This article characterizes RCF1, a cold-inducible DEAD box RNA helicase that is important for pre-mRNA splicing of genes. RCF1 regulates the expression of cold-regulated genes and is vital for cold tolerance in plants.
Abstract
Cold stress resulting from chilling and freezing temperatures substantially reduces crop production worldwide. To identify genes critical for cold tolerance in plants, we screened Arabidopsis thaliana mutants for deregulated expression of a firefly luciferase reporter gene under the control of the C-REPEAT BINDING FACTOR2 (CBF2) promoter (CBF2:LUC). A regulator of CBF gene expression1 (rcf1-1) mutant that is hypersensitive to cold stress was chosen for in-depth characterization. RCF1 encodes a cold-inducible DEAD (Asp-Glu-Ala-Asp) box RNA helicase. Unlike a previously reported DEAD box RNA helicase (LOW EXPRESSION OF OSMOTICALLY RESPONSIVE GENES4 [LOS4]) that regulates mRNA export, RCF1 does not play a role in mRNA export. Instead, RCF1 functions to maintain proper splicing of pre-mRNAs; many cold-responsive genes are mis-spliced in rcf1-1 mutant plants under cold stress. Functional characterization of four genes (PSEUDO-RESPONSE REGULATOR5 [PRR5], SHAGGY-LIKE SERINE/THREONINE KINASE12 [SK12], MYB FAMILY TRANSCRIPTION FACTOR CIRCADIAN1 [CIR1], and SPFH/PHB DOMAIN-CONTAINING MEMBRANE-ASSOCIATED PROTEIN [SPFH]) that are mis-spliced in rcf1-1 revealed that these genes are cold-inducible positive (CIR1 and SPFH) and negative (PRR5 and SK12) regulators of cold-responsive genes and cold tolerance. Together, our results suggest that the cold-inducible RNA helicase RCF1 is essential for pre-mRNA splicing and is important for cold-responsive gene regulation and cold tolerance in plants.
INTRODUCTION
Adaptation is the way of life for sessile and poikilothermic land plants, which must endure environmental stresses, including those caused by low/high temperatures, water deficit, and salinity. These abiotic stresses not only limit the geographical distribution of plants but also reduce the global productivity and quality of important agricultural crops. Although plant temperature changes with the ambient temperature, most temperate plants can acquire tolerance to freezing temperatures by a prior exposure to low nonfreezing temperatures, a process termed cold acclimation (Guy, 1990; Thomashow, 1999; Ruelland et al., 2009). Freezing tolerance is essential for temperate crops like winter wheat (Triticum aestivum) and canola (Brassica napus), but tropical and subtropical plants are incapable of cold acclimation. Thus, the productivity and quality of tropical crops (like rice [Oryza sativa], maize [Zea mays], soybean [Glycine max], cotton [Gossypium hirsutum], and tomato [Solanum lycopersicum]) are reduced even by nonfreezing low temperatures (i.e., chilling). Therefore, the engineering or breeding of chilling- and freezing-tolerant crop plants is an important goal in agriculture. Such engineering or breeding requires a thorough understanding of the molecular mechanisms of cold stress signal perception and transduction in plant cells that lead to chilling tolerance and/or cold acclimation.
The expression of many genes in plants is regulated by low temperature (Dhindsa and Monroy, 1994; Guy et al., 1994; Palva et al., 1994; Thomashow, 1994). Most of these genes maintain high levels of expression throughout cold treatment, but their expression decreases rapidly upon return from cold to normal growth temperatures (Thomashow, 1994). The cold-responsive genes encode a diverse array of proteins, including enzymes involved in respiration and in the metabolism of carbohydrates, lipids, phenylpropanoids, and antioxidants; molecular chaperones; antifreezing proteins; and many other proteins of unknown function (Guy et al., 1994).
Differential screening and cloning studies in plants have identified a core set of robustly cold-regulated genes (Thomashow, 1999). The promoters of many of these genes contain one or several copies of the dehydration-responsive element (DRE)/C-repeat (CRT) cis-element, which has the core sequence CCGAC (Yamaguchi-Shinozaki and Shinozaki, 1994; Stockinger et al., 1997). CRT binding factors (CBFs), also known as DRE binding proteins, are upstream transcription factors in the APETALA2/ETHYLENE RESPONSE FACTOR family that bind to the promoter cis-element and activate the expression of these cold-responsive genes (Stockinger et al., 1997; Liu et al., 1998). The CBF genes themselves are induced by low temperatures, and this induction is transient and precedes that of downstream cold-responsive genes with the DRE/CRT cis-element (Medina et al., 1999). Transgenic Arabidopsis thaliana plants ectopically expressing CBF3 contain elevated levels of Pro and soluble sugars, which correlate with increased freezing tolerance (Gilmour et al., 2000). INDUCER OF CBF EXPRESSION1 (ICE1), a basic helix-loop-helix protein, is an upstream transcription factor that binds to the CBF3 promoter and is required for activation of CBF3 expression upon cold stress (Chinnusamy et al., 2003). An R2R3-type MYB transcription factor, MYB15, interacts with ICE1 and negatively regulates the expression of CBF genes under cold stress (Agarwal et al., 2006). HOS1, a negative regulator of the CBF2 and CBF3 genes, was identified from a genetic screen for mutants with enhanced expression of CBF target genes (Ishitani et al., 1998; Lee et al., 2001). HOS1 encodes a RING finger protein that has ubiquitin E3 ligase activity (Lee et al., 2001; Dong et al., 2006). Both in vitro and in vivo ubiquitination assays showed that HOS1 mediates the polyubiquitination of ICE1 under cold conditions (Dong et al., 2006). SENSITIVE TO FREEZING6 plays a role in cold acclimation via the CBF pathway after CBF translation (Knight et al., 2009). Recently, Shi et al. (2012) reported that proteins in the ethylene signaling pathway negatively regulate freezing tolerance by downregulation of CBF and type-A Arabidopsis RESPONSE REGULATOR genes in Arabidopsis.
CBF2 negatively regulates CBF1 and CBF3 (Novillo et al., 2004), and Zn TRANSPORTER OF Arabidopsis thaliana12 (ZAT12; a C2H2 zinc finger protein) negatively regulates the expression of the CBF genes (Vogel et al., 2005). Novillo et al. (2007) reported that CBF1 and CBF3 function additively in cold acclimation and differently from CBF2; CBF2 defines different gene classes in the CBF regulon. Doherty et al. (2009) showed that members of calmodulin binding proteins of the CAMTA family of transcription factors can bind to one of the cis-elements in the CBF2 promoter in vitro and are important for freezing tolerance. Additional molecular factors modulating CBF2 expression remain to be identified.
To identify additional molecular factors that are critical for cold tolerance and cold-responsive gene regulation, we fused the CBF2 promoter to a firefly luciferase reporter gene (CBF2:LUC) and introduced this cold-inducible gene cassette into Arabidopsis plants. Screening of the ethyl methanesulfonate (EMS)–mutagenized M2 plants led to the isolation of mutants with altered CBF gene expression and/or cold stress tolerance. We designated these mutants as regulator of CBF gene expression (rcf). We report the in-depth characterization of one such mutant, rcf1-1. The rcf1-1 mutant plants are hypersensitive to chilling and freezing temperatures. Map-based cloning revealed that RCF1 encodes a cold-inducible DEAD (Asp-Glu-Ala-Asp) box RNA helicase. Unlike a previously reported DEAD box RNA helicase (LOS4) that regulates mRNA export, RCF1 maintains proper splicing of pre-mRNAs of nuclear-encoded genes. Loss-of-function and gain-of-function characterization of four genes that are mis-spliced in rcf1-1 under cold stress revealed that they are important regulators of cold-responsive genes and cold tolerance. Together, our results indicate that a cold-inducible nuclear-localized RNA helicase RCF1 is critical for pre-mRNA splicing and for cold-responsive gene regulation and cold tolerance in plants.
RESULTS
Isolation of the rcf1-1 Mutant
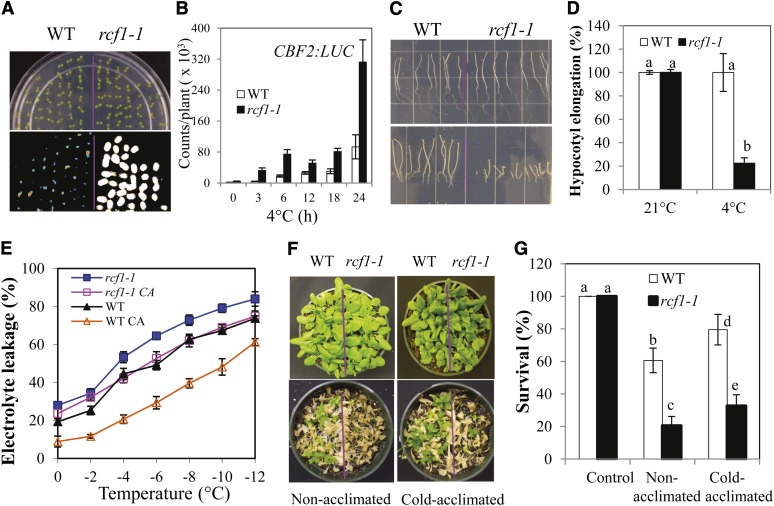
To identify novel molecular factors that regulate the expression of cold-responsive transcription factors and that have an essential role in cold tolerance, we generated transgenic Arabidopsis plants that express a firefly LUC reporter gene under the control of the cold-responsive CBF2 promoter (CBF2:LUC, referred to as the wild type) and subsequently screened the EMS-mutagenized M2 population for mutants with deregulated CBF2:LUC expression. We designated these mutants as rcf. The rcf1-1 mutant was chosen for in-depth characterization. Compared with wild-type plants, rcf1-1 plants have a much higher level of CBF2:LUC expression under cold stress (Figures 1A and 1B), suggesting that RCF1 might be a negative regulator of CBF2 gene expression.
Figure 1.
RCF1 Is a Positive Regulator of Cold Tolerance.
(A) CBF2:LUC expression in 7-d-old wild-type (WT) and rcf1-1 seedlings treated at 4°C for 24 h.
(B) Quantification of luminescence intensity in (A). CBF2:LUC expression was quantitatively measured as luminescence intensity (counts/plant). Data are also included for nonstressed and cold-treated seedlings at different time points.
(C) Chilling sensitivity of the wild type and rcf1-1 at 4°C in darkness. Top panel, 21°C for 15 d; bottom panel, 4°C for 52 d.
(D) Quantification of hypocotyl elongation of plants shown in (C). Hypocotyl elongation of the wild type at 21°C or 4°C was treated as 100%.
(E) Leakage of electrolytes from excised leaflets of rcf1-1 and wild-type plants when treated at temperatures below freezing. rcf1-1 CA, cold-acclimated rcf1-1 plants; WT CA, cold-acclimated wild-type plants.
(F) Whole-plant freezing tolerance of the wild type and rcf1-1. Plants were photographed 10 d after freezing treatments.
(G) Survival rates of rcf1-1 and the wild type in (F).
Error bars represent the sd (n = 20 to 40 in [B], 60 to 100 in [D], 8 to 16 in [E], and 100 to 140 in [G]). One-way analysis of variance (ANOVA; Tukey-Kramer test) was performed for data in (D) and (G), and statistically significant differences are indicated by different lowercase letters (P < 0.003). Experiments in were repeated at least three times with similar results, and values shown are from one experimental repetition.
We backcrossed rcf1-1 with the wild type. All F1 plants showed a wild-type phenotype in response to cold stress, and F2 plants segregated at ∼3:1 (wild type versus rcf1-1; see Supplemental Table 1 online). These results suggested that rcf1-1 is a recessive mutation in a single nuclear gene.
RCF1 Is Required for Plant Tolerance to Chilling and Freezing Stresses
We investigated the effect of the rcf1-1 mutation on plant sensitivity to chilling and freezing stresses. Chilling tolerance was assessed based on hypocotyl elongation in the dark. As shown in Figures 1C and 1D, hypocotyl elongation was similar for rcf1-1 and the wild type at 21°C but was dramatically less for rcf1-1 than for the wild type at 4°C, indicating that normal function of RCF1 is required for chilling stress tolerance. We subsequently determined the freezing tolerance of rcf1-1 by two methods: an electrolyte leakage assay (Sukumaran and Weiser, 1972; Ishitani et al., 1998) and a whole-plant freezing assay (Warren et al., 1996; Jaglo-Ottosen et al., 1998; Xin and Browse, 1998; Zhu et al., 2008). The rcf1-1 plants were hypersensitive to freezing temperatures before and after cold acclimation (Figures 1E to 1G), indicating that the ability to be fully acclimated is substantially reduced in rcf1-1. Together, these results suggest that RCF1 is a positive regulator of chilling and freezing tolerance in plants.
RCF1 Regulates Gene Expression under Cold Stress
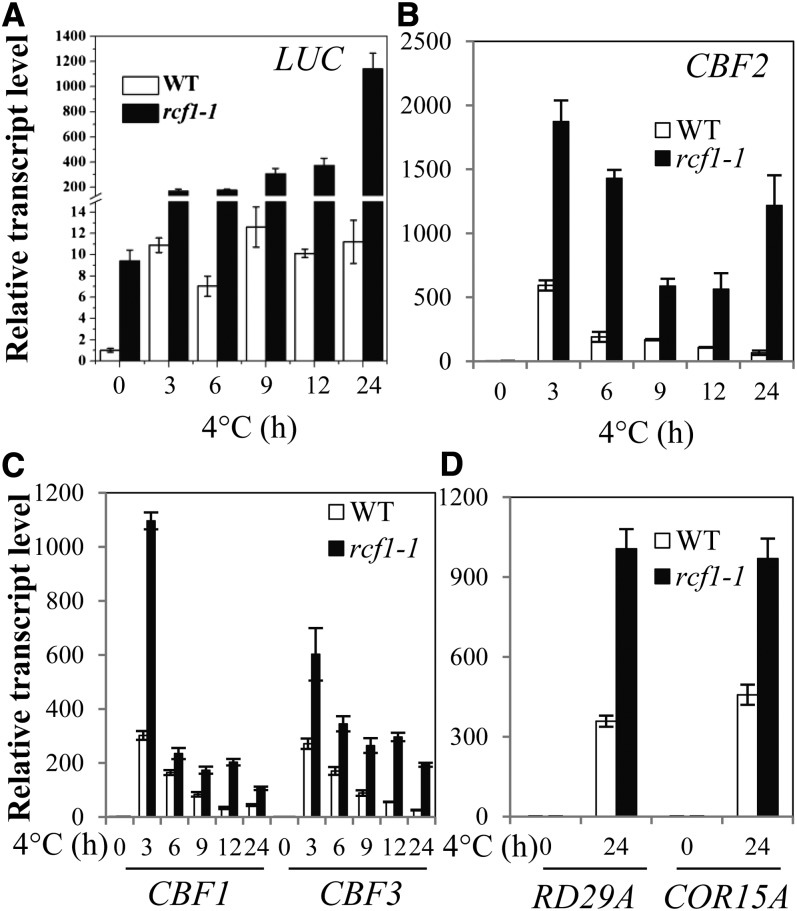
To gain insight into the molecular function of RCF1 in the cold stress tolerance pathway, we examined its role in gene regulation with quantitative real-time RT-PCR (qRT-PCR) analysis and whole-genome microarray analysis. Consistent with the increased CBF2:LUC expression in rcf1-1, qRT-PCR analysis revealed that transcripts of LUC and endogenous CBF2 were more abundant in rcf1-1 at all time points after cold treatment (Figures 2A and 2B). We then determined whether the rcf1-1 mutation affects other members in the CBF gene family. Expression of CBF1 and CBF3 was dramatically elevated in rcf1-1 compared with the wild type throughout cold treatment (Figure 2C). CBF members are known to regulate the expression of downstream genes that have the DRE/C-repeat cis-element in their promoters. Therefore, we examined whether rcf1-1 affects the expression of known CBF downstream genes, such as RD29A and COR15A. Cold induction of RD29A and COR15A was substantially greater in rcf1-1 than in wild-type plants (Figure 2D). These results, which sharply contrast with the reduced cold tolerance of the rcf1-1 mutant (Figures 1C to 1G), indicate that RCF1 is a negative regulator of the expression of CBF genes and their downstream target genes under cold stress. These results also suggest that CBF-independent factors may not function properly in the rcf1-1 mutant plants. Alternatively, increased expression of CBF genes in rcf1-1 may be a compensatory response to the severely reduced cold tolerance of rcf1-1.
Figure 2.
The rcf1-1 Mutation Causes Higher Induction of the LUC Gene and Endogenous Cold-Responsive Genes under Cold Stress.
Transcript levels of the LUC gene and of endogenous CBF2, CBF1, CBF3, RD29A, and COR15A in 14-d-old wild-type (WT) and rcf1-1 seedlings treated at 4°C for the indicated time periods. Error bars represent the sd (n = 4). Experiments in were repeated at least three times with similar results, and values shown are from one experimental repetition.
To detect changes in global gene expression in the rcf1-1 mutant plants, we performed a whole-genome microarray analysis with Affymetrix Arabidopsis ATH1 GeneChips. RNA was extracted from both wild-type and rcf1-1 seedlings that had been treated at 4°C for 0, 12, or 24 h. Analyses of the ATH1 microarray data indicated that, compared with their expression in wild-type seedlings, 35 genes were upregulated by at least threefold and five genes were downregulated by at least threefold in the rcf1-1 mutant under control conditions (see Supplemental Table 2 online). Among the 35 upregulated genes in rcf1-1, most encode proteins that are predicted to function in response to stress (see Supplemental Table 2A online). The ATH1 microarray data also showed that after a 12-h cold treatment and compared with the wild type, rcf1-1 contains 49 genes whose expression is significantly increased by at least threefold and 32 genes whose expression is significantly reduced by at least threefold (see Supplemental Data Set 1 online). After a 24-h cold treatment and compared with the wild type, expression in rcf1-1 was increased by at least threefold for 95 genes and decreased by at least threefold for 73 genes (see Supplemental Data Set 2 online). Compared with their expression in the wild type, the expression of five genes was significantly increased in rcf1-1 by at least threefold at all time points after cold treatment, while expression of one gene was significantly reduced in rcf1-1 by at least threefold at all time points after cold treatment (see Supplemental Tables 3A and 3B online). Microarray data analysis also revealed that transcripts of 20 genes were significantly increased in rcf1-1 by at least threefold after 12- and 24-h cold treatment, while transcripts of six genes were dramatically reduced in rcf1-1 by at least threefold after 12- and 24-h cold treatment (see Supplemental Tables 3C and 3D online). Although the genes in rcf1-1 that are differentially expressed in response to cold stress encode proteins involved in diverse biological processes, a relatively large proportion of the upregulated genes and some of the downregulated genes encode proteins that are predicted to function in biotic or abiotic stress response pathways (see Supplemental Table 3 and Supplemental Data Sets 1 and 2 online). Relative to publically available data concerning gene expression in response to cold stress in wild-type plants, gene expression substantially differs in rcf1-1 (see Supplemental Figures 1 to 5 online).
In our ATH1 microarray analysis, transcripts of CBF1, CBF2, and CBF3 were significantly increased in rcf1-1 after 12 or 24 h of cold stress, suggesting that our microarray data are reliable. Our microarray analysis was confirmed by qRT-PCR analysis of seven genes that showed altered expression patterns in rcf1-1 (Figure 2D; see Supplemental Figures 6A to 6D online). For example, both microarray analysis and qRT-PCR indicated that ZAT12 in rcf1-1 is upregulated after 12 and 24 h of cold treatment (see Supplemental Figure 6A online). Similarly, the detection of upregulation and downregulation was consistent for ATH1 microarray analysis and qRT-PCR for At5g59950, Stabilized 1, and At2g42270, which encode an RNA binding protein, a pre-mRNA splicing factor (STA1) (Lee et al., 2006), and a U5 small nuclear ribonucleoprotein helicase (Brr2b), respectively (see Supplemental Figures 6B and 6C online). Both ATH1 microarray and qRT-PCR analyses also indicated that one of the upstream transcriptional activators of CBF2, the CAMTA family protein CAMTA1, is downregulated in rcf1-1 (see Supplemental Data Set 1 and Supplemental Figure 6D online). These results suggest that elevated accumulation of the CBF2 gene in rcf1-1 is not due to the increased activity of CAMTA1. Together, these data indicate that RCF1 plays an important role in gene regulation under both normal and cold stress conditions.
RCF1 Encodes a Cold-Inducible DEAD Box RNA Helicase
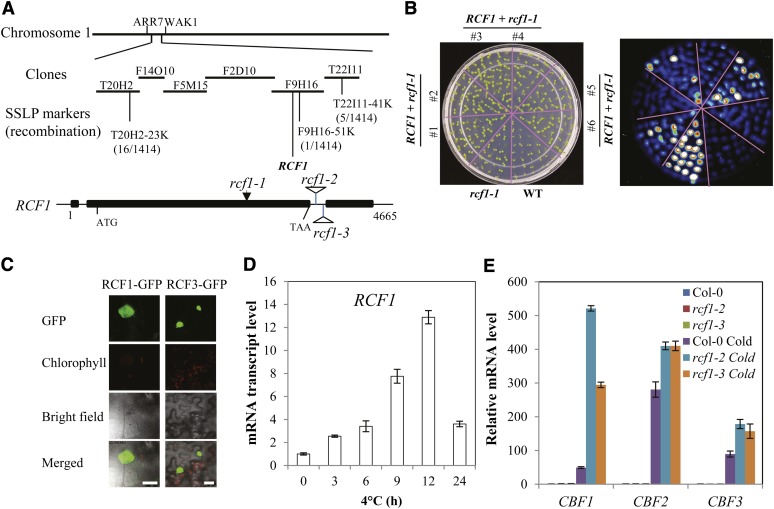
The rcf1-1 phenotypes indicate that RCF1 has an essential role in chilling and freezing tolerance and in the regulation of cold-responsive gene expression. We used a map-based cloning strategy to identify the RCF1 gene (Figure 3A). A segregating F2 population was generated from a cross between rcf1-1 (in the Columbia background) and the wild-type Landsberg erecta. A total of 1414 homozygous rcf1-1 mutant plants were selected from the F2 population, and genomic DNA was extracted from each plant for genetic mapping. The RCF1 locus is in the lower arm of chromosome 1 between BACs T20H2 and T22I11. The RCF1 locus was narrowed to BAC clone F9H16, and the candidate genes were sequenced. A single-nucleotide mutation from G to A at position 2650 from the transcription start site was found in At1g20920 of the rcf1-1 mutant. This mutation would change the amino acid Gly to Arg at position 808 of the decoded At1g20920 polypeptide. The At1g20920 gene encodes a putative DEAD box RNA helicase. We confirmed the identity of RCF1 with a gene complementation test using the wild-type RCF1 gene, including its own promoter and coding sequences (Figure 3B). We made an RCF1-GFP (for green fluorescent protein) fusion protein in transgenic tobacco (Nicotiana benthamiana) and Arabidopsis plants to determine the RCF1 subcellular localization, and confocal microscopy indicated that RCF1 is predominantly localized in the nucleus (Figure 3C; see Supplemental Figure 6E online). The RCF1-GFP fusion protein is able to fully restore the rcf1-1 mutant phenotype (see Supplemental Figure 6F online), indicating that the RCF1-GFP fusion protein is functional in planta. The expression of RCF1 appears to be upregulated by cold stress and reaches its peak level after 12 h of cold treatment (Figure 3D). qRT-PCR analysis indicated that rcf1-2 and rcf1-3 are null alleles of RCF1 and that the rcf1-2 and rcf1-3 mutations have the same effect as the rcf1-1 mutation on the expression of CBF genes (Figure 3E; see Supplemental Figure 6G online).
Figure 3.
Molecular Cloning of RCF1.
(A) Positional cloning of RCF1. Numbers of recombination are from 1414 F2 progeny seedlings that are homozygous for rcf1-1 phenotypes. The rcf1-1 mutation is caused by a single-nucleotide substitution (from G to A at position 2650, relative to transcription start site), and this mutation changes amino acid Gly-808 to Arg. Structure of the RCF1 gene and positions of rcf1-1, rcf1-2, and rcf1-3 mutations are indicated. Filled boxes indicate exons, and lines between boxes indicate introns.
(B) Molecular complementation of the rcf1-1 mutant by the wild-type (WT) RCF1 gene. Shown are seedlings on an MS agar plate (left) and the corresponding luminescence image after 4°C treatment for 24 h (right).
(C) RCF1-GFP is localized in the nucleus of tobacco leaf epidermal cells. RCF3-GFP fusion protein was used as a positive control (Guan et al., 2012). Bars = 25 µm.
(D) Time-course expression of RCF1 in 14-d-old wild-type seedlings.
(E) Transcript levels of CBF1, CBF2, and CBF3 in Col-0, rcf1-2, and rcf1-3 seedlings subjected to 0 or 12 h at 4°C. Error bars in (D) and (E) indicate the sd (n = 4).
RNA helicases are a class of enzymes that use energy derived from the hydrolysis of ATP to unwind double-stranded RNAs (de la Cruz et al., 1999). We produced recombinant RCF1 protein and tested its RNA helicase activity by measuring its ATPase activity. RCF1 indeed exhibits RNA-dependent ATPase activity (see Supplemental Figures 7A and 7B online). When the DEAD box domain (core amino acid sequence Asp-Glu-Ala-Asp [D-E-A-D]) is altered to DAAD, the RCF1 ATPase activity is abolished (see Supplemental Figure 7B online). We also found that RCF1 carrying this DAAD mutation under the control of the RCF1 native promoter failed to complement the rcf1-1 mutant (see Supplemental Figure 7C online), indicating that the DEAD box domain of RCF1 is critical for its function in planta.
RCF1 Is Not Involved in mRNA Export
The DEAD box RNA helicase LOS4 was previously identified through a genetic screen of deregulated expression of RD29A:LUC (Gong et al., 2005). LOS4 is localized in the cytoplasm in a nuclear rim–enriched pattern and is important for mRNA export at cold temperatures (Gong et al., 2005). We used the poly(A) in situ hybridization assay to determine whether RCF1 is also involved in mRNA export. As shown in Supplemental Figure 7D online, the different responses of los4-1 versus C24 (background of los4-1) to the presence and absence of cold stress suggest that our experimental conditions were similar to those reported by Gong et al. (2005), but differences were not detected in the responses of the wild type versus rcf1-1. These data indicate that RCF1 is not involved in mRNA export.
RCF1 Is Required for Proper Splicing of Pre-mRNAs for Cold-Responsive Genes Including Positive and Negative Regulators of CBFs and for Cold Tolerance
Database searches revealed that RCF1 coexpresses with a pre-mRNA splicing factor, STA1, and a U5 small nucleoprotein helicase, Brr2b. We isolated a knockdown mutant of Brr2b (see Supplemental Figure 8A online). We found that, like RCF1, STA1 and Brr2b also negatively regulate the expression of CBF2 (see Supplemental Figure 8B online). Double mutants of rcf1-1 sta1-2 and rcf1-1 brr2b did not show any additive effect on CBF2 expression (see Supplemental Figure 8B online), suggesting that RCF1, STA1, and Brr2b function in a common pathway to regulateCBF2 gene expression. We observed that Brr2b is cold inducible (see Supplemental Figure 8C online). Plants of sta1 and brr2b mutants were hypersensitive to chilling stress, as indicated by reduced hypocotyl growth (see Supplemental Figures 8D to 8G online; Lee et al., 2006). Both sta1-2 and brr2b mutant plants were more sensitive to freezing stress than wild-type plants before and after cold acclimation, as determined by an electrolyte leakage assay that indicated increased damage in the membranes of the mutant plants (see Supplemental Figure 8H online). Together, these data suggest that, like RCF1, STA1 and Brr2b are positive regulators of cold tolerance.
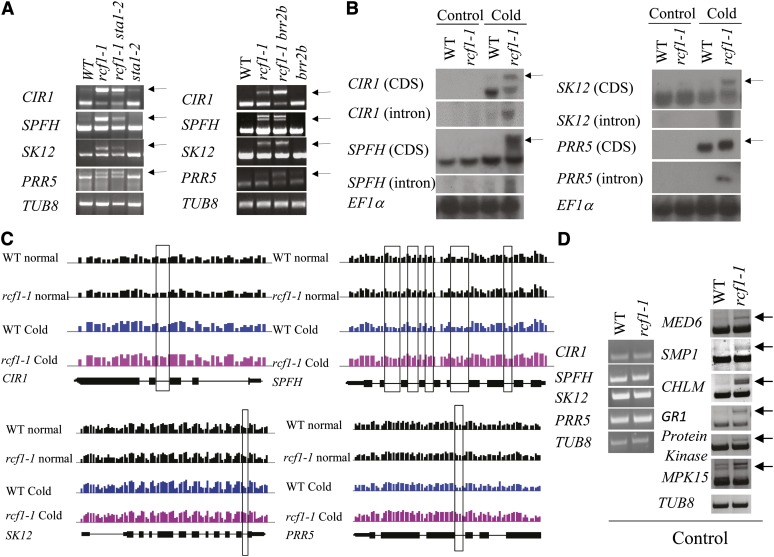
Because database searches suggested that RCF1, STA1, and Brr2b are part of a spliceosome containing more than 100 proteins (The Arabidopsis Information Resource) and because STA1 is involved in pre-mRNA splicing (Lee et al., 2006), we determined the potential role of RCF1 in pre-mRNA splicing. To identify candidate genes whose splicing events may be affected by the rcf1-1 mutation, we performed full-genome tiling arrays with Affymetrix Arabidopsis tiling array GeneChips (1.0R) with wild-type and rcf1-1 plants that had been treated at 4°C for 0 or 12 h. Statistical analysis of the 1.0R tiling array data detected intron retention events (false discovery rate < 0.05) for 204 unique genes in the rcf1-1 mutant plants subjected to cold treatment for 12 h, which is when RCF1 reaches its peak expression level (see Supplemental Data Set 3A online). These mis-spliced genes in rcf1-1 encode proteins with diverse functions in many biological processes, and the predicted roles of more than one-third (77 of 204) of these genes involve responses to abiotic or biotic stresses (see Supplemental Data Set 3A online). We designed primers and probes unique to the introns of genes that are retained in the rcf1-1 mutant, and we performed RT-PCR and RNA gel blot hybridization analyses to validate the 1.0R tiling array data. Six genes that displayed mis-spliced transcripts in rcf1-1 as revealed in the 1.0R tiling array experiments with cold-treated plants were selected for validation: At5g37260, At5g54100, At5g24470, At3g05840, At5g25350, and At1g27910. These genes encode MYB family transcription factor CIRCADIAN1 (CIR1), SPFH/PHB DOMAIN-CONTAINING MEMBRANE-ASSOCIATED PROTEIN (SPFH), PSEUDO-RESPONSE REGULATOR5 (PRR5), a SHAGGY-LIKE SERINE/THREONINE KINASE12 (SK12), EIN3 BINDING F BOX PROTEIN2 (EBF2), and PLANT U-BOX45 (PUB45), respectively. Transcripts of the six genes were found to contain at least one intron in the rcf1-1 mutant under cold treatment (Figures 4A to 4C; see Supplemental Figures 9A and 9B online). The retained introns were not detected in rcf1-1 plants without cold treatment (Figures 4B to 4D; see Supplemental Figures 9B and 9C online). The sta1-2 and brr2b mutations do not seem to suppress the mis-splicing effect of rcf1-1 on these six loci (Figure 4A; see Supplemental Figure 9A online), suggesting that RCF1 may function in a different pathway than STA1 and Brr2b for pre-mRNA splicing under cold stress.
Figure 4.
RCF1 Functions in Pre-mRNA Splicing under Cold Treatment.
(A) Mis-spliced transcripts of CIR1, SPFH, SK12, and PRR5 in rcf1-1, rcf1-1 sta1-2, and rcf1-1 brr2b, but not in sta1-2 or brr2b, under cold stress (4°C for 12 h) as determined by RT-PCR analysis. WT, the wild type.
(B) RNA gel blot hybridization analysis of CIR1, SPFH, SK12, and PRR5 genes in rcf1-1. Membranes were also hybridized with introns specific to CIR1, SPFH, SK12, and PRR5.
(C) Visualization of intron retention of CIR1, SPFH, SK12, and PRR5 in rcf1-1 with the integrated genome browser. Vertical bars represent averaged log2 expression values of unique probes in the gene region; introns that were retained in rcf1-1 under cold treatment are indicated with black boxes; gene structures on the bottom of each panel are based on TAIR10 annotation.
(D) Transcripts of genes tested in (A) and MED6 (At3g21350), SMP1 (At1g65660), CHLM (At4g25080), GR1 (At3g52115), protein kinase (At1g21590), and MPK15 (At1g73670) under unstressed condition. TUB8 and EF1α were used as loading controls.
Arrows in (A), (B), and (D) indicate intron-retained transcripts. The experiments in (except in [C]) were repeated at least three times with similar results, and data shown are from one experimental repetition.
[See online article for color version of this figure.]
Statistical analysis of the 1.0R tiling array data generated with RNA samples extracted from wild-type and rcf1-1 plants grown under normal conditions did not detect any genes with significant intron retention events (false discovery rate < 0.05) in rcf1-1 (Gene Expression Omnibus [GEO] accession number GSE41377). Although the 1.0R tiling array data produced from the unstressed wild-type and rcf1-1 plants did not pass the thresholds in our statistical analysis, we were able to confirm the intron retention events through RT-PCR analysis for the following six genes: At3g21350, At1g65660, At4g25080, At3g52115, At1g21590, and At1g73670. At3g21350 encodes MEDIATOR6 (MED6), which is an important component of the mediator complex in the regulation of RNA polymerase II transcription activity. At1g65660 encodes SWELLMAP1 (SMP1), which is a CCHC zinc finger protein that may function as a step II splicing factor. At4g25080 encodes a protein with methyltransferase activity responsible for the methylation of magnesium protoporphyrin IX (CHLM). At3g52115 encodes GAMMA RESPONSE GENE1 (GR1), which is induced in response to ionizing radiation and may be involved in DNA damage-induced growth arrest. At1g21590 encodes a protein kinase with an adenine nucleotide α-hydrolase–like domain. At1g73670 encodes MAP KINASE15 (MPK15) (see Supplemental Data Set 3B and Supplemental Figure 10 online; Figure 4D). These encoded proteins have diverse functions in cellular processes, including signal transduction (MPK15), gene regulation (SMP1 and MED6), and stress responses (GR1 and At1g21590) (see Supplemental Data Set 3B online).
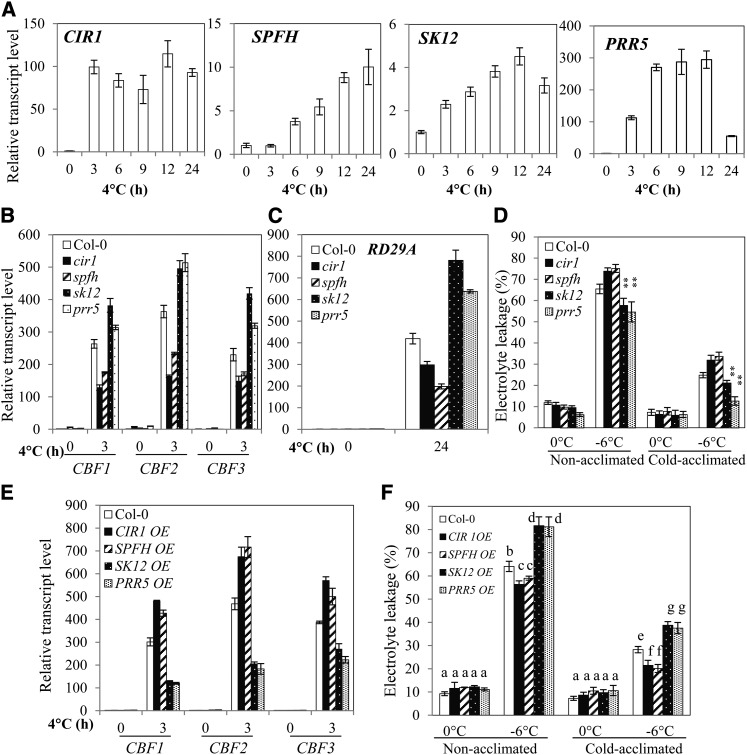
Searches of the publically available microarray databases revealed that many of the mis-spliced genes in rcf1-1 under cold stress are cold responsive in wild-type plants, suggesting that they play a role in the cold stress tolerance pathway (see Supplemental Figure 9D online). Indeed, CIR1, SPFH, SK12, and PRR5 are cold inducible (Figure 5A). To investigate possible functions of these four genes in cold stress responses, we isolated their T-DNA knockouts (see Supplemental Figures 11A to 11D online). As shown in Figures 5B to 5D, expression of CBFs and RD29A is substantially reduced in cir1 and spfh plants under cold stress, and these mutant plants are hypersensitive to freezing stress before and after cold acclimation. Furthermore, transgenic plants that overexpress CIR1 (CIR1 OE) and SPFH (SPFH OE) display increased expression of CBF genes and increased tolerance to freezing stress before and after cold acclimation (Figures 5E and 5F; see Supplemental Figure 11E online). These results indicate that CIR1 and SPFH are positive regulators for cold-responsive gene expression and cold tolerance. By contrast, sk12 and prr5 plants displayed elevated expression of CBFs and RD29A under cold and increased tolerance to freezing temperatures (Figures 5B to 5D). In addition, transgenic plants that overexpress SK12 (SK12 OE) and PRR5 (PRR5 OE) showed reduced expression of CBF genes and decreased tolerance to freezing stress (Figures 5E and 5F; see Supplemental Figure 11F online). These results suggest that SK12 and PRR5 are negative regulators of cold-responsive gene expression and cold tolerance.
Figure 5.
CIR1, SPFH, SK12, and PRR5 Regulate Expression of CBF Genes and Cold Tolerance.
(A) CIR1, SPFH, SK12, and PRR5 are cold inducible in the wild type.
(B) and (C) Relative transcript levels of CBF1, CBF2, CBF3, and RD29A in cir1, spfh, sk12, and prr5 mutant plants.
(D) Freezing tolerance of cir1, spfh, atsk12, and prr5 mutant plants determined by electrolyte leakage assays. **P < 0.01, as determined by Student’s t test.
(E) Relative transcript levels of CBF1, CBF2, and CBF3 in overexpression plants of CIR1 (CIR1 OE), SPFH (SPFH OE), At-SK12 (SK12 OE), and PRR5 (PRR5 OE).
(F) Freezing tolerance of CIR1 OE, SPFH OE, AtSK12 OE, and PRR5 OE plants. Cold acclimation in (D) and (F) was achieved by incubating plants at 4°C for 1 week. Data in (E) and (F) are from one representative transgenic line of two independent transgenic lines of CIR1 OE, SPFH OE, At-SK12 OE, and PRR5 OE plants (see Supplemental Figures 11E and 11F online).
Error bars indicate the sd (n = 4 in [B], [C], and [E]; n = 12 to 15 in [D] and [F]). One-way ANOVA (Tukey-Kramer test) was performed for data in (F), and statistically significant differences are indicated by different lowercase letters (P < 0.01). Experiments in were repeated at least four times with similar results, and values shown are from one experimental repetition.
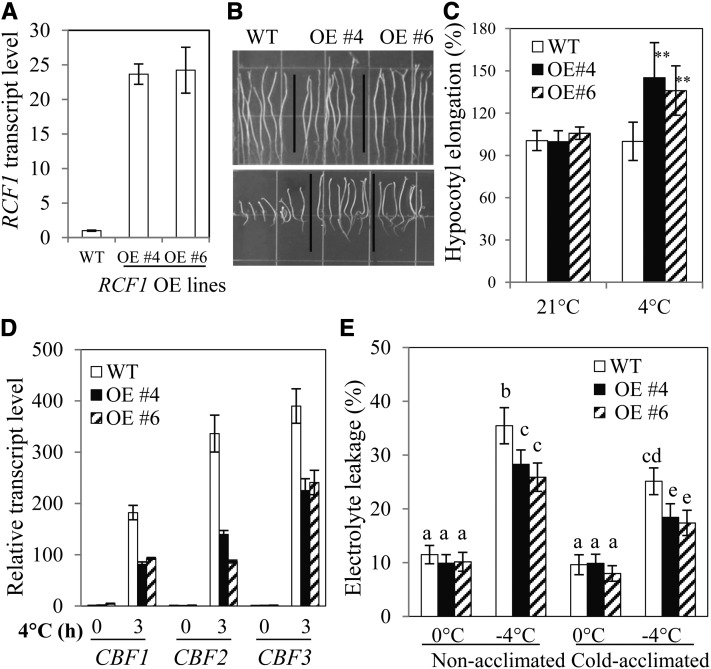
Overexpression of RCF1 in Arabidopsis Increases Tolerance to Chilling and Freezing Stresses
Because RCF1 is required for cold stress tolerance and is important for cold-responsive gene regulation, we investigated whether overproduction of RCF1 would improve the performance of plants under cold stress. We generated transgenic Arabidopsis plants that overexpress RCF1 under the control of the 35S promoter, and we selected two independent RCF1 overexpression lines (Figure 6A). The RCF1 overexpression lines developed longer hypocotyls than the wild type under chilling stress, indicating an increased resistance to chilling stress (Figures 6B and 6C). Expression of CBF genes is repressed in the RCF1 overexpression lines (Figure 6D). These results further support our conclusion that RCF1 is a negative regulator for these cold-responsive transcriptional activators. In an electrolyte leakage assay, the RCF1 overexpression lines displayed significantly increased tolerance to freezing temperature with or without cold acclimation (Figure 6E). Together, these results further support our conclusion that RCF1 is required for cold tolerance in plants.
Figure 6.
Overexpression of RCF1 Increases Cold Stress Tolerance.
(A) RCF1 expression levels in the wild type (WT) and two transgenic lines overexpressing RCF1 (OE).
(B) Chilling stress tolerance of wild-type and RCF1 OE plants. Top panel, 21°C for 13 d; bottom panel, 4°C in the dark for 32 d.
(C) Quantification of hypocotyl elongation of plants shown in (B). Hypocotyl elongation of the wild type (at 21°C or 4°C) was considered to be 100%. **P < 0.01, as determined by Student’s t test.
(D) Expression of CBF1, CBF2, and CBF3 in wild-type and RCF1 OE plants subjected to 0 or 3 h at 4°C.
(E) Freezing tolerance of RCF1 OE plants determined by electrolyte leakage assays.
Cold acclimated (4°C for 1 week). Error bars indicate the sd (n = 4 in [A] and [D]; n = 20 in [C]; n = 12 in [E]). One-way ANOVA (Tukey-Kramer test) was performed for data in (E), and statistically significant differences are indicated by different lowercase letters (P < 0.005). Experiments in were repeated at least three times with similar results, and values shown are from one experimental repetition.
DISCUSSION
We identified the cold-inducible DEAD box–containing RNA helicase RCF1 through a forward genetic screen for genes critical for cold tolerance. We confirmed that recombinant RCF1 protein produced in Escherichia coli has RNA helicase activity and that changing the functional DEAD box to DAAD abolishes its helicase activity (see Supplemental Figure 7 online). Similarly, the RCF1 gene carrying the DAAD mutation in the DEAD box failed to complement the rcf1-1 mutant, suggesting that the DEAD domain or the RNA helicase activity of RCF1 is essential for its biological function.
The RCF1 protein differs from a previously identified DEAD box RNA helicase, LOS4, in subcellular localization and function. Although both proteins are localized in the nucleus, LOS4 is enriched in the nuclear rims (Gong et al., 2005), but RCF1 is not (Figure 3C; see Supplemental Figure 6E online). The subcellular localization of LOS4 is correlated with its important role in mRNA export at cold temperatures (Gong et al., 2005), and the different location for RCF1 indicates that it is probably not involved in mRNA export. A role of RCF1 in mRNA export was ruled out by the poly(A) in situ hybridization assays (see Supplemental Figure 7D online).
DEAD box RNA helicases contribute to all aspects of RNA metabolism, including nuclear gene transcription, pre-mRNA splicing, nucleo-cytoplasmic transport, and gene expression (Rocak and Linder, 2004; Cordin et al., 2006). In this study, three lines of evidence demonstrate the role of RCF1 in pre-mRNA splicing. First, RCF1 coexpresses with another RNA helicase, Brr2b, and a pre-mRNA splicing factor, STA1. Second, database searches indicated that RCF1, STA1, and Brr2b are part of a spliceosome composed of over 100 proteins. Although there is no experimental evidence that Brr2b is involved in pre-mRNA splicing in Arabidopsis, a Brr2b homolog in yeast is involved in pre-mRNA splicing (Hahn et al., 2012). We therefore reasoned that RCF1 may function in some aspects of splicing. Third, full-genome tiling array analyses indicated that RCF1 regulates pre-mRNA splicing of six genes without cold stress and 204 unique genes under cold stress (see Supplemental Data Set 3 online). Because the mis-spliced genes in rcf1-1 under cold stress are not affected in sta1-2 or brr2b mutants, RCF1 seems to function in the pre-mRNA splicing pathway independently of STA1 and Brr2b. Based on our results, we believe that RCF1, Brr2b, and STA1 may be components of different pre-mRNA splicing complexes that regulate control the pre-mRNA splicing of different genes. That our 1.0R tiling array experiments detected only a relatively small number of mis-spliced genes in rcf1-1 under normal growth condition (Figure 4D; see Supplemental Figure 10 online; GEO accession number GSE41377) may be explained by the fact that the 1.0R tiling arrays used in this study may be insufficiently sensitive to detect all mis-splicing events.
Our data indicate that rcf1-1 mutant plants are defective in basal freezing tolerance (Figures 1E to 1G) and that the reduced basal freezing tolerance may be due to mis-splicing of the six genes in rcf1-1 under normal conditions (Figure 4D). Two of the six mis-spliced genes (ATGR1 and a protein kinase) in rcf1-1 under normal conditions have predicted roles in stress responses (see Supplemental Data Set 3B online). Mis-splicing of these two genes in rcf1-1 under normal conditions may reduce the ability of rcf1-1 to cope with freezing temperatures without cold acclimation. Two additional genes whose transcripts are mis-spliced in rcf1-1 under normal conditions (SMP1 and MED6) are involved in gene regulation. SMP1 encodes a CCHC zinc finger protein with step II splicing factor activity. The spliceosome catalyzes pre-mRNA splicing in two steps. After catalytic step I, a major remodeling of the spliceosome occurs to establish the active site for step II. Step II splicing factors are responsible for the correct selection of 3′ splicing sites (Chua and Reed, 1999). SMP1 is functionally redundant with SMP2. One of the functional targets of SMP1 and SMP2 is the transcript of STRUWWELPETER that resembles the subunits of the mediator complex required for transcriptional activation (Clay and Nelson, 2005). MED6 is one of the mediators of RNA polymerase II (Pol II). The mediator of Pol II is required for diverse aspects of the transcription process, including activation, repression, basal transcription, and phosphorylation of the C-terminal domain of the largest subunit of Pol II (Kim et al., 1994; Björklund and Kim, 1996). MED6 homologs in Drosophila melanogaster and yeast play important roles as transcriptional coactivators (Lee and Kim, 1998; Gim et al., 2001). The MED6 in Arabidopsis may function similarly to its homologs in yeast and Drosophila. Furthermore, splicing of MPK15 is defective in rcf1-1 under normal conditions, and MPK15 is likely involved in the activation of gene expression. As mentioned above, MPK15, SMP1, and MED6 are involved in gene regulation, and mis-splicing of these three genes in rcf1-1 may alter expression of genes, for example, those genes that are differentially expressed in rcf1-1 under nonstressed conditions (see Supplemental Table 2 online). The altered gene expression in rcf1-1 may contribute to reduced basal freezing tolerance.
Although rcf1-1 plants should theoretically display enhanced freezing tolerance when transcripts of cold-responsive genes accumulate to a higher level, the opposite phenotype was observed (Figures 1 and 2). Our results show that RCF1 is required for tolerance to chilling and freezing stresses in plants because rcf1-1 mutant plants are hypersensitive to cold stress, and overexpression of RCF1 increases cold tolerance (Figures 1 and 6). These results suggest that an essential regulator(s) of cold tolerance is defective in rcf1-1. CIR1 and SPFH are two good candidates for such essential regulators of cold tolerance, and transcripts of these two genes were mis-spliced in rcf1-1 under cold stress (Figure 4). CIR1 is involved in circadian-regulated developmental processes, and circadian rhythmic expression of CIR1 is regulated by the central oscillators CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), two MYB transcription factors that have partially redundant functions (Schaffer et al., 1998; Wang and Tobin, 1998; Mizoguchi et al., 2002; Zhang et al., 2007). The functions of CCA1 and LHY in the regulation of CBF expression have been reported (Dong et al., 2011), but the function of CIR1 in cold stress response has not been determined. SPFH is one of the SPFH/PHB (stomatin-prohibtin-flotillin-HflC/K) domain-containing superfamily proteins, and members of the SPFH protein superfamily are generally associated with plasma or mitochondrial membranes and are involved in many cellular processes, including protein turnover or oligomerization, cell proliferation, and ion channel regulation (Tavernarakis et al., 1999; Nadimpalli et al., 2000; Rivera-Milla et al., 2006). According to our investigation of the loss-of-function and gain-of-function of the CIR1 and SPFH genes, CIR1 and SPFH positively regulate expression of cold-responsive genes, including CBFs, and are required for cold tolerance (Figures 5B to 5F). CIR1 and SPFH appear to function in the cold stress tolerance pathway in a CBF-dependent manner. In addition, the list of RCF1 target genes as revealed by the ATH1 microarray and 1.0R tiling array analyses might include other positive and negative regulators that are essential for cold stress tolerance (for example, factors functioning in CBF-independent pathways). The molecular function of these positive and negative regulators for cold tolerance requires further study, but indications about these regulators were provided by the gene expression data generated in our microarray and tiling array analyses. Although the differentially expressed genes in rcf1-1 with or without cold stress encode proteins that are involved in diverse biological processes, many of the upregulated genes and some of the downregulated genes in rcf1-1 encode proteins that are predicted to function in the biotic or abiotic stress response pathways (see Supplemental Table 2 and Supplemental Data Sets 1 and 2 online). Gene expression data in rcf1-1 with and without cold treatment were quite different from publically available gene expression data from wild-type plants (see Supplemental Figures 1 to 5 online). In the latter case, genes are normally upregulated in wild-type plants after cold stress, but these genes failed to increase in rcf1-1 in response to cold (see Supplemental Figures 3 and 5 online). Similarly, genes that are repressed by cold stress in the wild type are turned on in rcf1-1 (see Supplemental Figures 1A, 2, and 4 online). Aberrant expression of these genes in rcf1-1 under cold stress potentially compromises the mutant’s ability to cope with freezing temperatures. Furthermore, many of the mis-spliced genes in rcf1-1 revealed by the 1.0R tiling array analyses are responsive to cold stress in wild-type plants, suggesting that they are involved in cold stress responses (see Supplemental Figure 9D online). The mis-splicing and resulting disruption of functions of some of these genes in rcf1-1 plants may contribute to the increased cold sensitivity of rcf1-1.
The increased accumulation of CBF genes and their downstream targets in rcf1-1 may be explained by the action of RCF1 target genes as revealed in the 1.0R tilling array experiments. PRR5 and SK12 appear to be negative regulators of CBFs and RD29A (Figures 5B and 5C). PRR5 is strongly induced by cold, and SK12 is moderately upregulated by cold (Figure 5A). PRR5 is a negative regulator of CBF genes and is partially redundant with PRR7 and PRR9 (Nakamichi et al., 2009). The null allele of prr5 used in this study (see Supplemental Figure 11D online) differed from those reported previously (Nakamichi et al., 2009), and this single mutation was sufficient to display phenotypes such as altered expression of CBF genes and sensitivity to freezing temperatures. Mis-spliced transcripts of PRR5 and SK12 in rcf1-1 presumably disrupt the normal function of these two genes, leading to increased induction of CBF genes in rcf1-1 in a similar way to that observed in the prr5 and sk12 mutant plants. Overexpression of PRR5 or SK12 results in reduced expression of CBF genes and reduced freezing tolerance (Figures 5E and 5F), confirming that PRR5 and SK12 are negative regulators of cold-responsive gene expression and cold tolerance.
There are additional examples of mutants that are more sensitive to cold stress than the wild type even though they accumulate higher levels of cold-responsive genes. The sta1-1, sta1-2, and brr2b mutant plants are hypersensitive to chilling and freezing stresses, although the CBF2 gene is expressed at a higher level in these three mutants (see Supplemental Figure 8 online). Thus, STA1 and Brr2b are required for cold tolerance. Because the cold response of rcf1-1 is similar to that of sta1 and brr2b, it is possible that RCF1, STA1, and Brr2b share a common subset of target genes critical for cold tolerance. Identification of these common target genes regulated by RCF1, STA1, and Brr2b requires further investigation. Nevertheless, STA1 and Brr2b are two examples (in addition to RCF1) of important proteins that are required for cold tolerance and that act as negative regulators of CBF gene expression. Increased expression of CBF2 in sta1-2 and brr2b can be a compensatory response to an increased sensitivity to cold stress, as was the case with sta1-2 and brr2b. The overaccumulated transcripts of CBF genes in rcf1-1 may be a compensatory response to the increased sensitivity of rcf1-1 to cold stress. The underlying mechanism for this type of compensatory response has been a mystery. Our results show that the defective splicing of SK12 and PRR5 explains at least part of the compensatory response in the rcf1-1 mutant. We have observed a similar phenomenon in hos9-1 and hos15 mutants, which accumulate higher levels of some cold-responsive genes but are hypersensitive to cold stress (Zhu et al., 2004, 2008). Unlike rcf1-1, however, hos9-1 and hos15 mutations do not affect cold induction of CBF genes.
The importance of RCF1 protein in cold stress responses is further supported by the consequences of RCF1 overexpression in Arabidopsis. Overexpression of RCF1 enhances the tolerance to chilling and freezing stresses (Figure 6), indicating that RCF1 and the splicing regulated by it are essential for cold tolerance. Furthermore, expression of CBF genes was downregulated in RCF1 overexpression lines, confirming that RCF1 is indeed a negative regulator of CBF genes. When RCF1 is overexpressed, positive regulators for cold tolerance are presumably produced at a higher level, and negative regulators are presumably suppressed to improve performance under cold stress. Although determining precisely how RCF1 functions in cold tolerance will require further investigation, our forward genetic analysis has demonstrated that by maintaining splicing of pre-mRNAs under cold conditions, RCF1 is a crucial regulator of cold tolerance in plants.
METHODS
Plant Materials and Growth Conditions
A firefly LUC reporter gene driven by the cold stress-responsive CBF2 promoter (−1500 to −1 bp upstream of the transcription start site) was introduced into Arabidopsis thaliana plants in the Columbia glabrous1 background. Seeds from one homozygous line expressing a single functional copy of the CBF2:LUC gene (referred to as the wild type) were mutagenized with EMS. The rcf1-1 mutant with altered CBF2:LUC gene expression was identified from M2 seedlings using a charge-coupled device camera imaging system (Ishitani et al., 1997).
Seeds of the following T-DNA insertion mutants were obtained from the ABRC: rcf1-2 (SALK_138032), rcf1-3 (SALK_062599), sta1-2 (SAIL_262-C08), brr2b (SALK_048780), prr5 (SALK_135000), sk12 (CS856017), cir1 (SALK_051843), and spfh (SALK_090074). The sta1-1 mutant was described previously (Lee et al., 2006). Arabidopsis seedlings on Murashige and Skoog (MS) medium agar plates (1× MS salts, 2% Suc, and 0.6% agar, pH 5.7) were routinely grown under cool, white light (∼120 μmol m−2 s−1) at 21 ± 1°C with a 16-h-light/8-h-dark photoperiod. Soil-grown plants were kept under cool, white light (∼100 μmol m−2 s−1) with a 16-h-light/8-h-dark photoperiod at 21 ± 1°C and with a 1:1 ratio of Metro Mix 360 and LC1 potting soil (Sun Gro Horticulture).
Chilling and Freezing Tolerance Assays
For chilling tolerance assays, seeds of relevant genotypes were sown side-by-side on agar plates containing MS medium. These agar plates were then wrapped with aluminum foil and kept vertically at 4°C or 21°C in growth chambers for the desired time. Reduction in hypocotyl elongation at 4°C relative to that of the wild type at 21°C was used as an indicator of sensitivity to chilling stress.
To evaluate freezing tolerance using electrolyte leakage assays, 3-week-old wild-type and mutant or transgenic plants were grown in soil at room temperature or at 4°C under a long-day photoperiod (16-h-light/8-h-dark) for 1 week. Fully developed rosette leaves were used for electrolyte leakage measurements as described (Sukumaran and Weiser, 1972; Ristic and Ashworth, 1993; Zhu et al., 2004). Whole-plant freezing tests were as described (Xiong et al., 2001) with modifications. Wild-type and rcf1-1 plants were grown in soil under a long-day photoperiod (16-h-light/8-h-dark) in a growth chamber for 3 weeks at 21°C and then at 4°C for 1 week for cold acclimation. The plants were then placed in a low-temperature chamber with the following freezing temperature regimen: from 4 to −1°C in 30 min, then hold at −1°C for 1 h (to initiate nucleation); then successive 2°C decreases at 30-min intervals to reach the next temperature (hold at the following temperatures for 3 h: −4, −6, −8, −10, and −12°C). Plant damage was scored 7 d later (Xiong et al., 2001).
Genetic Mapping and Complementation
The rcf1-1 mutant was crossed with the Landsberg erecta accession, and 1414 mutant plants were chosen from the F2 generation based on altered CBF2:LUC phenotype. Simple sequence length polymorphism markers were designed according to the information in the Cereon Arabidopsis Polymorphism Collection and were used to analyze recombination events (Jander et al., 2002). Initial mapping revealed that the rcf1-1 mutation is located on the upper arm of chromosome 1 between T20H2 and T22I11. Fine mapping within this chromosomal interval narrowed the RCF1 locus to the BAC clone F9H16. All candidate genes in this BAC were sequenced from the rcf1-1 mutant and compared with those in GenBank to find the rcf1-1 mutation.
For complementation of the rcf1-1 mutant, a 6975-bp genomic fragment of At1g20920 that included 2224 bp upstream of the translation initiation codon and 1251 bp downstream of the translation stop codon was amplified with F9H16 as a template (see Supplemental Data Set 4 online for primer sequences). The amplified fragment was first cloned through Gateway technology (Invitrogen) into the pENTR4 vector, resulting in plasmid pENTR4-RCF1. The pENTR4-RCF1 plasmid was then subjected to site-directed mutagenesis to change the DEAD domain into the DAAD mutant version at the amino acid level with the primer pair listed in Supplemental Data Set 4 online. The wild-type RCF1 gene and mutated RCF1 (DEAD to DAAD) were then introduced into pMDC99. The constructs were transferred into Agrobacterium tumefaciens (strain GV3101), and rcf1-1 plants were transformed by the floral dip method (Clough and Bent, 1998).
RCF1 Subcellular Localization and Overexpression of RCF1, PRR5, SK12, CIR1, and SPFH
The coding region of RCF1 was amplified by PCR and cloned into the pMDC83 vector to produce the RCF1-GFP fusion protein. This construct (pMDC83-RCF1) was then introduced into Arabidopsis wild-type (ecotype Columbia) and rcf1-1 plants by floral dip transformation with Agrobacterium strain GV3101. The pMDC83-RCF1 plasmid was also transformed into Agrobacterium strain C58C1 and coinfiltrated with 35S:p19 (p19 is a RNA silencing repressor protein from tomato bushy stunt virus; Voinnet et al., 2003) in Agrobacterium strain C58C1 into the 3-week-old leaves of tobacco (Nicotiana benthamiana) plants. The infiltrated tobacco plants were allowed to grow for an additional 3 d in a growth chamber under a 16-h-light/8-h-dark photoperiod at 21°C. The subcellular localization of RCF1-GFP protein in tobacco leaves or in root tissues of Arabidopsis transgenic plants (T2 generation) was determined with a Leica SP5X confocal microscope (Leica Microsystems). The rcf1-1 mutant plants transformed with pMDC83-RCF1 in the T2 generation were examined for CBF2:LUC expression to determine whether the pMDC83-RCF1 fusion protein is functional in planta. Nuclear localization of pMDC83-RCF3 fusion protein in tobacco leaves was used as a positive control. RCF3 protein was described previously (Guan et al., 2012).
For overexpression of RCF1, PRR5, SK12, CIR1, and SPFH, coding regions of these genes were amplified by PCR and cloned into the pMDC32 vector. The resulting plasmids (pMDC32-RCF1, pMDC32-PRR5, pMDC32-SK12, pMDC32-CIR1, and pMDC32-SPFH) were then transferred into Agrobacterium (strain GV3101), and wild-type Arabidopsis plants (ecotype Columbia) were transformed by the floral dip method. Transgenic plants resistant to hygromycin (50 µg/mL) in the T2 generation were tested for resistance to cold stress and for gene regulation under cold stress.
Microarray Analysis, Tilling Array Analysis, and Real-Time RT-PCR Analysis
Fourteen-day-old seedlings grown on MS medium were used for RNA isolation. Total RNA was extracted from the wild type, different mutants, and/or transgenic plants with Trizol reagent (Invitrogen) and treated with DNase I (New England Biolabs) to remove any genomic DNA contaminants.
For GeneChip Arabidopsis Genome (ATH1, Affymetrix) array analysis, total RNA was used to prepare biotin-labeled complementary RNA targets. Microarray analysis was performed at the School of Medicine, University of Maryland at Baltimore, as described (Breitling et al., 2004). Two biological replicates were used for each genotype. The data sets were subjected to the robust multiarray averaging normalization method. The robust multiarray averaging method for computing an expression measure is begun by computing background-corrected perfect match intensities for each perfect match cell on every GeneChip. The normalized data were further analyzed, and P values were generated by the affylmGUI component of Bioconductor in statistics environment R with the default parameters (Irizarry et al., 2003; Gentleman et al., 2004). Genes with statistically significant differences in expression between the rcf1-1 mutant and the wild type were selected by the RankProd method, which is a nonparametric method for identifying differentially expressed up- or downregulated genes based on the estimated percentage of false discoveries (P < 0.05) (Hong et al., 2006). RankProd results were summarized with the script written in PERL.
For GeneChip Arabidopsis Tiling (1.0R, Affymetrix) array analysis, total RNA extracted from cold-treated (4°C for 12 h, which is when RCF1 is expressed at its peak level) wild-type and rcf1-1 plants was used, and labeling, hybridization, and scanning were performed in the Genomics Core of the Institute for Integrative Genome Biology at the University of California at Riverside. Three biological replicates were used for each genotype. We first remapped the tiling array probes to the Arabidopsis genome (TAIR10) using SOAP2 (Li et al., 2009) and kept only probes that perfectly matched to a unique position in the genome for subsequent analyses. We then created a custom chip definition file using the probe mapping result and used the aroma.affymetrix framework (Bengtsson et al., 2008) to quantile normalize the raw tiling array data (three biological replicates each for the rcf1-1 mutant and wild-type control). To identify retained introns, we first calculated the log2 signal intensity for each annotated intron (TAIR10) based on the trimmed mean of signal intensities from all mapped probes. Introns with fewer than three mapped probes or with low expression (log2 expression value < 5 in all samples) were not further considered. The SAM algorithm (Tusher et al., 2001) was then used to identify introns with significantly elevated expression in the rcf1-1 mutant samples relative to the wild type. A false discovery rate of 0.05 was used as the significance cutoff. Intron retention in rcf1-1 of selected genes under both normal and cold stress conditions was visualized with the integrated genome browser (http://bioviz.org/igb/; Nicol et al., 2009).
For qRT-PCR analysis, 5 µg of total RNA was used to synthesize the first-strand cDNA with the Maxima first-strand cDNA synthesis kit (Fermentas) as described (Guan et al., 2012). Each experiment had three to five biological replicates (four technical replicates for each biological replicate), and each experiment was repeated at least three times. The comparative cycle threshold method was applied, and TUB8 was used as a reference gene.
RNA Gel Blot Hybridization Analysis
Fourteen-day-old seedlings grown on MS medium and subjected to 0- or 12-h cold treatment at 4°C were used for RNA isolation. Total RNA was extracted with Trizol reagent (Invitrogen) and treated with DNase I (New England Biolabs) for potential genomic DNA contamination. Total RNA (30 µg) was then separated on a formaldehyde-containing agarose (1.2%, [w/v]) gel, transferred to a nylon transfer membrane (GE Healthcare) overnight, and cross-linked. Blots were hybridized overnight (0.5 M phosphate buffer, pH 7.2, 7% [w/v] SDS, 1 mM EDTA, and 2 mM BSA) with 32P-labeled probe at 60°C. Washes (20 min each at 60°C) were performed first in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% SDS, then in 1× SSC and 0.1% SDS, and finally in 0.5× SSC and 0.1% SDS. The ELONGATION FACTOR1α (EF1α) gene was used as a loading control.
ATPase Activity Assay
A fragment of RCF1 cDNA (corresponding to amino acids 501 to 910 from the N-terminal region, spanning the ATPase motif) was amplified with the primer pair listed in Supplemental Data Set 3 online and cloned into pDEST15 to make a glutathione S-transferase–tagged GST-RCF1 fusion protein. The resulting plasmid pDEST15-RCF1 was used as a template to make a mutant version of RCF1 (DEAD domain changed to DAAD) through site-directed mutagenesis (pDEST15-RCF1 [DAAD]). The pDEST15-RCF1 and pDEST15-RCF1 (DAAD) plasmids were transformed into Escherichia coli. Rosetta (DE3)pLysS cells (EMD Millipore) and fusion proteins were purified with glutathione S-transferase affinity columns (GE Healthcare). The ATPase activities of RCF1 and its mutant version were determined as described by Iost et al. (1999) with minor modifications. This method uses pyruvate kinase and lactate dehydrogenase to link hydrolysis of ATP to oxidation of NADH, which results in a decrease in the absorbance at 338 nm. Briefly, assays were performed at 37°C in a reaction volume of 0.2 mL, in buffer containing 20 mM Tris-HCl, pH 8.0, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 1 mM ATP, 300 μM NADH, 2 mM phosphoenolpyruvate, 73.5 nM RCF1 (or RCF1 [DAAD]), and 3 units/mL of pyruvate kinase and lactate dehydrogenase with or without 50 μg/mL Arabidopsis total RNA.
Poly(A) RNA in Situ Hybridization Assay
Poly(A) RNA in situ hybridization was conducted essentially as described by Engler et al. (1994) and Gong et al. (2005). The samples were observed immediately using a Leica SP5X confocal microscope (Leica Microsystems) with a 488-nm excitation laser and a 522/DF35 emission filter. All samples were observed under the same conditions, including the use of the same ×60 objective lens and the same laser strength.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RCF1 (At1g20920), STA1 (At4g03430), Brr2b (At2g42270), CBF1 (At4g25490), CBF2 (At4g25470), CBF3 (At4g25480), RD29A (At5g52310), PRR5 (At5g24470), SK12 (At3g05840), PUB45 (At1g27910), EBF2 (At5g25350), CIR1 (At5g37260), SPFH (At5g54100), COR15A (At2g42540), MED6 (At3g21350), SMP1 (At1g65660), CHLM (At4g25080), GR1 (At3g52115), protein kinase (At1g21590), MPK15 (At1g73670), TUB8 (At5g23860), and EF1α (At1g07920). The microarray and tiling array data discussed in this article have been deposited in the National Center for Biotechnology Information’s GEO (Edgar et al., 2002) and can be accessed through GEO Series accession numbers GSE039090, GSE41377, and GSE41378.
Supplemental Data
The following is available in the online version of this article.
Supplemental Figure 1. Hierarchical Clustering Analysis of Genes in Wild-Type Plants in Response to Cold Stress Treatments Based on Publically Available Gene Expression Data; These Genes Showed Increased (A) and Reduced (B) Expression in rcf1-1 without Cold Treatment as Determined by the ATH1 Microarray Analysis.
Supplemental Figure 2. Hierarchical Clustering Analysis of Genes in Wild-Type Plants in Response to Cold Stress Treatments Based on Publically Available Gene Expression Data; These Genes Showed Increased Expression in rcf1-1 after 12-h Cold Treatment as Determined by the ATH1 Microarray Analysis.
Supplemental Figure 3. Hierarchical Clustering Analysis of Genes in Wild-Type Plants in Response to Cold Stress Treatments Based on Publically Available Gene Expression Data; These Genes Showed Reduced Expression in rcf1-1 after 12-h Cold Treatment as Determined by the ATH1 Microarray Analysis.
Supplemental Figure 4. Hierarchical Clustering Analysis of Genes in Wild-Type Plants in Response to Cold Stress Treatments Based on Publically Available Gene Expression Data; These Genes Showed Increased Expression in rcf1-1 after 24-h Cold Treatment as Determined by the ATH1 Microarray Analysis.
Supplemental Figure 5. Hierarchical Clustering Analysis of Genes in Wild-Type Plants in Response to Cold Stress Treatments Based on Publically Available Gene Expression Data; These Genes Showed Reduced Expression in rcf1-1 after 24-h Cold Treatment as Determined by the ATH1 Microarray Analysis.
Supplemental Figure 6. Expression of Cold-Responsive Genes in Wild-Type and rcf1-1 Seedlings, Subcellular Localization of RCF1, and RCF1 Expression in rcf1-2 and rcf1-3 Plants.
Supplemental Figure 7. RCF1 Is an RNA-Dependent RNA Helicase That Is Not Involved in mRNA Export.
Supplemental Figure 8. STA1 and Brr2b Negatively RegulateCBF2 Expression and Are Positive Regulators of Cold Tolerance.
Supplemental Figure 9. Mis-Spliced Transcripts of EBF2 and PUB45 in rcf1-1 as Determined by RT-PCR and Hierarchical Clustering Analysis of Genes in Wild-Type Plants in Response to Cold Stress Treatments Based on Publically Available Gene Expression Data.
Supplemental Figure 10. Visualization of Intron Retention of MED6 (At3g21350), SMP1 (At1g65660), CHLM (At4g25080), GR1 (At3g52115), Protein Kinase (At1g21590), and MPK15 (At1g73670) under Unstressed Condition in rcf1-1 with the Integrated Genome Browser.
Supplemental Figure 11. Expression of CIR1, SPFH, SK12, and PRR5 in T-DNA knockouts of CIR1, SPFH, SK12, and PRR5 (cir1, spfh, prr5, and sk12), CIR1 Overexpression (CIR1 OE), SPFH Overexpression (SPFH OE), SK12 Overexpression (SK12 OE), and PRR5 Overexpression (PRR5OE) Seedlings Determined by qRT-PCR Analysis.
Supplemental Table 1. Genetic Analysis of rcf1-1 (Wild Type [Female] × rcf1-1 [Male] Cross).
Supplemental Table 2. Genes with Increased and Reduced Expression in rcf1-1 without Cold Treatment as Determined by Microarray Analysis.
Supplemental Table 3. Common Genes with Altered Expression Levels in rcf1-1 after 0-, 12-, and 24-h Cold Treatment and Common Genes with Altered Expression Levels in rcf1-1 after 12- and 24-h Cold Treatment as Determined by Microarray Analysis.
Supplemental Data Set 1. Genes with Increased and Reduced Expression in rcf1-1 after 12-h Cold Treatment as Determined by Microarray Analysis.
Supplemental Data Set 2. Genes with Increased and Reduced Expression in rcf1-1 after 24-h Cold Treatment as Determined by Microarray Analysis.
Supplemental Data Set 3. Mis-Spliced Genes in rcf1-1 as Determined by Tiling Array Analysis.
Supplemental Data Set 4. Primers Used in This Study.
Acknowledgments
We thank Jheesoo Ahn, Gary Coleman, Lixin Li, and Xiule Yue for their technical assistance. This work was supported by a USDA hatch fund (CA-R*-BPS-7754 H) to R.L. and by National Science Foundation Grants IOS0919745 and MCB0950242 to J.Z.
AUTHOR CONTRIBUTIONS
Q.G., J.W., and J.Z. designed the research. Q.G., J.W., Y.Z., C.C., C.J., and J.Z. performed the research. R.L. analyzed the tiling array data. Q.G. and J.Z. analyzed the remaining data. Q.G. and J.Z. wrote the article.
Glossary
- CBF
CRT binding factor
- EMS
ethyl methanesulfonate
- qRT-PCR
quantitative real-time RT-PCR
- GEO
Gene Expression Omnibus
- Pol II
RNA polymerase II
- MS
Murashige and Skoog
- ANOVA
analysis of variance
References
- Agarwal M., Hao Y., Kapoor A., Dong C.H., Fujii H., Zheng X., Zhu J.-K. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281: 37636–37645 [DOI] [PubMed] [Google Scholar]
- Bengtsson H., Simpson K., Bullard J., Hansen K. (2008). aroma.affymetrix: A Generic Framework in R for Analyzing Small to Very Large Affymetrix Data Sets in Bounded Memory. Tech Report #745. (Berkeley, CA: University of California)
- Björklund S., Kim Y.J. (1996). Mediator of transcriptional regulation. Trends Biochem. Sci. 21: 335–337 [DOI] [PubMed] [Google Scholar]
- Breitling R., Armengaud P., Amtmann A., Herzyk P. (2004). Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X., Agarwal M., Zhu J.-K. (2003). ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K., Reed R. (1999). The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature 402: 207–210 [DOI] [PubMed] [Google Scholar]
- Clay N.K., Nelson T. (2005). The recessive epigenetic swellmap mutation affects the expression of two step II splicing factors required for the transcription of the cell proliferation gene STRUWWELPETER and for the timing of cell cycle arrest in the Arabidopsis leaf. Plant Cell 17: 1994–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cordin O., Banroques J., Tanner N.K., Linder P. (2006). The DEAD-box protein family of RNA helicases. Gene 367: 17–37 [DOI] [PubMed] [Google Scholar]
- de la Cruz J., Kressler D., Linder P. (1999). Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24: 192–198 [DOI] [PubMed] [Google Scholar]
- Dhindsa R.S., Monroy A.F. (1994). Low temperature signal transduction, gene expression, and cold acclimation: Multiple roles of low temperature. In Biochemical and Cellular Mechanisms of Stress Tolerance in Plants, J.H. Cherry, ed (Berlin: Springer-Verlag), pp. 501–514
- Doherty C.J., Van Buskirk H.A., Myers S.J., Thomashow M.F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.-K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M.A., Farré E.M., Thomashow M.F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 7241–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler J.A., Van Montagu M., Engler G. (1994). Hybridization in situ of whole-mount messenger RNA in plants. Plant Mol. Biol. Rep. 12: 321–331 [Google Scholar]
- Gentleman R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S.J., Sebolt A.M., Salazar M.P., Everard J.D., Thomashow M.F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gim B.S., Park J.M., Yoon J.H., Kang C., Kim Y.J. (2001). Drosophila Med6 is required for elevated expression of a large but distinct set of developmentally regulated genes. Mol. Cell. Biol. 21: 5242–5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Dong C.H., Lee H., Zhu J., Xiong L., Gong D., Stevenson B., Zhu J.-K. (2005). A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q., Wen C., Zeng H., Zhu J. (November 21, 2012). A KH domain-containing putative RNA-binding protein is critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Mol. Plant http://dx.doi.org/10.1093/mp/sss119 [Google Scholar]
- Guy C.L. (1990). Cold acclimation and freezing stress tolerance: Role of protein metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41: 187–223 [Google Scholar]
- Guy C.L., Anderson J.V., Haskell D.W., Li Q.-B. (1994). Caps, cors, dehydrins, and molecular chaperones: Their relationship with low temperature responses in spinach. In Biochemical and Cellular Mechanisms of Stress Tolerance in Plants, J.H. Cherry, ed (Berlin: Springer-Verlag), pp. 479–499
- Hahn D., Kudla G., Tollervey D., Beggs J.D. (2012). Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev. 26: 2408–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F., Breitling R., McEntee C.W., Wittner B.S., Nemhauser J.L., Chory J. (2006). RankProd: A bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22: 2825–2827 [DOI] [PubMed] [Google Scholar]
- Iost I., Dreyfus M., Linder P. (1999). Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 274: 17677–17683 [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Ishitani M., Xiong L., Lee H.J., Stevenson B., Zhu J.-K. (1998). HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 10: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M., Xiong L., Stevenson B., Zhu J.-K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9: 1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen K.R., Gilmour S.J., Zarka D.G., Schabenberger O., Thomashow M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Björklund S., Li Y., Sayre M.H., Kornberg R.D. (1994). A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77: 599–608 [DOI] [PubMed] [Google Scholar]
- Knight H., Mugford S.G., Ulker B., Gao D., Thorlby G., Knight M.R. (2009). Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 58: 97–108 [DOI] [PubMed] [Google Scholar]
- Lee B.H., Kapoor A., Zhu J., Zhu J.-K. (2006). STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18: 1736–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., Zhu J.-K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.C., Kim Y.J. (1998). Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell. Biol. 18: 5364–5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. (2009). SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.R., Carré I.A., Coupland G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Medina J., Bargues M., Terol J., Pérez-Alonso M., Salinas J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadimpalli R., Yalpani N., Johal G.S., Simmons C.R. (2000). Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. J. Biol. Chem. 275: 29579–29586 [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kusano M., Fukushima A., Kita M., Ito S., Yamashino T., Saito K., Sakakibara H., Mizuno T. (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50: 447–462 [DOI] [PubMed] [Google Scholar]
- Nicol J.W., Helt G.A., Blanchard S.G., Jr, Raja A., Loraine A.E. (2009). The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F., Alonso J.M., Ecker J.R., Salinas J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 3985–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F., Medina J., Salinas J. (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 104: 21002–21007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palva E.T., Welin B., Vahala T., Olson A., Nordin-Henriksson K., Mantyla E., Lang V. (1994). Regulation of low temperature-induced genes during cold acclimation of Arabidopsis thaliana. In Biochemical and Cellular Mechanisms of Stress Tolerance in Plants, J.H. Cherry, ed (Berlin: Springer-Verlag), pp. 527–542
- Rivera-Milla E., Stuermer C.A.O., Málaga-Trillo E. (2006). Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: Convergent evolution of the SPFH domain. Cell. Mol. Life Sci. 63: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic Z., Ashworth E.N. (1993). Changes in leaf ultrastructure and carbohydrates in Arabidopsis thaliana (Heynh) cv. Columbia during rapid cold acclimation. Protoplasma 172: 111–123 [Google Scholar]
- Rocak S., Linder P. (2004). DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5: 232–241 [DOI] [PubMed] [Google Scholar]
- Ruelland E.V.M., Zachowski A., Vaughn H. (2009). Cold signaling and cold acclimation in plants. Adv. Bot. Res. 49: 36–54 [Google Scholar]
- Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carré I.A., Coupland G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Shi Y., Tian S., Hou L., Huang X., Zhang X., Guo H., Yang S. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24: 2578–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E.J., Gilmour S.J., Thomashow M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran N.P., Weiser C.J. (1972). An excised leaflet test for evaluation potato frost tolerance. HortScience 7: 467–468 [Google Scholar]
- Thomashow M.F. (1994). Arabidopsis thaliana as a model for studying mechanisms of plant cold tolerance. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 807–834
- Thomashow M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Tavernarakis N., Driscoll M., Kyrpides N.C. (1999). The SPFH domain: Implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 24: 425–427 [DOI] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J.T., Zarka D.G., Van Buskirk H.A., Fowler S.G., Thomashow M.F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Warren G., McKown R., Marin A.L., Teutonico R. (1996). Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Tobin E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Xin Z., Browse J. (1998). Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl. Acad. Sci. USA 95: 7799–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Ishitani M., Lee H., Zhu J.-K. (2001). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen Y., Wang Z.-Y., Chen Z., Gu H., Qu L.-J. (2007). Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 51: 512–525 [DOI] [PubMed] [Google Scholar]
- Zhu J., Jeong J.C., Zhu Y., Sokolchik I., Miyazaki S., Zhu J.-K., Hasegawa P.M., Bohnert H.J., Shi H., Yun D.J., Bressan R.A. (2008). Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. USA 105: 4945–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Shi H., Lee B.H., Damsz B., Cheng S., Stirm V., Zhu J.-K., Hasegawa P.M., Bressan R.A. (2004). An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc. Natl. Acad. Sci. USA 101: 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]