It has been assumed that only seed plants are able to sense far-red light as PHYA, which is essential for far-red light perception, specifically evolved in seed plants. Here, we show that also cryptogams, such as mosses and ferns, respond to far-red light and have phytochromes with PHYA-like properties. This suggests that far-red light sensing is an evolutionarily ancient trait.
Abstract
Phytochromes are plant photoreceptors important for development and adaptation to the environment. Phytochrome A (PHYA) is essential for the far-red (FR) high-irradiance responses (HIRs), which are of particular ecological relevance as they enable plants to establish under shade conditions. PHYA and HIRs have been considered unique to seed plants because the divergence of seed plants and cryptogams (e.g., ferns and mosses) preceded the evolution of PHYA. Seed plant phytochromes translocate into the nucleus and regulate gene expression. By contrast, there has been little evidence of a nuclear localization and function of cryptogam phytochromes. Here, we identified responses to FR light in cryptogams, which are highly reminiscent of PHYA signaling in seed plants. In the moss Physcomitrella patens and the fern Adiantum capillus-veneris, phytochromes accumulate in the nucleus in response to light. Although P. patens phytochromes evolved independently of PHYA, we have found that one clade of P. patens phytochromes exhibits the molecular properties of PHYA. We suggest that HIR-like responses had evolved in the last common ancestor of modern seed plants and cryptogams and that HIR signaling is more ancient than PHYA. Thus, other phytochromes in seed plants may have lost the capacity to mediate HIRs during evolution, rather than that PHYA acquired it.
INTRODUCTION
Plants do not only gain energy from light, they also use light as a source of information in order to adapt growth and development to environmental conditions. They have a range of photoreceptors to detect different aspects of their light environment, such as the light intensity and spectral composition, the direction of the light gradient, or temporal light patterns (Kami et al., 2010). Cryptochromes, phototropins, members of the ZEITLUPE (ZTL) family, and UV RESISTANCE LOCUS 8 (UVR8) monitor the blue (B) and UV-B range of the light spectrum, whereas phytochromes (PHYs) are essential for the perception of red (R) and far-red (FR) light (Kami et al., 2010; Heijde and Ulm, 2012). Phytochromes can exist in two different states, the inactive Pr form with maximal absorption in R light, and the active or Pfr form of phytochromes, which has an absorption peak in FR light. By absorption of light, these forms reversibly convert into each other, resulting in an equilibrium with a wavelength-specific Pfr/Ptot ratio (Ptot = Pfr + Pr [inactive form of phytochromes]) (Mancinelli, 1994).
Phytochromes have been most intensively studied in seed plants, but they are also present in ferns, mosses, and green algae (i.e., in cryptogams) (Mathews, 2006). They are encoded by small gene families, which are the result of independent gene duplication events in the different lineages (Mathews, 2006; Mittmann et al., 2009). In seed plants, the phytochrome gene lineage split into type I and type II phytochromes that are represented in Arabidopsis thaliana by PHYTOCHROME A (PHYA) and PHYTOCHROME B-E (PHYB-E), respectively. PHYB is the most abundant phytochrome in light-grown seedlings and adult plants. It translates the R/FR light ratio in the environment into a corresponding Pfr/Ptot ratio, which is the molecular basis of the shade avoidance response. Under light conditions resulting in high Pfr/Ptot levels, such as in strong R or white (W) light, PHYB also plays an important role in seed germination, deetiolation, and induction of flowering (Li et al., 2011; Kami et al., 2012). PHYA is the only type I phytochrome. It is rapidly degraded in light but highly abundant in dark-grown (i.e., etiolated) seedlings. In contrast with type II phytochromes, PHYA mediates germination or deetiolation in response to low levels of Pfr/Ptot, which are typically achieved by light pulses of any wavelength (very low fluence responses) or by continuous irradiation with FR light (high-irradiance responses [HIRs]) (Li et al., 2011).
Cryptogam phytochromes evolved independently of seed plant phytochromes and cannot be assigned to either the type I or type II clade of seed plant phytochromes (Mathews, 2006). Fern (Pteridophyta), moss (Bryophyta), and liverwort (Marchantiophyta) phytochromes are involved in R/FR light-reversible spore germination and deetiolation, but there are hardly any reports of HIR-like responses in cryptogams, perhaps none outside of vascular plants (i.e., seed plants and ferns) (Mathews, 2006). A number of studies have shown that cryptogam phytochromes also regulate phototropic growth of protonema filaments and light-induced chloroplast movement (Mittmann et al., 2004; Kanegae and Wada, 2006; Lamparter, 2006). As these responses are rapidly induced and depend on the orientation of the E vector in polarized light, it has been concluded that cryptogam phytochromes localize to the cytosol or associate with the plasma membrane by interacting with phototropins (Wada et al., 1983; Jaedicke et al., 2012). Indeed, in contrast with seed plants, there is only very limited evidence for a function of cryptogam phytochromes in the nucleus (Rösler et al., 2010). Experiments based on microbeam irradiation suggested that nuclear-localized phytochromes play a role in branch formation in the moss Physcomitrella patens or in spore germination in the fern Adiantum capillus-veneris, and a recent report indicated the presence of Ac-PHY2 in nuclei of transiently transformed A. capillus-veneris gametophytes (Uenaka et al., 2005; Tsuboi et al., 2012). However, spectroscopic methods and transient expression assays do not support the existence of nuclear-localized phytochromes in mosses but rather point to a localization and function at the plasma membrane or in the cytosol (Böse et al., 2004; Uenaka and Kadota, 2007).
By contrast, the vast majority of phytochrome-mediated responses in seed plants depend on nuclear-localized phytochromes, and only a few examples of cytosolic phytochrome functions have been reported (Rösler et al., 2010). In seed plants, light-activated phytochromes translocate from the cytosol into the nucleus (Kami et al., 2010). They interact with PHYTOCHROME INTERACTING FACTORS (PIFs), which are a subgroup of the basic helix-loop-helix transcription factors important for the regulation of elongation growth and photomorphogenesis (Leivar and Quail, 2011). In total, several hundred genes in Arabidopsis are regulated by phytochromes at the transcriptional level (Li et al., 2011).
Interestingly, type I and type II phytochromes from seed plants employ different mechanisms for translocation into the nucleus. The main type II phytochrome, PHYB, possibly does not rely on a specific transport protein and may enter the nucleus bound to transcription factors, such as PIFs, or using a nuclear localization signal (NLS) of its own (Chen et al., 2005; Pfeiffer et al., 2012). By contrast, light-regulated nuclear accumulation of PHYA depends on the two functional homologs FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and FHY1-LIKE (FHL), which contain a NLS and physically interact with PHYA (Hiltbrunner et al., 2006; Rösler et al., 2007).
Based on the different subcellular localization and site of action of seed plant and cryptogam phytochromes, it has been assumed that in the two plant lineages phytochrome signaling relies on fundamentally different molecular mechanisms and mediates different response modes (Rösler et al., 2010). Type I and type II phytochromes have virtually identical photophysical properties, based on which they are expected to have an action peak in R light, where the Pfr/Ptot ratio is maximal (Mancinelli, 1994; Eichenberg et al., 2000). While R light indeed is the most efficient trigger for responses depending on PHYB and other type II phytochromes, we have recently shown that PHYA-specific properties, such as the rapid degradation of the Pfr form and the interaction with the nuclear transport proteins FHY1 and FHL, shift the action peak of PHYA from R to FR light (Rausenberger et al., 2011). This shift is typical of HIRs, which exclusively depend on PHYA and have been considered unique to seed plants. Under FR light conditions, seed plants depend on PHYA for germination and deetiolation (Yanovsky et al., 1995; Botto et al., 1996). Thus, HIRs have been hypothesized to be essential for survival in light environments dominated by FR light, such as in canopy shade (Yanovsky et al., 1995). HIRs are ubiquitous among angiosperms, and rudimentary HIRs have also been described in some gymnosperms (Burgin et al., 1999). By contrast, there has been little evidence for HIR-like responses in cryptogams, and the emergence of HIRs has been associated with the evolution of seed plant PHYA and PHYA orthologs (Mathews, 2006).
However, data presented in this report support the idea that HIR-like responses to FR light are not restricted to seed plants and do not specifically require PHYA. We describe the light-dependent nuclear accumulation of phytochromes in the moss P. patens. Furthermore, we show that P. patens FHY1 (Pp-FHY1) is functionally equivalent to Arabidopsis FHY1 and plays a key role in phytochrome nuclear transport and HIR signaling in P. patens.
RESULTS
Light Induces Rapid Nuclear Accumulation of Cryptogam Phytochromes
We have chosen the moss P. patens as a model system to investigate the subcellular localization of PHYs in cryptogams. By homologous recombination, we generated independent transgenic P. patens lines expressing endogenous Pp-PHY1, Pp-PHY2, Pp-PHY3, Pp-PHY4, Pp-PHY5a, or Pp-PHY5b with a C-terminal yellow fluorescent protein (YFP) tag. In dark-adapted gametophore leaves and protonema filaments exposed to microscope light, we observed a clear nuclear localization of Pp-PHY1:YFP (Figures 1A and 1B). Pp-PHY3:YFP and Pp-PHY4:YFP showed a similar localization, and after chlorophyll bleaching using the herbicide norflurazone, we also found a nuclear accumulation of Pp-PHY2:YFP and Pp-PHY5a:YFP, which were only weakly expressed (see Supplemental Figures 1A to 1D online). In an immunoblot analysis, full-length phytochrome YFP fusions were detected (see Supplemental Figure 1E online). We could not detect Pp-PHY5b:YFP by microscopy or immunoblot.
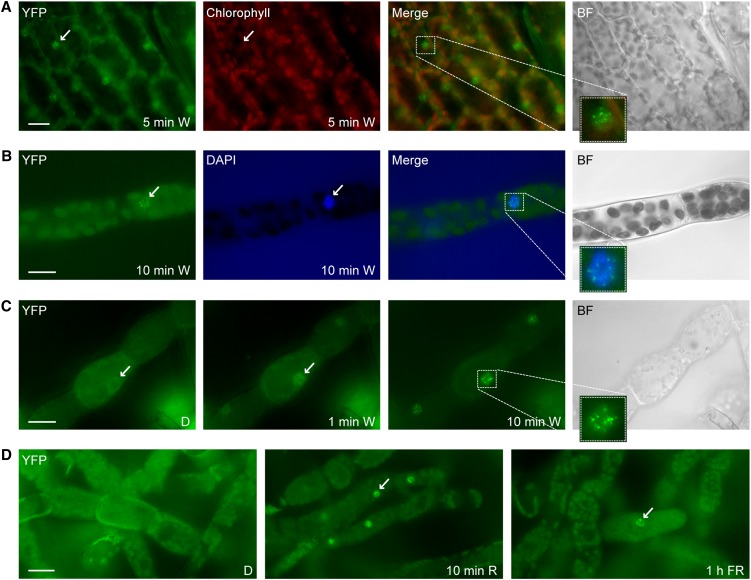
Figure 1.
Rapid Light-Induced Nuclear Transport of P. patens PHY1.
(A) Light-regulated nuclear accumulation of Pp-PHY1 in gametophores. Dark-adapted gametophores of transgenic P. patens plants expressing Pp-PHY1:YFP were exposed to W light for 5 min and used for fluorescence microscopy.
(B) DAPI staining. Dark-adapted protonema filaments of Pp-PHY1:YFP expressing P. patens plants were exposed to W light for 10 min, fixed with formaldehyde, stained with DAPI, and analyzed by fluorescence microscopy.
(C) Rapid light-induced nuclear transport of Pp-PHY1 in protonema filaments. Dark-adapted protonema filaments of P. patens plants expressing YFP-tagged Pp-PHY1 were used for time series fluorescence microscopy. Images were acquired before (dark control [D]) and 1 and 10 min after the onset of irradiation with W light.
(D) Nuclear accumulation of P. patens PHY1 is induced by R and FR light. Protonema filaments of P. patens plants expressing Pp-PHY1:YFP were dark adapted and used for fluorescence microscopy. Images were acquired before and after irradiation with R light (10 min, 22 μmol m−2 s−1) or FR light (1 h, 18 μmol m−2 s−1). The samples were fixed with formaldehyde before microscopy analysis.
Arrows indicate nuclei; insets show enlargements of nuclei. Merge, merge of YFP and chlorophyll/DAPI channels; BF, bright field. Bars = 20 μm.
Time series experiments confirmed the light dependency of P. patens phytochrome nuclear accumulation (Figure 1C). Pp-PHY1:YFP did not accumulate to detectable levels in nuclei of dark-adapted protonema filaments, whereas 1 min of irradiation with microscope light was sufficient to induce nuclear translocation. Although we detected Pp-PHY1:YFP in the nucleus after both R and FR light treatments, the nuclear signal was stronger in R light than in FR light (Figure 1D). We also observed Pp-PHY3:YFP in the nucleus after R light treatment but possibly due to the weaker YFP signal it was hardly detectable in the nucleus after irradiation with FR light (see Supplemental Figure 2B online). All nuclear-localized P. patens phytochromes formed nuclear bodies, which are also observed for seed plant phytochromes and which have been implicated in signal transduction (Figure 1; see Supplemental Figure 1 online) (Van Buskirk et al., 2012).
Using particle bombardment, we also found that PHY2 from the moss Ceratodon purpureus (Cp-PHY2) accumulated in the nucleus of P. patens protonema cells (see Supplemental Figure 3A online). Moreover, phytochromes from the fern A. capillus-veneris (Ac-PHY1, Ac-PHY2, and Ac-PHY3) localized in the nucleus of A. capillus-veneris gametophore cells, although a fraction may remain in the cytosol (see Supplemental Figure 3B online). In summary, our findings in cryptogams support the hypothesis that nuclear accumulation of phytochromes is not restricted to seed plants.
To investigate if nuclear transport of cryptogam phytochromes depends on a mechanism similar to seed plants, we expressed YFP-tagged Cp-PHY2 and Ac-PHY1 in Arabidopsis plants. In etiolated seedlings irradiated for 6 h with either R or FR light, Cp-PHY2:YFP and Ac-PHY1:YFP accumulated in the nucleus (see Supplemental Figure 4A online). Moreover, transiently expressed Arabidopsis PHYA:YFP localized to the nucleus in P. patens protonema cells (see Supplemental Figure 4B online), confirming recent results by Jaedicke et al. (2012). This suggests that phytochrome nuclear transport is mediated by a similar mechanism in seed plants and cryptogams. However, despite light-dependent nuclear accumulation, cryptogam phytochromes are not functional in Arabidopsis but interfere with proper light perception (see Supplemental Figures 4C and 4D online).
P. patens FHY1 Is Essential for Light-Regulated Phytochrome Nuclear Transport and Gene Expression
FHY1-like proteins consist of an NLS and a PHYA binding motif, linked by a spacer of roughly 150 amino acid residues (Genoud et al., 2008). Using the consensus sequence of the PHYA binding motif to search protein, genome, and EST databases, we found potential FHY1-like proteins from different cryptogam species. In EST databases for P. patens, Selaginella moellendorffii (spikemoss), and Ceratopteris richardii (fern), we identified clones that code for proteins, which contain an NLS, a spacer, and a C-terminal PHYA binding motif (Figure 2A). For A. capillus-veneris (fern) and Closterium sp (green alga), our search identified partial cDNA clones coding for the PHYA binding motif and part of the spacer, but lacking the 5′ end of the coding sequence (Figure 2A). Using RT-PCR, we amplified the Pp-FHY1 coding sequence predicted in the database (Phypa_446283). Pp-FHY1 contains two introns, similar to FHY1 from Arabidopsis, and codes for a protein of 402 amino acids.
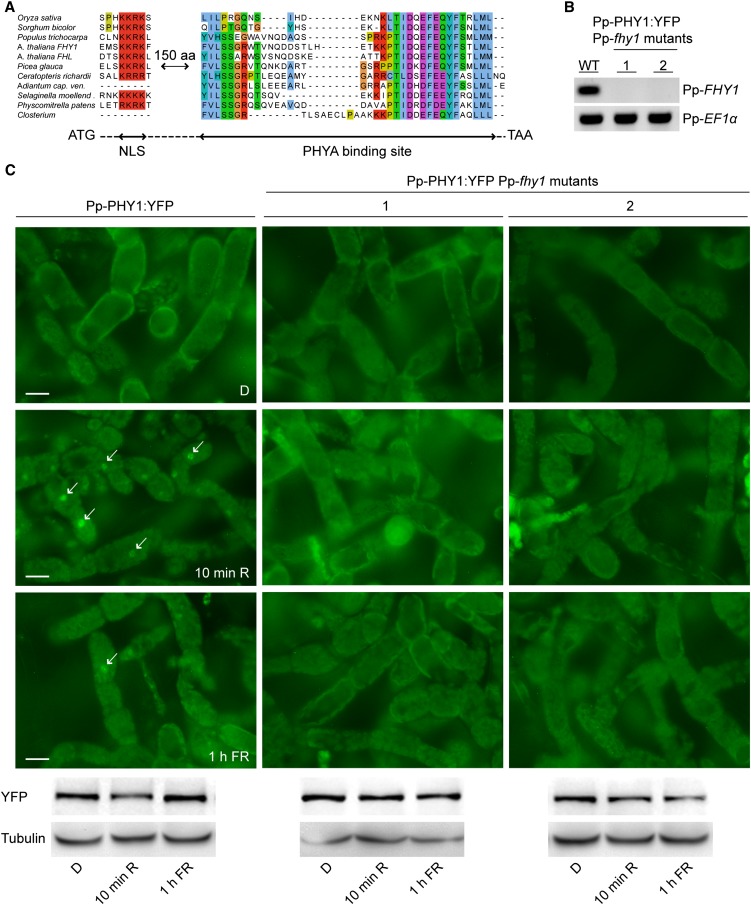
Figure 2.
Pp-FHY1 Is Essential for Pp-PHY1 Nuclear Transport in P. patens.
(A) Cryptogams contain FHY1-like proteins. Sequence alignment of FHY1-like proteins from monocots, dicots, gymnosperms, and cryptogams. Only regions of high sequence similarity are shown. The dashed line indicates nonaligned regions. aa, amino acids.
(B) RT-PCR analysis of Pp-fhy1 mutants. Pp-FHY1 was deleted in Pp-PHY1:YFP-expressing lines. Two independent Pp-fhy1 mutants were used for RT-PCR analysis and Pp-EF1α was used as a control. WT, the wild type.
(C) Light-induced nuclear transport of Pp-PHY1 depends on Pp-FHY1. Dark-adapted protonema filaments of P. patens wild type or Pp-fhy1 mutants expressing Pp-PHY1:YFP were fixed before microscopy analysis. Images were acquired before (dark control [D]) and after irradiation with either R light (10 min, 22 μmol m−2 s−1) or FR light (1 h, 18 μmol m−2 s−1). Arrows indicate nuclei. Immunoblot analysis shows Pp-PHY1:YFP levels in the different light conditions. Bars = 20 μm.
In Arabidopsis, FHY1 (At-FHY1) is essential for PHYA nuclear translocation and signaling. To test cryptogam FHY1-like proteins for a function in phytochrome nuclear transport, we knocked out P. patens FHY1 in Pp-PHY1:YFP lines using gene targeting (Figure 2B). Nuclear accumulation of Pp-PHY1:YFP was strongly reduced in two independent Pp-FHY1 KO lines (Figure 2C; see Supplemental Figure 5 online), suggesting that Pp-FHY1 functions as a nuclear transport factor for phytochromes, similar to seed plant FHY1.
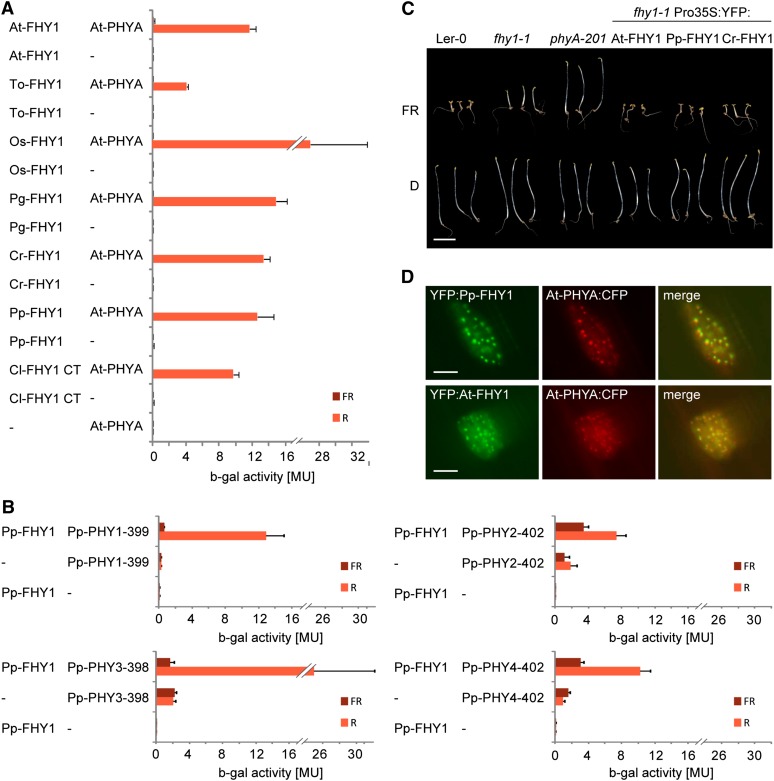
Using yeast two-hybrid assays, we found that FHY1-like proteins from ferns, mosses, and algae interact with Arabidopsis PHYA in a Pfr-dependent manner, similar to FHY1-like proteins from gymnosperms, monocots, and dicots (Figure 3A). Moreover, Pp-FHY1 interacted predominantly with the Pfr form of Pp-PHY1, Pp-PHY2, Pp-PHY3, and Pp-PHY4 fragments corresponding to the minimal At-PHYA fragment (PHYA 1–406), which binds to At-FHY1 (Figure 3B). Testing further cryptogam FHY1-like proteins and phytochromes, we also found Pfr-specific interactions between Cr-FHY1 and both Ac-PHY1 and Ac-PHY2 as well as between Pp-FHY1 and Cp-PHY2 (see Supplemental Figures 6A and 6B online). Furthermore, the expression of Pp-FHY1 and Cr-FHY1 in the Arabidopsis fhy1-1 mutant restored hypocotyl growth inhibition in FR light (Figure 3C), suggesting that FHY1-like proteins from cryptogams and seed plants are functionally equivalent. In line with this notion, we found that Pp-FHY1 colocalizes with At-PHYA in light-induced nuclear bodies in mustard (Sinapis alba; Figure 3D), similar to seed plant FHY1 (Hiltbrunner et al., 2005, 2006). Moreover, transiently expressed Pro35S:Pp-FHY1:YFP was detected in the nucleus of P. patens protonema cells (see Supplemental Figure 6C online).
Figure 3.
Cryptogam and Seed Plant FHY1 Are Functional Homologs.
(A) Cryptogam FHY1 proteins contain a PHYA binding motif. AD plasmids containing the coding sequence for the C-terminal phytochrome binding motif of FHY1 from Closterium sp (Cl; green algae) or full-length FHY1 from Arabidopsis (At), dandelion (Taraxacum officinale; To), rice (Oryza sativa; Os), white spruce (Picea glauca; Pg), Ceratopteris richardii (Cr; fern), or P. patens (Pp) fused to the GAL4 activation domain were used for yeast two-hybrid analysis with Arabidopsis PHYA fused to the GAL4 DNA binding domain. To convert PHYA to the Pfr or Pr form, yeast cultures were irradiated for 5 min with R (12 μmol m−2 s−1) or FR light (12 μmol m−2 s−1) and incubated for 4 h in the dark before measuring the β-galactosidase activity. MU, Miller Units; Error bars represent se; n = 3.
(B) P. patens phytochromes interact with Pp-FHY1 in a light regulated fashion. N-terminal fragments of P. patens phytochromes fused to the binding domain were used for yeast two-hybrid assays with AD:Pp-FHY1 as described in (A). MU, Miller Units; Error bars represent se; n = 3.
(C) P. patens and Ceratopteris FHY1 are functional in Arabidopsis. Landsberg erecta-0 (Ler-0), fhy1-1, and phyA-201 as well as fhy1-1 seedlings expressing 35S promoter–driven YFP:At-FHY1, YFP:Pp-FHY1, or YFP:Cr-FHY1 were grown for 5 d in darkness (D) or FR (12 μmol m−2 s−1). Bar = 5 mm.
(D) Pp-FHY1 and At-PHYA colocalize in light-induced nuclear bodies. Etiolated mustard seedlings were transformed by particle bombardment with constructs coding for Pro35S:PHYA:CFP and either Pro35S:YFP:At-FHY1 or Pro35S:YFP:Pp-FHY1. After transformation, the seedlings were incubated for 2 d in darkness and used for microscopy. The images were acquired after 5 min irradiation with W light. Bars = 10 μm.
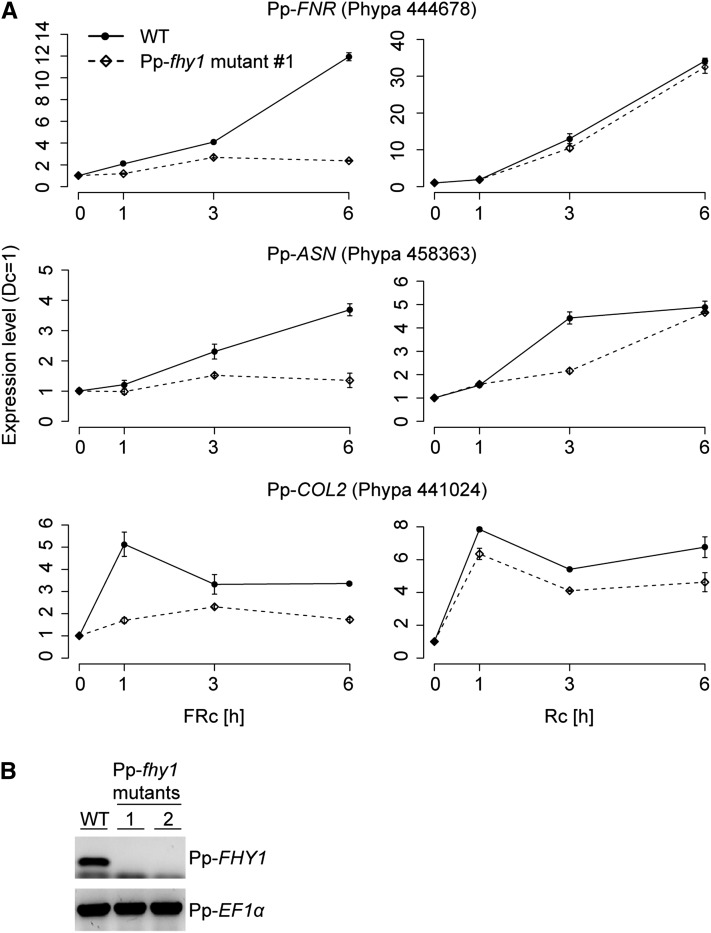
Considering a potential nuclear function of cryptogam phytochromes, we analyzed the transcriptional activity of P. patens genes that are homologous to different R and FR light–regulated Arabidopsis genes (Tepperman et al., 2001; Hare et al., 2003; Jang et al., 2007). Quantitative RT-PCR analyses showed that in wild-type P. patens protonemata, the expression of CONSTANS-LIKE2 (COL2) (Phypa_441024), Asparagine Synthetase (ASN) (Phypa_458363), and Ferredoxin NADP+ Reductase-like Protein (FNR) (Phypa_444678) is induced by R and FR light (Figure 4A; see Supplemental Figure 7 online). Interestingly, in Pp-fhy1 knockout mutants, FR-dependent transcription of COL2, ASN, and FNR is strongly impaired, whereas the transcript levels in R light were only slightly changed (Figures 4A and 4B; see Supplemental Figure 7 online).
Figure 4.
Pp-FHY1 Is Essential for FR Light–Induced Gene Expression.
(A) Protonemata cultures of P. patens wild type (WT) and Pp-fhy1 mutant lines were dark adapted and exposed to either R light (28 μmol m−2 s−1) or FR light (16 μmol m−2 s−1). Samples for quantitative RT-PCR analyses were harvested after 1, 3, and 6 h of light treatment or darkness. The expression levels of FNR, ASN, and COL2 were normalized to the levels of 26S rRNA. Expression levels in darkness were set to 1. Error bars represent se of technical replicates, n = 3. An independent biological replicate is shown in Supplemental Figure 7 online.
(B) RT-PCR analysis of Pp-fhy1 mutants. Pp-fhy1 knockout lines were generated using gene targeting. Two independent Pp-fhy1 mutant lines were used for RT-PCR analysis with primers specific for either Pp-FHY1 or Pp-EF1α.
P. patens Shows HIR-Like Responses
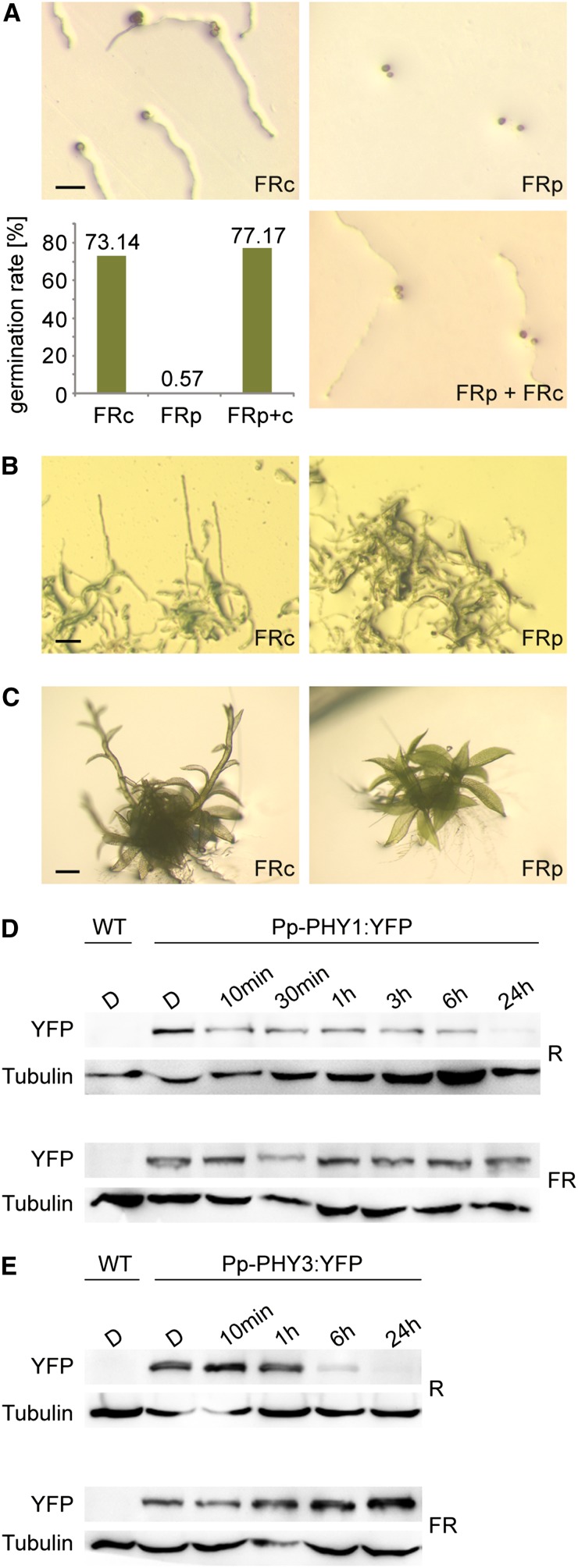
Next, we investigated whether FR light–dependent regulatory processes affected adaptive and developmental responses. P. patens spore germination as well as protonemata and gametophore growth were induced in continuous FR light (Figures 5A to 5C), whereas no growth was observed in darkness (see Supplemental Figure 8 online). This response mode was reminiscent of HIRs in seed plants. A distinctive feature of HIRs in seed plants is that they do not follow the reciprocity law (i.e., FR light pulses with the same total fluence cannot substitute for continuous FR irradiation) (Li et al., 2011). Indeed, in P. patens, FR light–induced spore germination as well as growth of protonemata and gametophores depended on continuous irradiation (Figures 5A to 5C) and therefore may be classified as HIR-like responses.
Figure 5.
HIR-Like Responses to High Fluence Rate FR Light in P. patens.
(A) Spore germination in FR light requires continuous irradiation. P. patens spores were irradiated for 3 d with continuous FR light (FRc; 3.5 μmol m−2 s−1) or with 3-min FR light pulses (FRp; 70 μmol m−2 s−1) of the same total fluence, interrupted by 57-min dark periods. To ensure that spores irradiated with FR pulses were viable, they were irradiated for an additional 3 d with continuous FR light (FRp + FRc). The bar plot shows significantly reduced germination rate in FRp (Fisher’s exact test P < 2.2e-16). Bar = 100 μm.
(B) Protonemata growth in FR light depends on continuous irradiation. P. patens cultures were grown for 20 d in continuous FR light (3.5 μmol m−2 s−1) or irradiated with 3-min FR light pulses (70 μmol m−2 s−1) of the same total fluence, interrupted by 57-min dark periods. For dark controls, see Supplemental Figure 8B online. Bar = 100 μm.
(C) Continuous irradiation is essential for FR light–induced gametophore growth. P. patens gametophores were grown for 9 d in continuous FR light (3.5 μmol m−2 s−1) or irradiated with 3-min FR light pulses (70 μmol m−2 s−1) of the same total fluence, interrupted by 57-min dark periods. For dark controls, see Supplemental Figure 8C online. Bar = 500 μm.
(D) Pfr-dependent degradation of Pp-PHY1. Dark-adapted protonemata cultures of P. patens lines expressing YFP-tagged Pp-PHY1 were irradiated with R light (22 μmol m−2 s−1) or FR light (18 μmol m−2 s−1) for different time periods. Total protein was isolated and analyzed by SDS-PAGE and immunoblotting with anti-YFP antibody. Protein extracts from dark-adapted wild-type P. patens cultures were used as negative controls. Tubulin is shown as a loading control. D, darkness; WT, the wild type.
(E) Pfr-dependent degradation of Pp-PHY3. Dark-adapted protonemata cultures of P. patens lines expressing Pp-PHY3:YFP were irradiated with either R or FR light and used for SDS-PAGE and immunoblot analysis as described in (D).
[See online article for color version of this figure.]
Pp-FHY1 Is Required for HIR-Like Responses of P. patens
Rapid degradation of PHYA after its conversion into the light-activated Pfr form is essential for HIRs in seed plants (Rausenberger et al., 2011). We analyzed the stability of P. patens phytochromes in lines expressing endogenous phytochromes tagged with YFP. Immunoblot analyses with YFP-specific antibodies and microscopy analyses demonstrated that Pp-PHY1:YFP and Pp-PHY3:YFP were degraded in the Pfr form (R light) but were stable in Pr (FR light) (Figures 5D and 5E; see Supplemental Figures 2A and 2B online). Pp-PHY1 and Pp-PHY3 are therefore potential photoreceptors for the HIR-like responses in P. patens.
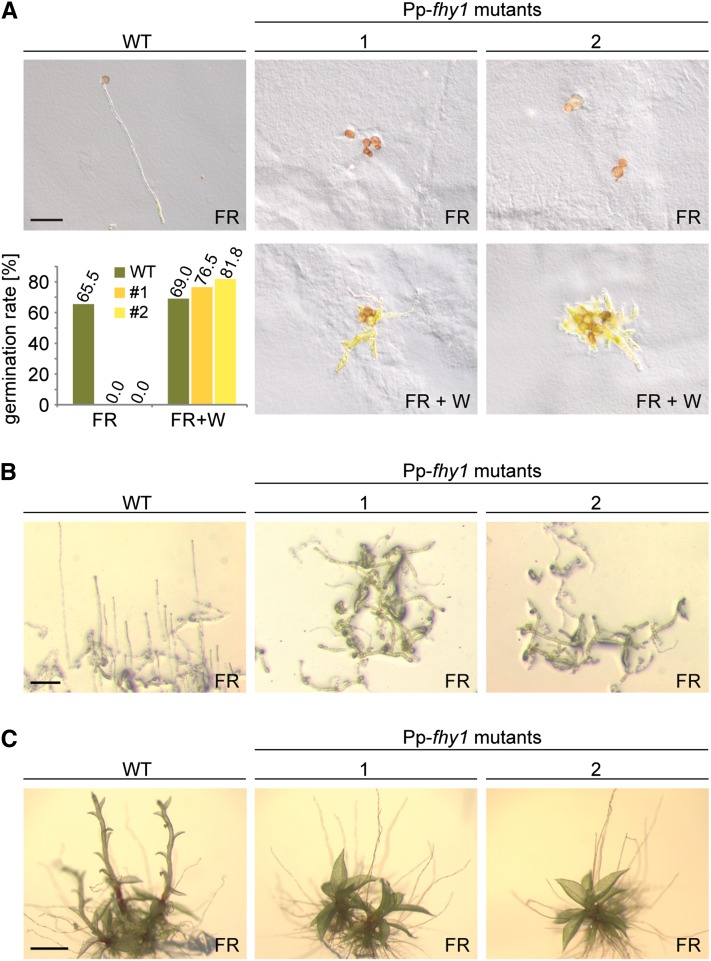
Another essential determinant for Arabidopsis HIRs is continuous photocycling and nuclear accumulation of PHYA, which depends on the nuclear transport proteins At-FHY1 and At-FHL (Rausenberger et al., 2011). Pp-fhy1 knockout interfered with the nuclear accumulation of Pp-PHY1:YFP (Figure 2C; see Supplemental Figure 5 online), which is in line with the idea that Pp-PHY1 and its closest homolog, Pp-PHY3, are receptors for HIR-like responses in P. patens. Analogous to At-FHY1 in Arabidopsis, Pp-FHY1 would then constitute an upstream prerequisite for phytochrome-mediated HIR-like responses in P. patens. Indeed, FR light–dependent spore germination as well as protonemata and gametophore growth were severely inhibited in two independent Pp-fhy1 mutant lines (Figures 6A to 6C). Furthermore, protonemata side branching was reduced in light with a low R:FR ratio (see Supplemental Figure 9 online). These effects of Pp-fhy1 knockout were specific for FR light and low Pfr/Ptot ratios, as we did not observe any differences in other light qualities or in darkness (see Supplemental Figures 8A to 8C online). Thus, Pp-FHY1 is essential for HIR-like responses in P. patens.
Figure 6.
Pp-FHY1 Is Essential for HIR-Like Responses to High Fluence Rate FR Light.
(A) Spore germination in FR light depends on Pp-FHY1. Spores from wild-type (WT) P. patens plants and two independent Pp-fhy1 mutant lines were irradiated for 8 d with continuous FR light (18 μmol m−2 s−1). To ensure that Pp-fhy1 mutant spores irradiated with FR light were viable, they were irradiated for an additional 5 d with W light after the FR light treatment. The bar plot shows significantly reduced germination rate in Pp-fhy1 mutants (Fisher’s exact test P < 3.4e-05). Bar = 100 μm.
(B) Pp-FHY1 is essential for protonemata growth in FR light. Wild-type and Pp-fhy1 mutant P. patens cultures were grown for 13 d in FR light (18 μmol m−2 s−1). Bar = 100 μm.
(C) FR light–induced gametophore growth requires Pp-FHY1. Wild-type and Pp-fhy1 mutant P. patens gametophores were grown for 11 d in FR (18 μmol m−2 s−1). Bar = 500 μm.
[See online article for color version of this figure.]
DISCUSSION
Localization and Function of Cryptogam Phytochromes in the Nucleus
Using stable transgenic P. patens lines, we visualized the dynamics of the subcellular localization of cryptogam phytochromes. We found that P. patens phytochromes accumulated in the nucleus upon light-dependent conversion to the active Pfr form, which is reminiscent of the first steps in phytochrome signaling in seed plants. Moreover, we showed that cryptogam phytochromes translocated into the nucleus in Arabidopsis seedlings and that, in agreement with Jaedicke et al. (2012), Arabidopsis PHYA accumulated in the nucleus of P. patens protonema cells. Thus, it seems that seed plant and cryptogam phytochrome nuclear transport rely on an evolutionarily conserved mechanism. Interestingly, previous studies have described cryptogam phytochromes as mainly localizing and acting in the cytosol and/or at the plasma membrane, and there has been only limited evidence for a function in the nucleus (Rösler et al., 2010; Tsuboi et al., 2012). In contrast with this, phytochromes from Arabidopsis and other seed plants were shown to translocate to the nucleus after activation by light, where they mediate changes in gene expression (Li et al., 2011). Consistent with our observations of a light-dependent nuclear localization of P. patens phytochromes, R and FR light also induced changes in gene expression in P. patens. However, moss and fern phytochromes interfered with phytochrome signaling in Arabidopsis, which might reflect differences between seed plants and cryptogams regarding downstream signal transduction. Considering these novel aspects of phytochrome localization and signaling in cryptogams, one can assume a dual localization and spatial function of P. patens phytochromes, with one phytochrome pool in the cytosol and/or at the plasma membrane and one pool translocating to the nucleus. In summary, our data show that nuclear localization and function of phytochromes are not exclusive to seed plants but are common to all land plants.
FHY1 Proteins from Seed Plants and Cryptogams Are Functional Homologs
The split of the phytochrome gene lineage into PHYA- and PHYB-like phytochromes only occurred in seed plants. Nevertheless, even though cryptogams do not have PHYA-like phytochromes, we identified FHY1-like proteins in several cryptogams. In Arabidopsis, the nuclear translocation of light-activated At-PHYA depends on At-FHY1 and its homolog At-FHL and is about 10-fold faster than At-PHYB nuclear transport (Kircher et al., 2005; Hiltbrunner et al., 2006; Rösler et al., 2007). The rapid nuclear accumulation we observed for P. patens phytochromes is reminiscent of At-PHYA nuclear transport, suggesting a similar transport mechanism for cryptogams. Indeed, in the P. patens fhy1 KO mutant, the nuclear accumulation of Pp-PHY1:YFP was strongly reduced in R and FR light and FR-induced gene expression was inhibited. Furthermore, FHY1-like proteins from cryptogams were functional in Arabidopsis. We therefore postulate the existence of an FHY1-dependent phytochrome nuclear transport system in cryptogams, similar to the transport system essential for PHYA signaling in seed plants. Interestingly, recent studies have shown that growth of P. patens mutants deficient in phytochrome chromophore biosynthesis is strongly reduced in R light (Chen et al., 2012), whereas the phenotype of Pp-fhy1 knockout mutants is mostly FR light specific. Thus, we assume the existence of alternative nuclear transport systems for phytochromes in P. patens as present in seed plants (Chen et al., 2005; Pfeiffer et al., 2012).
HIR-Like Responses Are Not Restricted to Seed Plants
According to the current assumption, seed plants acquired the ability to respond to FR light along with the evolution of PHYA. It has been shown that PHYA is essential for germination and seedling establishment in environments enriched in FR light, such as in canopy shade (Yanovsky et al., 1995; Botto et al., 1996). In contrast with seed plants, cryptogams do not contain PHYA-like phytochromes, and there has been little evidence that FR light has major effects on growth and development of cryptogams (Mathews, 2006). Thus, it has been suggested that PHYA-like FR light signaling is restricted to seed plants. However, we observed that FR light triggered several adaptive and developmental responses in the moss P. patens. Importantly, these responses failed to obey the reciprocity law (i.e., FR light pulses with the same total fluence could not substitute for continuous FR irradiation). This is a hallmark of the HIRs in seed plants, which strictly depend on PHYA (Mathews, 2006; Li et al., 2011). We have recently shown that rapid degradation of the Pfr form and FHY1-dependent nuclear transport are key features of PHYA that are required for the HIRs (Rausenberger et al., 2011). Interestingly, in P. patens, nuclear transport of Pp-PHY1 depends on Pp-FHY1, and Pp-PHY1 as well as Pp-PHY3 are rapidly degraded after conversion to the Pfr form. Thus, our data suggest that Pp-PHY1 and Pp-PHY3 are potential receptors for HIRs in P. patens and that HIR-like responses do not require PHYA. In general, any phytochrome that meets the requirements defined by Rausenberger et al. (2011) (i.e., rapid degradation of the Pfr form and FHY1-dependent nuclear transport) may work as a sensor for HIRs. We conclude that FR light signaling and HIR-like responses are not restricted to seed plants but had already evolved in the last common ancestor of modern seed plants and cryptogams. The capacity to mediate HIRs might have been intrinsic to all phytochromes and could have been lost in the course of evolution by PHYB and other type II phytochromes from seed plants. In seed plants, PHYB works as sensor for the R/FR light ratio, which is important to detect potential competitors at an early stage. As a function in HIR signaling is likely to interfere with the perception of R/FR light ratios, plants may have been under ecological pressure driving the subfunctionalization of phytochromes specialized for either detecting the R/FR light ratio or mediating HIRs. Although the mechanistic similarity of FR light sensing in seed plants and cryptogams strongly suggests a common evolutionary origin, we cannot rule out that they are the result of convergent evolution, leading to similar FR light signaling and response mechanisms.
Interestingly, FR light–induced protonemata and gametophore growth in P. patens were reminiscent of etiolation and shade avoidance growth in seed plants. In Arabidopsis, these responses can be considered as default development at low levels of activated phytochromes, which are thus positive regulators of deetiolation and photomorphogenesis. Under FR light, PHYA antagonizes the shade avoidance response by promoting deetiolation. In contrast with this, we found that P. patens etiolation- and shade avoidance–like growth were impaired in Pp-fhy1 knockout lines. They therefore do not constitute a default response but rather depend on phytochromes acting as positive regulators. This indicates that in seed plants and cryptogams the adaptation to shade conditions relies on phytochrome-dependent FR light sensing but follows different strategies.
In terms of physiological relevance, it is conceivable that FR light signaling is advantageous not only to seed plants, which had to adapt to increased shade conditions, but also to cryptogams. Grassland habitats of P. patens might equally require adaptation of growth and development to micro environments enriched in FR light. Due to its short height, P. patens has to cope with shade conditions induced by neighboring plants typical for grasslands and could thus rely on the ability to perceive and integrate this information. Moreover, other cryptogams typically grow in forest habitats, where the ability to sense FR light might be equally important as for seed plants. As we also identified a potential FHY1 homolog in the green algae Closterium sp (Figure 2A), one could even assume a more general role of FR light signaling, which would not only be important for land plants but also for green algae and which might already have been relevant in their last common ancestor.
Similar to seed plants, the phytochrome gene lineage in P. patens split into two branches, with Pp-PHY1 and Pp-PHY3 on one branch and Pp-PHY2, Pp-PHY4, and Pp-PHY5a/b/c on the other branch (Mittmann et al., 2009). Thus, it is tempting to speculate that Pp-PHY1 and Pp-PHY3 have PHYA-like functions, whereas Pp-PHY2, Pp-PHY4, and Pp-PHY5a/b/c may have functions similar to PHYB. Future work will have to resolve the subfunctionalization of phytochromes from cryptogams and even more ancient branches of the plant kingdom in order to increase our knowledge on the evolution of phytochrome signaling.
METHODS
Cloning of Constructs
A detailed description of DNA constructs and a list including all primers used in this work can be found in Supplemental Methods 1 online. A schematic representation of targeting constructs and genomic loci is given in Supplemental Figure 10 online.
Plant Material and Growth Conditions
The Arabidopsis thaliana wild type used was either the Columbia-0 or Landsberg erecta-0 ecotype. The fhy1-1 and phyA-201 mutants as well as a transgenic line expressing YFP:At-FHY1 (fhy1-1 Pro35S:YFP:FHY1) have been described previously (Whitelam et al., 1993; Quail et al., 1994; Hiltbrunner et al., 2005). Arabidopsis seeds were stratified for at least 2 d at 4°C and grown on 0.5× Murashige and Skoog medium (Duchefa)/0.7% (w/v) agar at 22°C in continuous R (12 μmol m−2 s−1, 656 nm, 24 nm full width at half maximum [FWHM]) or FR light (15 μmol m−2 s−1, 730 nm, 128 nm FWHM).
Cultivation of Physcomitrella patens on solid Knop medium or in liquid Knop culture was performed according to Frank et al. (2005). To induce sporophyte development, gametophores were grown at 16°C and 8/16-h light/dark photoperiod (20 μmol m−2 s−1 PAR). After 4 to 6 weeks, gametophores were flooded with autoclaved tap water and cultivated for another 6 weeks. The sporophytes were opened by mechanical disruption, distributed on solid Knop medium, and incubated in R (670 nm), FR (740 nm), and B light (473 nm) or in darkness. For analysis of protonemata growth, protonemata liquid culture was spotted on solid Knop medium and vertically incubated in R, FR, and B light or darkness. For testing gametophore growth, P. patens gametophores were pregrown in white light and then incubated in R, FR, or B light or darkness.
Adiantum capillus-veneris gametophyte cultures (provided by M. Wada, Kyushu University, Fukuoka, Japan) were grown on White’s Basal Salt Mixtures (Sigma-Aldrich)/0.8% (w/v) agar at 25°C and 16/8-h light/dark photoperiod (50 to 70 μmol m−2 s−1 PAR). Fragmentation of the culture was performed every 2 weeks using a T18 Ultra Turrax disperser (IKA).
Transformation of Arabidopsis and P. patens
Arabidopsis lines expressing Pro35S:Ac-PHY1:YFP:TerRbcS, Pro35S:Cp-PHY2:YFP:TerRbcS, Pro35S:YFP:Pp-FHY1:TerRbcS, or Pro35S:YFP:Cr-FHY1:TerRbcS were obtained by Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998). The selection for transgenic plants using the herbicides BASTA (Hoechst Schering AgrEvo) and Butafenacil/Inspire (Syngenta Agro) was performed as previously described (Block et al., 1987; Rausenberger et al., 2011).
P. patens transformation was performed according to Frank et al. (2005) using liquid cell culture for protoplast preparation. The selection was done on 12.5 mg/mL G418 or hygromycin. Positively selected gametophores were tested with PCR using primers derived from the sequence of the gene of interest and the integrated YFP (p050/p051 for Pp-PHY1:YFP, p028/ah070 for Pp-PHY2:YFP, p062/ah070 for Pp-PHY3:YFP, p023/ah070 for Pp-PHY4:YFP, p030/ah070 for Pp-PHY5a:YFP, and p026/ah070 for Pp-PHY5b:YFP) or with primers p046/p047 and p223/p225 for Pp-FHY1:HPT (see Supplemental Table 1 online for primer sequences). PCR products were ligated blunt-end into pJET1.2 (Fermentas) and sequenced to verify sequences and borders. The molecular characterization of Pp-fhy1 mutant lines is shown in Supplemental Figure 11 online.
Fluorescence Microscopy
Arabidopsis seedlings used for microscopy were grown for 4 d on 0.5× Murashige and Skoog medium/0.7% (w/v) agar as described above and irradiated with either R light (12 μmol m−2 s−1, 656 nm, 24 nm FWHM) or FR light (15 μmol m−2 s−1, 730 nm, 128 nm FWHM) prior to microscopy analyses. A Zeiss Axioscope 2 equipped with YFP-, cyan fluorescent protein (CFP-), and mCherry-specific filter sets (AHF Analysentechnik) was used for image acquisition and ImageJ (version 1.44k; National Institute of Health) and Photoshop (version 10.0.0.1; Adobe) software for image processing.
P. patens protonema liquid culture was transferred to Knop plates with or without 5 μM norflurazone and grown for 2 to 6 d at 25°C and 16/8-h light/dark photoperiod (50 to 70 μmol m−2 s−1 PAR). The cultures were then adapted to darkness for 3 to 6 d. The preparation of samples was done in safety green light (526 nm); for preparation of fixed probes, the protonemata samples were transferred to 1.2% formaldehyde/MTSB (7.5 g PIPES, 0.95 g EGTA, 0.66 g MgSO4•7 H2O, and 2.5 g KOH in 500 mL, pH 7.0) directly after light treatment and incubated in darkness for 10 to 15 min. For 4′,6-diamidino-2-phenylindole (DAPI) staining, P. patens samples were exposed to white light for 3 to 10 min and then imbibed in 0.1 mg/L DAPI in 3% p-formaldehyde/MTSB and incubated in darkness for 15 min. Image acquisition of P. patens and A. capillus-veneris cultures was done on an Axiovert 200M MAT system (Zeiss) or an Axioscope 2 equipped with YFP-, CFP-, chlorophyll-, and mCherry-specific filter sets. All images were acquired using Metamorph (version 6.2r4). ImageJ (version 1.44k) and Photoshop (version 10.0.0.1) software was used for image processing.
Transient Expression in Mustard, P. patens, and A. capillus-veneris
The transient transformation of mustard (Sinapis alba) seedlings has been described previously (Stolpe et al., 2005). After cotransformation with pUC1940:At-PHYA and either pCHF70:At-FHY1 or pCHF70:Pp-FHY1, the mustard seedlings were grown overnight in darkness and used for microscopy analysis as described for Arabidopsis seedlings.
P. patens and A. capillus-veneris cultures were inoculated onto cellophane sheets placed on solid medium, covered with another sheet of cellophane to prevent upright growth, and incubated under standard growth conditions for 4 to 6 d. pCHF70:Pp-FHY1, pPPO30:Ac-PHY1, pUC1930:Ac-PHY2, pUC1930:Ac-PHY3, pPPO30:Cp-PHY2, and pUC1930:At-PHYA, either with or without pCHF150myc or pUC1942, were transiently transformed into P. patens or A. capillus-veneris cultures by particle bombardment with the Biolistic Particle Delivery System (Bio-Rad). Gold particle diameter was 1 μm, helium pressure 7 bar, chamber vacuum pressure 0.8 bar, and target distance 5 cm. After bombardment, the samples were kept in darkness for 1 to 5 d before microscopy analysis.
RNA Extraction and RT-PCR Analyses
P. patens protonemata liquid cultures were inoculated onto cellophane sheets placed on solid Knop medium and incubated for 7 d under standard conditions. At the end of the light period on the 7th day, the samples were transferred to darkness for 5 d. After incubation in R or FR light or continuing darkness, the cultures were harvested and RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). Total RNA (0.5 to 1 µg) was treated with DNaseI (Fermentas) and reverse transcribed using the First-Strand cDNA Synthesis Kit (Fermentas). One to two microliters of the resulting cDNA solution was used for PCR or quantitative PCR (Maxima SYBR Green qPCR Master Mix, Fermentas; Bio-Rad CFX384 real-time system) using standard protocols and primers as follows: p065/p066 for Pp-EF1α, p069/p070 for Pp-FHY1, p158/p159 for 26S rRNA, p118/p119 for Pp-COL2, p186/p187 for Pp-ASN, and p188/p189 for Pp-FNR (see Supplemental Table 1 online for primer sequences).
Immunoblot Analyses
P. patens protonemata liquid cultures were inoculated onto cellophane sheets placed on solid Knop medium and incubated in darkness for 5 d. After light treatment, the plant material was harvested, ground in liquid nitrogen, and resuspended in protein extraction buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% [v/v] glycerol, 1 mM DTT, and complete Protease Inhibitor [Roche]). After centrifugation at 4°C, the cleared protein samples were used for SDS-PAGE and immunoblot transfer according to standard protocols. Immunodetection was performed using protein-specific antibodies (GFP, Abcam; tubulin, Sigma-Aldrich).
Yeast Two-Hybrid Analyses
Yeast two-hybrid analyses and o-Nitrophenyl-β-D-galactopyranoside (ONPG) assays were performed according to previously published protocols (Hiltbrunner et al., 2005). For all yeast two-hybrid assays, the yeast strain Y187 (Clontech) was used. The growth medium was supplemented with phycocyanobilin purified from Spirulina (final concentration 10 μM) (Kunkel et al., 1993).
Database Searches
We searched the databases available at http://www.ncbi.nlm.nih.gov/ and http://www.cosmoss.org/ for proteins from cryptogams containing the consensus PHYA binding motif of FHY1/FHY1-like proteins from seed plants (YVLSSGRWXVNQDKPTIDQEFEQYFSMLML). As suggested by the National Center for Biotechnology Information BLAST program selection guide (http://www.ncbi.nlm.nih.gov/blast/producttable.shtml), we used settings optimized to identify short, nearly exact matches for this search. Sequences were aligned using MAFFT v6.717b and Jalview 2.7 (Waterhouse et al., 2009).
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource, GenBank, or Cosmoss (www.cosmoss.org) databases under the following accession numbers: DK958635 (Ac-FHY1), AB016168 (Ac-PHY1), AB016232 (Ac-PHY2), AB012082 (Ac-PHY3), AT5G02200 (At-FHL), AT2G37678 (At-FHY1), AT1G09570 (At-PHYA), BW647715 (Cl-FHY1), U56698 (Cp-PHY2), BE640872 (Cr-FHY1), AK070454 (Os-FHY1), BT111284 (Pg-FHY1), Phypa_458363 (Pp-ASN), Phypa_441024 (Pp-COL2), Phypa_424011 (Pp-EF1α), Phypa_446283 (Pp-FHY1), Phypa_444678 (Pp-FNR), AB275304 (Pp-PHY1), AB275305 (Pp-PHY2), XM_001765983 (Pp-PHY3), AB275307 (Pp-PHY4), XM_001761093 (Pp-PHY5a), XM_001767172 (Pp-PHY5b), HM751653 (Pp-26S rRNA), XM_002309031 (Pt-FHY1), XM_002468063 (Sb-FHY1), XM_002992728 (Sm-FHY1), and DY831102 (To-FHY1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Nuclear Localization of Physcomitrella Phytochromes.
Supplemental Figure 2. Pfr-Dependent Degradation of PHY1 and PHY3 in Physcomitrella.
Supplemental Figure 3. Nuclear Localization of Ceratodon and Adiantum Phytochromes.
Supplemental Figure 4. Conserved Nuclear Transport Mechanisms for Cryptogam and Seed Plant Phytochromes.
Supplemental Figure 5. Light-Induced Nuclear Transport of Pp-PHY1 Depends on Pp-FHY1.
Supplemental Figure 6. Light-Regulated Interaction of FHY1 and Phytochromes from Mosses and Ferns.
Supplemental Figure 7. Pp-FHY1 Is Essential for FR Light–Induced Gene Expression.
Supplemental Figure 8. Phenotype of the Pp-fhy1 Mutant Lines under Different Light Conditions.
Supplemental Figure 9. Pp-FHY1 Is Required for Branching of Protonema Filaments in a R-FR Light Mixture.
Supplemental Figure 10. Schematic Representation of Genomic Loci (Pp-PHY1-5b and Pp-FHY1) and Targeting Cassettes (Pp-PHY1-5b:YFP and Pp-FHY1 KO).
Supplemental Figure 11. Molecular and Phenotypical Characterization of Independent Pp-fhy1 Mutant Lines.
Supplemental Table 1. Primer List.
Supplemental Methods 1. Cloning of Constructs.
Acknowledgments
We thank Wolfgang Frank (University of Munich, Germany), Ralf Reski (University of Freiburg, Germany), Masamitsu Wada (Kyushu University, Fukuoka, Japan), Tilman Lamparter (Karlsruhe Institute of Technology, Karlsruhe, Germany), Jerzy Paszkowski (University of Geneva, Switzerland), Stanley J. Roux (University of Texas, Austin, TX), Hiroyuki Sekimoto (Japan Women's University, Tokyo, Japan), Andreas Wachter (University of Tübingen, Germany), The Rice Genome Resource Center (Tsukuba, Japan), and the Center for Forest Research (Université Laval, Canada) for providing cDNA clones and plasmids. We thank Wolfgang Frank, Ralf Reski, and Masamitsu Wada for technical advice and for providing P. patens and A. capillus-veneris plants. We thank Claudia König (University of Tübingen, Germany) for excellent technical work. We also acknowledge support by Eberhard Schäfer (University of Freiburg, Germany), in whose lab this project started. We thank Claude Becker (Max Planck Institute for Developmental Biology, Tuebingen, Germany) and Sascha Laubinger (Centre for Plant Molecular Biology, University of Tuebingen, Germany) for helpful discussions and critical reading of the article. This work was supported by a grant (HI 1369/2-1) from the German Research Foundation (Deutsche Forschungsgemeinschaft) to A.H.
AUTHOR CONTRIBUTIONS
A.P. and A.H. designed the research, did the experimental work, analyzed the data, and wrote the article.
Glossary
- B
blue
- R
red
- FR
far-red
- W
white
- HIR
high-irradiance responses
- NLS
nuclear localization signal
- YFP
yellow fluorescent protein
- FWHM
full width half maximum
- CFP
cyan fluorescent protein
- DAPI
4′,6-diamidino-2-phenylindole
References
- Block M.D., Botterman J., Vandewiele M., Dockx J., Thoen C., Gosselé V., Movva N.R., Thompson C., Montagu M.V., Leemans J. (1987). Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 6: 2513–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böse G., Schwille P., Lamparter T. (2004). The mobility of phytochrome within protonemal tip cells of the moss Ceratodon purpureus, monitored by fluorescence correlation spectroscopy. Biophys. J. 87: 2013–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto J.F., Sanchez R.A., Whitelam G.C., Casal J.J. (1996). Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol. 110: 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin M.J., Casal J.J., Whitelam G.C., Sanchez R.A. (1999). A light-regulated pool of phytochrome and rudimentary high-irradiance responses under far-red light in Pinus elliottii and Pseudotsuga menziesii. J. Exp. Bot. 50: 831–836 [Google Scholar]
- Chen M., Tao Y., Lim J., Shaw A., Chory J. (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 15: 637–642 [DOI] [PubMed] [Google Scholar]
- Chen Y.R., Su Y.S., Tu S.L. (2012). Distinct phytochrome actions in nonvascular plants revealed by targeted inactivation of phytobilin biosynthesis. Proc. Natl. Acad. Sci. USA 109: 8310–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Eichenberg K., Bäurle I., Paulo N., Sharrock R.A., Rüdiger W., Schäfer E. (2000). Arabidopsis phytochromes C and E have different spectral characteristics from those of phytochromes A and B. FEBS Lett. 470: 107–112 [DOI] [PubMed] [Google Scholar]
- Frank W., Decker E.L., Reski R. (2005). Molecular tools to study Physcomitrella patens. Plant Biol. (Stuttg.) 7: 220–227 [DOI] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. (2008). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare P.D., Moller S.G., Huang L.F., Chua N.H. (2003). LAF3, a novel factor required for normal phytochrome A signaling. Plant Physiol. 133: 1592–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M., Ulm R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17: 230–237 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Tscheuschler A., Viczián A., Kunkel T., Kircher S., Schäfer E. (2006). FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczián A., Bury E., Tscheuschler A., Kircher S., Tóth R., Honsberger A., Nagy F., Fankhauser C., Schäfer E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15: 2125–2130 [DOI] [PubMed] [Google Scholar]
- Jaedicke K., Lichtenthäler A.L., Meyberg R., Zeidler M., Hughes J. (2012). A phytochrome-phototropin light signaling complex at the plasma membrane. Proc. Natl. Acad. Sci. USA 109: 12231–12236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang S.W., Yang J.Y., Chua N.H. (2007). Independent and interdependent functions of LAF1 and HFR1 in phytochrome A signaling. Genes Dev. 21: 2100–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C., Hersch M., Trevisan M., Genoud T., Hiltbrunner A., Bergmann S., Fankhauser C. (2012). Nuclear phytochrome A signaling promotes phototropism in Arabidopsis. Plant Cell 24: 566–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C., Lorrain S., Hornitschek P., Fankhauser C. (2010). Light-regulated plant growth and development. In Current Topics in Developmental Biology, M.C.P. Timmermans, ed (Oxford, UK: Academic Press), pp. 29–66 [DOI] [PubMed]
- Kanegae T., Wada M. (2006). Photomorphogenesis of ferns. In Photomorphogenesis in Plants and Bacteria., E. Schäfer and F. Nagy, eds (Dordrecht, The Netherlands: Springer), pp. 515–534
- Kircher S., Merkle T., Schäfer E., Nagy F. (2005). Nuclear import of plant proteins. In Nuclear Import and Export, T. Tzfira and V. Citovsky, eds (New York: Kluwer Academic/Plenum Publishers), pp. 100–117
- Kunkel T., Tomizawa K., Kern R., Furuya M., Chua N.H., Schäfer E. (1993). In vitro formation of a photoreversible adduct of phycocyanobilin and tobacco apophytochrome B. Eur. J. Biochem. 215: 587–594 [DOI] [PubMed] [Google Scholar]
- Lamparter T. (2006). Photomorphogenesis in mosses. In Photomorphogenesis in Plants and Bacteria, E. Schäfer and F. Nagy, eds (Dordrecht, The Netherlands: Springer), pp. 537–565
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Wang Deng X. (2011). Phytochrome signaling mechanisms. The Arabidopsis Book 9: e0148, doi/10.1199/tab.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli A.L. (1994). The physiology of phytochrome action. In Photomorphogenesis in Plants, R.E. Kendrick and G.M.H. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 211–269
- Mathews S. (2006). Phytochrome-mediated development in land plants: Red light sensing evolves to meet the challenges of changing light environments. Mol. Ecol. 15: 3483–3503 [DOI] [PubMed] [Google Scholar]
- Mittmann F., Brücker G., Zeidler M., Repp A., Abts T., Hartmann E., Hughes J. (2004). Targeted knockout in Physcomitrella reveals direct actions of phytochrome in the cytoplasm. Proc. Natl. Acad. Sci. USA 101: 13939–13944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittmann F., Dienstbach S., Weisert A., Forreiter C. (2009). Analysis of the phytochrome gene family in Ceratodon purpureus by gene targeting reveals the primary phytochrome responsible for photo- and polarotropism. Planta 230: 27–37 [DOI] [PubMed] [Google Scholar]
- Pfeiffer A., Nagel M.K., Popp C., Wüst F., Bindics J., Viczián A., Hiltbrunner A., Nagy F., Kunkel T., Schäfer E. (2012). Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc. Natl. Acad. Sci. USA 109: 5892–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H., Briggs W.R., Chory J., Hangarter R.P., Harberd N.P., Kendrick R.E., Koornneef M., Parks B., Sharrock R.A., Schäfer E., Thompson W.F., Whitelam G.C. (1994). Spotlight on phytochrome nomenclature. Plant Cell 6: 468–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. (2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825 [DOI] [PubMed] [Google Scholar]
- Rösler J., Jaedicke K., Zeidler M. (2010). Cytoplasmic phytochrome action. Plant Cell Physiol. 51: 1248–1254 [DOI] [PubMed] [Google Scholar]
- Rösler J., Klein I., Zeidler M. (2007). Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc. Natl. Acad. Sci. USA 104: 10737–10742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolpe T., Süsslin C., Marrocco K., Nick P., Kretsch T., Kircher S. (2005). In planta analysis of protein-protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma 226: 137–146 [DOI] [PubMed] [Google Scholar]
- Tepperman J.M., Zhu T., Chang H.S., Wang X., Quail P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi H., Nakamura S., Schäfer E., Wada M. (2012). Red light-induced phytochrome relocation into the nucleus in Adiantum capillus-veneris. Mol. Plant 5: 611–618 [DOI] [PubMed] [Google Scholar]
- Uenaka H., Kadota A. (2007). Functional analyses of the Physcomitrella patens phytochromes in regulating chloroplast avoidance movement. Plant J. 51: 1050–1061 [DOI] [PubMed] [Google Scholar]
- Uenaka H., Wada M., Kadota A. (2005). Four distinct photoreceptors contribute to light-induced side branch formation in the moss Physcomitrella patens. Planta 222: 623–631 [DOI] [PubMed] [Google Scholar]
- Van Buskirk E.K., Decker P.V., Chen M. (2012). Photobodies in light signaling. Plant Physiol. 158: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Kadota A., Furuya M. (1983). Intracellular localization and dichroic orientation of phytochrome in plasma membrane and/or ectoplasm of a centrifuged protonema of fern Adiantum capillus-veneris L. Plant Cell Physiol. 24: 1441–1447 [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. (2009). Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam G.C., Johnson E., Peng J., Carol P., Anderson M.L., Cowl J.S., Harberd N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M.J., Casal J.J., Whitelam G.C. (1995). Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth-responses to natural radiation in Arabidopsis: Weak de-etiolation of the phyA mutant under dense canopies. Plant Cell Environ. 18: 788–794 [Google Scholar]