Abstract
Background
The Staphylococcus aureus RecU protein is homologous to a Bacillus subtilis Holliday junction resolvase. Interestingly, RecU is encoded in the same operon as PBP2, a penicillin-binding protein required for cell wall synthesis and essential for the full expression of resistance in Methicillin Resistant S. aureus strains. In this work we have studied the role of RecU in the clinical pathogen S. aureus.
Results
Depletion of RecU in S. aureus results in the appearance of cells with compact nucleoids, septa formed over the DNA and anucleate cells. RecU-depleted cells also show increased septal recruitment of the DNA translocase SpoIIIE, presumably to resolve chromosome segregation defects. Additionally cells are more sensitive to DNA damaging agents such as mitomycin C or UV radiation. Expression of RecU from the ectopic chromosomal spa locus showed that co-expression of RecU and PBP2 was not necessary to ensure correct cell division, a process that requires tight coordination between chromosome segregation and septal cell wall synthesis.
Conclusions
RecU is required for correct chromosome segregation and DNA damage repair in S. aureus. Co-expression of recU and pbp2 from the same operon is not required for normal cell division.
Keywords: Staphylococcus aureus, recU, Chromosome segregation, DNA repair
Background
All cells have to repair DNA lesions caused not only by DNA damaging agents but also under normal growth conditions. Chromosome replication is not a continuous process and a series of barriers such as tightly bound proteins, abnormal DNA structures and DNA damage can cause replication fork arrest, which is a major source of genome instability [1-3]. In order to surpass these obstacles, bacteria have developed mechanisms to grant faithful inheritance of genomic information. One example is the process of homologous recombination, required to re-establish stalled and collapsed replication forks and to repair double strand breaks (DSBs) [4,5]. DSB repair is initiated by recognition of the damaged DNA, followed by processing of its ends, leaving a 3’ overhanging strand. The RecA protein associates with these overhanging strands, strand invasion occurs and a Holliday junction is formed and extended unidirectionally by branch migrating proteins such as RuvAB [6]. Holliday junction resolvases, such as Bacillus subtilis RecU, have multiple roles during this process as they promote RecA-mediated strand invasion, associate with the branch migrating proteins and resolve the Holliday junction through DNA cleavage [7-9]. The replication fork can then be re-established, generating either crossover or non-crossover products [10,11]. Importantly, B. subtilis RecU biases homologous recombination towards non-crossover products, therefore avoiding the formation of dimeric chromosomes that cannot be segregated to daughter cells in the absence of a compensating recombination reaction [11].
In agreement with the role of RecU in homologous recombination and DNA damage repair, B. subtilis recU mutants show several chromosome segregation defects. These include nucleoids that are bisected by the division septa, abnormal nucleoid position and anucleate cells [11,12], as well as an increased susceptibility to DNA damaging agents such as mitomycin C (MMC), methyl methanesulfonate (MMS) and UV light [13,14].
Homologous recombination is involved in the transfer of DNA within and, occasionally, between species, which can lead to acquisition of new traits including increased virulence or antibiotic resistance [15,16]. It is therefore of particular relevance to study this process in clinical pathogens. In this work, we focus on Staphylococcus aureus, an important clinical pathogen responsible for high mortality rates in hospitals, mainly due to the presence of methicillin-resistant S. aureus (MRSA) strains [17,18]. The study of RecU in S. aureus is relevant not only because of its putative role in homologous recombination, but also because it is encoded by the same operon as PBP2. This penicillin-binding protein is required for cell wall synthesis and essential for the full expression of resistance in MRSA strains [19,20]. It is interesting that a protein involved in homologous recombination and a protein involved in cell wall synthesis, two biochemically independent processes, are part of the same operon. Importantly, this genetic organization is conserved in other gram-positive bacteria, such as Streptococcus pneumoniae[21] and B. subtilis[22].
During cell division, the processes of chromosome replication and septum synthesis have to be tightly coordinated to avoid the disastrous consequences of DNA guillotining by a septum forming over the DNA. Since PBP2 is required for septum synthesis and RecU is apparently involved in chromosome segregation, we wondered if the regulation of this operon could constitute a possible checkpoint for cell division coordination in S. aureus. Here we show that recU absence causes cell growth defects due to an inability of the mutant to repair damaged DNA and to properly segregate the chromosomes. We also show that co-expression of recU and pbp2 from the same operon is not required for normal cell division.
Methods
Bacterial strains and growth conditions
All strains and plasmids used in this study are listed in Table 1 and primer sequences are listed in Table 2. S. aureus strains were grown in tryptic soy broth (TSB, Difco) or on tryptic soy agar (TSA, Difco) at 37°C with aeration. The medium was supplemented when required with appropriate antibiotics (erythromycin 10 μg/ml, chloramphenicol 10 μg/ml), with 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside 100 μg/ml (X-Gal; BDH Prolabo) or with isopropyl-β-D-thiogalactopyranoside 0.5 mM (IPTG; VWR).
Table 1.
Strains and plasmids used in this study
| Strain/Plasmid | Relevant characteristics | Source/ Reference |
|---|---|---|
|
E. coli |
|
|
| DH5α |
Cloning strain, recA endA1 gyrA96 thi-1 hsdR17 supE44 relA1 ϕφ80 ΔlacZΔM15 |
Gibco-BRL |
|
S. aureus |
|
|
| NCTC8325-4 |
MSSA strain |
R. Novick |
| BCBHV008 |
NCTC8325-4Δspa::Pspac-MCS-lacI lacImc, Cmr |
[23] |
| 8325-4ΔrecU |
NCTC8325-4 recU mutant lacking initial 165 codons |
This study |
| 8325-4recUspaL |
NCTC8325-4 Δspa::Pspac-recU-lacI |
This study |
| BCBRP001 |
NCTC8325-4 ΔrecU Δspa::Pspac-recU-lacI |
This study |
| 8325-4recUi |
NCTC8325-4 ΔrecU Δspa::Pspac-recU-lacI lacImc, Cmr |
This study |
| BCBHV017 |
BCBHV008 strain expressing spoIIIE-yfp from the native chromosomal locus, Cmr |
This study |
| BCBRP002 |
8325-4recUi mutant strain expressing spoIIIE-yfp, Cmr |
This study |
| Plasmids |
|
|
| pMAD |
E. coli – S. aureus shuttle vector with the bgaB gene encoding a ß-galactosidase and thermosensitive origin of replication for Gram-positive bacteria, Ampr Eryr |
[24] |
| pBCB13 |
pMAD derivative with Pspac-lacI between up- and downstream regions of the spa gene, Ampr Eryr |
[25] |
| pMGPII |
Plasmid encoding lacI gene; Cmr |
[26] |
| pMADrecUKO |
pMAD derivative used for deletion of the first 165 codons of recU, Ampr Eryr |
This study |
| pBCB13recUspaL |
pBCB13 derivative containing Pspac-recU-lacI Ampr Eryr |
This study |
| pMUTINYFPKan |
Integrative vector for C-termini YFP fusions; Ampr, Kanr |
[27] |
| pBCBHV007 |
pMUTINYFPKan containing spoIIIE–yfp Ampr, Kanr |
This study |
| pMADspoIIIEyfp |
pMAD derivative containing 3’ end spoIIIE –linker-yfp, Ampr Eryr |
This study |
| pBCBHV008 | pMAD derivative containing 3’ end spoIIIE–linker-yfp-3’ 64 bp and downstream region of spoIIIE, Ampr Eryr | This study |
lacImc – cells expressing multicopies of the lacI gene (encoded by pMGPII).
Table 2.
Primers used in this study
| Primer name | Sequence (5’-3’) |
|---|---|
| recUp1 |
ATCGAGATCTATGTACTTCAGGTGCGT |
| recUp2 |
TAGACTTTTTAAAATTTCACCACACAAGTTTGGTAG |
| recUp3 |
ACTTGTGTGGTGAAATTTTAAAAAGTCTATAAC |
| recUp4 |
ATCGGGATCCCAATGTTTTGACGTTC |
| recUp5 |
TGGTGTATTGTGTCTTTCG |
| recUp6 |
TTCCCACCATTATTACCG |
| recUp7 |
ATCTGCATGCTTAATTATGTTGGC |
| recUp8 |
ATACCCGGGTGTGTGGTGAAATTTATG |
| recUp9 |
TATGCTCGAGTCATACGCGGTCC |
| spoIIIEp1 |
GCTGCGGTACCGTCATAGCTATTTTAGTAGTTG |
| spoIIIEp2 |
GCTGCGGTACCGGAGGCGCCGCAGGACACCTCGTCATTATTAAGATC |
| spoIIIEp3 |
TGAGGATCCGATGAAAAATTCCCGTCT |
| spoIIIEp4 |
TACTCCCCGGGTTACTTGTACAGCTCGTCC |
| spoIIIEp5 |
TACTCCCCGGGCGGTCCACAAAAAGGAAG |
| spoIIIEp6 | TGCATTCCATGGGACATGCTGATCTTTGAATTTTGAAATTG |
Underlined sequences correspond to the restriction site. Bold sequences correspond to the five codon linker.
Construction of a RecU null mutant
To construct a S. aureus recU mutant lacking the initial 165 codons we amplified two 1 Kb DNA fragments, one containing the upstream region of recU up to its start codon (using primers recUp1 and recUp2), and the other containing the 3’end of recU including promoter P2 (see Figure 1A) [19] and the 5’ region of pbp2 (using primers recUp3 and recUp4). The resulting PCR products were joined by overlap PCR using primers recUp1 and recUp4. The PCR product was digested with BamHI and BglII and cloned into the thermosensitive pMAD plasmid [24], resulting in plasmid pMADrecUKO. The insert was sequenced and the plasmid was electroporated into the transformable S. aureus strain RN4220 as previously described [28]. The plasmid was subsequently transduced to strain NCTC8325-4 using phage 80α [29] and insertion and excision of pMADrecUKO into the chromosome was performed as previously described [24]. Deletion of recU was confirmed by two different PCR reactions using the primers recUp5/recUp6 and recUp7/recUp6 and the resulting strain was named 8325-4ΔrecU.
Figure 1.
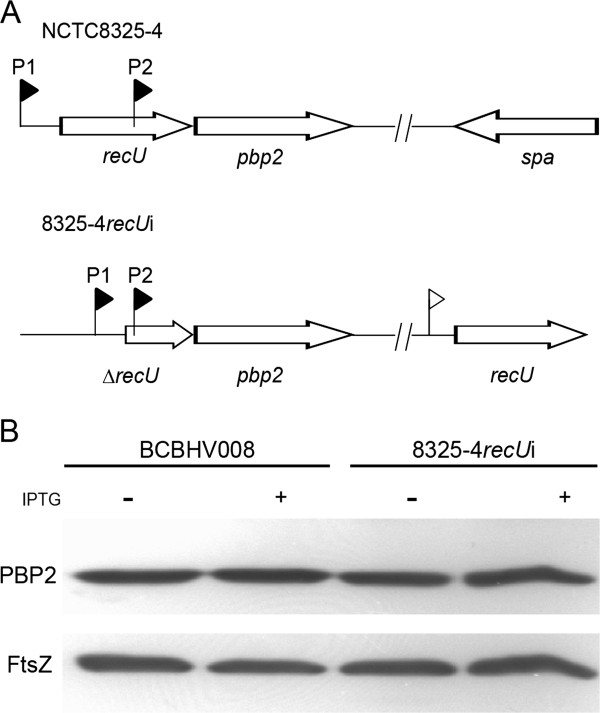
RecU and PBP2 are encoded in the same operon. A – Schematic representation of the recU-pbp2 operon in the NCTC8325-4 wild-type strain (top) and the 8325-4recUi mutant strain (bottom) where the recU gene, including the RBS, was placed in the spa locus under the control of the IPTG inducible Pspac promoter (white flag). Subsequently, the first 165 codons of the native copy of recU were deleted. Black flags represent the promoters (P1 and P2) of the recU-pbp2 operon. B – Western blot analysis of PBP2 levels in control strain BCBHV008 and recU inducible mutant 8325-4recUi grown in the presence or absence of IPTG showing that PBP2 levels were not affected by recU deletion. FtsZ was used as an internal control of total protein loaded.
Construction of a recU inducible mutant
To generate an inducible recU mutant, a full copy of the recU coding sequence was first placed in the spa locus under the control of the IPTG inducible-Pspac promoter using the pBCB13 plasmid [25]. The sequence encoding the first 165 amino acids of RecU was subsequently deleted from the native chromosomal locus using plasmid pMADrecUKO as described above. To clone recU into the spa locus, the entire recU coding sequence and the RBS was amplified by PCR using primers recUp8 and recUp9. The PCR product was digested with XmaI and XhoI restriction enzymes and cloned into pBCB13 generating the plasmid pBCB13recUspaL. The insert was sequenced, the plasmid was introduced into RN4220 by electroporation and subsequently transduced into NCTC8325-4. Integration and excision of the plasmid in the chromosome was performed as previously described [24] and the resulting strain, which contains two copies of recU in the chromosome, one in the native locus and another in the spa locus, was named 8325-4recUspaL. In order to delete recU from its normal locus in the background of strain 8325-4recUspaL, the pMADrecUKO plasmid was transduced into this strain and deletion of the recU gene was performed and verified as described in the previous paragraph, but in the presence of IPTG, resulting in the strain BCBRP001. In order to ensure tight regulation of the expression of recU from the Pspac promoter [30] we transduced the pMGPII plasmid, which encodes the lacI gene [26], into BCBRP001 and the resulting strain was named 8325-4recUi.
SpoIIIE-YFP localization
To study SpoIIIE localization in BCBHV008 [23] and 8325-4recUi strains, derivatives of these strains expressing a C-terminal SpoIIIE-YFP fusion from its native locus were constructed. For that purpose, a DNA fragment encompassing a copy of the spoIIIE gene without its STOP codon and encoding a five amino acid linker was cloned, in frame with the yfp gene, in the pMUTINYFPKan plasmid [27]. This fragment was amplified from NCTC8325-4 genomic DNA using primers spoIIIEp1 and spoIIIEp2, digested with KpnI and cloned into pMUTINYFPKan, giving rise to pBCBHV007. The insert in pBCBHV007 was sequenced and this plasmid was used as a template to amplify a DNA fragment containing the 3’ end of the spoIIIE gene (1065 bp) connected to the linker and the yfp gene, using primers spoIIIEp3 and spoIIIEp4. This fragment was digested and cloned into the BamHI and XmaI restriction sites of the pMAD vector [24], generating plasmid pMADspoIIIEyfp. A second PCR product, encompassing the last 64 bp of spoIIIE (containing the Shine-Dalgarno sequence of the downstream gene) and the 1 Kb region downstream of spoIIIE, was amplified from NCTC8325-4 genomic DNA using primers spoIIIEp5 and spoIIIEp6. The PCR product was digested with XmaI and NcoI and subsequently cloned into pMADspoIIIEyfp generating the plasmid pBCBHV008. The two inserts in pBCBHV008 were sequenced and the plasmid was electroporated into RN4220 and subsequently transduced to strains BCBHV008 and 8325-4recUi (selection with erythromycin, chloramphenicol and IPTG), using phage 80α. Integration and excision of pBCBHV008 from the genome was performed as previously described [24] and colonies in which spoIIIE had been replaced by the spoIIIE-yfp (with the last 64 bp of spoIIIE duplicated after the yfp gene), were selected by PCR. The BCBHV008 and 8325-4recUi strains expressing spoIIIE-yfp were named BCBHV017 and BCBRP002, respectively. Functionality of spoIIIE-yfp was confirmed by introduction of the fusion protein into a spoIIIE null mutant that resulted in complementation of the defective phenotype typical of this strain (data not shown).
Growth analysis of S. aureus strains
Growth of S. aureus in liquid culture was analyzed by diluting overnight cultures 1/500 into fresh media, incubating them at 37°C with aeration and following the optical density at 600 nm (OD600nm). Strains encoding an inducible recU gene and the corresponding control strains were grown overnight in TSB containing chloramphenicol and IPTG. Cells were harvested, washed three times with TSB, and re-inoculated into fresh media supplemented or not with IPTG.
Western blot analysis
Expression levels of PBP2 were analyzed by western blotting, using a polyclonal anti-PBP2 antibody [31]. A polyclonal anti-FtsZ antibody was used as an internal control. Samples were taken from cultures of BCBHV008 and 8325-4recUi supplemented or not with IPTG, grown until an OD600nm 0.5. Cells were broken with glass beads in a Fast Prep FP120 (Thermo Electro Corporation) and unbroken cells were removed by centrifugation. The total protein content of the extracts was quantified by the Bradford method, using bovine serum albumin as a standard (BCA protein assay kit, Pierce). Equal amounts of protein from each sample were loaded onto an 8% SDS-PAGE gel and separated at 120 V. Proteins were then transferred to a Hybond-P Polyvinylidene fluoride (PVDF) membrane (GE Healthcare) using a semidry transfer cell (Bio-Rad). The membranes were cut to separate the region containing PBP2 and FtsZ. Each half of the membrane was blocked with blocking buffer (PBS, 5% milk, 0.5% Tween 20) for 1 hour and incubated with either a polyclonal anti-PBP2 antibody (1/1000 dilution in blocking buffer) or with an anti-FtsZ antibody (1/5000 dilution in blocking buffer) for 16 hours at 4°C. Membranes were washed three times with PBS-T (PBS containing 0.5% Tween 20) and incubated with secondary antibodies (anti-rabbit for PBP2, ECL; anti-sheep for FtsZ, Pierce) diluted 1/100,000 in blocking buffer. The detection was performed using ECL Plus Western blotting detection system (Amersham) according to the manufacturer’s guidelines.
Fluorescence microscopy
Strains were incubated overnight at 37°C in TSB supplemented with the appropriate antibiotics and IPTG. Cultures were washed three times with fresh TSB and diluted 1/500 in fresh TSB and supplemented with IPTG when required. During exponential phase (O.D600nm 0.5) 1 ml of 8325-4recUi culture was taken and incubated with membrane dye Nile Red (5 μg/ml, Invitrogen), DNA dye Hoechst 33342 (1 μg/ml, Invitrogen) and the cell wall dye Van-FL (Invitrogen) mixed in a 1:1(v:v) proportion with non-fluorescent vancomycin (4 μg/ml, Sigma), at room temperature for 5 minutes with shaking. The same protocol was followed for strains BCBHV017 and BCBRP002 but the incubation was performed with the membrane dye FM 5–95 (1 μg/ml, Invitrogen) and with the DNA dye Hoechst 33342 (1 μg/ml, Invitrogen). The cultures were then centrifuged, re-suspended in PBS and 1 μl was placed on a thin layer of 1.2% agarose in PBS. Fluorescence microscopy was performed using a Zeiss Axio Observer.Z1 microscope equipped with a Photometrics CoolSNAP HQ2 camera (Roper Scientific), using Metamorph software (Molecular devices). Analysis of fluorescence images was performed using Metamorph and ImageJ software.
Determination of mitomycin C minimum inhibitory concentration (MIC)
Determination of the MIC to mitomycin C of 8325-4recUi and BCBHV008 strains was performed in liquid medium by micro-dilution. Overnight cultures containing IPTG and chloramphenicol were washed three times with fresh TSB and added at a final cell density of 5×105 CFU/ml to wells containing 2-fold dilutions of mitomycin C in TSB supplemented or not with 0.5 mM IPTG. The 96-well plates were incubated for 24 hours at 37°C and the MIC was recorded as the lowest concentration of mitomycin C that inhibited bacterial growth. All MIC determinations were performed in triplicate.
UV survival assays
BCBHV008 and 8325-4recUi strains were incubated overnight at 37°C with aeration, in TSB supplemented with chloramphenicol and IPTG. These cultures were washed three times with TSB and then diluted 1/500 into fresh TSB, supplemented or not with IPTG and incubated at 37°C until O.D600nm 0.5. Serial dilutions (100 to 10-6) were made in TSB and 10 μl of each dilution was spotted on TSA plates containing chloramphenicol and supplemented or not with IPTG. Plates were then irradiated with UV light (Vilber Lourmat, VL-6.LC model, 254 nm) at a dose of 4 J/m2 for 0, 10, 20, 30, 40 and 60 seconds and incubated overnight at 37°C in the dark. CFUs were counted and the fraction surviving was determined with reference to an unirradiated control plate.
Results
S. aureus RecU is required for optimal growth
In order to functionally characterize the RecU homologue in S. aureus we deleted the 5’ region of the recU gene (encoding the first 165 amino acids) in the background of NCTC8325-4 generating strain 8325-4ΔrecU. The recU gene is encoded upstream of pbp2, in the same operon (Figure 1A). This operon contains two promoters, one upstream of recU (P1) and the other contained within the recU coding sequence (P2) [19]. In order not to affect pbp2 expression in the recU mutant, the last 43 recU codons (which contain P2) were not deleted. Growth analysis of the 8325-4ΔrecU strain indicated that RecU is not essential, but it is required for optimal growth of S. aureus since the deletion mutant had a two-fold increase in the doubling time when compared to the parental strain NCTC8325-4 (40 versus 22 minutes, respectively). Given that mutants with very slow growth rates may accumulate suppressor mutations that increase fitness, we generated a recU inducible mutant, to be used for further studies. For the construction of this mutant a full copy of recU was placed under the control of the IPTG-inducible Pspac promoter in the ectopic spa locus (which encodes for the non-essential Protein A), and subsequently the first 165 codons of recU were deleted from the native locus, while in the presence of IPTG (Figure 1A). In order to achieve strong repression of the Pspac promoter, we introduced the pMGPII plasmid [26], which encodes the lacI repressor, generating strain 8325-4recUi. Although the two promoters driving expression of pbp2 are present in this strain, deletion of recU decreased the spacing between P1 and P2 promoters. To exclude the possibility that expression of pbp2 was altered in the 8325-4recUi strain, and to ensure that the phenotypes observed in further studies were due only to the absence of RecU and not to low PBP2 levels, we analyzed PBP2 levels in strain 8325-4recUi cultured in the presence or absence of IPTG. Figure 1B shows that PBP2 levels are similar in 8325-4recUi and the control strain BCBHV008 (where the spa gene was replaced by the construct Pspac-MCS-lacI and the pMGPII plasmid was introduced), indicating that mutation of recU does not affect PBP2 production.
RecU depletion leads to defects in DNA repair and in chromosome morphology and segregation
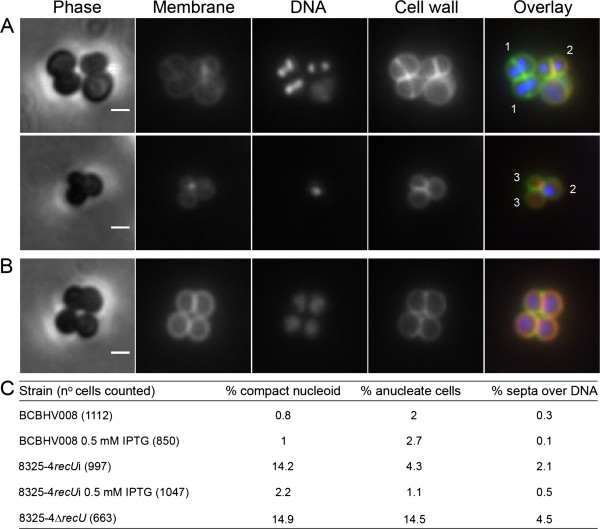
In order to study the effects of RecU depletion, strain 8325-4recUi was incubated in the absence of IPTG for three hours and then observed by fluorescence microscopy (Figure 2). Approximately 14% of the RecU-depleted cells (n = 1046) showed compact nucleoids, while 4% had no DNA (anucleate cells) and 2% presented septa over a compact nucleoid. These phenotypes were shown to be due to the lack of RecU, as they were complemented by ectopic expression of RecU from the spa locus (Figure 2B, C). Importantly these phenotypes were also found in cells from the recU null mutant strain 8325-4ΔrecU (Figure 2C) but at a higher frequency. This difference may result from prolonged growth in the absence of RecU in the null mutant or from residual RecU protein present in the inducible strain.
Figure 2.
RecU depletion in S. aureus leads to chromosome segregation defects. The fluorescence microscopy images show cells of recU inducible strain 8325-4recUi incubated in the absence (A) or presence (B) of IPTG. Panels from left to right show phase-contrast images, cells stained with membrane dye Nile Red, DNA dye Hoechst 33342, cell wall dye Van-FL and the overlay of the three fluorescence images showing the membrane in red, the DNA in blue and the cell wall in green. The absence of RecU (A) led to the formation of cells with septa bisecting the DNA (1), compact nucleoids (2) and anucleate cells (3). Ectopic expression of RecU in 8325-4recUi strain, through the addition of IPTG, resulted in the disappearance of the aberrant phenotypes (B). Scale bars 1 μm. Panel (C) shows a comparison of the phenotypes of control strain BCBHV008; 8325-4recU inducible mutant, incubated in the presence or absence of IPTG and 8325-4ΔrecU mutant.
The presence of anucleate cells can be associated with chromosome segregation defects that result in one sister cell with two chromosomes and another with none. However, they could also arise as a result of DNA degradation caused by DNA guillotining by the septum or due to decreased DNA damage repair. We therefore tested the susceptibility of recU mutants to UV light and mitomycin C, both of which cause DNA lesions [32,33]. Depletion of recU in the strain 8325-4recUi resulted in a 2-fold decrease in mitomycin C MIC (from 0.8 to 0.4 ng/ml), compared to the same strain grown in the presence of IPTG or to the control strain BCBHV008. Importantly, addition of IPTG recovered the MIC to wild-type levels. Similar results were obtained for the null mutant strain 8325-4ΔrecU which had a 6-fold decrease in the mitomycin MIC compared to the parental strain. RecU depletion also caused S. aureus to become more sensitive to UV damage, since 10 sec of exposure time to UV light were sufficient to kill approximately 99% of the 8325-4recUi cells grown in the absence of ITPG but had no significant effect on BCBHV008 cells or 8325-4recUi cells grown in the presence of the inducer, which required 20 sec of UV exposures for similar decrease in cell viability (Figure 3). Taken together, these results indicate that RecU is required for DNA damage repair in S. aureus and that its ectopic expression from the spa locus was sufficient to fully recover UV and mitomycin C resistance to wild type levels.
Figure 3.
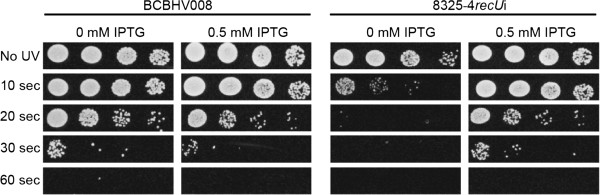
RecU depletion in 8325-4recUi strain leads to increased susceptibility to UV damage. Cultures of control strain BCBHV008 and recU inducible mutant 8325-4recUi showing serial dilutions from 10-2 (left) to 10-5 (right). 10 μl spots were placed on TSA agar, containing or not IPTG, and irradiated with a UV dose of 4 J/m2/sec for 0, 10, 20, 30 and 60 seconds. Plates were then incubated overnight and the number of CFU’s was counted.
Absence of RecU leads to increased recruitment of the SpoIIIE DNA pump to the division septum
SpoIIIE is a DNA pump crucial for moving DNA into the forespore of B. subtilis during sporulation [34]. During vegetative growth of B. subtilis this protein plays an important backup role when the chromosome fails to segregate prior to septum formation [35-37]. The presence of SpoIIIE foci localized near the center of the septum in a small fraction (~6%) of vegetatively growing B. subtilis cells is thought to reflect its role in post-septational chromosome partioning [38]. Therefore, we have used SpoIIIE recruitment to the septum as a marker for the requirement to resolve chromosome segregation defects. For that purpose we fused SpoIIIE to the yellow fluorescent protein YFP and expressed this fusion protein in the 8325-4recUi background, generating the strain BCBRP002 (Figure 4). SpoIIIE-YFP foci were present in 10% (n = 580) of the cells cultured in the presence of inducer. However, when the same strain was cultured in the absence of IPTG, the number of cells with SpoIIIE-YFP foci increased to 44% (n = 536). In a control experiment, addition of IPTG did not change the fraction of cells exhibiting SpoIIIE foci in the control strain BCBHV017, a strain identical to BCBRP002 but lacking the recU mutations (data not shown). These results suggest that RecU is required for correct segregation of the S. aureus chromosome as its absence increases the need for SpoIIIE-mediated post-septational chromosome partitioning.
Figure 4.
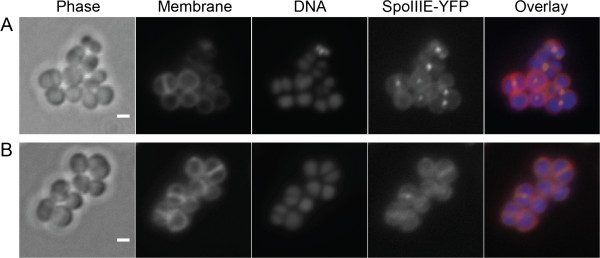
RecU-depleted cells show increased frequency of SpoIIIE-YFP foci. The figure shows SpoIIIE-YFP localization in recU inducible strain BCBRP002 incubated in the absence (A) or presence (B) of IPTG. SpoIIIE-YFP foci are present in 44% of BCBRP002 RecU-depleted cells in comparison with 10% of the cells of the same strain when expressing RecU. Panels from left to right show phase-contrast image, membrane labeled with FM 5–95, DNA stained with Hoechst 33342, SpoIIIE-YFP localization, and the overlay of the three fluorescence images showing the membrane in red, DNA in blue and SpoIIIE-YFP in yellow. Scale bars 1 μm.
Discussion
The role of RecU in homologous recombination and in DNA repair has been well studied in a small number of organisms [39-41]. However DSB repair mechanisms studied in one bacterial species cannot be directly extrapolated to other species since the phenotypes that arise from the same mutations in different bacteria are not always the same [42]. Furthermore, homologous recombination has an important role in the evolution of antibiotic resistance and acquisition of virulence determinants [15,16], emphasizing the relevance of studying this mechanism in pathogenic bacteria.
We have now studied the role of RecU in the clinical pathogen S. aureus and found that the major phenotypes observed in RecU depleted S. aureus cells were compatible with defects in chromosome segregation and DNA repair. These phenotypes include: (i) The presence of anucleate cells, which can result from deficient chromosome partioning causing one of the daughter cells to inherit the two copies of the genome and the other none. Alternatively, anucleate cells can arise from DNA degradation resulting from DNA breaks due to chromosome guillotining by septum placement over the nucleoid [12,23] or from DNA damage that is not repaired [43]. (ii) Compaction of the nucleoid, a phenotype that has already been observed in B. subtilis and E. coli under DNA damaging conditions, such as UV irradiation. Interestingly, these observations led to the suggestion that a dramatic alteration of nucleoid morphology may be part of an active mechanism to protect the cell’s genome when DNA repair is required [43-45], a mechanism which we now suggest also occurs in S. aureus. (iii) Increased sensitivity to UV irradiation and mitomycin C, a phenotype in agreement with a role of RecU in DNA damage repair. (iv) Increased recruitment of the DNA translocase SpoIIIE. In B. subtilis, RecU has been shown to bias homologous recombination towards non-crossover products [7,11], decreasing the formation of chromosome dimers that would not be properly segregated into the daughter cells [46-48]. When present, chromosome dimers can be resolved by dedicated recombinases in a process that requires the presence of at least one of the two DNA translocases, SpoIIIE or SftA [49]. Furthermore, the presence of septal SpoIIIE foci was proposed to be associated with its role in post-septational chromosome partitioning [38]. Therefore, the fact that approximately half of the S. aureus cells grown in the absence of RecU had SpoIIIE-YFP foci (compared to 10% of the cells grown in its presence), suggests that RecU has a major role in chromosome segregation, maybe through biasing recombination towards non-crossover products. (v) The presence of septa placed over the DNA, a phenotype that could be caused by segregation defects or, alternatively, by the lack of a cell division checkpoint required to prevent septum formation over the DNA (see below). Together, the phenotypes observed for RecU depleted cells strongly point to an important role of this protein in DNA repair and chromosome segregation, in agreement with what would be expected for a Holliday junction resolvase.
In the course of S. aureus cell division, the synthesis of cell wall occurs at the septum, which progressively closes to originate the two daughter cells. During this process the chromosome is replicated and the two resulting DNA molecules are segregated. Tight coordination between chromosome segregation (which requires RecU) and septum synthesis (which requires PBP2, encoded in the same operon as RecU), two biosynthetically unrelated events, is therefore essential for proper division, to ensure that the septum does not form over the nucleoid, which would result in DNA damage. Given that the genetic organization of the recU-pbp2 operon is maintained in other gram-positive bacteria [19,21,22], we hypothesized that co-regulation of the expression of these two proteins could be central for the coordination of cell division events. We have abolished this co-regulation (but maintained the presence of RecU in the cell) in strain 8325-4recUi by placing an inducible copy of recU in the distant spa locus, under the control of the Pspac promoter and deleting the native gene from the recU-pbp2 operon. When this mutant is incubated with IPTG, RecU is produced from the ectopic spa locus while PBP2 is expressed from its native locus, under the control of its native promoters. If recU/pbp2 co-regulation constituted a checkpoint for cell division, we should detect a subpopulation of cells with cell division defects when the 8325-4recUi strain was incubated with IPTG. This is not what we have observed, since ectopic expression of recU led to a reversal of the phenotypes observed in the absence of RecU, namely the presence of anucleate cells and cells with septa over DNA (Figure 2A-C). This indicates that although RecU may have a role in preventing chromosome trapping by the septum, co-regulation of recU and pbp2 expression from the same operon is not required during cell division.
Conclusions
We have shown that lack of S. aureus RecU protein has important consequences in the cells, doubling the duplication time, increasing the susceptibility to DNA damage and leading to the appearance of a large population of cells with compact nucleoids, lacking a nucleoid or with septa placed over the chromosome. This shows that the role of RecU in chromosome segregation and DNA repair is crucial for normal growth of S. aureus cells. RecU is encoded in the same operon as the cell wall synthesis protein PBP2 and consequently the two proteins are overexpressed under certain conditions, such as in the presence of cell wall targeting antibiotics [50]. We have shown that this genetic organization is not required for correct cell division in rich medium, but it remains to be determined if it becomes advantageous under other, more clinically relevant, conditions.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ARP, PR and MGP designed research, analyzed data and wrote the paper, HV contributed with new genetic constructs, ARP performed research. All authors read and approved the final manuscript.
Contributor Information
Ana R Pereira, Email: arpereira@itqb.unl.pt.
Patricia Reed, Email: preed@itqb.unl.pt.
Helena Veiga, Email: hveiga@itqb.unl.pt.
Mariana G Pinho, Email: mgpinho@itqb.unl.pt.
Acknowledgements
This work was funded by grants PTDC/BIA-BCM/66449/2006, PTDC/BIA-BCM/099152/2008 and PEst-OE/EQB/LA0004/2011 from Fundação para a Ciência e Tecnologia. P.R. and H.V. were supported by fellowships SFRH/BPD/23812/2005 and SFRH/BD/38732/2007, respectively. The anti-FtsZ antibody was kindly provided by Dr. E.J. Harry (University of Technology, Sydney, Australia).
References
- Kuzminov A. Instability of inhibited replication forks in E. coli. Bioessays. 1995;17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R. Recombination proteins and rescue of arrested replication forks. DNA Repair. 2007;6:967–980. doi: 10.1016/j.dnarep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Wyman C, Ristic D, Kanaar R. Homologous recombination-mediated double-strand break repair. DNA Repair. 2004;3:827–833. doi: 10.1016/j.dnarep.2004.03.037. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell. 2001;8:1163–1174. doi: 10.1016/S1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- Ayora S, Carrasco B, Doncel-Perez E, Lurz R, Alonso JC. Bacillus subtilis RecU protein cleaves Holliday junctions and anneals single-stranded DNA. Proc Natl Acad Sci U S A. 2004;101:452–457. doi: 10.1073/pnas.2533829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canas C, Carrasco B, Ayora S, Alonso JC. The RecU Holliday junction resolvase acts at early stages of homologous recombination. Nucleic Acids Res. 2008;36:5242–5249. doi: 10.1093/nar/gkn500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco B, Ayora S, Lurz R, Alonso JC. Bacillus subtilis RecU Holliday-junction resolvase modulates RecA activities. Nucleic Acids Res. 2005;33:3942–3952. doi: 10.1093/nar/gki713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Leach DR. Control of crossing over. Mol Cell. 2000;6:815–826. doi: 10.1016/S1097-2765(05)00095-X. [DOI] [PubMed] [Google Scholar]
- Carrasco B, Cozar MC, Lurz R, Alonso JC, Ayora S. Genetic recombination in Bacillus subtilis 168: contribution of Holliday junction processing functions in chromosome segregation. J Bacteriol. 2004;186:5557–5566. doi: 10.1128/JB.186.17.5557-5566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Setlow P. Penicillin-binding protein-related factor A is required for proper chromosome segregation in Bacillus subtilis. J Bacteriol. 2000;182:1650–1658. doi: 10.1128/JB.182.6.1650-1658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez H, Carrasco B, Cozar MC, Alonso JC. Bacillus subtilis RecG branch migration translocase is required for DNA repair and chromosomal segregation. Mol Microbiol. 2007;65:920–935. doi: 10.1111/j.1365-2958.2007.05835.x. [DOI] [PubMed] [Google Scholar]
- Sanchez H, Kidane D, Reed P, Curtis FA, Cozar MC, Graumann PL, Sharples GJ, Alonso JC. The RuvAB branch migration translocase and RecU Holliday junction resolvase are required for double-stranded DNA break repair in Bacillus subtilis. Genetics. 2005;171:873–883. doi: 10.1534/genetics.105.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson CG, Barcus V, King S, Pickerill P, Whatmore A, Yeo M. Horizontal gene transfer and the evolution of resistance and virulence determinants in Streptococcus. Soc Appl Bacteriol Symp Ser. 1997;26:42S–51S. [PubMed] [Google Scholar]
- Spratt BG, Zhang QY, Jones DM, Hutchison A, Brannigan JA, Dowson CG. Recruitment of a penicillin-binding protein gene from Neisseria flavescens during the emergence of penicillin resistance in Neisseria meningitidis. Proc Natl Acad Sci U S A. 1989;86:8988–8992. doi: 10.1073/pnas.86.22.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM. et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- Boucher C. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008;46(Suppl 5):S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- Pinho MG, de Lencastre H, Tomasz A. Transcriptional analysis of the Staphylococcus aureus penicillin binding protein 2 gene. J Bacteriol. 1998;180:6077–6081. doi: 10.1128/jb.180.23.6077-6081.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho MG, de Lencastre H, Tomasz A. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci U S A. 2001;98:10886–10891. doi: 10.1073/pnas.191260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Briese T, Hakenbeck R. Nucleotide sequences of genes encoding penicillin-binding proteins from Streptococcus pneumoniae and Streptococcus oralis with high homology to Escherichia coli penicillin-binding proteins 1a and 1b. J Bacteriol. 1992;174:4517–4523. doi: 10.1128/jb.174.13.4517-4523.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham DL, Setlow P. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. J Bacteriol. 1995;177:326–335. doi: 10.1128/jb.177.2.326-335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga H, Jorge AM, Pinho MG. Absence of nucleoid occlusion effector Noc impairs formation of orthogonal FtsZ rings during Staphylococcus aureus cell division. Mol Microbiol. 2011;80:1366–1380. doi: 10.1111/j.1365-2958.2011.07651.x. [DOI] [PubMed] [Google Scholar]
- Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira PM, Veiga H, Jorge AM, Pinho MG. Fluorescent reporters for studies of cellular localization of proteins in Staphylococcus aureus. Appl Environ Microbiol. 2010;76:4346–4353. doi: 10.1128/AEM.00359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho MG, Filipe SR, de Lencastre H, Tomasz A. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J Bacteriol. 2001;183:6525–6531. doi: 10.1128/JB.183.22.6525-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilano ML, Pereira PM, Yates J, Reed P, Veiga H, Pinho MG, Filipe SR. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2010;107:18991–18996. doi: 10.1073/pnas.1004304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga H, Pinho MG. Inactivation of the SauI type I restriction-modification system is not sufficient to generate Staphylococcus aureus strains capable of efficiently accepting foreign DNA. Appl Environ Microbiol. 2009;75:3034–3038. doi: 10.1128/AEM.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshida T, Tomasz A. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J Bacteriol. 1992;174:4952–4959. doi: 10.1128/jb.174.15.4952-4959.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana M, Luong TT, Komatsuzawa H, Shigeta M, Lee CY. A method for demonstrating gene essentiality in Staphylococcus aureus. Plasmid. 2000;44:100–104. doi: 10.1006/plas.2000.1473. [DOI] [PubMed] [Google Scholar]
- Reed P, Veiga H, Jorge AM, Terrak M, Pinho MG. Monofunctional transglycosylases are not essential for Staphylococcus aureus cell wall synthesis. J Bacteriol. 2011;193:2549–2556. doi: 10.1128/JB.01474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VN, Szybalski W. A molecular mechanism of mitomycin action: linking of complementary DNA strands. Proc Natl Acad Sci U S A. 1963;50:355–362. doi: 10.1073/pnas.50.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peak MJ, Peak JG, Moehring MP, Webb RB. Ultraviolet action spectra for DNA dimer induction, lethality, and mutagenesis in Escherichia coli with emphasis on the UVB region. Photochem Photobiol. 1984;40:613–620. doi: 10.1111/j.1751-1097.1984.tb05349.x. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- Sharpe ME, Errington J. Postseptational chromosome partitioning in bacteria. Proc Natl Acad Sci U S A. 1995;92:8630–8634. doi: 10.1073/pnas.92.19.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Grossman AD. Synthetic lethal phenotypes caused by mutations affecting chromosome partitioning in Bacillus subtilis. J Bacteriol. 1999;181:5860–5864. doi: 10.1128/jb.181.18.5860-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaimer C, Gonzalez-Pastor JE, Graumann PL. SpoIIIE and a novel type of DNA translocase, SftA, couple chromosome segregation with cell division in Bacillus subtilis. Mol Microbiol. 2009;74:810–825. doi: 10.1111/j.1365-2958.2009.06894.x. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez S, Sorokin A, Alonso JC. Genetic recombination in Bacillus subtilis 168: effects of recU and recS mutations on DNA repair and homologous recombination. J Bacteriol. 1998;180:3405–3409. doi: 10.1128/jb.180.13.3405-3409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SJ, Li J, Setlow P, Jedrzejas MJ. Structure, flexibility, and mechanism of the Bacillus stearothermophilus RecU holliday junction resolvase. Proteins. 2007;68:961–971. doi: 10.1002/prot.21418. [DOI] [PubMed] [Google Scholar]
- Sluijter M, Aslam M, Hartwig NG, van Rossum AM, Vink C. Identification of amino acid residues critical for catalysis of Holliday junction resolution by Mycoplasma genitalium RecU. J Bacteriol. 2011;193:3941–3948. doi: 10.1128/JB.00247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayora S, Carrasco B, Cardenas PP, Cesar CE, Canas C, Yadav T, Marchisone C, Alonso JC. Double-strand break repair in bacteria: a view from Bacillus subtilis. FEMS Microbiol Rev. 2011;35:1055–1081. doi: 10.1111/j.1574-6976.2011.00272.x. [DOI] [PubMed] [Google Scholar]
- Smith BT, Grossman AD, Walker GC. Localization of UvrA and effect of DNA damage on the chromosome of Bacillus subtilis. J Bacteriol. 2002;184:488–493. doi: 10.1128/JB.184.2.488-493.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Zaidman S, Frenkiel-Krispin D, Shimoni E, Sabanay I, Wolf SG, Minsky A. Ordered intracellular RecA-DNA assemblies: a potential site of in vivo RecA-mediated activities. Proc Natl Acad Sci U S A. 2000;97:6791–6796. doi: 10.1073/pnas.090532397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odsbu I, Morigen, Skarstad K. A reduction in ribonucleotide reductase activity slows down the chromosome replication fork but does not change its localization. PLoS One. 2009;4:e7617. doi: 10.1371/journal.pone.0007617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre FX, Soballe B, Michel B, Aroyo M, Robertson M, Sherratt D. Circles: the replication-recombination-chromosome segregation connection. Proc Natl Acad Sci U S A. 2001;98:8189–8195. doi: 10.1073/pnas.111008998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Recchia GD, Penel-Colin M, Ehrlich SD, Sherratt DJ. Resolution of Holliday junctions by RuvABC prevents dimer formation in rep mutants and UV-irradiated cells. Mol Microbiol. 2000;37:180–191. doi: 10.1046/j.1365-2958.2000.01989.x. [DOI] [PubMed] [Google Scholar]
- Hendricks EC, Szerlong H, Hill T, Kuempel P. Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC and xerD) of Escherichia coli. Mol Microbiol. 2000;36:973–981. doi: 10.1046/j.1365-2958.2000.01920.x. [DOI] [PubMed] [Google Scholar]
- Kaimer C, Schenk K, Graumann PL. Two DNA translocases synergistically affect chromosome dimer resolution in Bacillus subtilis. J Bacteriol. 2011;193:1334–1340. doi: 10.1128/JB.00918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle-Vavra S, Yin S, Challapalli M, Daum RS. Transcriptional induction of the penicillin-binding protein 2 gene in Staphylococcus aureus by cell wall-active antibiotics oxacillin and vancomycin. Antimicrob Agents Chemother. 2003;47:1028–1036. doi: 10.1128/AAC.47.3.1028-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]