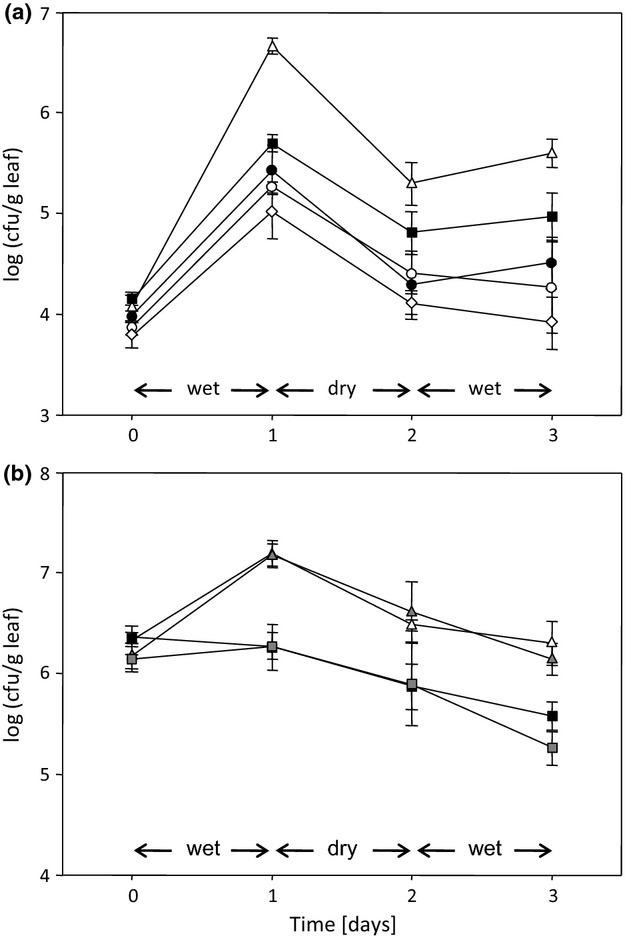
(a) Phyllosphere performance of Arthrobacter isolates cp10 (open circles), cp12 (open diamond), and cp15 (open triangles), compared to that of soil bacterium Arthrobacter chlorophenolicus A6 (closed circles), and phyllosphere bacterium Pantoea agglomerans 299R (closed squares). (b) Phyllosphere performance of Arthrobacter isolate cp15 (triangles) and P. agglomerans 299R (squares) at high densities, inoculated either alone (cp15 = open symbols; 299R = closed symbols) or in competition with each other (gray symbols). The graphs show bacterial numbers on bean leaves after 1 day at 97% relative humidity (“wet”), 1 day at 50% relative humidity (“dry”), and another day at 97% relative humidity (“wet”). Error bars represent standard deviations, n = 4.
We conclude then that the Arthrobacter strains that were isolated from orchard ground vegetation were good phyllosphere colonizers. The same was true for strain A6, which was originally recovered from soil. It is likely that many of the traits that generally make Arthrobacter species excellent survivors in soil (Mongodin et al. 2006), for example desiccation tolerance (Labeda et al. 1976), contributed to the ability of A6 and the cp isolates to deal with and rebound from the imposed stress of low relative humidity on leaf surfaces (Fig. 3). Remarkable is our finding that strains of Arthrobacter excelled at reproducing on leaf surfaces under growth-conducive conditions. It is unlikely that this ability depended on the capacity to catabolize 4-CP. In culture, isolate cp15 had the lowest yield on 4-CP, relative to other Arthrobacter strains (Fig. 2), yet it showed superior epiphytic growth during the first 24 h on bean leaves. Also, we were not able to recover 4-CP from the surface of bean leaves at concentrations that were detectable by GC-MS (not shown) or that would allow population increases such as those that were seen on leaves for cp10, cp12, and cp15 during the first 24 h on the bean leaf surface (Fig. 3). Thus, foliar growth by Arthrobacter under these conditions must be attributed to the acquisition of other carbon and energy sources on the leaf surface, most likely photosynthates such as fructose, glucose, and sucrose, which are among the most abundant sources of carbon on leaf surfaces (Leveau 2006). This does not preclude the possibility that on other plant species, in particular those that are known to harbor phenolic compounds on their leaf surfaces (Yadav et al. 2005), Arthrobacter would benefit from the possession of cph genes and the ability to catabolize substituted phenols. Moreover, as it is likely that phyllosphere Arthrobacter bacteria spend part of their life cycle in the soil environment, carrying cph genes might be advantageous for degradation of other aromatic compounds in soil, for example those formed during degradation of lignin and humic acids.
In conclusion, our findings show that members of the genus Arthrobacter fit the definition of residual epiphytes and that the presence of Arthrobacter on leaf surfaces should be interpreted in light of the demonstrated capacity to reproduce epiphytically. This capacity, together with the notions that Arthrobacter species (1) exhibit high levels of resistance to desiccation stress (Labeda et al. 1976), (2) have a wide range of pollutant degradation capabilities (Alvarez and Vogel 1991; Keuth and Rehm 1991; Westerberg et al. 2000; Kotouckova et al. 2004), and (3) can be retrieved as culturable bacteria from leaf surfaces (this study; Ercolani 1991; Thompson et al. 1995; Heuer and Smalla 1999; Krimm et al. 2005; Jackson et al. 2006), makes the Arthrobacter genus a promising group for further development as a model for the study of phyllosphere-based bioremediation (Sandhu et al. 2007). Using other bacterial species, such phylloremediation has been demonstrated for a number of pollutants such as toluene, phenol, and phenanthrene (De Kempeneer et al. 2004; Sandhu et al. 2007, 2009; Waight et al. 2007; Yutthammo et al. 2010), as well as for agrochemicals such as dichlorvos and acetamiprid (Ning et al. 2010; Zhou et al. 2011). Phyllosphere isolates of Arthrobacter strains may have practical utility as foliar sprays for the initial or accelerated attrition of pesticide residue associated with the use of atrazine, cyanazine, phenylurea herbicides, glyphosate, and malathion, all of which have been reported to be targets for destruction by Arthrobacter (Kertesz et al. 1994; Strong et al. 2002; Tixier et al. 2002). Application of biodegradation-capable, phyllosphere-competent strains of Arthrobacter to ground cover or buffer-zone vegetation may be a sustainable strategy to mitigate and reduce levels of environmental contamination associated with runoff of pesticides (Reichenberger et al. 2007).
Acknowledgments
We thank Bart Heijne from Applied Plant Research (PPO), Randwijk, The Netherlands, for his help and advice on the sampling site. We thank Robin Tecon for his comments on an early version of the manuscript. This work was supported as part of the BACSIN project within the 7th framework program of the European Union. This is NIOO-KNAW publication 5386.
Conflict of Interest
None declared.
References
- Al-Awadhi H, El-Nemr I, Mahmoud H, Sorkhoh NA, Radwan SS. Plant-associated bacteria as tools for the phytoremediation of oily nitrogen-poor soils. Int. J. Phytorem. 2009;11:11–27. [Google Scholar]
- Alvarez PJJ, Vogel TM. Substrate interactions of benzene, toluene, and para-xylene during microbial degradation by pure cultures and mixed culture aquifer slurries. Appl. Environ. Microbiol. 1991;57:2981–2985. doi: 10.1128/aem.57.10.2981-2985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl M, Clark EM, Lindow SE. Characterization of the indole-3 acetic acid (IAA) biosynthetic pathway in an epiphytic strain of Erwinia herbicola and IAA production in vitro. Can. J. Microbiol. 1996;42:586–592. [Google Scholar]
- Da Silva JP, Vieira Ferreira LF. Surface photochemistry of pesticides: an approach using diffuse reflectance and chromatography techniques. Environ. Sci. Technol. 2004;38:2849–2856. doi: 10.1021/es0348501. [DOI] [PubMed] [Google Scholar]
- De Kempeneer L, Sercu B, Vanbrabant W, Verstraete H, Van Langenhove W. Bioaugmentation of the phyllosphere for the removal of toluene from indoor air. Appl. Microbiol. Biotechnol. 2004;64:284–288. doi: 10.1007/s00253-003-1415-3. [DOI] [PubMed] [Google Scholar]
- Ercolani GL. Distribution of epiphytic bacteria on olive leaves and the influence of leaf age and sampling time. Microb. Ecol. 1991;21:35–48. doi: 10.1007/BF02539143. [DOI] [PubMed] [Google Scholar]
- Heuer H, Smalla K. Bacterial phyllosphere communities of Solanum tuberosum L. and T4-lysozyme-producing transgenic variants. FEMS Microbiol. Ecol. 1999;28:357–371. [Google Scholar]
- Hua NP, Kobayashi F, Iwasaka Y, Shi GY, Naganuma T. Detailed identification of desert-originated bacteria carried by Asian dust storms to Japan. Aerobiologia. 2007;23:291–298. [Google Scholar]
- Iesce MR, Graziano ML, Cermola F, Montella S, Stasio L, di Gioia C. Effects of sensitizers on the photodegradation of the systemic fungicide triadimenol. Chemosphere. 2003;51:163–166. doi: 10.1016/S0045-6535(02)00823-8. [DOI] [PubMed] [Google Scholar]
- Jackson EF, Echlin HL, Jackson CR. Changes in the phyllosphere community of the resurrection fern, Polypodium polypodioides, associated with rainfall and wetting. FEMS Microbiol. Ecol. 2006;58:236–246. doi: 10.1111/j.1574-6941.2006.00152.x. [DOI] [PubMed] [Google Scholar]
- Kageyama A, Morisaki K, Omura S, Takahashi Y. Arthrobacter oryzae sp. nov. and Arthrobacter humicola sp. nov. Int. J. Syst. Evol. Microbiol. 2008;58:53–56. doi: 10.1099/ijs.0.64875-0. [DOI] [PubMed] [Google Scholar]
- Kertesz MA, Cook AM, Leisinger T. Microbial metabolism of sulfur-containing and phosphorus-containing xenobiotics. FEMS Microbiol. Rev. 1994;15:195–215. doi: 10.1111/j.1574-6976.1994.tb00135.x. [DOI] [PubMed] [Google Scholar]
- Keuth S, Rehm HJ. Biodegradation of phenanthrene by Arthrobacter polychromogenes isolated from a contaminated soil. Appl. Microbiol. Biotechnol. 1991;34:804–808. [Google Scholar]
- Kotouckova L, Schumann P, Durnova E, Sproer C, Sedlacek I, Neca J, et al. Arthrobacter nitroguajacolicus sp. nov., a novel 4-nitroguaiacol-degrading actinobacterium. Int. J. Syst. Evol. Microbiol. 2004;54:773–777. doi: 10.1099/ijs.0.02923-0. [DOI] [PubMed] [Google Scholar]
- Krimm U, Abanda-Nkpwatt D, Schwab W, Schreiber L. Epiphytic microorganisms on strawberry plants (Fragaria ananassa cv. Elsanta): identification of bacterial isolates and analysis of their interaction with leaf surfaces. FEMS Microbiol. Ecol. 2005;53:483–492. doi: 10.1016/j.femsec.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Labeda DP, Liu KC, Casida LE. Colonization of soil by Arthrobacter and Pseudomonas under varying conditions of water and nutrient availability as studied by plate counts and transmission electron microscopy. Appl. Environ. Microbiol. 1976;31:551–561. doi: 10.1128/aem.31.4.551-561.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematic. New York, NY: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- Leveau JHJ. Microbial communities in the phyllosphere. In: Riederer M, Müller M, editors. Biology of the plant cuticle. Oxford, UK: Blackwell Publishing; 2006. pp. 334–367. [Google Scholar]
- Leveau JHJ, Tech JJ. Grapevine microbiomics: bacterial diversity on grape leaves and berries revealed by high-throughput sequence analysis of 16S rRNA amplicons. Acta Hortic. (ISHS) 2011;905:31–42. [Google Scholar]
- Lindow SE. Novel method for identifying bacterial mutants with reduced epiphytic fitness. Appl. Environ. Microbiol. 1993;59:1586–1592. doi: 10.1128/aem.59.5.1586-1592.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduell P, Armengol G, Llagostera M, Orduz S, Lindow S. B. thuringiensis is a poor colonist of leaf surfaces. Microb. Ecol. 2008;55:212–219. doi: 10.1007/s00248-007-9268-4. [DOI] [PubMed] [Google Scholar]
- Manulis S, Haviv-Chesner A, Brandl MT, Lindow SE, Barash I. Differential involvement of indole-3-acetic acid biosynthetic pathways in pathogenicity and epiphytic fitness of Erwinia herbicola pv. gypsophilae. Mol. Plant Microbe Interact. 1998;11:634–642. doi: 10.1094/MPMI.1998.11.7.634. [DOI] [PubMed] [Google Scholar]
- Mercier J, Lindow SE. Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl. Environ. Microbiol. 2000;66:369–374. doi: 10.1128/aem.66.1.369-374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KM, Leveau JHJ. Microbiology of the phyllosphere: a playground for testing ecological concepts. Oecologia. 2012;168:621–629. doi: 10.1007/s00442-011-2138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongodin EF, Shapir N, Daugherty SC, Deboy RT, Emerson JB, Shvartzbeyn A, et al. Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens TC1. PLoS Genet. 2006;2:2094–2106. doi: 10.1371/journal.pgen.0020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier JM, Lindow SE. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl. Environ. Microbiol. 2004;70:346–355. doi: 10.1128/AEM.70.1.346-355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning JY, Bai ZH, Gang G, Jiang D, Hu Q, He JZ, et al. Functional assembly of bacterial communities with activity for the biodegradation of an organophosphorus pesticide in the rape phyllosphere. FEMS Microbiol. Lett. 2010;306:135–143. doi: 10.1111/j.1574-6968.2010.01946.x. [DOI] [PubMed] [Google Scholar]
- Nordin K, Unell M, Jansson JK. Novel 4-chlorophenol degradation gene cluster and degradation route via hydroxyquinol in Arthrobacter chlorophenolicus A6. Appl. Environ. Microbiol. 2005;71:6538–6544. doi: 10.1128/AEM.71.11.6538-6544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RDM. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Polymenakou PN, Mandalakis M, Stephanou EG, Tselepides A. Particle size distribution of airborne microorganisms and pathogens during an intense African dust event in the eastern Mediterranean. Environ. Health Perspect. 2008;116:292–296. doi: 10.1289/ehp.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL, Leveau JHJ. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012;6:1812–1822. doi: 10.1038/ismej.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenberger S, Bach M, Skitschak A, Frede HG. Mitigation strategies to reduce pesticide inputs into ground- and surface water and their effectiveness: a review. Sci. Total Environ. 2007;384:1–35. doi: 10.1016/j.scitotenv.2007.04.046. [DOI] [PubMed] [Google Scholar]
- Ruinen J. The phyllosphere. I. An ecologically neglected mileu. Plant Soil. 1961;15:81–109. [Google Scholar]
- Sabaratnam S, Beattie GA. Differences between Pseudomonas syringae pv. syringae B728a and Pantoea agglomerans BRT98 in epiphytic and endophytic colonization of leaves. Appl. Environ. Microbiol. 2003;69:1220–1228. doi: 10.1128/AEM.69.2.1220-1228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu A, Halverson LJ, Beattie GA. Bacterial degradation of airborne phenol in the phyllosphere. Environ. Microbiol. 2007;9:383–392. doi: 10.1111/j.1462-2920.2006.01149.x. [DOI] [PubMed] [Google Scholar]
- Sandhu A, Halverson LJ, Beattie GA. Identification and genetic characterization of phenol-degrading bacteria from leaf microbial communities. Microb. Ecol. 2009;57:276–285. doi: 10.1007/s00248-008-9473-9. [DOI] [PubMed] [Google Scholar]
- Schippers-Lammertse AF, Muijsers AO, Klatser-Oedekerk KB. Arthrobacter polychromogenes nov. spec., its pigments, and a bacteriophage of this species. Antonie Van Leeuwenhoek. 1963;29:1–15. doi: 10.1007/BF02046034. [DOI] [PubMed] [Google Scholar]
- Sguros PL. Microbial transformations of the tobacco alkaloids I. Cultural and morphological characteristics of a nicotinophile. J. Bacteriol. 1955;69:28–37. doi: 10.1128/jb.69.1.28-37.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong LC, Rosendahl C, Johnson G, Sadowsky MJ, Wackett LP. Arthrobacter aurescens TC1 metabolizes diverse s-triazine ring compounds. Appl. Environ. Microbiol. 2002;68:5973–5980. doi: 10.1128/AEM.68.12.5973-5980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson IP, Bailey MJ, Ellis RJ, Lilley AK, McCormack PJ, Purdy KJ, et al. Short-term community dynamics in the phyllosphere microbiology of field-grown sugar beet. FEMS Microbiol. Ecol. 1995;16:205–211. [Google Scholar]
- Tixier C, Sancelme M, Ait-Aissa S, Widehem P, Bonnemoy F, Cuer A, et al. Biotransformation of phenylurea herbicides by a soil bacterial strain, Arthrobacter sp. N2: structure, ecotoxicity and fate of diuron metabolite with soil fungi. Chemosphere. 2002;46:519–526. doi: 10.1016/s0045-6535(01)00193-x. [DOI] [PubMed] [Google Scholar]
- Unell M, Abraham PE, Shah M, Zhang B, Ruckert C, VerBerkmoes NC, et al. Impact of phenolic substrate and growth temperature on the Arthrobacter chlorophenolicus proteome. J. Proteome Res. 2009;8:1953–1964. doi: 10.1021/pr800897c. [DOI] [PubMed] [Google Scholar]
- Waight K, Pinyakong O, Luepromchai E. Degradation of phenanthrene on plant leaves by phyllosphere bacteria. J. Gen. Appl. Microbiol. 2007;53:265–272. doi: 10.2323/jgam.53.265. [DOI] [PubMed] [Google Scholar]
- Wang QQ, Lemley AT. Competitive degradation and detoxification of carbamate insecticides by membrane anodic Fenton treatment. J. Agric. Food Chem. 2003;51:5382–5390. doi: 10.1021/jf034311f. [DOI] [PubMed] [Google Scholar]
- Westerberg K, Elvang AM, Stackebrandt E, Jansson JK. Arthrobacter chlorophenolicus sp. nov., a new species capable of degrading high concentrations of 4-chlorophenol. Int. J. Syst. Evol. Microbiol. 2000;50:2083–2092. doi: 10.1099/00207713-50-6-2083. [DOI] [PubMed] [Google Scholar]
- Whipps JM, Hand P, Pink D, Bending GD. Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 2008;105:1744–1755. doi: 10.1111/j.1365-2672.2008.03906.x. [DOI] [PubMed] [Google Scholar]
- Yadav RKP, Karamanoli K, Vokou D. Bacterial colonization of the phyllosphere of Mediterranean perennial species as influenced by leaf structural and chemical features. Microb. Ecol. 2005;50:185–196. doi: 10.1007/s00248-004-0171-y. [DOI] [PubMed] [Google Scholar]
- Yashiro E, Spear RN, McManus PS. Culture-dependent and culture-independent assessment of bacteria in the apple phyllosphere. J. Appl. Microbiol. 2011;110:1284–1296. doi: 10.1111/j.1365-2672.2011.04975.x. [DOI] [PubMed] [Google Scholar]
- Yutthammo C, Thongthammachat N, Pinphanichakarn P, Luepromchai E. Diversity and activity of PAH-degrading bacteria in the phyllosphere of ornamental plants. Microb. Ecol. 2010;59:357–368. doi: 10.1007/s00248-009-9631-8. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Qiao XW, Li WJ, Xu JF, Wang W, Chen XY. Phyllosphere bacterial communities associated with the degradation of acetamiprid in Phaseolus vulgaris. Afr. J. Biotechnol. 2011;10:3809–3817. [Google Scholar]
- Zwielehner J, Handschur M, Michaelsen A, Irez S, Demel M, Denner EBM, et al. DGGE and real-time PCR analysis of lactic acid bacteria in bacterial communities of the phyllosphere of lettuce. Mol. Nutr. Food Res. 2008;52:614–623. doi: 10.1002/mnfr.200700158. [DOI] [PubMed] [Google Scholar]


