Abstract
Multiple myeloma (MM) is a largely incurable plasma cell malignancy with a poorly understood and heterogeneous clinical course. To identify potential, functionally relevant somatic mutations in MM, we performed whole-exome sequencing of five primary MM, corresponding germline DNA and six MM cell lines, and developed a bioinformatics strategy that also integrated published mutational data of 38 MM patients. Our analysis confirms that identical, recurrent mutations of single genes are infrequent in MM, but highlights that mutations cluster in important cellular pathways. Specifically, we show enrichment of mutations in adhesion molecules of MM cells, emphasizing the important role for the interaction of the MM cells with their microenvironment. We describe an increased rate of mutations in receptor tyrosine kinases (RTKs) and associated signaling effectors, for example, in EGFR, ERBB3, KRAS and MAP2K2, pointing to a role of aberrant RTK signaling in the development or progression of MM. The diversity of mutations affecting different nodes of a particular signaling network appears to be an intrinsic feature of individual MM samples, and the elucidation of intra- as well as interindividual redundancy in mutations that affect survival pathways will help to better tailor targeted therapeutic strategies to the specific needs of the MM patient.
Keywords: multiple myeloma, somatic mutations, whole-exome sequencing, adhesion, receptor tyrosine kinases
Introduction
Multiple myeloma (MM) is characterized by the expansion of plasma cells in the bone marrow (BM), causing hematopoietic insufficiency, osteolytic bone lesions and renal failure.1 The introduction of the novel therapeutic agents thalidomide, lenalidomide and bortezomib, in conjunction with autologous stem cell transplantation protocols, has led to a considerable improvement of overall survival times of MM patients.2 However, the disease remains largely incurable and, moreover, shows a remarkably heterogeneous clinical course, suggesting the existence of different biologically defined subgroups. As a prerequisite for novel targeted approaches, more insights into the genetic and molecular pathogenesis of MM are therefore warranted, which will lead to the identification of crucially activated or inactivated survival pathways.
MM is thought to evolve through accumulation of a multitude of genetic aberrations in a multi-step transformation process initiating from a post-germinal center B-cell via the premalignant state of monoclonal gammopathy of undetermined significance to overt clinical disease. The occurrence of hyperdiploidy with chromosomal trisomies or of genetic translocations involving the immunoglobulin heavy chain locus and certain oncogenes represent rather early pathogenetic events, whereas the progression to symptomatic MM is frequently accompanied, if not facilitated by mutations of RAS, TP53, genetic rearrangements or amplifications of MYC, mutations affecting the nuclear factor-κB pathway and dysregulation of the phosphoinositide-3-kinase/AKT pathway.2, 3, 4
Recent high-throughput genomic and whole-exome sequencing efforts have set the stage for the description of the genomic landscape of MM.5, 6, 7 Sequencing of 38 MM tumor genomes/exomes identified novel, recurrent somatic mutations that—despite their low frequency at the single gene level—appear to cluster in important cellular pathways. For example, the RNA-processing machinery and protein homeostasis mechanisms are affected by mutations (DIS3, FAM46C), as are histone-modifying enzymes (MLL, MLL2, MLL3). Frequent somatic mutations of genes involved in the nuclear factor-κB pathway point to a more potent role of this signaling cascade in MM than was previously appreciated.5 BRAF mutations were detected in 4% of MM cases,5 suggesting a potential therapeutic role for recently developed BRAF inhibitors for such patients. Finally, sequencing has also revealed differences in the mutational landscape between t(4;14)- and t(11;14)-positive MM.7
Our own whole-exome sequencing efforts, including five patient-derived (primary) MM samples, their matched normal DNA and six MM cell lines, provide—in concert with previously published data of 38 MM patients5—information on two additional, functionally related groups of genes that show an enrichment of mutations in MM, namely adhesion- and receptor tyrosine kinase (RTK)-associated molecules. The pivotal role of the BM microenvironment in the maintenance and proliferation of the tumor cell clone and in the response to therapy2, 8, 9, 10, 11, 12 is well established in MM. However, our findings suggest that MM cells themselves are being selected for clones with increased numbers of somatically mutated adhesion molecules that may modulate their interaction with bystander cells of the BM microenvironment, to promote anti-apoptotic or pro-proliferative properties, or to decrease dependency during tumor progression. The finding of an increased rate of mutations in genes involved in RTK function underlines the importance of these signaling networks in the pathogenesis of MM, and should stimulate efforts to interfere with these signaling events therapeutically.
Material and methods
MM specimens and cell lines
BM aspirates and corresponding peripheral blood mononuclear cells (PBMCs) of five patients with MM were obtained after informed consent. Two patients presented with an extramedullary involvement, one of whom also had plasma cell leukemia and one MM showed anaplastic morphology. Three patients were untreated at the time the biopsy was taken, including the patient with anaplastic MM, whereas two patients had received multiple prior lines of therapy. Tumor cells were isolated using CD138 microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany) as previously described.13 Fluorescence in situ hybridizations for the detection of losses in 13q14 and 17p13, gains in 1q21, 9q34 and chr11, and for the translocations (4;14), 14;16) and (11;14) were performed according to standard protocols (Supplementary Table S1).14, 15 The human MM cell lines JJN3, AMO1, U266, OPM2 and L363 were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany), and MM.1S cells were purchased from LGC Biolabs (Wesel, Germany). MM cells were cultured as previously described.13 The study was approved by the Ethics Committee of the Medical Faculty, University of Würzburg (reference number 18/09).
Whole-exome resequencing
Sample preparation of the primary MM and corresponding PBMC samples and of the MM cell lines was performed using the Paired End DNA Sample Preparation Kit from Illumina (Illumina, San Diego, CA, USA) according to the standard protocol. Amplification and exome enrichment was performed according to the V1 and V2 SeqCap EZ Exome Library SR user guides from Nimblegen (Roche Diagnostics, Mannheim, Germany). Clonal cluster generation and 76 bp paired-end sequencing was performed on the GAIIx (Illumina).
Microarray analysis
RNA and DNA were isolated from six MM cell lines, using the Allprep Kit, and then hybridized to HG-U133 plus 2.0 arrays and SNP 6.0 arrays (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's protocol.
Bioinformatic evaluation of sequencing data
File conversion and alignment
File conversion was performed using the off-line base caller software (Illumina) and GERALD (Illumina), and the alignment to the reference genome hg19 was performed using the Burrows Wheeler Aligner. Furthermore, the sequences were processed using the Picard tool and R/Bioconductor. For details see Supplementary Material and Methods.
Single Nucleotide_Variant calling, filtering and biological interpretation
Unpaired Single Nucleotide_Variant (SNV) calling was performed using the Genome Analysis Toolkit pipeline as described,16 followed by the SeattleSeq annotation and filtering steps (Figure 1). In addition, we matched the readily filtered mutational data of 38 MM, published in the supplements of Chapman et al.5 with the mutation data of our cases. This increased the number of primary MM to 43 and helped us to efficiently extract the tumor-relevant SNVs from our six cell lines (Figure 1). Specifically, for our discovery approaches, we focused on genes that were affected by a mutation in at least 1 of our 5 primary MM, or 1 of our 6 cell lines, plus at least 1 of the 38 primary MM.5
Figure 1.
SNV-filtering strategy. After alignment and SNV calling (see Material and Methods), SNVs were filtered using different databases and annotation tools. In a first step, SNVs were excluded that appeared in the databases 1000 genomes or dbSNP, as they were described to occur in healthy individuals. Next, SNVs were excluded, which did not lead to an amino acid exchange (synonymous mutations) and which were benign according to the predictor tool Polyphen 1, using the SeattleSeq annotation tool. Of note, this annotation refers to all transcripts of a gene, which leads to a higher amount of total SNVs. In detail, it might occur that the SNV is located to the same position, but might affect different transcripts. Thus, the amount of detected SNVs after annotation does not equal the amount of SNVs before SeattleSeq annotation. Moreover, SNVs were excluded that occurred in the corresponding blood samples, as we were interested in tumor-related SNVs. In addition, we decided to focus on genes that were affected in at least one of the primary MM sample. To extract the tumor-relevant SNVs efficiently from the 6 cell lines, we integrated the previously published mutation data of 38 MM.5 This allowed us to exclude SNVs that affected genes in only a cell line but not in a primary sample (including Chapman et al.5). Finally, we applied three functional predictors to increase the possibility that the detected amino acid exchange leads to a functional change.
The resulting gene lists (Figure 1) were then used to discover new tumor-relevant pathways using the GSEA and String analysis.17, 18 Information on protein domains was obtained using the graphical view of the nucleotide database from NCBI and the String database. For details, see Supplementary Material and Methods.
PCR and Sanger sequencing
For technical validation, PCR and Sanger sequencing were performed using standard protocols. All primers (Supplementary Table S3) were purchased from Integrated DNA Technologies (Leuven, Belgium).
Results
Sequencing output and quality assessment
According to the sequencing chemistry and respective software version used, we obtained 17–120 million reads per lane and 56–192 million reads per sample, performing a 36- or 76-bp paired end run on a GAIIx. Of those reads, approximately 90% could be mapped to the reference genome. On average, 10% of those reads were not unique or mapped to more than one position. Mapping of those reads to the target region revealed 50–80% of uniquely mapped unique reads ‘on target,' dependent on the enrichment kit and the read length used. With the SBS v5 chemistry (Illumina) and the SCS 2.9 software (Illumina), we obtained an average coverage ‘on target' of ∼100 × per lane. Uniformity at 1 × coverage was, on average, 97%. One sample pair showed a worse uniformity for unknown reasons and was, thus, not included in this calculation. The transition/transversion ratios were around 3.0 in all samples, indicating a low rate of false-positive calls and no significant technical bias. All technical parameters and a more detailed description of this Results section can be found in Supplementary Table S2 and Supplementary Results.
SNV filtering and validation
To detect somatic mutations that might be relevant for MM pathogenesis, we developed a bioinformatics pipeline. This included the extraction of tumor-associated SNVs using the databases dbSNP and 1000 genomes, and the corresponding normal sample of each primary MM, SeattleSeq annotation to extract the non-synonymous SNVs, as well as the application of functional predictors (Polyphen, phastCons, GERP) to determine SNVs at conservative sites or SNVs that may lead to structural changes. By this strategy, 55 363 raw SNVs that were called in a total of 16 samples (6 MM cell lines and 5 primary MM, plus corresponding PBMC samples) were narrowed down to 330 putative cancer-relevant SNVs (Figure 1, Supplementary Table S4 and Table S5). Moreover, we integrated published mutation data of 38 primary MM5 to efficiently extract tumor-relevant SNVs from the 6 MM cell lines. Interestingly, 480/1429 of the mutated genes that were published by Chapman et al.,5 were also mutated in our data set. Thus, for our discovery approach we were able to focus on those genes that were mutated in at least 1 sample of our data set (including cell lines), but also in at least 1 primary MM (5 own and 38 published) (Figure 1). Validations by Sanger sequencing were performed randomly after SeattleSeq annotation (4130 SNVs) at various steps of the analysis. In total, 199 of 213 SNVs (94.3%) could be confirmed (primers listed in Supplementary Table S3), indicating only a very low technical bias as already evidenced by the good transition/transversion ratio. For more details, see legend of Figure 1 and Supplementary Results.
MM is characterized by a low frequency of recurrent gene mutations
In line with the previous report,5 our results confirm that recurrent gene mutations are rare in MM. Combining both data sets, only 8 genes (KRAS, FAM46C, WHSC1, DIS3, LRP1B, PKHD1, NRAS, ALOX12B) were affected in at least 4 out of 43 primary MM and at least 1 MM cell line, and passed the thresholds for at least 2 functional predictors (Supplementary Table S6). Of note, all mutations were validated by Sanger sequencing in the cell lines and the primary MM samples.
Higher mutational load in MM with anaplastic morphology
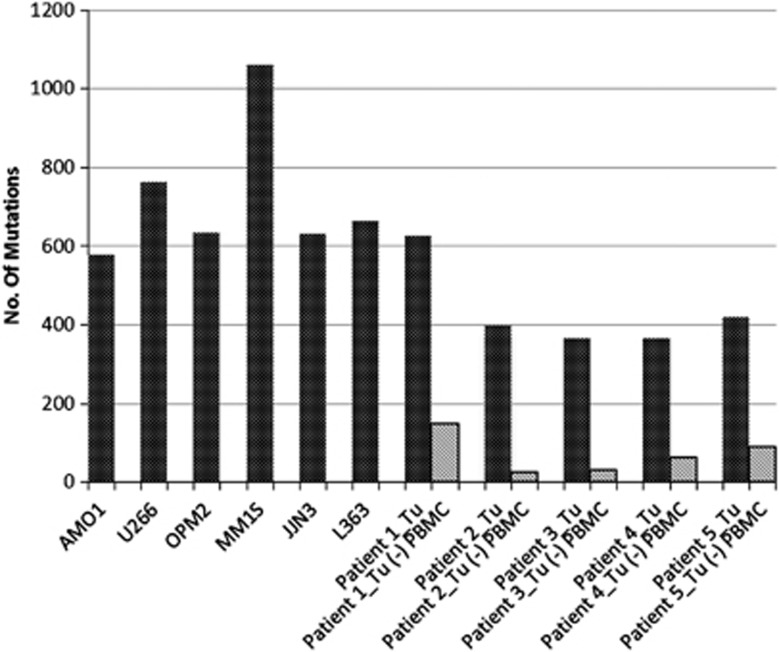
After SeattleSeq annotation and before the exclusion of SNVs detected in the corresponding blood (Figure 1), MM cell lines showed an overall higher mutational load than the primary MM as expected (Figure 2). Only one primary MM (Patient 1, Figure 2) showed a mutational load equivalent to the one observed in MM cell lines. Interestingly, this MM, studied at primary diagnosis, showed an anaplastic morphology. Of note, the number of total SNVs per primary MM was dramatically reduced (up to 16 × ) after exclusion of the SNVs, which also occurred in the corresponding PBMC sample, emphasizing the advantage of the inclusion of matched normal DNA for an unbiased SNV-filtering approach and supporting our strategy to focus on mutations that occurred in at least one primary MM.
Figure 2.
Mutation load of the six MM cell lines and the five primary MM samples. MM cell lines showed an overall higher mutation load, with the exception of Patient 1. The bars in dark gray reflect the mutation load after SeattleSeq annotation and before the exclusion of SNVs detected in the corresponding blood (see Figure 1). The bars in light gray reflect the mutation load of the primary sample after the exclusion of SNVs that occurred in the corresponding blood.
The mutational profile of MM is enriched for genes associated with growth factor receptor and adhesion molecule signaling
Pathway annotation of the 79 genes (137 SNVs, Supplementary Table S7) affected in at least 1 of our 5 primary MM or in 1 of our 5 primary MM and in any of the 6 cell lines revealed an enrichment of 9 growth factor receptor signaling pathways and 1 adhesion pathway among the top 20 gene sets (Table 1), and 15 growth factor receptor signaling pathways and 1 adhesion pathway among the 50 most significant gene sets (Supplementary Table S8). Moreover, annotation of the 193 genes (330 SNVs, Supplementary Table S5) that are affected in at least 1 of our 5 primary MM or in 1 MM cell line, plus at least 1 of the 38 primary MM of the Chapman data set, revealed an enrichment of 14 pathways related to specific cancers (for example, liver, melanoma) or cancer in general, and 3 adhesion pathways among the first 20 gene sets (Supplementary Table S9A). Among the top 50 gene sets, we detected 4 adhesion-associated pathways and 4 growth factor receptor signaling pathways (Supplementary Table S9B). In an independent validation approach, pathway analysis was performed on genes that were mutated in the published 38 primary MM cases (1429), revealing an enrichment of 2 adhesion-associated pathways among the 20 most significant gene sets, and 4 adhesion-associated pathways among the top 50 most significant gene sets (Supplementary Table S10). Moreover, an enrichment of two gene sets associated with histone modification was observed, which confirms the findings of Chapman et al.5 that this pathway might have an important role in MM pathogenesis.
Table 1. GSEA pathway annotation.
| Gene set name | Genes in gene set | Genes in overlap | P-value |
|---|---|---|---|
| REACTOME SIGNALING BY EGFR | 48 | 4 | 1.59E-05 |
| ST_MYOCYTE_AD_PATHWAY | 23 | 3 | 5.09E-05 |
| KEGG_LONG_TERM_DEPRESSION | 70 | 4 | 7.12E-05 |
| KEGG_LONG_TERM_POTENTATION | 70 | 4 | 7.12E-05 |
| VERRECCHIA_DELAYED_RESPONSE_TO_TGFB1 | 36 | 3 | 1.99E-04 |
| ST_B_CELL_ANTIGEN_RECEPTOR | 39 | 3 | 2.53E-04 |
| KEGG_BLADDER_CANCER | 42 | 3 | 3.16E-04 |
| BENPORATH_SUZ12_TARGETS | 1037 | 11 | 3.82E-04 |
| NAKAMURA_METASTASIS | 47 | 3 | 4.42E-04 |
| VERRECCHIA_RESPONSE_TO_TGFB1_C4 | 11 | 2 | 5.30E-04 |
| KEGG_ENDOMETRIAL_CANCER | 52 | 3 | 5.95E-04 |
| REACTOME_SHC_MEDIATED_SIGNALING | 12 | 2 | 6.34E-04 |
| KEGG_NON_SMALL_CELL_LUNG_CANCER | 54 | 3 | 6.65E-04 |
| KEGG_NEUROTROPHIN_SIGNALING_PATHWAY | 126 | 4 | 6.81E-04 |
| KEGG_AXON_GUIDANCE | 129 | 4 | 7.44E-04 |
| REACTOME_GRB2_EVENTS_IN_EGFR_SIGNALING | 13 | 2 | 7.48E-04 |
| REACTOME_SOS_MEDIATED_SIGNALING | 13 | 2 | 7.48E-04 |
| KEGG_TIGHT_JUNCTION | 134 | 4 | 8.57E-04 |
| AMIT_EGF_RESPONSE_20_MCF10A | 14 | 2 | 8.71E-04 |
| REACTOME_SHC_RELATED_EVENTS | 14 | 2 | 8.71E-04 |
Abbreviation: GSEA, gene set enrichment analysis. Twenty most significant gene sets; 79 genes, mutated in at least 1 of the 5 primary MM (Figure 1) were annotated to pathways, using the c2 collection of the GSEA annotation database. Red label: growth factor receptor signaling pathways, yellow label: adhesion-associated pathway. For the top 50 gene sets see Supplementary Table S8.
Mutations in RTKs and their downstream effectors
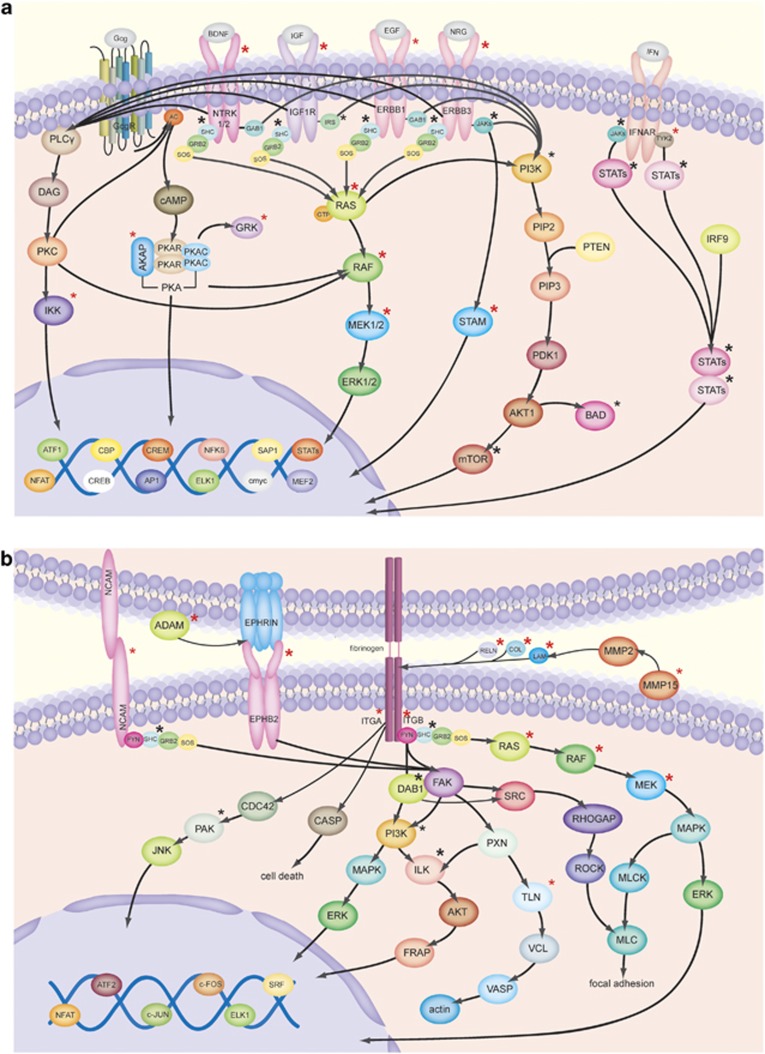
Given the importance of tyrosine kinase signaling in the pathogenesis of MM and its therapeutic relevance in other cancers, we screened our data set (after exclusion of SNVs detected in the corresponding PBMC samples, Figure 1) for mutations in 91 known RTKs (Supplementary Table S11 A) and discovered that 14 out of 91 RTKs (∼15%) were mutated in our data set. Compared with the reported average mutation rate of 35 non-synonymous SNVs in MM,5 which is similar to our results in primary MM (Patient 1–5, bars in light gray, Figure 2), this represents a high enrichment. Specifically, we found somatic mutations in some well-known RTKs, such as IGF1R, EGFR, ERBB3, NTRK1 and NTRK2, as well as in several potential downstream effectors, such as KRAS, BRAF, MAP2K2, AKAP1, IKBKE, STAM2 and GRK. All of these mutations occurred in at least one primary MM and all were designated as ‘damaging' (mutations occur at conservative sites or lead to structural changes) by three different functional predictors (Figure 3a and Table 2). Mutations in NTRK1, NTRK2, AKAP1, IKBKE, STAM2, GRK2/ADRBK1 and MAP2K2 have not previously been reported in MM. All but one mutation labeled in Figure 3a were validated by Sanger sequencing in our data set.
Figure 3.
(a) RTK signaling and (b) adhesion molecule signaling. Asterisks indicate a mutation. Asterisks in red (bold): mutated in at least one primary MM (including Chapman et al.5) and ‘damaging' according to three functional predictors. Asterisks in red: mutated in 1 of the 43 primary MM (including Chapman et al.5). Asterisks in black (bold): mutated in a cell line and ‘damaging' according to three functional predictors. Asterisks in black: mutated in at least one cell line. All mutations were confirmed by Sanger sequencing in our samples, except for MMp15. Moreover, sequence information of GRK/ADBRK1 was not evaluable. Please note that not all members of a family were validated (for details see Supplementary Table S12).
Table 2. Mutation profile of six cell lines and five primary MM samples with respect to adhesion and RTK signaling.
| Gene | AMO1 | U266 | OPM2 | MM1S | JJN3 | L363 | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MMP15 | x(homoz.) | x | x | ||||||||
| MUC4 | xx (<2) | x | |||||||||
| COL10A1 | x | x | |||||||||
| SDK1 | x | x | |||||||||
| LRFN5 | x | x | x | ||||||||
| THBS1 | x | x | |||||||||
| SLAMF1 | x | ||||||||||
| COL6A3 | xx | x | |||||||||
| VCAN | x | ||||||||||
| THSD7A | x | ||||||||||
| ALOX12B | x (<2) | x | |||||||||
| HSD17B1 | x | ||||||||||
| NCAM2 | x | ||||||||||
| ADAM29 | x | x | |||||||||
| ADAMTS1 | x | ||||||||||
| ADAMTSL1 | x(homoz.) | ||||||||||
| BCAN | x | x | |||||||||
| COL17A1 | x | ||||||||||
| COL7A1 | x | ||||||||||
| DCHS1 | x | x | |||||||||
| DCHS2 | x | ||||||||||
| ITGA10 | x | x | |||||||||
| ODZ1 | x | ||||||||||
| SIGLEC1 | x | ||||||||||
| WHSC1 | x | ||||||||||
| LAMB2 | x | ||||||||||
| PCDHGA7 | x | xx | x | ||||||||
| ADAMTS19 | x (<2) x | ||||||||||
| ADAMTS9 | x | ||||||||||
| CAMTA1 | x | ||||||||||
| ADAMTS2 | x (<2) | x | |||||||||
| COL12A1 | x | ||||||||||
| COL16A1 | x | ||||||||||
| HS6ST3 | x | x | |||||||||
| LAMA5 | x | x | |||||||||
| NRXN1 | x | ||||||||||
| ODZ4 | x | ||||||||||
| PCDH10 | x | ||||||||||
| ADAMTS18 | x | ||||||||||
| CDH11 | x | ||||||||||
| CELSR2 | x | ||||||||||
| GPC6 | x | x | |||||||||
| ITGB1 | x | ||||||||||
| LAMB1 | x | ||||||||||
| PCDH15 | x(homoz.) | ||||||||||
| PCDH17 | x | ||||||||||
| PCDH20 | x | ||||||||||
| RELN | xx | ||||||||||
| SPOCK1 | x | ||||||||||
| TLN2 | x | ||||||||||
| TNC | x | x (<2) | |||||||||
| TGFBR2 | x | ||||||||||
| CHIA | x(homoz.) | ||||||||||
| NTRK1 | x | x | |||||||||
| EPHB2 | x | ||||||||||
| EPHA8 | x | x | x | ||||||||
| NTRK2 | x | xx | |||||||||
| EGFR | x | x | |||||||||
| ERBB3 | x | x | |||||||||
| IGF1R | x | ||||||||||
| TTN | x (<2) | ||||||||||
| HK3 | x | ||||||||||
| STAM2 | x | ||||||||||
| IKBKE | x | ||||||||||
| ADRBK1 | x | ||||||||||
| AKAP1 | x | ||||||||||
| TYK2 | x | ||||||||||
| KRAS | x | x | x | ||||||||
| NRAS | x | ||||||||||
| BRAF | x | ||||||||||
| MAP2K2 | x | x |
Abbreviations: MM, multiple myeloma; RTK, receptor tyrosine kinase; x, mutated; xx, mutated twice in the same sample; homoz., homozygous
Adhesion molecules, RTKs and some downstream molecules are listed that were mutated in at least 1 of the5 primary MM or in at least 1 cell line, plus at least 1 of the 38 published primary MM.5 All mutations mentioned (x, xx, homoz.) were damaging in at least two functional predictors, if not otherwise specified (<2=less than 2 predictors). Genes that belonged to the same signaling network according to the STRING database or manual literature search were highlighted in bold and italics (bold: medium confidence; italics: low confidence). The mutation details for the 38 primary MM5 are shown in Supplementary Table S12.
Mutations in adhesion molecules and their downstream effectors
The finding that the pathway annotation of genes mutated in MM revealed the biological feature of ‘adhesion' caught our interest, as the microenvironment is known to have an important role in MM pathogenesis. Therefore, we screened 653 known adhesion molecules (Supplementary Table S11 B) and found that 201 of those (∼31%) were mutated in our data set. These mutations affected integrins (for example, ITGB1, ITGB5, ITGA7 and TGFBR2), several cadherins (for example, CDH9, CDH11, CDH18 and CDH23) and protocadherins (for example, PCDH9, PCDHGA3, PCDH10 and PCDH20), as well as ICAM and NCAM2, extracellular matrix molecules (for example, COL10A1 and LAMB2), proteoglycans (for example, VCAN and BCAN), metalloproteases (for example, ADAM29 and ADAMTS1), as well as different ephrin receptors (for example, EPHB2 and EPHA2) and several downstream effectors (partially shown in Figure 3b and Table 2). Of note, there was no prevalence of mutations affecting adhesion molecules in the two MM patients with extramedullary involvement. Mutations marked in Figure 3 and also present in cell line(s) are detailed in Supplementary Table S12. All but one mutation labeled in Figure 3b were validated by Sanger sequencing.
Approximately 50% of MMs are affected by more than one mutation in the same signaling network
We noticed that some MM cell lines carried multiple mutations within the same signaling network. For example, in the cell line AMO1, mutations potentially affecting the mitogen-activated protein kinase pathway were found for EGFR, ERBB3, KRAS (A146T) and MAP2K2. This prompted us to investigate this surprising finding in MM cell lines and primary MM in more detail.
All 6 MM cell lines and all primary MM samples, as well as almost all 38 primary MM (36/38 of MM reported in Chapman et al.5) carried mutations in genes that encode parts of the same signaling network containing adhesion molecules, RTKs and their downstream effectors (Table 2, Supplementary Table S13 and 14). Moreover, all 6 MM cell lines and approximately 50% of primary MM (including Chapman et al.5) showed mutations in more than 1 gene of the same signaling network, and approximately 30% showed more than 2 mutations within this network. Of note, the second largest number of mutations (n=16) within this network, affecting the adhesion molecules SDK1, LRFN5, THBS1, SLAMF1, VCAN, ALOX12B, NCAM2 and TGFBR2, the RTK EPHB2 and EPHA8, and the effector molecules HK3, STAM2, IKBKE, ADRBK1, AKAP1 and MAP2K2, was identified in 1 primary MM showing anaplastic features (Table 2). Thus, this biological feature does not only seem to be a culture artefact of established MM cell lines, but is probably also a genuine trait of malignant plasma cell clones in vivo.
Correlation of somatic mutations with copy number, gene expression and copy neutral loss of heterozygosity
In the six MM cell lines, selected mutations in RTKs and adhesion molecules were correlated with mRNA expression, chromosomal copy number changes and copy neutral loss of heterozygosity (Tables 3a and b, for mutation details see Supplementary Table S14). The EGFR mutation in MM.1S showed a weak, positive correlation with gene expression (Table 3). Moreover, IGF1R, NTRK1 and ERBB3 mutations were accompanied by increased copy numbers. Although we observed only a weak correlation of somatic mutations and gene expression for RTKs, we noticed a remarkable correlation for mutations in adhesion molecules with gene expression (Table 3). Specifically, in the presence of the respective mutation, we observed a fourfold upregulation of TNC, a fourfold downregulation of LAMA5 and a ninefold downregulation of both LAMB2 and RELN. Moreover, LAMB2 was affected by cn-loss of heterozygosity in MM.1S, pointing to a possible suppressor function. From four primary MM samples, cytogenetic information was available (Supplementary Table S1). Of note, the MM sample with p53 deficiency carried the least number of mutations in adhesion- and RTK-associated molecules, suggesting a dominant pathogenetic event for the p53 alteration that might make additional mutations in the adhesion/RTK network dispensable.
Table 3. Correlation of selected mutations in RTKs and adhesion molecules with gene expression, CN changes and copy neutral LOH.
| Gene | GE | CN | LOH | CL | Predictors |
|---|---|---|---|---|---|
| (A) | |||||
| IGF1R | — | + | L363 | 3 | |
| NTRK2 | — | — | L363 | 3 | |
| NTRK1 | — | + | MM1S | 2 | |
| EGFR | — | — | AMO1 | 2 | |
| EGFR | ↑ | — | MM1S | 2 | |
| ERBB3 | — | + | AMO1 | 3 | |
| ERBB3 | — | — | L363 | 3 | |
| (B) | |||||
| SDK1 | — | + | JJN3 | 3 | |
| LRFN5 | — | — | X | MM1S | 3 |
| LRFN5 | — | — | U266 | 3 | |
| DCHS2 | — | + | OPM2 | 3 | |
| DCHS2 | — | — | MM1S | 3 | |
| SIGLEC1 | — | + | U266 | 2 | |
| SIGLEC1 | — | — | MM1S | 3 | |
| PCDHGA7 | — | — | JJN3 | 2 | |
| PCDH10 | — | — | AMO1 | 3 | |
| THBS1 | — | — | AMO1 | 2 | |
| BCAN | — | ++ | X | AMO1 | 3 |
| BCAN | ↓ | ++ | OPM2 | 3 | |
| LAMB2 | ↓↓↓ | — | X | MM1S | 3 |
| CAMTA1 | — | — | OPM2 | 3 | |
| ADAMTS2 | — | — | OPM2 | 3 | |
| ADAMTS2 | — | — | AMO1 | 2 | |
| LAMA5 | ↓↓ | + | OPM2 | 2 | |
| LAMA5 | — | — | MM1S | 3 | |
| TNC | ↑↑ | — | AMO1 | 3 | |
| ADAMTS19 | ↓ | — | U266 | 3 | |
| COL12A1 | — | — | L363 | 3 | |
| CDH11 | — | — | L363 | 3 | |
| CELSR2 | — | + | JJN3 | 3 | |
| ITGB1 | ↓↓ | — | X | U266 | 3 |
| PCDH20 | ↑ | + | AMO1 | 3 | |
| RELN | ↓↓↓ | + | U266 | 3 | |
Abbreviations: CL, cell line; CN, copy number; GE, gene expression; LOH, loss of heterozygosity; MM, multiple myeloma; RTK, receptor tyrosine kinase.
One arrow represents a twofold up- or downregulation, one and a half arrows a threefold up- or downregulation, two arrows a fourfold up- or downregulation, and three arrows a ninefold up- or downregulation in mRNA expression compared with the median mRNA expression of all six cell lines; ± represents a simple copy number gain (three copies) and ++ represents four copies of that gene. Copy neutral LOH or LOH that was accompanied by a chromosomal gain is depicted by x.
aRTKs and adhesion molecules that were mutated in at least 1 of the 43 primary MM (including Chapman et al.5) and 1 cell line, that were ‘damaging' by at least 2 functional predictors and that were validated by Sanger sequencing. These conditions apply to all genes of Table A and B.
Tyrosine kinase catalytic domains of RTKs are frequently affected by somatic mutations in MM
To gain more information on the potential biological impact of the non-synonymous mutations in RTKs listed in Table 3, we searched for the affected domains using the Nucleotide database from NCBI and the String database. Of note, three out of five RTKs (NTRK2, IGF1R and EGFR) were affected by mutations in the tyrosine kinase domain in at least one of the samples investigated, and the remaining mutations mostly affected the furin-like domains or the region between two furin-like domains, close to the ligand-binding domain. For example, the somatic point mutations in NTRK2 affected the tyrosine kinase catalytic domain in one primary MM of our discovery set (Supplementary Figure S1); IGF1R showed a somatic point mutation in L363 in the tyrosine kinase catalytic domain, and the somatic point mutations in EGFR that were detected in MM.1S and in a primary MM5 also appeared to target the tyrosine kinase catalytic domain. For more details see Supplementary Results.
Discussion
With the use of novel drugs, such as thalidomide, lenalidomide and bortezomib, and risk-adapted strategies, including autologous stem cell transplantation in eligible patients, 10-year overall survival rates in the small subset of MM patients younger than 60 years have reached up to 30%.2, 3, 19 Patients who carry MM clones with hyperdiploidy or a t(11;14) were shown to have a relatively better outcome compared with those whose tumors harbor a t(4;14), t(14;16) or del 17p.20 Interestingly, MM patients carrying the t(4;14) translocation that often leads to deregulation of FGFR3 seem to benefit from the inclusion of bortezomib into their first-line treatment21 in that the adverse prognosis may be overcome. This suggests future concepts in which the detection of molecular alterations may ultimately lead to refinement of therapeutic approaches. Nevertheless, MM currently remains largely incurable and efforts are underway to develop more individualized, molecular therapies that target critical growth and survival pathways in the tumor cells.
Novel sequencing technologies nowadays allow for the identification of somatically mutated genes in malignant tumors on a global scale. Initial genome sequencing of MM5 and published literature suggest that recurrent genetic translocations (t(11;14), t(4;14), t(14;16)) or oncogene mutations, for example, in KRAS or NRAS, with frequencies of at best 15–20%.3 Novel mutations are detected at even lower frequencies. BRAF, for example, is mutated in only 4% of all samples,5 in sharp contrast to the scenario in hairy cell leukemia that harbors BRAF mutations in most, if not all tumors.22 Thus, it appears unlikely that the development of a targeted therapy against a single molecule will prove to be a successful strategy in MM. Rather, there has been a recent focus in clinical phase I/II trials to test the hypothesis whether the modulation of molecular pathways, for example, by inhibiting mitogen-activated protein kinase or AKT, or by using epigenetic agents, could represent a promising way forward.23, 24 Such an approach is supported by a recent sequencing analysis of 38 MM tumor genomes/exomes that showed clustering of somatic mutations in important cellular pathways, including the RNA-processing machinery, histone-modifying enzymes and genes involved in nuclear factor-κB signaling.5
Our discovery and validation approach, integrating altogether 43 primary MM samples (including published MM samples) and 6 MM cell lines, provides evidence that the neoplastic cells in MM are affected by multiple and heterogeneous somatic mutations in adhesion- and RTK-associated signaling molecules, which is a hitherto undescribed finding. Notably, 147 out of 201 mutated adhesion molecules were newly detected in our analysis, including the integrins ITGA6, ITGA7 and ITGB5, the cadherins CDH23, CDH18 and CDH9, and the protocadherin clusters (for example, PCDH9 and PCDHGA3) among others. Twenty mutations in adhesion molecules were found to be affected in at least 1 of the 43 primary MM samples and 1 cell line, were ‘damaging' by at least 2 functional predictors and were validated by Sanger sequencing. Available gene expression and copy number data in six investigated MM cell lines allowed us to correlate the presence of specific mutations in adhesion-associated molecules with accompanying alterations on the mRNA expression and genomic copy number level. From this analysis, it remains unclear whether mutations in adhesion molecules might increase or decrease their function, that is, whether they might act as oncogenes or as tumor suppressors.
However, the frequent occurrence of somatic mutations in adhesion molecules in primary MM emphasizes their presumably important biological role for the interaction of the neoplastic plasma cells with their BM microenvironment. For example, the interaction between MM cells and BM stromal cells was shown to provide adhesion-mediated drug resistance.8, 25, 26 In addition, this interaction may lead to the activation of signal transduction pathways that promote cell cycle progression and cell survival via expression of growth factors such as insulin-like growth factor and vascular endothelial growth factor, as well as chemokines and cytokines.8 Our results support the notion that MM cells are being selected for clones with increased numbers of mutated adhesion molecules during tumor evolution. As a consequence, therapeutic efforts to more effectively target the interaction between MM cells and their surrounding stroma appear reasonable, a strategy that is currently tested in chronic lymphocytic leukemia, in which the stromal cell-derived factor-1/C-X-C chemokine receptor type 4 axis in BM and lymph node infiltrates is targeted by the C-X-C chemokine receptor type 4antagonists, such as plerixafor and T140.27
Another important finding of our study is the enrichment of somatic mutations in RTKs in MM cells. The well-known RTKs EGFR, IGF1R, ERBB3, NTRK1 and NTRK2 were affected in at least 1 of the 43 primary MM samples and in at least 1 MM cell line, and mutations in these genes were predicted to be ‘damaging' according to at least 2 functional predictors. This finding is of interest for two reasons. First, RTKs likely have a role in the pathogenesis of MM, as evidenced by the presence of FGFR3 translocations in a considerable subset of MM,2 and signaling cascades downstream of RTKs that involve the aberrant activation or deregulation of the RAS/mitogen-activated protein kinase and phosphoinositide-3-kinase/AKT pathways are frequently altered in MM.10, 28, 29 Second, these receptors may represent interesting targets for novel therapeutic approaches reinforcing initial concepts in MM to develop small-molecule inhibitors that block anti-apoptotic or pro-proliferative effects of RTK activation. RTK inhibition is already a widely used strategy for treating various types of solid cancers, including epidermal growth factor receptor inhibitors (erlotinib, gefitinib) in non-small cell lung cancer or the monoclonal antibody trastuzumab and the small-molecule inhibitor lapatinib in patients with ERBB2/HER2-neu positive breast and gastric cancer, respectively.30, 31, 32 More recently, insulin-like growth factor-1 receptor inhibitors have been under investigation in clinical trials of non-small cell lung cancer, Ewing sarcoma and prostate cancer.33, 34, 35, 36, 37 Given the increased frequency of RTK mutations in MM, therapeutic intervention using RTK inhibitors could prove useful in subsets of MM patients.
Our data, in line with published results,5 suggest that the pathogenesis and progression of MM is accompanied by the development of multiple and heterogeneous somatic mutations in various important cellular signaling cascades (RNA-processing machinery, histone-modifying enzymes, nuclear factor-κB system, adhesion- and RTK-associated molecules), leading to a profound ‘interindividual pathway redundancy'. For example, all 6 MM cell lines and approximately 95% of 43 primary MM samples carried mutations in at least 1 adhesion molecule, RTK or RTK downstream-signaling effector that all cluster within the same signaling network, indicating that pathways belonging to this network are of importance in MM pathogenesis. Approximately 50% of primary MM were affected by mutations in more than one gene of this network, and almost one third of primary MM samples carried multiple mutations (>2) in this signaling network, suggesting an ‘intra-individual pathway redundancy'. Of note, 1 MM with anaplastic morphology carried 16 mutations (including, for example, ALOX12B, NCAM2, EPHB2 and MAP2K2). This observed pathway redundancy' underlines the necessity of comprehensive genetic approaches to make informed decisions on how to employ future targeted molecular therapies most effectively for the benefit of individual MM patients.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (KFO 216). Moreover, we would like to acknowledge the expert technical assistance from Theodora Nedeva.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri S, Stein H, et al. IARC: Lyon; 2008. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. [Google Scholar]
- Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- Chesi M, Bergsagel PL. Many multiple myelomas: making more of the molecular mayhem. Hematology Am Soc Hematol Educ Program. 2011;2011:344–353. doi: 10.1182/asheducation-2011.1.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan JB, Shi CX, Tembe W, Christoforides A, Kurdoglu A, Sinari S, et al. Whole genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution and clonal tides. Blood. 2012;120:1060–1066. doi: 10.1182/blood-2012-01-405977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BA, Wardell CP, Melchor L, Hulkki S, Potter NE, Johnson DC, et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120:1077–1086. doi: 10.1182/blood-2012-03-412981. [DOI] [PubMed] [Google Scholar]
- Bommert K, Bargou RC, Stühmer T. Signalling and survival pathways in multiple myeloma. Eur J Cancer. 2006;42:1574–1580. doi: 10.1016/j.ejca.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Klein B, Tarte K, Jourdan M, Mathouk K, Moreaux J, Jourdan E, et al. Survival and proliferation factors of normal and malignant plasma cells. Int J Hematol. 2003;78:106–113. doi: 10.1007/BF02983377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentzsch S, Chatterjee M, Gries M, Bommert K, Gollasch H, Dorken B, et al. PI3-K/AKT/FKHR and MAPK signaling cascades are redundantly stimulated by a variety of cytokines and contribute independently to proliferation and survival of multiple myeloma cells. Leukemia. 2004;18:1883–1890. doi: 10.1038/sj.leu.2403486. [DOI] [PubMed] [Google Scholar]
- Tai YT, Podar K, Catley L, Tseng YH, Akiyama M, Shringarpure R, et al. Insulin-like growth factor-1 induces adhesion and migration in human multiple myeloma cells via activation of beta1-integrin and phosphatidylinositol 3'-kinase/AKT signaling. Cancer Res. 2003;63:5850–5858. [PubMed] [Google Scholar]
- Yaccoby S, Wezeman MJ, Henderson A, Cottler-Fox M, Yi Q, Barlogie B, et al. Cancer and the microenvironment: myeloma-osteoclast interactions as a model. Cancer Res. 2004;64:2016–2023. doi: 10.1158/0008-5472.can-03-1131. [DOI] [PubMed] [Google Scholar]
- Stühmer T, Arts J, Chatterjee M, Borawski J, Wolff A, King P, et al. Preclinical anti-myeloma activity of the novel HDAC-inhibitor JNJ-26481585. Br J Haematol. 2010;149:529–536. doi: 10.1111/j.1365-2141.2010.08126.x. [DOI] [PubMed] [Google Scholar]
- Liebisch P, Dohner H. Cytogenetics and molecular cytogenetics in multiple myeloma. Eur J Cancer. 2006;42:1520–1529. doi: 10.1016/j.ejca.2005.12.028. [DOI] [PubMed] [Google Scholar]
- Ross F, Avet-Loiseau H, Ameye G, Gutierrez NC, Liebisch P, OC S, et al. Report from the european myeloma network on interphase FISHin multiple myeloma and related disorders. Haematologica. 2012;97:1272–1277. doi: 10.3324/haematol.2011.056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39 (Database issue:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsagel PL. Impressions of the myeloma landscape. Blood. 2010;116:2403–2404. doi: 10.1182/blood-2010-07-294009. [DOI] [PubMed] [Google Scholar]
- Kumar SK, Mikhael JR, Buadi FK, Dingli D, Dispenzieri A, Fonseca R, et al. Management of newly diagnosed symptomatic multiple myeloma: updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines. Mayo Clin Proc. 2009;84:1095–1110. doi: 10.4065/mcp.2009.0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda-Roman M, Zangari M, Haessler J, Anaissie E, Tricot G, van Rhee F, et al. Sustained complete remissions in multiple myeloma linked to bortezomib in total therapy 3: comparison with total therapy 2. Br J Haematol. 2008;140:625–634. doi: 10.1111/j.1365-2141.2007.06921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiacci E, Trifonov V, Schiavoni G, Holmes A, Kern W, Martelli MP, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocio EM, Mateos MV, Maiso P, Pandiella A, San-Miguel JF. New drugs in multiple myeloma: mechanisms of action and phase I/II clinical findings. Lancet Oncol. 2008;9:1157–1165. doi: 10.1016/S1470-2045(08)70304-8. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Barlogie B, Shaughnessy JD. The molecular characterization and clinical management of multiple myeloma in the post-genome era. Leukemia. 2009;23:1941–1956. doi: 10.1038/leu.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Stühmer T, Herrmann P, Bommert K, Dorken B, Bargou RC. Combined disruption of both the MEK/ERK and the IL-6R/STAT3 pathways is required to induce apoptosis of multiple myeloma cells in the presence of bone marrow stromal cells. Blood. 2004;104:3712–3721. doi: 10.1182/blood-2004-04-1670. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Bergsagel PL, Kuehl WM, Anderson KC. Advances in biology of multiple myeloma: clinical applications. Blood. 2004;104:607–618. doi: 10.1182/blood-2004-01-0037. [DOI] [PubMed] [Google Scholar]
- Burger JA. Chemokines and chemokine receptors in chronic lymphocytic leukemia (CLL): from understanding the basics towards therapeutic targeting. Semin Cancer Biol. 2010;20:424–430. doi: 10.1016/j.semcancer.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Steinbrunn T, Stühmer T, Gattenlöhner S, Rosenwald A, Mottok A, Unzicker C, et al. Mutated RAS and constitutively activated Akt delineate distinct oncogenic pathways, which independently contribute to multiple myeloma cell survival. Blood. 2011;117:1998–2004. doi: 10.1182/blood-2010-05-284422. [DOI] [PubMed] [Google Scholar]
- Zöllinger A, Stühmer T, Chatterjee M, Gattenlöhner S, Haralambieva E, Müller-Hermelink HK, et al. Combined functional and molecular analysis of tumor cell signaling defines 2 distinct myeloma subgroups: Akt-dependent and Akt-independent multiple myeloma. Blood. 2008;112:3403–3411. doi: 10.1182/blood-2007-11-119362. [DOI] [PubMed] [Google Scholar]
- Awada A, Bozovic-Spasojevic I, Chow L. New therapies in HER2-positive breast cancer: A major step towards a cure of the disease. Cancer Treat Rev. 2012;38:494–504. doi: 10.1016/j.ctrv.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364:1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begnami MD, Fukuda E, Fregnani JH, Nonogaki S, Montagnini AL, da Costa WL, et al. Prognostic implications of altered human epidermal growth factor receptors (HERs) in gastric carcinomas: HER2 and HER3 are predictors of poor outcome. J Clin Oncol. 2011;29:3030–3036. doi: 10.1200/JCO.2010.33.6313. [DOI] [PubMed] [Google Scholar]
- Friedlander TW, Weinberg VK, Huang Y, Mi JT, Formaker CG, Small EJ, et al. A phase II study of insulin-like growth factor receptor inhibition with nordihydroguaiaretic acid in men with non-metastatic hormone-sensitive prostate cancer. Oncol Rep. 2012;27:3–9. doi: 10.3892/or.2011.1487. [DOI] [PubMed] [Google Scholar]
- Juergens H, Daw NC, Geoerger B, Ferrari S, Villarroel M, Aerts I, et al. Preliminary efficacy of the anti-insulin-like growth factor type 1 receptor antibody figitumumab in patients with refractory Ewing sarcoma. J Clin Oncol. 2011;29:4534–4540. doi: 10.1200/JCO.2010.33.0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki RG. Small is beautiful: insulin-like growth factors and their role in growth, development, and cancer. J Clin Oncol. 2010;28:4985–4995. doi: 10.1200/JCO.2009.27.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappo AS, Patel SR, Crowley J, Reinke DK, Kuenkele KP, Chawla SP, et al. R1507, a monoclonal antibody to the insulin-like growth factor 1 receptor, in patients with recurrent or refractory Ewing sarcoma family of tumors: results of a phase II Sarcoma Alliance for Research through Collaboration study. J Clin Oncol. 2011;29:4541–4547. doi: 10.1200/JCO.2010.34.0000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam SS, Spigel DR, Chen D, Steins MB, Engelman JA, Schneider CP, et al. Randomized phase II study of erlotinib in combination with placebo or R1507, a monoclonal antibody to insulin-like growth factor-1 receptor, for advanced-stage non-small-cell lung cancer. J Clin Oncol. 2011;29:4574–4580. doi: 10.1200/JCO.2011.36.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.