Abstract
Type II topoisomerases are required for the management of DNA superhelicity and chromosome segregation, and serve as frontline targets for a variety of small-molecule therapeutics. To better understand how these enzymes act in both contexts, we determined the 2.9-Å-resolution structure of the DNA cleavage core of human topoisomerase IIα (TOP2A) bound to a doubly nicked, 30-bp duplex oligonucleotide. In accord with prior biochemical and structural studies, TOP2A significantly bends its DNA substrate using a bipartite, nucleolytic center formed at an N-terminal dimerization interface of the cleavage core. However, the protein also adopts a global conformation in which the second of its two inter-protomer contact points, one at the C-terminus, has separated. This finding, together with comparative structural analyses, reveals that the principal site of DNA engagement undergoes highly quantized conformational transitions between distinct binding, cleavage, and drug-inhibited states that correlate with the control of subunit–subunit interactions. Additional consideration of our TOP2A model in light of an etoposide-inhibited complex of human topoisomerase IIβ (TOP2B) suggests possible modification points for developing paralog-specific inhibitors to overcome the tendency of topoisomerase II-targeting chemotherapeutics to generate secondary malignancies.
Keywords: type IIA topoisomerase, allostery, double-strand DNA breaks, protein–drug interactions, chemotherapeutics
Introduction
Topoisomerases are essential enzymes that manage the higher-order structural state of DNA. By selectively cleaving, rearranging, and religating DNA strands, topoisomerases can both alter the superhelical state of DNA and help disentangle interlinked chromosomes.1 Topoisomerases fall into two general categories, termed type I or type II, depending on whether one or both DNA strands of a single duplex are cut during a catalytic cycle, respectively. Although necessary for activity, the DNA breaks generated by topoisomerases can be subverted to give rise to mutagenic or cytotoxic lesions.2 A variety of small-molecule agents capable of inducing such effects have been identified to date, many of which serve as frontline antibiotics or anticancer agents.3
Type II topoisomerases operate by a complex mechanism that involves the controlled association and dissociation of subunit dimerization elements.4–8 In this reaction, one DNA duplex (termed the “G-segment”) is bound and cleaved by the enzyme, while a second double-stranded DNA (the “T-segment”) is transported through the break. G-segment breakage is catalyzed by a pair of symmetrically related tyrosines9–11 forming a transient, but covalently linked, topoisomerase·DNA assembly termed the cleavage complex. Strand passage is typically dependent upon ATP,12–15 a cofactor that not only promotes T-segment capture and stimulates G-segment cleavage but also triggers the successive opening and closing of inter-protomer interfaces, or “gates”.4–6,16–19 Type IIA topoisomerases, which comprise one of two type II topoisomerase subfamilies found predominantly in bacteria and eukaryotes,20 possess three such regions, termed the N-gate, DNA-gate, and C-gate.1,21 Although a general structural framework for type IIA topoisomerase action has been established for some time,22 many mechanistic questions, such as the role of metal ions in DNA cleavage, or how specific catalytic events are coordinated with gate actuation, remain ill defined.
Humans express two type IIA topoisomerase paralogs, or isoforms, termed topoisomerase IIα (TOP2A)23 and topoisomerase IIβ (TOP2B).24,25 Type IIA topoisomerases share significant amino acid sequence homology between species, with the human type II topoisomerases being ~ 40% identical with Saccharomyces cerevisiae topoisomerase II (Top2) (Fig. S1). Overall, the two human enzymes are 68% identical with each other,26 with the catalytic portions of the proteins sharing ~ 78% identity. TOP2A and TOP2B have different expression patterns in mouse,27 with TOP2A produced in proliferating tissues and TOP2B expressed more systemically. Expression of both isoforms are cell cycle dependent; levels for both are highest in dividing cells, but TOP2B is the predominant paralog in non-dividing and post-mitotic cells.28–31 In general, TOP2A plays a more central role in chromosome segregation and DNA replication,32,33 whereas TOP2B seems to be involved primarily in transcriptional regulation.34–36
Due to its role in cell proliferation, TOP2A expression has been used as a cancer cell marker.37–40 Both human type II topoisomerases are targets of highly effective anticancer drugs such as epipodophyllotoxins (e.g., etoposide); however, in some human patients, these agents also increase the likelihood of therapy-related malignancies, such as acute myeloid leukemia.41–43 Several studies have suggested that these secondary cancers may be linked to the undesired action of such drugs on TOP2B,44–47 raising the possibility that TOP2A-specific inhibitors (e.g., NK31448) might be useful for improving therapeutic outcomes. One challenge to identifying such compounds arises from the fact that the principal drug-binding locus, the DNA-gate, is one of the most highly conserved regions between the two enzymes. Structural insights from both drug-bound and drug-free states of DNA-bound TOP2A and TOP2B complexes are ultimately needed to aid efforts aimed at improving specificity. Currently, only an etoposide-inhibited form of the human TOP2B DNA cleavage core has been imaged crystallographically.49
To better understand both the type IIA topoisomerase mechanism and to identify features that can distinguish between closely related epipodophyllotoxin-binding sites, we have crystallized and solved the structure of the central catalytic region of human TOP2A bound noncovalently to a doubly nicked DNA substrate. The complex not only exhibits the now-canonical DNA bend imparted by type IIA topoisomerase family members49–55 but also fosters comparative studies that provide new insights into how the relative configuration of the active site for DNA cleavage correlates with both metal-ion occupancy and the opening and closing of a distal C-terminal dimer interface. Analysis of the TOP2A structure in light of the etoposide-inhibited state of human TOP2B confirms the close relationship between the two isoforms that favors inhibitor cross-reactivity, while also highlighting two positions in the drug-binding pocket that could serve as differentiating features for developing more selective anti-topoisomerase agents.
Results and Discussion
Overview of the human TOP2A–DNA complex
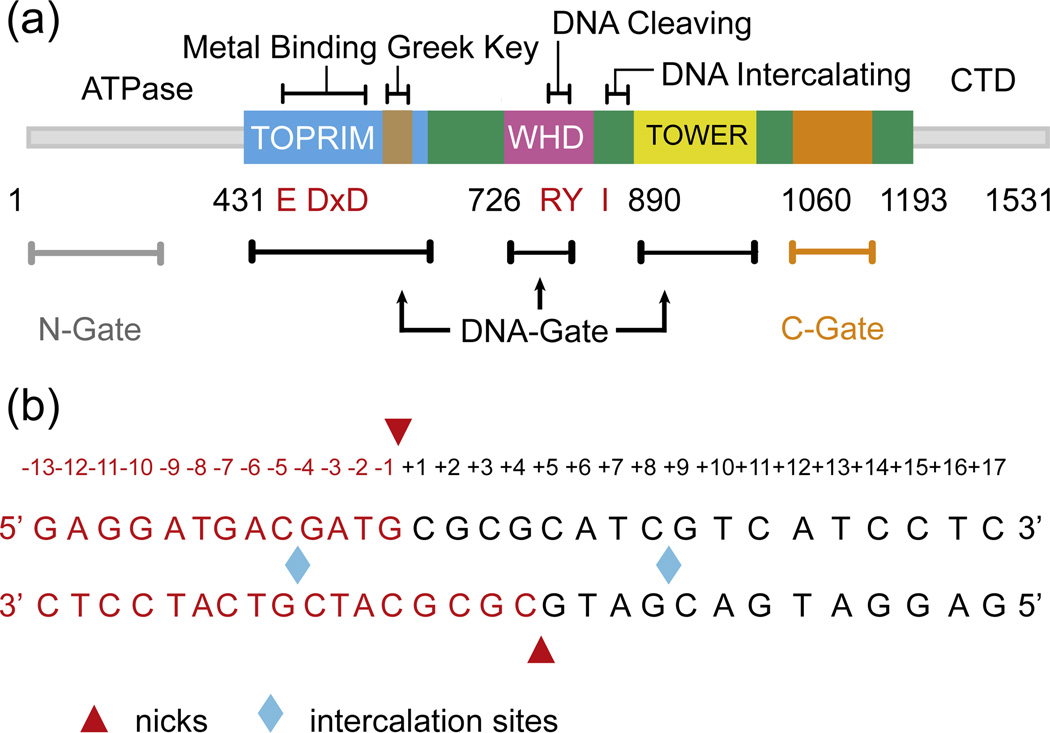
As no structure of the human TOP2A DNA binding and cleavage core had been determined previously, we used sequence homology and existing type IIA topoisomerase structures as a guide for constructing a minimal, crystallizable fragment (residues 431–1193, Fig. 1a). This protein was expressed and purified (Materials and Methods) and subsequently co-crystallized with a duplex DNA 13mer containing complementary four-base, 5′ overhangs (Fig. 1b). Following data collection and phasing by molecular replacement (Materials and Methods), the resultant electron density maps readily permitted modeling of the entire DNA substrate and essentially all of the protein (Fig. 2a and b). The final model was refined to 2.9 Å resolution with an Rwork/Rfree of 22.7%/27.0% and excellent stereochemistry (Table 1).
Fig. 1.
Crystallization constructs. (a) Primary structure of human TOP2A. The region crystallized (residues 431–1193) is shown in color. Specific domains (TOPRIM, WHD, tower) and dimerization gates are labeled with important residues and features highlighted: catalytic tyrosine and associated arginine, RY; intercalating isoleucine, I; metal-binding triad, DxD and E. (b) Schematic of the doubly nicked, 30-bp DNA substrate. Nicks and intercalation sites are highlighted.
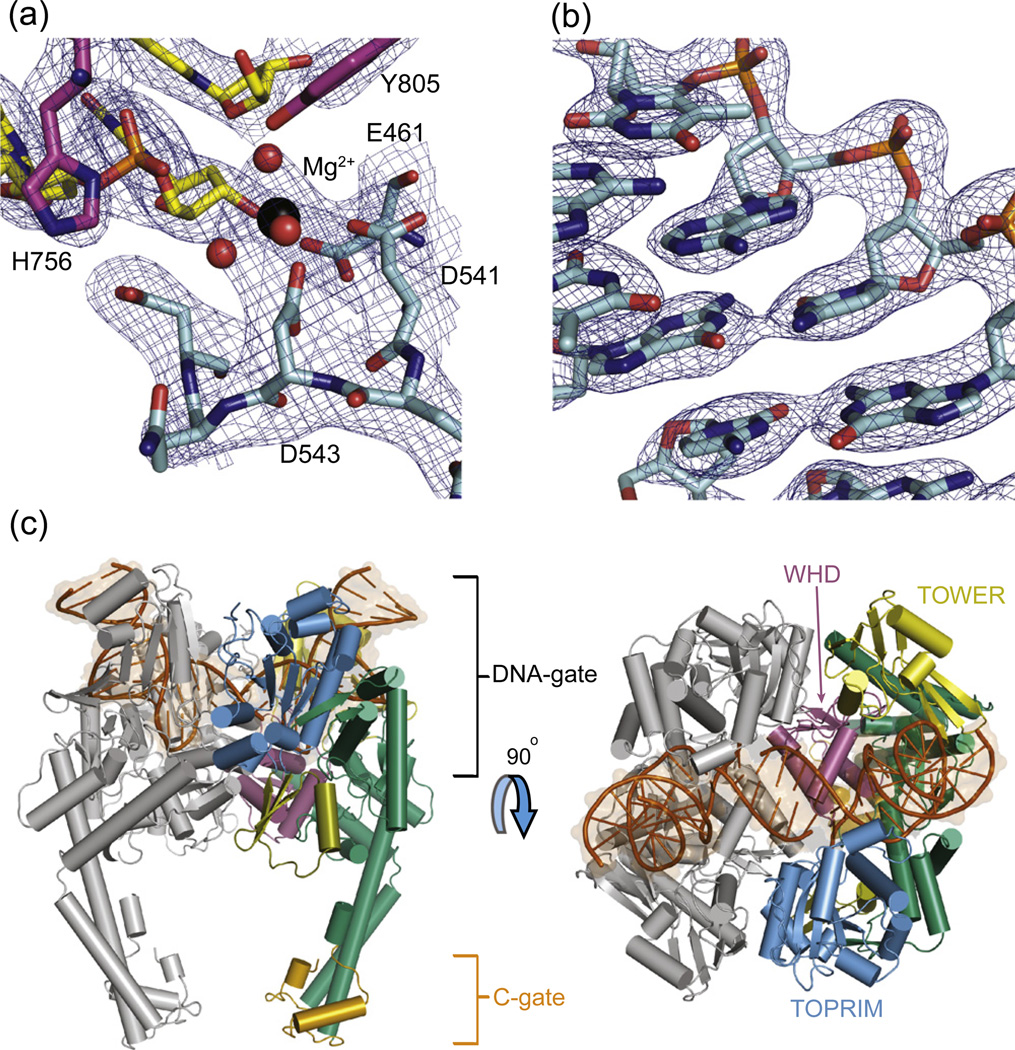
Fig. 2.
Structure of the human TOP2A DNA binding and cleavage core bound to DNA. (a) Refined 2Fo − Fc map around the nucleolytic center (1.4 σ contour). The active site is formed around DNA (yellow) at the interface between the TOPRIM domain (cyan) and WHD (magenta) of opposing protomers. (b) Refined 2Fo − Fc electron density map (2 σ contour) clearly shows the bound DNA substrate. (c) The human TOP2A·DNA complex as viewed from the front (left) and above (right). Domains are colored as in Fig. 1a, with DNA in dark orange. The G-segment is bound in the DNA-gate, and the C-gate dimerization interface is dissociated.
Table 1.
Diffraction and refinement statistics for the DNA-bound human TOP2A complex
| Structure | Human topo IIα–DNA complex |
|---|---|
| Space group | C2221 |
| Unit cell dimensions | |
| a, b, c (Ǻ) | 85.91, 215.08, 128.65 |
| α, β, γ (°) | 90, 90, 90 |
| Data collection | |
| Wavelength (Å) | 1.1159 |
| Resolution (highest shell) | 49.6–2.9 (3.00–2.90) |
| Observed reflections | 552,602 |
| Unique reflections | 25,373 |
| Rsym (%) (3.00–2.90 Å)a | 9.3 (71.2) |
| I/σI (3.00–2.90 Å) | 15.4 (2.0) |
| Completeness (3.00–2.90 Å) | 95.3 (91.6) |
| Redundancy (3.00–2.90 Å) | 7.2 (4.5) |
| Refinement | |
| Rworkb (%) | 22.7 |
| Rfreeb (%) | 27.0 |
| Reflections in test set (% of total) | 1285 (5.1) |
| No. of atoms | |
| Protein/DNA | 6538 |
| Solvent | 44 |
| Average B-factor (Å2) | |
| Protein/DNA | 80.2 |
| Solvent | 37.6 |
| Bond length deviations from ideal (Å) | 0.006 |
| Bond angle deviations from ideal (°) | 0.847 |
| Ramachandran outliersc (%) | 0 |
| Ramachandran favoredc (%) | 96.4 |
Rsym = Σ|I − Iσ|/ΣI, where I is the integrated intensity for a given reflection.
Rwork = Σ|Fo − Fc|/ΣFo, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree is calculated as for Rwork, but from a subset of the data (5.1%) that was withheld from crystallographic refinement.
As defined by MolProbity.56
The TOP2A·DNA complex forms a homodimer (Fig. 2c), in which the two protomers derive from a pair of asymmetric units related by crystal symmetry. One DNA oligo associates with each protomer, forming a doubly nicked, pseudo-continuous duplex in which the complementary overhangs pair with one another across the molecular dyad. Dimerization at the site of DNA binding (corresponding to the DNA-gate) buries 2560 Å2 of surface area between three elements—a metal-ion binding TOPRIM (TOpoisomerase/PRIMase) domain, a winged-helix domain (WHD) bearing the active-site tyrosine, and the “tower” domain—which reside on both halves of the dimer (Figs. 1a and 2c). Two isoleucines, one on each protomer (Ile856), intercalate into the minor groove of DNA, together bending the duplex by 130°; additional DNA contacts are manifest by numerous nonspecific, electrostatic interactions within the large (~ 3130 Å2), positively charged DNA-binding surface of the protein. The degree of bending seen in the structure coincides with that reported for TOP2A by fluorescence resonance energy transfer (136 ± 17°)55 and with structures of other type IIA topoisomerase·DNA complexes.50,52,53,57
Within each protomer, a cluster of invariant amino acids forms the active site for DNA cleavage (Fig. 2a). The catalytic tyrosine (Tyr805), responsible for nucleophilic attack on the DNA, resides ~ 6 Å from the backbone, opposite the nick; the two aspartates (Asp541 and Asp543) that form the conserved DxD motif of the TOPRIM region sit nearby, coordinating a single Mg2+. A third invariant acidic moiety of the TOPRIM domain, Glu461, hydrogen bonds to the 3′ OH of the DNA strand, while Arg804, which ligands the scissile phosphate in covalent cleavage complexes determined for type IIA topoisomerases, coordinates the − 1 phosphate in this structure. In general, these contacts comport well with the active-site environments seen in other structures of type IIA topoisomerases complexed with DNA.49–53,57,58
Implications for inhibition of TOP2A by exogenous agents
Type IIA topoisomerases are targets of both small-molecule inhibitors and posttranslational regulatory events that can alter enzyme activity. The structure of the TOP2A·DNA complex affords a new understanding of two such strategies. One is SUMOylation, which recently has emerged as an important, albeit poorly understood, modification of topo II.59,60 Most posttranslational modification sites, SUMOylation loci included,61 reside in the highly divergent C-terminal region of eukaryotic type IIA topoisomerases. However, a recent study found that a catalytic core residue, Lys660 of Xenopus laevis TOP2A, could be SUMOylated by Protein Inhibitor of Activated STAT-gamma (PIASγ), resulting in loss of decatenation activity.62 In human TOP2A, this residue (Lys662) is directly involved in binding G-segment DNA (Fig. S2). This contact indicates that SUMOylation likely inhibits TOP2A by interfering with G-segment binding or by preventing the TOPRIM domain from properly forming a functional DNA-gate. Curiously, SUMOylation of Xenopus TOP2A in vitro was observed only in the presence of DNA, even though the reactive lysine is buried in the DNA binding interface. Moreover, Lys662 does not reside in a typical φ-K-X-E/D SUMOylation target motif (where φ is a hydrophobic residue) recognized by the Xenopus or human enzyme.63,64 These peculiarities suggest that PIASγ may be SUMOylating Lys662 in an unusual fashion, perhaps capturing a state of the enzyme where the G-segment binding site has undergone a marked rearrangement from the conformations seen crystallographically.
Our TOP2A model also provides useful insights into epipodophyllotoxin inhibition, particularly with respect to efforts aimed at engineering compounds that discriminate between the two human isoforms. A recent landmark structure of human TOP2B in complex with etoposide establishes the binding pocket and modality for this class of topoisomerase II poisons.49 This pocket is solvent accessible in our TOP2A structure, but insufficiently large to accommodate drug. In TOP2B, the expanded pocket derives from changes in global protein conformation: association of the relatively nonplanar compound alters the disposition of the two WHDs in the dimer interface with respect to each other, deforming the DNA and pushing each TOPRIM domain away from the WHD of its partner protomer. These observed structural changes comprise rigid-body, en bloc movements between the WHDs and TOPRIM domains in the dimer, suggesting that following DNA cleavage, drug binding occurs during transient opening or breathing of the DNA-gate.
To investigate how etoposide might interact with human TOP2A, we individually superposed both the WHD and TOPRIM domain from our structure onto that of TOP2B. A single superposition using both domains simultaneously was not feasible due to the aforementioned conformational changes in the region (Fig. 3a); however, the backbones of both domains individually align well with those of TOP2B (WHD Cα RMSD = 0.70 Å, TOPRIM Cα RMSD = 0.51 Å). Of the four TOP2B residues that make side-chain contacts with the drug, the only residue that is different in the corresponding TOP2A binding site is the substitution of a methionine (Met762) for glutamine (Gln778 in TOP2B) (Fig. 3b and c). The sulfur atom in methionine is unlikely to participate in the hydrogen-bonding interaction seen to occur between the glutamine amide of TOP2B and the O12 moiety on the “D” ring of etoposide. Nevertheless, etoposide is known to act in vitro against both human paralogs65 and targets both proteins in cells.66,67 Since Gln778 adopts two conformations in the TOP2B structure,49 its observed contacts with etoposide likely are not crucial for inhibitor activity.
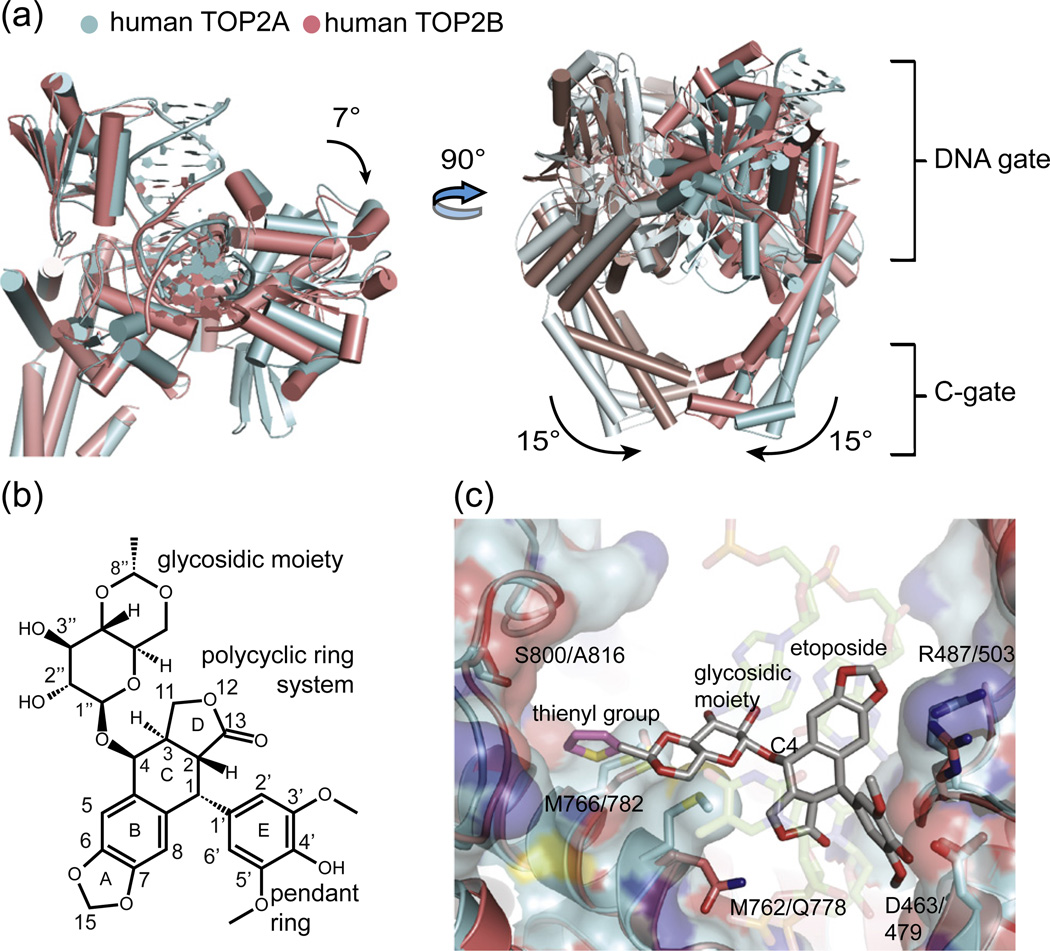
Fig. 3.
Comparison of TOP2A and etoposide-bound TOP2B. (a) The global conformation of TOP2A and TOP2B differs. Superposition of the two human isoforms shows that the TOPRIM domain in the etoposide-bound TOP2B cleavage complex (PDB ID 3QX3) is rotated relative to the WHD and tower domain in the TOP2A structure (left panel, only one protomer shown). The dimeric TOP2B cleavage complex has a closed C-gate compared to TOP2A (right panel). (b) Structure and ring numbering of etoposide. (c) Only two amino acids differ in the etoposide-binding pocket of TOP2A and TOP2B. The WHD and TOPRIM of TOP2A (cyan sticks and colored surface) are shown individually superposed on the TOP2B (salmon sticks) drug-binding site. Two residues that differ—Met762 in TOP2A versus Gln778 in TOP2B, and Ser800 versus Ala816—are labeled, as are three amino acids that are preserved (Arg487/503, Asp463/479, and Met766/782). Etoposide is shown as gray/red sticks; the additional thienyl group of the related anticancer drug teniposide is modeled in magenta to show its relation to the Ser800/Ala816 region.
A comparison of the two structures suggests a few possible avenues by which etoposide might be modified to better discriminate between TOP2A and TOP2B. Adding different substituents to the lactone group of etoposide to take advantage of the different chemistries, steric bulk, and rotamer preferences of glutamine versus methionine could constitute one approach. The reactivity of methionine might also be exploited, for instance, through platinum conjugates,68,69 to select for TOP2A reactivity. Our modeling suggests that the glycosidic moiety on the etoposide “C” ring is another possible candidate for modification (Fig. 3c). In the TOP2B complex, this group resides within a large binding pocket on the enzyme but makes few contacts with the protein surface. NMR studies have corroborated this paucity of contacts,70 and removal of the sugar is known to cause little change in drug activity in vitro.70,71 Comparison of this region between human TOP2A and TOP2B reveals a single point difference between the two isoforms, with Ser800 of TOP2A replacing Ala816 of TOP2B (Fig. 3c). This substitution suggests that the addition of a hydrogen-bonding or serine-reactive extension to the 8″ carbon position of the glycosidic group of etoposide might be useful for increasing TOP2A selectivity. Interestingly, teniposide, an etoposide analog that shows a fourfold greater inhibitor effect against TOP2A as compared with TOP2B in vitro,72 may already employ such an approach: teniposide possesses a thienyl sulfur in lieu of the methyl group found on etoposide's glycosidic C-ring, which modeling suggests could make van der Waals contacts with Ser800.
Distinguishing features between TOP2A and other eukaryotic type II topoisomerases
Outside of the active site, various structures have captured a range of conformational states for the DNA binding and cleavage core of type IIA topoisomerases as a whole. Our present TOP2A model bears greatest resemblance to a noncovalent complex between a doubly nicked, duplex DNA and the corresponding region of S. cerevisiae topoisomerase II (Fig. 4a).50 Both models retain a closed DNA-gate and exhibit similar DNA-bend angles and catalytic-tyrosine positions relative to the DNA backbone. More strikingly, the C-terminal dimerization interface that regulates DNA transport through type IIA topoisomerases (the “C-gate”) is separated in both structures.
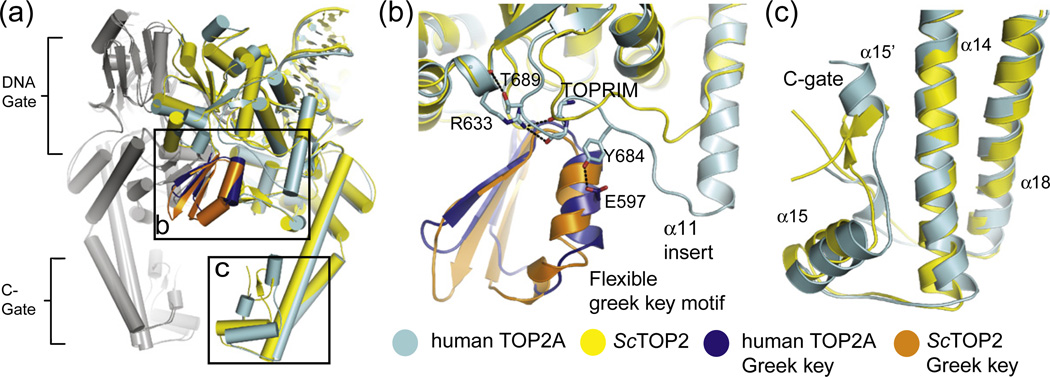
Fig. 4.
Comparison of TOP2A and S cerevisiae topoisomerase II. (a) The quaternary conformation of the human TOP2A·DNA complex most closely resembles the yeast Top2–DNA complex with a doubly nicked substrate. Architectural differences between the two structures are most pronounced in the Greek key motif and in the C-gate (boxed regions). (b) Differences in the TOPRIM insert region. A tyrosine (Tyr684) in the 670–686 insert of TOP2A forms hydrogen bonds with Glu597 in the Greek key element of the TOPRIM domain. Interactions seen in TOP2A, but not yeast topoisomerase II, are shown as sticks/broken lines and labeled. (c) Differences in the C-gate. A β-hairpin in the yeast topoisomerase II C-gate is replaced by an α-helix (A′α15′) in TOP2A.
Aside from single amino acid substitutions, the most significant differences between human TOP2A and yeast Top2 map to two regions of secondary structure. One is an insertion in helix α11 (residues 670–686 of human TOP2A, Fig. S1) of the three-helix bundle that both connects the TOPRIM domain to the WHD and forms part of the DNA binding interface (Fig. 4b). This insertion packs against a flexible “Greek-key” element, which constitutes a distinct augmentation to the TOPRIM fold of type IIA topoisomerases, pinning this subdomain against the WHD of the partner subunit. The second difference maps to the C-gate (Fig. 4c), where a short α-helix in human TOP2A (residues 1086–1092) replaces a β-hairpin in the yeast model, thereby forming an interface with one of the two long α-helical arms (residues 1015–1060) that extend from the DNA-gate. The functional significance of these modifications, if any, is not known at present.
By contrast to these respective similarities and differences with yeast Top2, the human TOP2A·DNA complex is more akin to human TOP2B in terms of local structure, but distinct insofar as global conformation. Like TOP2A, TOP2B contains the extended helix in the three-helix bundle between the TOPRIM and WHD, as well as the extra helix near the C-gate. However, the TOPRIM-associated Greek-key domain is not visible in the TOP2B structure, despite the high resolution of the model (2.2 Å).49 This element may be disordered in TOP2B due to natural substitutions of Tyr684 for phenylalanine and Thr689 for alanine, which together would abrogate interactions between the α11 extension and the Greek-key motif seen in TOP2A (Fig. 4b). Alternatively, this subdomain may become disordered due to conformational differences in the DNA-gate between the two enzymes. In particular, in accommodating etoposide, the TOPRIM domain of TOP2B is rotated by ~ 7° toward the dimer interface compared to TOP2A (Fig. 3a), concomitant with a lateral displacement of ~ 3 Å between its dyad-related WHDs. This movement leads the DNA-gate to crack open, decreasing the buried surface area in this region of TOP2B by ~ 2000 Å2. Even more strikingly, the C-gate of TOP2B is fully dimerized, compared to the separated, or “open”, state seen in TOP2A (Fig. 3a).
Inter-subunit conformational states correlate with DNA cleavage status
The TOP2A·DNA complex presented here joins a growing number of structures for the DNA binding and cleavage core of type IIA topoisomerases. This collection in turn provides a wealth of data for comparative analyses. While considering our TOP2A model in light of these related systems, we noted that the majority of structures (22 out of 25) retained an associated, or “closed”, C-gate.22,49,51–53,57,58,73–78 However, three completely distinct complexes—S. cerevisiae Top2,50 Acinetobacter baumannii topoisomerase IV,52 and human TOP2A (the present study)—have also been obtained in which the C-gate interface is dissociated. The existence of multiple models that derive from different topoisomerase homologs and crystal packing environments, yet still exhibit an “open” C-gate, strongly argues against suggestions that this conformation represents a nonphysiologic state.51
The factors that control C-gate status have been the subject of debate. One proposal has suggested that the formation of a covalent protein·DNA cleavage complex favors C-gate association, whereas G-segment binding alone promotes (but does not strictly enforce) C-gate opening.53 Support for this argument centers on the existence of a physical linkage between the active-site tyrosine and the α-helical arms that extend into the C-terminal interface (α14 and α18 in human TOP2A); this connection would appear to allow DNA-gate movements, triggered by entry into a cleavage-competent conformational state, to lever the arms toward each other and shut the gate. By contrast, the imaging of certain noncovalent, DNA-bound type IIA topoisomerases with a closed C-gate57,58 has raised the possibility that an alternative mechanism may account for the opening of this region. Entrapment of the T-segment in the cavity between the DNA-gate and C-gate could constitute one such approach.79
A close inspection between all 22 published type IIA topoisomerase structures bearing a closed DNA-gate helps resolve this question.49–53,57,58,73,76–78 In particular, superposition between the WHDs of these models reveals that the DNA-gate samples only a limited number of conformational states as the region engages a duplex substrate. For example, in comparing apo and substrate-bound forms, the presence of DNA correlates with a single, quantized sliding motion of the WHDs with respect to each other, along a vector that is both perpendicular to the dyad axis of the topoisomerase dimer and roughly parallel with the axes of the two α-helices that abut each other at the dimer interface (α3 in bacterial GyrA, and A′α3 in eukaryotic topo II) (Fig. 5a and b). This movement corresponds to a translational shift of the helix–turn–helix motif of each WHD past each other by a distance corresponding to single helical turn, clamping the TOPRIM and tower domains around the G-segment DNA. Interestingly, the etoposide-bound TOP2B·DNA structure exhibits an analogous shift of two helical turns, along with a rotational reorientation of the TOPRIM domain (Fig. 5b), as its DNA-gate cracks open.
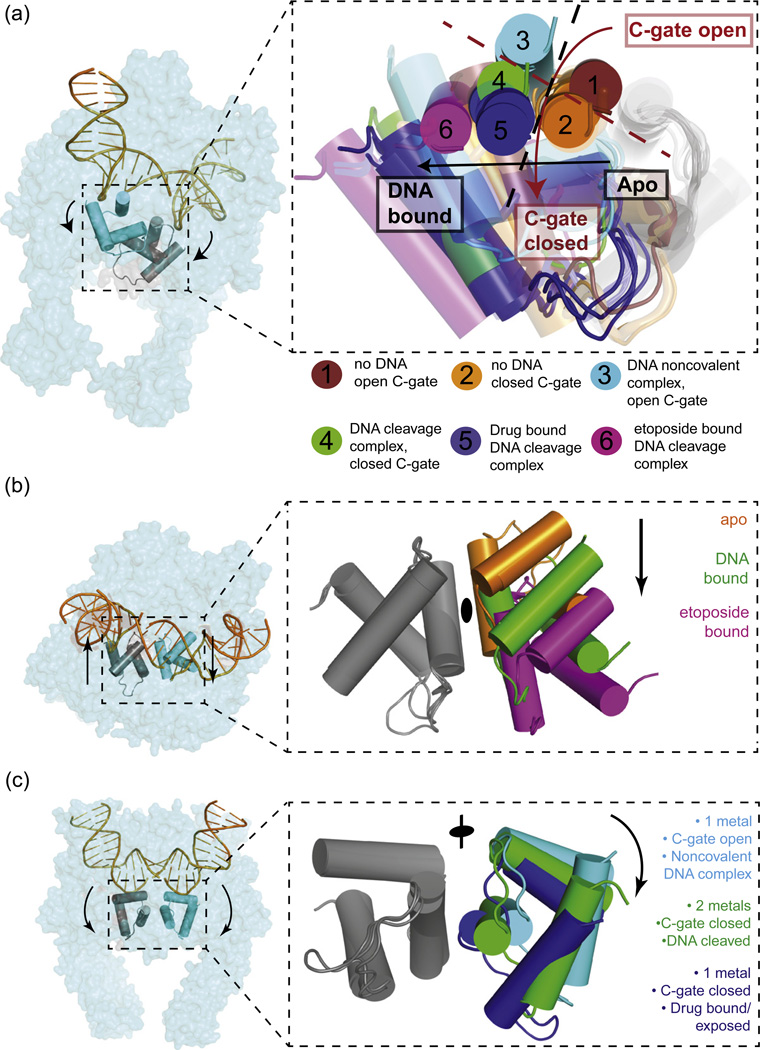
Fig. 5.
Substrate-dependent movements in the type IIA topoisomerase DNA-gate are quantized. This figure depicts the superposition of 22 type IIA topoisomerase structures, aligned using the WHD of one protomer to reveal the relative displacements of its partner WHD. The juxtapositions fall into six groups. Cartoons depict just the three-helix bundles of the two WHDs; surface representations of TOP2A are provided for orientation. (a) Overview of the six juxtapositions: (1) no DNA bound, C-gate open [A. baumannii topo IV (2XKJ)]; (2) no DNA bound, C-gate closed [Escherichia coli gyrase (3NUH), also E. coli gyrase (1AB4), M. tuberculosis gyrase (3ILW)]; (3) DNA noncovalently bound, C-gate open [human TOP2A (4FM9), also S. cerevisiae topoisomerase II (2RGR)]; (4) DNA cleavage complex, C-gate closed [S. cerevisiae topoisomerase II (3L4K)]; (5) DNA cleavage complex, C-gate closed, initial crystallization with drug bound [A. baumannii topo IV bound to moxifloxacin (2XKK), also S. pneumoniae topo IV cleavage complex bound to moxifloxacin (3FOF), clinafloxacin (3FOE), levofloxacin (3K9F), a dione inhibitor (3LTN), the dione complex back soaked with EDTA (3KSA), and the EDTA-soaked complex resealed by a MgCl2 soak (3KSB). The S. aureus gyrase noncovalent complex bound to GSK299943 (2XCR, 2XCS) and cleavage complex bound to ciprofloxacin (2XCT) further map to this group]; (6) DNA cleavage complex, C-gate closed, etoposide bound [human TOP2B (3QX3)]. The red broken line indicates the demarcation in relative WHD rotation angles between open and closed C-gate states. The black broken line indicates the demarcation of lateral WHD displacement between DNA-bound and free forms. (b) Binding of DNA causes the WHDs in opposing protomers to slide past each other by one helical turn along the α3 helix (A′α3 helix in eukaryotic topoisomerase IIs). This sliding can be quantized into juxtapositions taken by the apoenzyme [conformations 1 and 2 in (a)] and juxtapositions taken by DNA-bound enzymes [conformations 3–5 in (a)]. A more extreme lateral shift is seen in the etoposide-bound TOP2B complex where the DNA-gate interface has been disrupted [conformation 6 in (a)]. (c) DNA cleavage and C-gate dimerization are correlated to a rocking of the WHDs against each other. This rotation is quantized into juxtapositions with an open C-gate and noncovalently bound DNA [conformations 1 and 3 in (a)], a closed C-gate and covalently bound DNA [conformations 2 and 4 in (a)], and a closed C-gate, covalently bound DNA, and drug binding [conformations 5 and 6 in (a)].
The relative rotational status of the two WHDs and their associated active-site tyrosines is similarly quantized and correlates with both DNA cleavage status and the dimerization state of the C-gate (Fig. 5a and c). An open C-gate is observed only at one end of the rotational range, which is populated by two noncovalent topoisomerase·DNA complexes, as well as one apoenzyme structure.50,52 By contrast, all other apo and DNA-bound complexes occupy a distinct set of alternate WHD·WHD juxtapositions that coincide with C-gate closure. For instance, all DNA-cleavage complexes stabilized by small-molecule poisons not only have an associated C-gate but also exhibit a consistent counterclockwise rotation of the two WHDs with respect to each other.51,52,57,58 This conformational change is preserved in the etoposide-inhibited structure between DNA and human TOP2B,49 where an extra translational shift between WHDs has also occurred (Fig. 5b). A less-extreme, but still evident, counterclockwise movement is seen in every DNA-free structure that displays a closed C-gate73,77,78 and in an S. cerevisiae Top2 cleavage complex that was obtained using a phosphorothiolated suicide substrate.53
Overall, in instances where DNA is present, these comparisons reveal a tight correlation between a relative movement on the part of the WHDs to elicit DNA cleavage, and the predisposition of the C-gate to be closed. Indeed, out of the 15 topoisomerase II·DNA co-crystal structures in the database, only 2 appear to run counter to this correspondence. However, a closer consideration of these outliers (which represent noncovalent, DNA-bound states) demonstrates that neither structure in fact violates the dependence of C-gate dimerization on DNA-gate status proposed above: both structures actually derive from topoisomerase/DNA complexes that were exposed to a small-molecule agent that interferes with the normal cleavage cycle of the enzyme. In one example, the cleavage core of Staphylococcus aureus gyrase was co-crystallized with an intact DNA duplex and a non-intercalative inhibitor of strand scission (GSK299943). Interestingly, this inhibitor binds at the interface between the two apposed WHDs, in a pocket that materializes when the domains adopt a cleavage-like orientation relative to each other.58 This concurrence suggests that GSK299943 traps the DNA-gate of bacterial type IIA topoisomerases in a conformation similar to that adopted after the action of fluoroquinolone poisons, which are known to impede normal phosphotransfer chemistry.51,52,57,58 In the other instance, the core region of Streptococcus pneumoniae topo IV was first crystallized with the dione PD 0305970 to stabilize a cleavage complex, after which religation was promoted in situ by first soaking out the drug in the presence of ethylenediaminetetraacetic acid (EDTA) and then resoaking the crystals with Mg2+.57 Thus, this latter structure derives from a state in which the topoisomerase·DNA complex had already been captured with a rotated WHD configuration and closed C-gate. An inspection of the contacts arising from the crystal lattice indicates that molecular packing interactions would prevent subsequent WHD/WHD rearrangements or C-gate opening upon removal of the drug.
These findings have additional intriguing implications for the mechanism of DNA cleavage. Based on biochemical data, there is a general consensus that type IIA topoisomerases rely on two metal ions for DNA cleavage.80–82 Crystallographic efforts have refined this picture to suggest that only one metal ion (metal A) actively assists with the chemistry of strand scission and religation, whereas the other (metal B) serves a more structural role, anchoring the DNA at the − 1 phosphate position.49–53,57,58 The use of a single metal for phosphodiester bond breakage and reformation parallels the mechanism of strand cleavage employed by ββα-Me and HUH nucleases.83 However, there is debate as to whether the two metals found in the type II topoisomerase active site bind simultaneously during DNA breakage and religation, or if a single “dynamic” ion hops between the two coordination sites at different stages of the reaction.
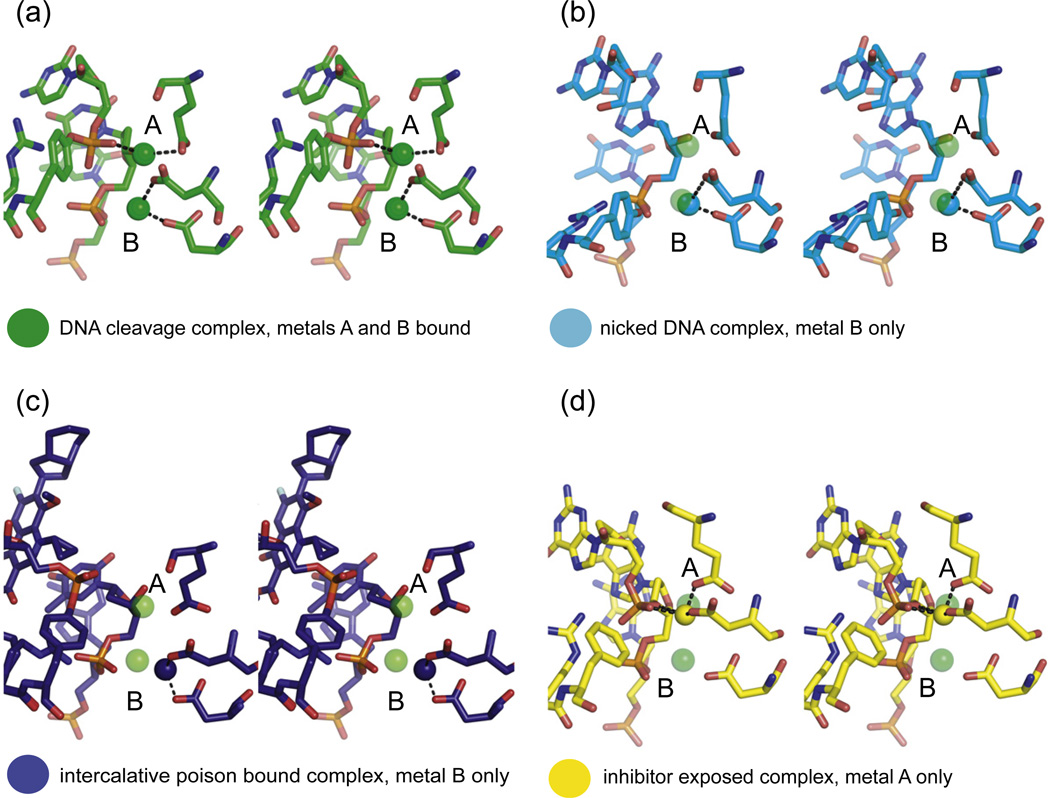
Support for the simultaneous two-metal mechanism has derived from thiol-rescue studies,81,82 and from a structure of S. cerevisiae Top2 captured in a covalent product complex with a phosphorothiolated DNA that was soaked with Zn2+ (Fig. 6a).53 By comparison, the dynamic metal concept derives from a number of type IIA topoisomerase·DNA complex structures that exhibit only a single metal in either one position or the other.49–52,57,58 However, each of these one-metal structures corresponds to, or derives from, either drug-inhibited states of the enzyme or complexes in which the DNA substrate is not competent to support cleavage. For instance, in all structures where the protein is bound to an intercalative poison, or where the DNA is nicked at the scissile position (as in the present TOP2A model), only metal B is visible (Fig. 6b and c; Fig. S3a). Alternatively, in the two topoisomerase·DNA complexes where the duplex backbone is intact and a poison is not present—either because the protein is bound to a non-intercalative drug (e.g., GSK29994) or because the enzyme was first co-crystallized with a poison before removal of the drug—only site A is filled (Fig. 6d; Fig. S3b and c). The inability of poisoned or nicked DNAs to support metal binding at the A-position is readily understandable, as the phosphate that coordinates this metal ion is either shifted out of the active-site pocket (in drug-bound states) or missing altogether. Similarly, the physical impediment to metal binding at site B in the GSK29994-associated or dione-pretreated complexes coincides with a misalignment of the coordinating residues in the DxD motif. This alteration likely is coupled to the more extreme WHD juxtaposition exhibited by these structures, which matches that of other cleavage-impaired drug-bound complexes (Fig. 5). These shifts may also explain why metal A remains present in the enzyme active site even after EDTA treatment of the dione-soaked crystals and why no other divalent species appear following the addition of Mg2+: this behavior suggests that the presence of drug reconfigures the active site to tightly bind only a single cofactor, which is sufficient to promote religation in a nonideal crystalline environment once drug is removed. Overall, the most parsimonious interpretation of these data is that under normal circumstances, two metals are present in the active site during strand scission, whereas inhibitor binding introduces new conformational constraints that allow for occupancy of only a single metal ion. This latter mode of action would be expected to perturb the cleavage/religation equilibrium of the enzyme, a well-established property of type IIA topoisomerase poisons in general.
Fig. 6.
Comparison of metal occupancy between type IIA topoisomerase structures. (a) The structure of S. cerevisiae topoisomerase II trapped in a cleavage intermediate by a phosphorothiolate suicide substrate has two metals, A and B, bound in the active site (PDB 3L4K shown). (b) In structures solved with nicked substrates, only metal B is present, as metal A binding requires coordination by the missing scissile phosphate (PDB 4FM9 shown). (c) Structures bound to a type IIA topoisomerase poison bind only metal B, due to a shift of the phosphotyrosine out of the metal A binding site (PDB 2XKK shown). (d) In structures bound to a non-intercalative drug (e.g., GSK29994), or where poison has been removed following co-crystallization, only metal A is present. In these structures, residues in the DxD motif do not form appropriate geometry for metal B coordination, due possibly to the retention of an altered DNA-gate state (cf. Fig. 5) (PDB 2XCS shown).
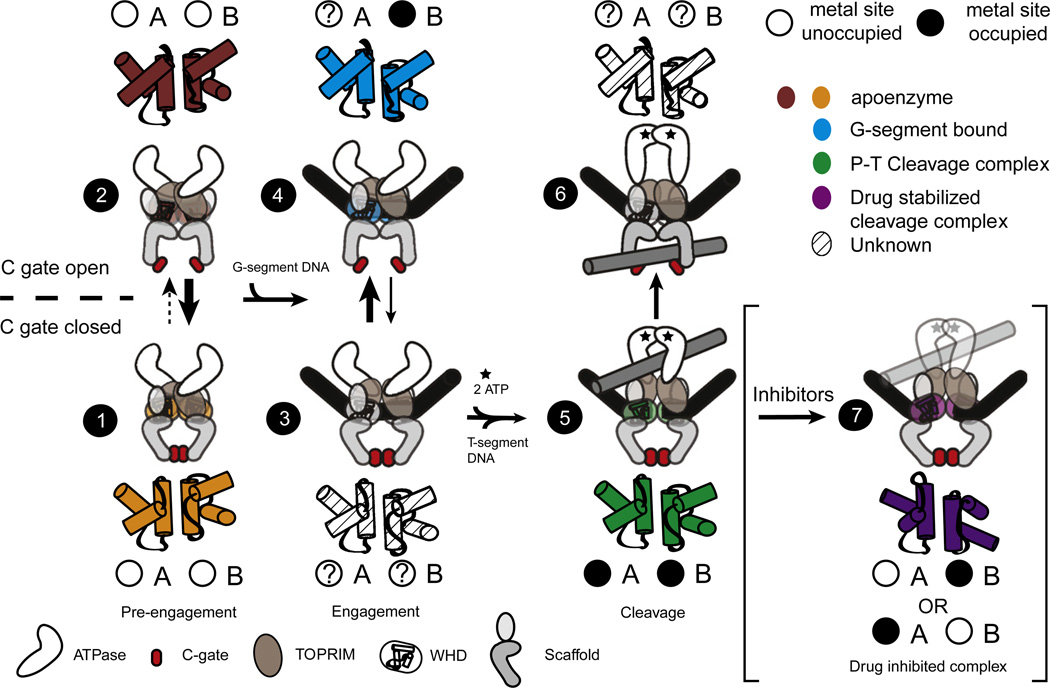
Together, the observed pattern of coupled conformational changes and DNA binding/cleavage status not only supports the idea that a direct allosteric connection exists between the state of the DNA-gate and C-gate actuation but also highlights probable substeps in the type IIA topoisomerase reaction (Fig. 7; Movie S1). Prior to G-segment binding, the DNA-gate can oscillate between dissociated and associated conformers that are accompanied by a dimerized (closed) C-gate (1).22,73–78 Occasionally, when the DNA-gate adopts a closed configuration, the C-gate can sample an open state (2), as is seen in A. baumannii topo IV.52 We propose that binding of an intact G-segment promotes the transition from this “pre-engagement” state to an “engagement” configuration (3), in which subunit-related TOPRIM and WHD elements shift with respect to each other to both clamp around DNA; this structural change simultaneously imposes a WHD/WHD juxtaposition that modestly destabilizes the C-gate and more readily allows for separation (4), possibly by introducing strain into the coiled-coil arms that link the DNA-gate and C-gate together. ATP- and/or T-segment binding would subsequently promote entry into a new physical state—one that promotes DNA cleavage—by inducing the WHDs to rotate with respect to each other and bring the active-site tyrosines in-line to attack DNA (5). This movement also would serve to relieve any strain imposed by the engagement state and thereby maintain C-gate closure. Resealing of the G-segment would return the DNA-gate to a conformation promoting C-gate dissociation and allow for T-segment release (6). Interestingly, our comparative studies indicate that the binding of either small-molecule poisons (which push apart the 3′ and 5′ ends of a cleaved DNA) or other interfacial inhibitors that associate with the DNA-gate interface can promote the formation of a more extreme rotation of the WHDs with respect to each other (7). This action may help such drugs trap the enzyme in an off-pathway state, thereby impeding recovery to a religation-competent form. Consistent with this model, the binding of etoposide, which is substantially nonplanar compared to its fluoroquinolone counterparts, promotes an even more pronounced WHD rotation and partial separation of the DNA-gate. An interesting secondary feature of this model is that it suggests that the energy landscape between conformational transitions controlling DNA cleavage and gate opening is relatively flat, with only subtle energetic differences needed to shift from one structural intermediate to another.
Fig. 7.
Model relating DNA-gate status to C-gate dimerization and metal occupancy in the context of the type IIA topoisomerase reaction. Topoisomerase IIA dimers exist in an equilibrium between different combinations of associated or dissociated proteinaceous interfaces. Where double arrows are shown, the relative thickness of the arrows denotes the likely weighted distribution of the two states. Prior to DNA binding, DNA-gate closure can occur with either a closed (1) or an open (2) C-gate. G-segment binding promotes the TOPRIM domain, WHD, and tower domain of both protomers to clamp around the DNA (3); this event may promote C-gate opening (4). ATP binding and capture of a second duplex, the T-segment, promote cleavage of the G-segment (5), favoring C-gate closure. The T-segment exits through an open C-gate following its passage through the G-segment and resealing of the cleaved DNA (6). Binding of topoisomerase II poisons and inhibitors such as GSK299943 perturb the DNA-gate interface to a more extended rotation that is impaired for religation and may help maintain the C-gate in a closed conformation (7). Relative WHD displacements as observed between different ligand-bound and/or free states are shown as cartoon cylinders and colored as per Fig. 5. The relative occupancies of the two metal-binding sites are also shown, with question marks denoting states that have yet to be imaged. The lone question mark associated with site A, state (4), reflects the fact that the DNAs found in these models lack a scissile phosphate and, hence, may not capable of binding a metal ion accordingly.
Concluding remarks
In summary, this work presents the first structure of the DNA binding and cleavage core of human TOP2A bound to DNA. Consideration of the model in light of existing structures of homologous enzymes has enabled us to better understand several important facets of type IIA topoisomerase mechanism and inhibition. In particular, we find that the two WHDs of a topoisomerase dimer, which form part of the region responsible for cleaving and opening DNA (the DNA-gate), adopt only a small set of positional configurations with respect to each other. These quantized conformations appear to couple DNA cleavage status with the allosteric control of interactions at a distal subunit interface, the C-gate, which sits more than 50 Å away from the enzyme active site (Movie S1). Anti-topoisomerase agents that bind within the DNA-gate appear to subvert these movements, altering the position and occupancy of reactive elements to either impede or stabilize cleavage and religation events. Comparisons of our model with an etoposide-inhibited state of the second human type IIA topoisomerase, TOP2B, not only highlight the similarities between the two isoforms but also suggest avenues for promoting epipodophyllotoxin selectivity that might be of use in countering secondary malignancies. Future efforts aimed at imaging additional intermediates and drug-bound states will be needed to investigate these ideas further.
Materials and Methods
A truncation of the DNA binding and cleavage core from the human gene TOP2A23 (residues 431–1193, Fig. 1a) was cloned into a GAL1-based yeast shuttle vector, in frame with an N-terminal tobacco etch virus protease-cleavable His6-MBP tag. Protein was expressed in S. cerevisiae strain BCY123 (MATα pep3::HIS3 prb1::LEU2 bar1::HISG lys2::GAL1/10-GAL4 can1 ade2 trp1 ura3 his3 leu2-3, 112,84 originally from the laboratory of R. Kornberg) grown in CSM-Ura− media with a 2% lactic acid and 1.5% glycerol carbon source at 30 °C. Overexpression was induced by the addition of 2% galactose at A600 = 0.9. Six hours after induction, cells were centrifuged (4500 rpm, 4 °C, 15 min), resuspended in 1 mM EDTA and 250 mM NaCl (1 mL per liter of liquid culture), and flash frozen dropwise in liquid nitrogen.
For purification, frozen cells were first lysed under liquid nitrogen by grinding with a mortar and pestle, and the resultant powder was thawed and resuspended in 20 mM Tris–HCl (pH 8.5), 300 mM KCl, 20 mM imidazole, and 10% glycerol (20 mL per liter of liquid culture) supplemented with protease inhibitors (1 mM PMSF, 2.34 µM leupeptin, and 1.45 µM pepstatin). Lysate was clarified by centrifugation (15,000 rpm, 4 °C, 20 min) and run over a nickel-chelating Sepharose column (GE) at 4 °C. Following a column wash in resuspension buffer, tagged TOP2A was eluted stepwise with resuspension buffer plus 200 mM imidazole. Eluted protein was concentrated using a 100-kDa cutoff centrifugal filter (Amicon), buffer exchanged with resuspension buffer, and then incubated overnight with His-tagged tobacco etch virus protease at 4 °C.85 Protease and uncleaved protein were removed by running over a second nickel column, and the eluted cleaved protein was run over a gel-filtration column (S300, GE) equilibrated in 500 mM KCl, 20 mM Tris–HCl (pH 7.9), and 10% glycerol. Purity was estimated by SDS-PAGE, and peak fractions were combined and concentrated to ~ 15.8 mg/mL protein using a 100-kDa cutoff filter.
Oligonucleotides were ordered from Integrated DNA Technologies. After receipt, DNAs were purified by urea-formamide PAGE, resuspended in 200 mM KCl and 10 mM Tris–HCl (pH 7.9), and annealed to form a 13-bp duplex with a complementary four-base, 5′ overhang. This construct results in the formation of a doubly nicked, 30-bp DNA substrate (Fig. 1b) when bound to a TOP2A homodimer. DNA and protein were combined in a 1.1:1 molar ratio of DNA 30mer:TOP2A dimer and dialyzed stepwise against 20 mM Tris–HCl (pH 7.9) and 10 mM MgCl2 and decreasing concentrations of KCl (500 mM, 250 mM, 100 mM). Dialysis ran over the course of 16 h at 4 °C to a final condition of 20 mM Tris–HCl (pH 7.9), 10 mM MgCl2, and 100 mM KCl using a 1000-Da cutoff mini-capillary dialyzer (Harvard Apparatus). Crystals grew within a day of setting hanging drops by mixing 150 nL of a 5-mg/mL protein/DNA sample with 150 nL 10% (w/v) polyethylene glycol 3000, 100 mM Na-cacodylate (pH 6.5), and 200 mM MgCl2 solution at 18 °C. Crystals were looped and cryoprotected with well solution plus 29% glycerol before being flash frozen in liquid nitrogen.
Diffraction experiments were performed at the 8.3.1 beamline of the Advanced Light Source at Lawrence Berkeley National Laboratory.86 Data were collected on an ADSC Q210 CCD detector using a wavelength of 1.1159 Å, and crystals were maintained at 100 K by a gaseous N2 cyrostat. Data were integrated and scaled using HKL200087 and reduced using CCP4 software.88 Based on BLAST analysis of type IIA topoisomerase homologs at the time (the human TOP2B model had not yet been published),89,90 a molecular replacement search model comprising the DNA-gate (residues 431–1014 and 1165–1189, TOP2A numbering) of a single protomer bound to half the DNA substrate but excluding the α-helical arms and C-gate was prepared from the crystal structure of the yeast topoisomerase II DNA binding and cleavage core bound to DNA [residues 419–989 and 1153–1177, Protein Data Bank (PDB) ID: 2RGR].50 Side-chain lengths in the search model were reduced to the last common atom using CHAINSAW.91 From this model, PHASER was used to find a positional solution and determine initial phases;92 molecular replacement was performed with the same initial search model, but without DNA, and yielded an identical solution with strong positive difference density confirming that the DNA substrate was present. A second molecular replacement search using only the globular domains of the C-gate was performed against the first solution to determine placement of this region, again using the yeast topoisomerase II DNA structure as a search model (residues 1029–1136, PDB ID: 2RGR). The resultant asymmetric unit consists of a single protomer and half the DNA substrate; the protein dimer is generated by a crystallographic 2-fold axis. After a round of rigid-body refinement using PHENIX,93 the model was improved by building in Coot94 and refinement in PHENIX using individual site refinement, individual ADP refinement, and TLS. The TLS Server95,96 was used to inform decisions for TLS groups. The final model included the entire DNA substrate and the entire protein construct, excluding residues 1092–1124, the N-terminus (residues 431–432), and the C-terminus (residues 1190–1193). Model validation was performed by MolProbity;56 comparisons between different topoisomerase models were conducted using PyMOL.97
Supplementary Material
Acknowledgements
The authors would like to thank members of the Berger group for helpful discussions. This work was supported by a National Science Foundation graduate research fellowship to T.J.W. (DGE 1106400), an Leukaemia Lymphoma Research grant to C.A.A. (07038), and a National Cancer Institute-sponsored RO1 to J.M.B. (CA077373).
Abbreviations used
- TOP2A
topoisomerase IIα
- TOP2B
topoisomerase IIβ
- TOPRIM
TOpoisomerase/PRIMase
- WHD
winged-helix domain
- EDTA
ethylenediaminetetraacetic acid
- PDB
Protein Data Bank
Footnotes
Accession numbers
Structure factors and atomic coordinates have been deposited in the PDB with accession number 4FM9.
Supplementary Data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2012.07.014
References
- 1.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;1:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 2.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 5.Roca J, Wang JC. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 6.Roca J, Berger JM, Harrison SC, Wang JC. DNA transport by a type II two-gate mechanism topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl Acad. Sci. USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams NL, Maxwell A. Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry. 1999;38:13502–13511. doi: 10.1021/bi9912488. [DOI] [PubMed] [Google Scholar]
- 8.Wigley DB, Davies GJ, Dodson EJ, Maxwell A, Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 9.Morrison A, Cozzarelli NR. Site-specific gyrase cleavage of DNA by E. coli DNA. Cell. 1979;17:175–184. doi: 10.1016/0092-8674(79)90305-2. [DOI] [PubMed] [Google Scholar]
- 10.Tse YC, Kirkegaard K, Wang JC. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J. Biol. Chem. 1980;255:5560–5565. [PubMed] [Google Scholar]
- 11.Buhler C, Lebbink JH, Bocs C, Ladenstein R, Forterre P. DNA topoisomerase VI generates ATP-dependent double-strand breaks with two-nucleotide overhangs. J. Biol. Chem. 2001;276:37215–37222. doi: 10.1074/jbc.M101823200. [DOI] [PubMed] [Google Scholar]
- 12.Gellert M, Mizuuchi K, O'Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl Acad. Sci. USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu LF, Liu C-C, Alberts BM. T4 DNA topoisomerase: a new ATP-dependent enzyme essential for initiation of T4 bacteriophage DNA replication. Nature. 1979;281:456–461. doi: 10.1038/281456a0. [DOI] [PubMed] [Google Scholar]
- 14.Baldi MI, Benedetti P, Mattoccia E, Tocchini-Valentini GP. In vitro catenation and decatenation of DNA and a novel eucaryotic ATP-dependent topoisomerase. Cell. 1980;20:461–467. doi: 10.1016/0092-8674(80)90632-7. [DOI] [PubMed] [Google Scholar]
- 15.Goto T, Wang J. Yeast DNA topoisomerase II. An ATP-dependent type II topoisomerase that catalyzes the catenation, decatenation, unknotting, and relaxation of double-stranded DNA rings. J. Biol. Chem. 1982;257:5866–5872. [PubMed] [Google Scholar]
- 16.Baird CL, Harkins TT, Morris SK, Lindsley JE. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl Acad. Sci. USA. 1999;96:13685–13690. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harkins TT, Lewis TJ, Lindsley JE. Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 2. Kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry. 1998;37:7299–7312. doi: 10.1021/bi9729108. [DOI] [PubMed] [Google Scholar]
- 18.Corbett KD, Benedetti P, Berger JM. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat. Struct. Mol. Biol. 2007;14:611–619. doi: 10.1038/nsmb1264. [DOI] [PubMed] [Google Scholar]
- 19.Graille M, Cladiere L, Durand D, Lecointe F, Gadelle D, Quevillon-Cheruel S, et al. Crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms. Structure. 2008;16:360–370. doi: 10.1016/j.str.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 20.Forterre P, Gribaldo S, Gadelle D, Serre M-C. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Bates AD, Berger JM, Maxwell A. The ancestral role of ATP hydrolysis in type II topoisomerases: prevention of DNA double-strand breaks. Nucleic Acids Res. 2011;39:6327–6339. doi: 10.1093/nar/gkr258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 23.Tsai-Pflugfelder M, Liu LF, Liu AA, Tewey KM, Whang-Peng J, Knutsen T, et al. Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21–22. Proc. Natl Acad. Sci. USA. 1988;85:7177–7181. doi: 10.1073/pnas.85.19.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung TD, Drake FH, Tan KB, Per SR, Crooke ST, Mirabelli CK. Characterization and immunological identification of cDNA clones encoding two human DNA topoisomerase II isozymes. Proc. Natl Acad. Sci. USA. 1989;86:9431–9435. doi: 10.1073/pnas.86.23.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Austin CA, Fisher LM. Isolation and characterization of a human cDNA clone encoding a novel DNA topoisomerase II homologue from HeLa cells. FEBS Lett. 1990;266:115–117. doi: 10.1016/0014-5793(90)81520-x. [DOI] [PubMed] [Google Scholar]
- 26.Austin CA, Sng J-H, Patel S, Fisher LM. Novel HeLa topoisomerase II is the II beta isoform: complete coding sequence and homology with other type II topoisomerases. Biochim. Biophys. Acta. 1993;1172:283–291. doi: 10.1016/0167-4781(93)90215-y. [DOI] [PubMed] [Google Scholar]
- 27.Capranico G, Tinelli S, Austin CA, Fisher ML, Zunino F. Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim. Biophys. Acta. 1992;1132:43–48. doi: 10.1016/0167-4781(92)90050-a. [DOI] [PubMed] [Google Scholar]
- 28.Padget K, Pearson A, Austin C. Quantitation of DNA topoisomerase IIalpha and beta in human leukaemia cells by immunoblot. Leukemia. 2005;14:1997–2005. doi: 10.1038/sj.leu.2401928. [DOI] [PubMed] [Google Scholar]
- 29.Tsutsui K, Tsutsui K, Okada S, Watanabe M, Shohmori T, Seki S, Inoue Y. Molecular cloning of partial cDNAs for rat DNA topoisomerase II isoforms and their differential expression in brain development. J. Biol. Chem. 1993;268:19076–19083. [PubMed] [Google Scholar]
- 30.Tsutsui K, Sano K, Kikuchi A, Tokunaga A. Involvement of DNA topoisomerase IIbeta in neuronal differentiation. J. Biol. Chem. 2001;276:5769–5778. doi: 10.1074/jbc.M008517200. [DOI] [PubMed] [Google Scholar]
- 31.Lyu YL, Wang JC. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIbeta. Proc. Natl Acad. Sci. USA. 2003;100:7123–7128. doi: 10.1073/pnas.1232376100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grue P, Grasser A, Sehested M, Jensen PB, Uhse A, Straub T, et al. Essential mitotic functions of DNA topoisomerase IIalpha are not adopted by topoisomerase IIbeta in human H69 cells. J. Biol. Chem. 1998;273:33660–33666. doi: 10.1074/jbc.273.50.33660. [DOI] [PubMed] [Google Scholar]
- 33.Niimi A, Suka N, Harata M, Kikuchi A, Mizuno S. Co-localization of chicken DNA topoisomerase IIα, but not β, with sites of DNA replication and possible involvement of a C-terminal region of α through its binding to PCNA. Chromosoma. 2001;110:102–114. doi: 10.1007/s004120100140. [DOI] [PubMed] [Google Scholar]
- 34.Ju B-G, Lunyak W, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 35.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 36.McNamara S, Wang H, Hanna N, Miller WH. Topoisomerase IIbeta negatively modulates retinoic acid receptor alpha function: a novel mechanism of retinoic acid resistance. Mol. Cell. Biol. 2008;28:2066–2077. doi: 10.1128/MCB.01576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dingemans AM, Pinedo HM, Giaccone G. Clinical resistance to topoisomerase-targeted drugs. Biochim. Biophys. Acta. 1998;1400:275–288. doi: 10.1016/s0167-4781(98)00141-9. [DOI] [PubMed] [Google Scholar]
- 38.Fry AM, Chresta CM, Davies SM, Walker MC, Harris AL, Hartley JA, et al. Relationship between topoisomerase II Level and chemosensitivity in human tumor cell lines relationship between topoisomerase II level and chemosensitivity in human. Cancer. 1991:6592–6595. [PubMed] [Google Scholar]
- 39.Kasahara K, Fujiwara Y, Sugimoto Y, Nishio K, Tamura T, Matsuda T, Saijo N. Determinants of response to the DNA topoisomerase II inhibitors doxorubicin and etoposide in human lung cancer cell lines. J. Natl Cancer Inst. 1992;84:113–118. doi: 10.1093/jnci/84.2.113. [DOI] [PubMed] [Google Scholar]
- 40.Boege F, Andersen A, Jensen S, Zeidler R, Kreipe H. Proliferation-associated nuclear antigen Ki-S1 is identical with topoisomerase IIalpha. Am. J. Pathol. 1995;146:1302–1308. [PMC free article] [PubMed] [Google Scholar]
- 41.Felix C. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim. Biophys. Acta. 1998;1400:233–255. doi: 10.1016/s0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 42.Felix C. Leukemias related to treatment with DNA topoisomerase II inhibitors. Med. Pediatr. Oncol. 2001;36:525–535. doi: 10.1002/mpo.1125. [DOI] [PubMed] [Google Scholar]
- 43.Pedersen-Bjergaard J. Genetic pathways in therapy-related myelodysplasia and acute myeloid leukemia. Blood. 2002;99:1909–1912. doi: 10.1182/blood.v99.6.1909. [DOI] [PubMed] [Google Scholar]
- 44.Azarova AM, Lyu YL, Lin C-P, Tsai Y-C, Lau JY-N, Wang JC, Liu LF. Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc. Natl Acad. Sci. USA. 2007;104:11014–11019. doi: 10.1073/pnas.0704002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyu YL, Kerrigan JE, Lin CP, Azarova AM, Tsai YC, Ban Y, Liu LF. Topoisomerase IIbeta mediated DNA double-strand breaks: implications in doxorubicin cardiotoxicity and prevention by dexrazoxane. Cancer Res. 2007;67:8839–8846. doi: 10.1158/0008-5472.CAN-07-1649. [DOI] [PubMed] [Google Scholar]
- 46.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat. Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowell IG, Sondka C, Smith K, Lee KC, Manville CM, Sidorczuk-Lesthuruge M, et al. A model for MLL translocations in therapy-related leukemia involving topoisomerase II-beta mediated DNA strand breaks and gene proximity. Proc. Natl Acad. Sci. USA. 2012;109:8989–8994. doi: 10.1073/pnas.1204406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toyoda E, Kagaya S, Cowell IG, Kurosawa A, Kamoshita K, Nishikawa K, et al. NK314, a topoisomerase II inhibitor that specifically targets the alpha isoform. J. Biol. Chem. 2008;283:23711–23720. doi: 10.1074/jbc.M803936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu C-C, Li T-K, Farh L, Lin L-Y, Lin T-S, Yu Y-J, et al. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011;333:459–462. doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 50.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 51.Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, et al. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 52.Wohlkonig A, Chan PF, Fosberry AP, Homes P, Huang J, Kranz M, et al. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol. 2010;17:1152–1153. doi: 10.1038/nsmb.1892. [DOI] [PubMed] [Google Scholar]
- 53.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charvin G, Strick TR, Bensimon D, Croquette V. Topoisomerase IV bends and overtwists DNA upon binding. Biophys. J. 2005;89:384–392. doi: 10.1529/biophysj.105.060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hardin AH, Sarkar SK, Seol Y, Liou GF, Osheroff N, Neuman KC. Direct measurement of DNA bending by type IIA topoisomerases: implications for non-equilibrium topology simplification. Nucleic Acids Res. 2011;39:5729–5743. doi: 10.1093/nar/gkr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM, Sanderson MR. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One. 2010;5:e11338. doi: 10.1371/journal.pone.0011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bax BD, Chan PF, Eggleston DS, Fosberry A, Gentry DR, Gorrec F, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 59.Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 60.Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee M-T, Bachant J. SUMO modification of DNA topoisomerase II: trying to get a CENse of it all. DNA Repair. 2009;8:557–568. doi: 10.1016/j.dnarep.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryu H, Furuta M, Kirkpatrick D, Gygi SP, Azuma Y. PIASy-dependent SUMOylation regulates DNA topoisomerase IIalpha activity. J. Cell Biol. 2010;191:783–794. doi: 10.1083/jcb.201004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 64.Hay RT. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 65.Gentry AC, Pitts SL, Jablonsky MJ, Bailly C, Graves DE, Osheroff N. Interactions between the etoposide derivative F14512 and human type II topoisomerases: implications for the C4 spermine moiety in promoting enzyme-mediated DNA cleavage. Biochemistry. 2011;50:3240–3249. doi: 10.1021/bi200094z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Willmore E, Frank AJ, Padget K, Tilby MJ, Austin CA. Etoposide targets topoisomerase IIalpha and IIbeta in leukemic cells: isoform-specific cleavable complexes visualized and quantified in situ by a novel immunofluorescence technique. Mol. Pharmacol. 1998;54:78–85. doi: 10.1124/mol.54.1.78. [DOI] [PubMed] [Google Scholar]
- 67.Bandele OJ, Osheroff N. The efficacy of topoisomerase II-targeted anticancer agents reflects the persistence of drug-induced cleavage complexes in cells. Biochemistry. 2008;47:11900–11908. doi: 10.1021/bi800981j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dedon PC, Borch RF. Characterization of the reactions of platinum antitumor agents with biological and nonbiological sulfur-containing nucleophiles. Biochem. Pharmacol. 1987;36:1955–1964. doi: 10.1016/0006-2952(87)90494-1. [DOI] [PubMed] [Google Scholar]
- 69.Fuertes MA, Alonso C, Pérez JM. Biochemical modulation of cisplatin mechanisms of action: enhancement of antitumor activity and circumvention of drug resistance. Chem. Rev. 2003;103:645–662. doi: 10.1021/cr020010d. [DOI] [PubMed] [Google Scholar]
- 70.Bender RP, Jablonsky MJ, Shadid M, Romaine I, Dunlap N, Anklin C, et al. Substituents on etoposide that interact with human topoisomerase IIalpha in the binary enzyme–drug complex: contributions to etoposide binding and activity. Biochemistry. 2009;47:4501–4509. doi: 10.1021/bi702019z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilstermann AM, Bender RP, Godfrey M, Choi S, Anklin C, Berkowitz DB, et al. Topoisomerase II–drug interaction domains: identification of substituents on etoposide that interact with the enzyme. Biochemistry. 2007:8217–8225. doi: 10.1021/bi700272u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drake FH, Hofmann GA, Bartus HF, Mattern MR, Crooke ST, Mirabelli CK. Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II. Biochemistry. 1989;28:8154–8160. doi: 10.1021/bi00446a029. [DOI] [PubMed] [Google Scholar]
- 73.Morais Cabral JH, Jackson AP, Smith CV, Shikotra N, Maxwell A, Liddington RC. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 74.Fass D, Bogden CE, Berger JM. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat. Struct. Mol. Biol. 1999;6:322–326. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 75.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 76.Laponogov I, Veselkov DA, Sohi MK, Pan XS, Achari A, Yang C, et al. Breakage-reunion domain of Streptococcus pneumoniae topoisomerase IV: crystal structure of a gram-positive quinolone target. PLoS One. 2007;2:e301. doi: 10.1371/journal.pone.0000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tretter EM, Schoeffler AJ, Weisfield SR, Berger JM. Crystal structure of the DNA gyrase GyrA N-terminal domain from Mycobacterium tuberculosis. Proteins: Struct. Funct. Bioinf. 2010;78:492–495. doi: 10.1002/prot.22600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schoeffler AJ, May AP, Berger JM. A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic Acids Res. 2010;38:7830–7844. doi: 10.1093/nar/gkq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roca J. The path of the DNA along the dimer interface of topoisomerase II. J. Biol. Chem. 2004;279:25783–25788. doi: 10.1074/jbc.M402555200. [DOI] [PubMed] [Google Scholar]
- 80.West KL, Meczes EL, Thorn R, Turnbull RM, Marshall R, Austin CA. Mutagenesis of E477 or K505 in the B′ domain of human topoisomerase II beta increases the requirement for magnesium ions during strand passage. Biochemistry. 2000;39:1223–1233. doi: 10.1021/bi991328b. [DOI] [PubMed] [Google Scholar]
- 81.Noble CG, Maxwell A. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two metal-ion mechanism. J. Mol. Biol. 2002;318:361–371. doi: 10.1016/S0022-2836(02)00049-9. [DOI] [PubMed] [Google Scholar]
- 82.Deweese JE, Guengerich FP, Burgin AB, Osheroff N. Metal ion interactions in the DNA cleavage/ligation active site of human topoisomerase IIalpha. Biochemistry. 2009;48:8940–8947. doi: 10.1021/bi900875c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang W. An equivalent metal ion in one- and two-metal-ion catalysis. Nat. Struct. Mol. Biol. 2008;15:1228–1231. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wasserman RA, Wang JC. Mechanistic studies of amsacrine-resistant derivatives of DNA topoisomerase II. Implications in resistance to multiple antitumor drugs targeting the enzyme. J. Biol. Chem. 1994;269:20943–20951. [PubMed] [Google Scholar]
- 85.Tropea JE, Cherry S, Waugh DS. High throughput protein expression and purification. Methods Mol. Biol. 2009;498:297–307. doi: 10.1007/978-1-59745-196-3_19. [DOI] [PubMed] [Google Scholar]
- 86.MacDowell A, Celestre RS, Howells M, McKinney W, Krupnick J, Cambie D, et al. Suite of three protein crystallography beamlines with single super-conducting bend magnet as the source. J. Synchrotron Radiat. 2004;11:447–455. doi: 10.1107/S0909049504024835. [DOI] [PubMed] [Google Scholar]
- 87.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:306–315. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 88.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK. Protein database searches using compositionally adjusted substitution matrices. FEBS, J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stein N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Crystallogr. 2008;41:641–643. [Google Scholar]
- 92.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 96.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 2006;39:109–111. [Google Scholar]
- 97.The PyMOL Molecular Graphics System, Version 1.2r3pre. Schödinger, LLC; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.