Abstract
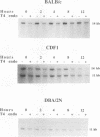
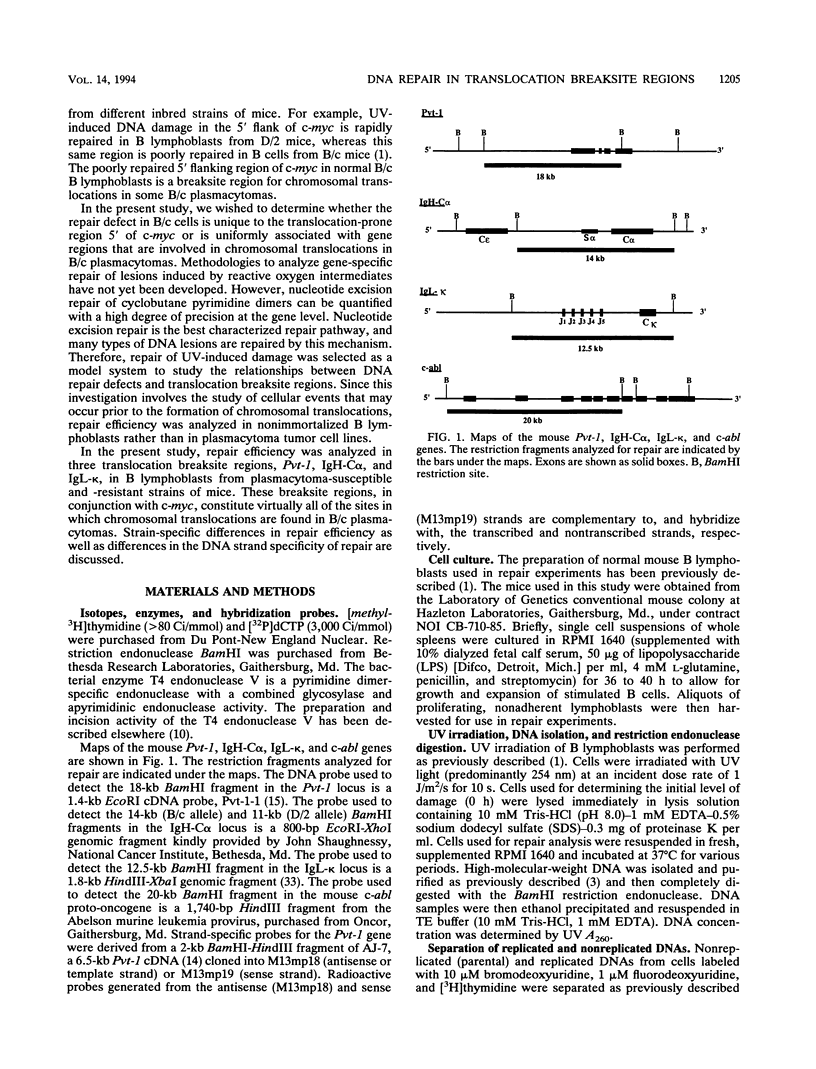
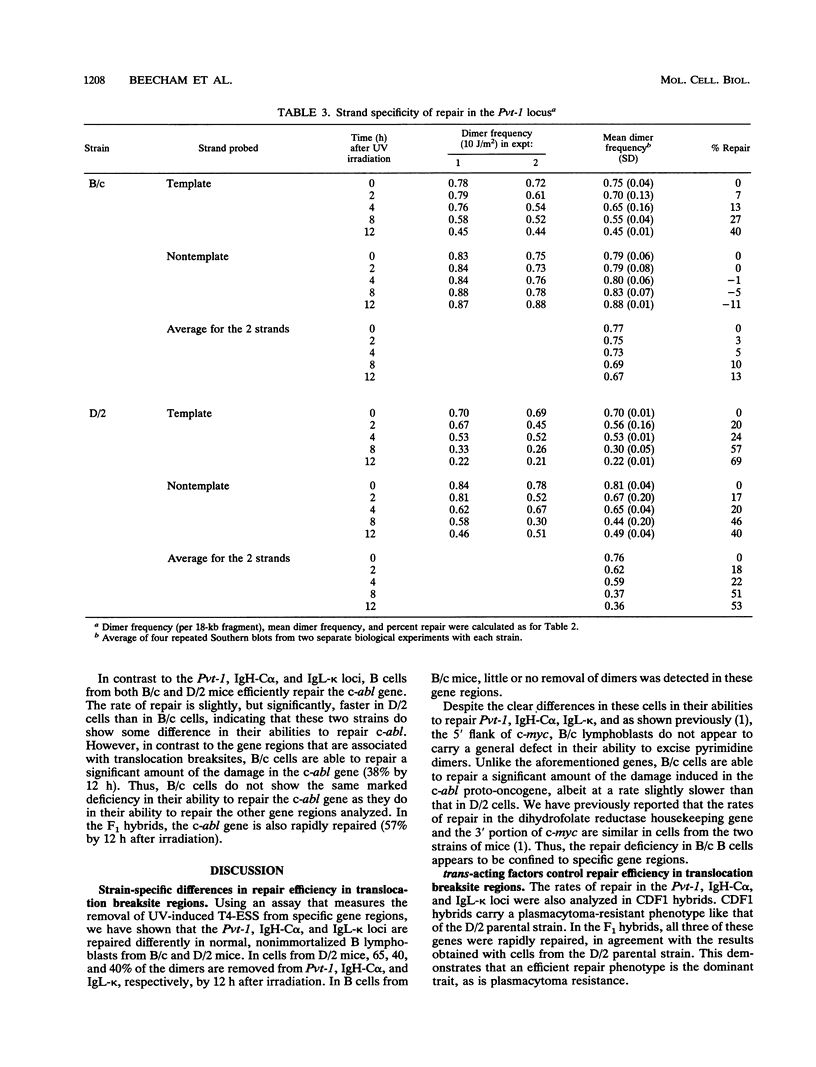
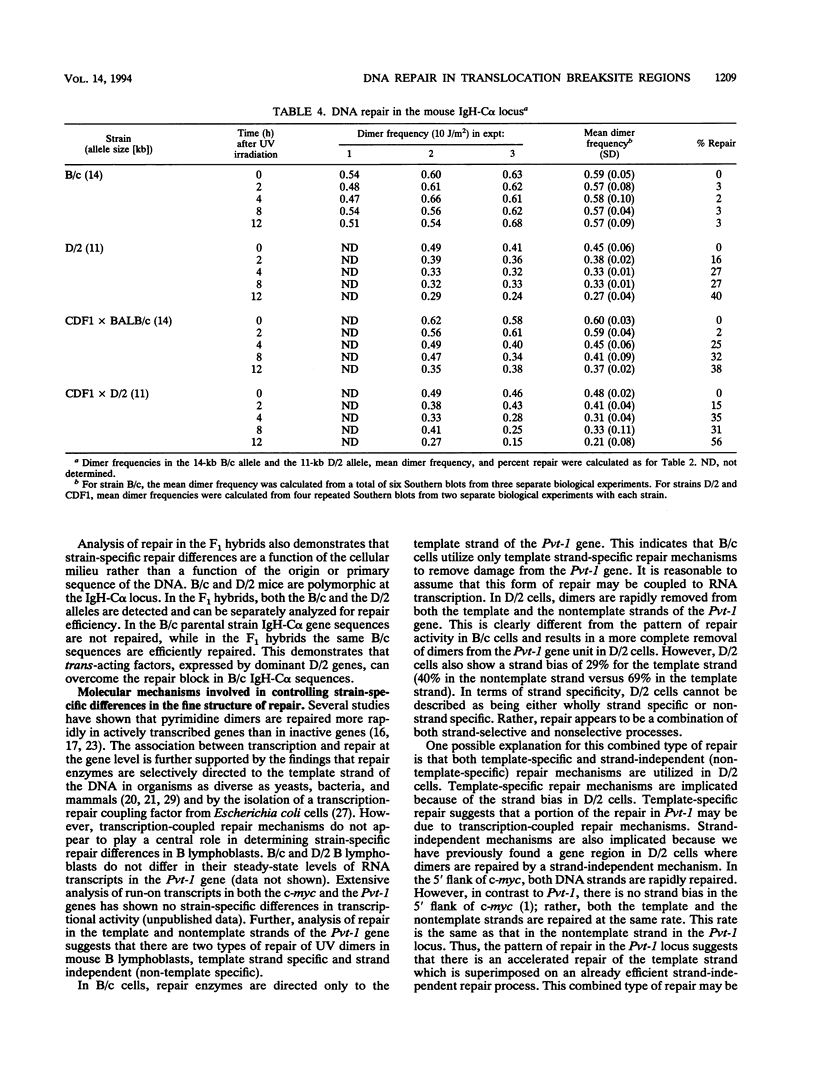
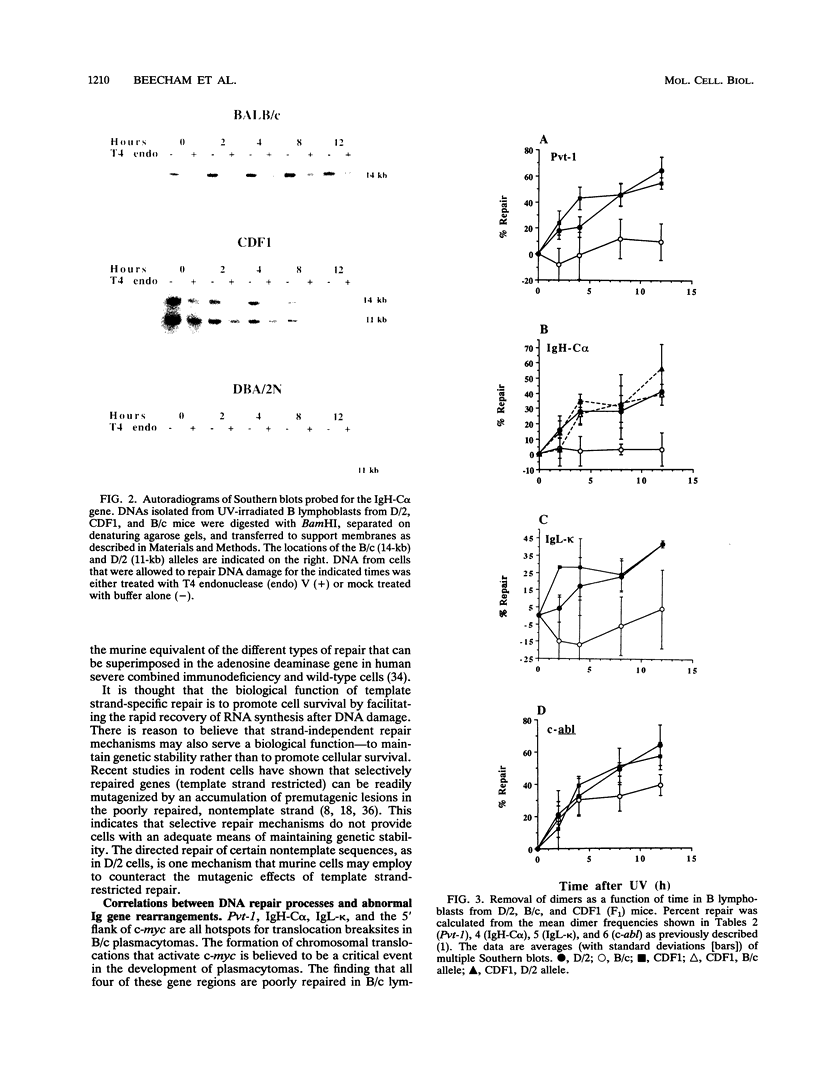
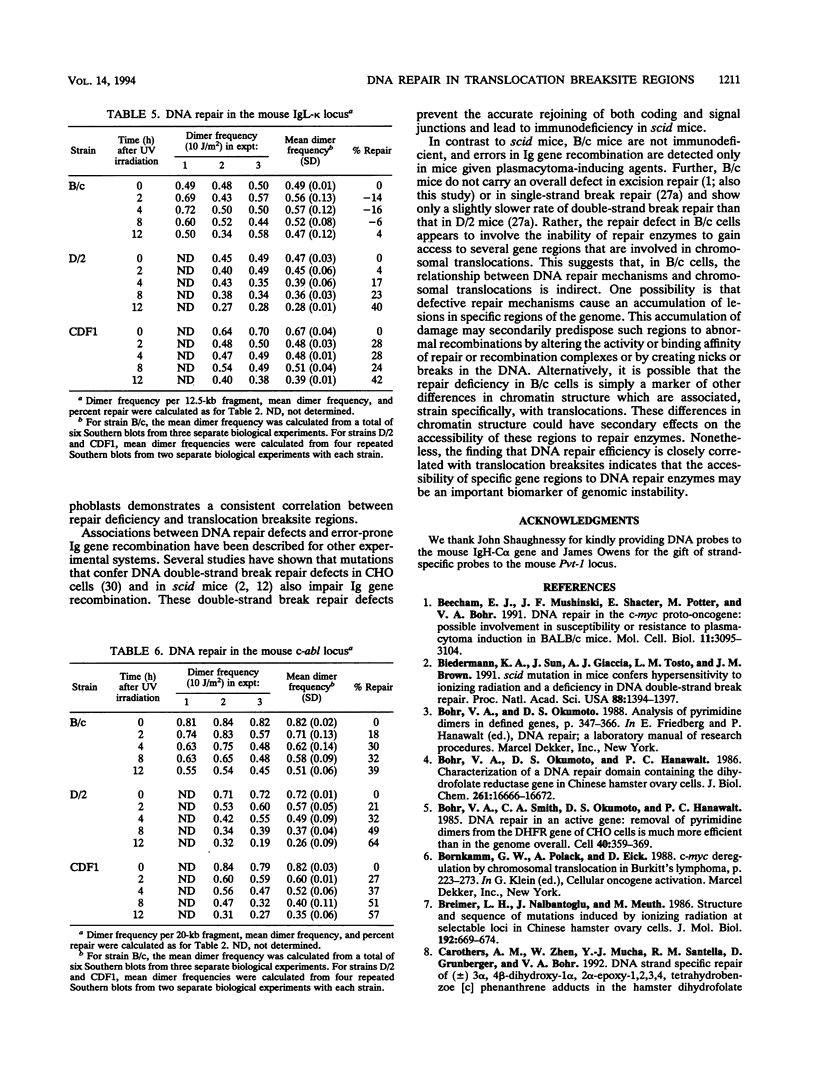
Using an assay that measures the removal of UV-induced pyrimidine dimers in specific DNA sequences, we have found that the Pvt-1, immunoglobulin H-C alpha (IgH-C alpha), and IgL-kappa loci are poorly repaired in normal B lymphoblasts from plasmacytoma-susceptible BALB/cAnPt mice. Breaksites in these genes are associated with the chromosomal translocations that are found in > 95% of BALB/cAnPt plasmacytomas. In contrast to those from BALB/cAnPt mice, B lymphoblasts from plasmacytoma-resistant DBA/2N mice rapidly repair Pvt-1, IgH-C alpha, and IgL-kappa. Further, (BALB/cAnPt x DBA/2N)F1 hybrids, which are resistant to plasmacytoma development, carry an efficient (DBA/2N-like) repair phenotype. Analysis of allele-specific repair in the IgH-C alpha locus indicates that efficient repair is controlled by dominant, trans-acting factors. In the F1 heterozygotes, these factors promote efficient repair of BALB/cAnPt IgH-C alpha gene sequences. The same sequences are poorly repaired in the BALB/cAnPt parental strain. Analysis of the strand specificity of repair indicates that both strand-selective and nonselective forms of repair determine repair efficiency at the gene level in nonimmortalized murine B lymphoblasts.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beecham E. J., Mushinski J. F., Shacter E., Potter M., Bohr V. A. DNA repair in the c-myc proto-oncogene locus: possible involvement in susceptibility or resistance to plasmacytoma induction in BALB/c mice. Mol Cell Biol. 1991 Jun;11(6):3095–3104. doi: 10.1128/mcb.11.6.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann K. A., Sun J. R., Giaccia A. J., Tosto L. M., Brown J. M. scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr V. A., Okumoto D. S., Ho L., Hanawalt P. C. Characterization of a DNA repair domain containing the dihydrofolate reductase gene in Chinese hamster ovary cells. J Biol Chem. 1986 Dec 15;261(35):16666–16672. [PubMed] [Google Scholar]
- Bohr V. A., Smith C. A., Okumoto D. S., Hanawalt P. C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985 Feb;40(2):359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Breimer L. H., Nalbantoglu J., Meuth M. Structure and sequence of mutations induced by ionizing radiation at selectable loci in Chinese hamster ovary cells. J Mol Biol. 1986 Dec 5;192(3):669–674. doi: 10.1016/0022-2836(86)90284-6. [DOI] [PubMed] [Google Scholar]
- Cory S. Activation of cellular oncogenes in hemopoietic cells by chromosome translocation. Adv Cancer Res. 1986;47:189–234. doi: 10.1016/s0065-230x(08)60200-6. [DOI] [PubMed] [Google Scholar]
- Evans M. K., Robbins J. H., Ganges M. B., Tarone R. E., Nairn R. S., Bohr V. A. Gene-specific DNA repair in xeroderma pigmentosum complementation groups A, C, D, and F. Relation to cellular survival and clinical features. J Biol Chem. 1993 Mar 5;268(7):4839–4847. [PubMed] [Google Scholar]
- Grosovsky A. J., Drobetsky E. A., deJong P. J., Glickman B. W. Southern analysis of genomic alterations in gamma-ray-induced aprt- hamster cell mutants. Genetics. 1986 Jun;113(2):405–415. doi: 10.1093/genetics/113.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson E. A., Qin X. Q., Bump E. A., Schatz D. G., Oettinger M., Weaver D. T. A link between double-strand break-related repair and V(D)J recombination: the scid mutation. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4061–4065. doi: 10.1073/pnas.88.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Bohr V. A., Hanawalt P. C. Demethylation enhances removal of pyrimidine dimers from the overall genome and from specific DNA sequences in Chinese hamster ovary cells. Mol Cell Biol. 1989 Apr;9(4):1594–1603. doi: 10.1128/mcb.9.4.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppi K., Siwarski D., Skurla R., Klinman D., Mushinski J. F. Pvt-1 transcripts are found in normal tissues and are altered by reciprocal(6;15) translocations in mouse plasmacytomas. Proc Natl Acad Sci U S A. 1990 Sep;87(18):6964–6968. doi: 10.1073/pnas.87.18.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadon S. A., Snowden M. M. Differential repair of DNA damage in the human metallothionein gene family. Mol Cell Biol. 1988 Dec;8(12):5331–5338. doi: 10.1128/mcb.8.12.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H. D., Bohr V. A., Hanawalt P. C. Differential DNA repair in transcriptionally active and inactive proto-oncogenes: c-abl and c-mos. Cell. 1986 May 9;45(3):417–423. doi: 10.1016/0092-8674(86)90327-2. [DOI] [PubMed] [Google Scholar]
- McGregor W. G., Chen R. H., Lukash L., Maher V. M., McCormick J. J. Cell cycle-dependent strand bias for UV-induced mutations in the transcribed strand of excision repair-proficient human fibroblasts but not in repair-deficient cells. Mol Cell Biol. 1991 Apr;11(4):1927–1934. doi: 10.1128/mcb.11.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Bohr V. A., Smith C. A., Hanawalt P. C. Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Hanawalt P. C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989 Nov 2;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987 Oct 23;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Okumoto D. S., Bohr V. A. DNA repair in the metallothionein gene increases with transcriptional activation. Nucleic Acids Res. 1987 Dec 10;15(23):10021–10030. doi: 10.1093/nar/15.23.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter M. Neoplastic development in B-lymphocytes. Carcinogenesis. 1990 Jan;11(1):1–13. doi: 10.1093/carcin/11.1.1. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Witkin E. M., Sancar A. Escherichia coli mfd mutant deficient in "mutation frequency decline" lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shacter E., Beecham E. J., Covey J. M., Kohn K. W., Potter M. Activated neutrophils induce prolonged DNA damage in neighboring cells. Carcinogenesis. 1988 Dec;9(12):2297–2304. doi: 10.1093/carcin/9.12.2297. [DOI] [PubMed] [Google Scholar]
- Sweder K. S., Hanawalt P. C. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10696–10700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taccioli G. E., Rathbun G., Oltz E., Stamato T., Jeggo P. A., Alt F. W. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993 Apr 9;260(5105):207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- Thacker J. The nature of mutants induced by ionising radiation in cultured hamster cells. III. Molecular characterization of HPRT-deficient mutants induced by gamma-rays or alpha-particles showing that the majority have deletions of all or part of the hprt gene. Mutat Res. 1986 May;160(3):267–275. doi: 10.1016/0027-5107(86)90137-5. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Kas E., Chasin L. A., Funanage V. L., Myoda T. T., Hamlin J. Effect of gamma rays at the dihydrofolate reductase locus: deletions and inversions. Somat Cell Mol Genet. 1986 Nov;12(6):555–566. doi: 10.1007/BF01671941. [DOI] [PubMed] [Google Scholar]
- Van Ness B. G., Coleclough C., Perry R. P., Weigert M. DNA between variable and joining gene segments of immunoglobulin kappa light chain is frequently retained in cells that rearrange the kappa locus. Proc Natl Acad Sci U S A. 1982 Jan;79(2):262–266. doi: 10.1073/pnas.79.2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J., Bartosová Z., Natarajan A. T., van Zeeland A. A., Mullenders L. H. Transcription affects the rate but not the extent of repair of cyclobutane pyrimidine dimers in the human adenosine deaminase gene. J Biol Chem. 1992 May 5;267(13):8852–8856. [PubMed] [Google Scholar]
- Vrieling H., Simons J. W., Arwert F., Natarajan A. T., van Zeeland A. A. Mutations induced by X-rays at the HPRT locus in cultured Chinese hamster cells are mostly large deletions. Mutat Res. 1985 Dec;144(4):281–286. doi: 10.1016/0165-7992(85)90065-x. [DOI] [PubMed] [Google Scholar]
- Vrieling H., Venema J., van Rooyen M. L., van Hoffen A., Menichini P., Zdzienicka M. Z., Simons J. W., Mullenders L. H., van Zeeland A. A. Strand specificity for UV-induced DNA repair and mutations in the Chinese hamster HPRT gene. Nucleic Acids Res. 1991 May 11;19(9):2411–2415. doi: 10.1093/nar/19.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]