Abstract
Sal4 is a monoclonal polymeric IgA antibody directed against the O antigen (O-Ag) of Salmonella enterica serovar Typhimurium (S. Typhimurium), which is sufficient to protect mice against intestinal infections from S. Typhimurium. We recently reported that the exposure of S. Typhimurium to Sal4 results in the immediate loss of flagellum-based motility, in alterations to the outer membrane (OM) integrity, and in the concomitant appearance of a mucoid phenotype that is reminiscent of cells in the earliest stages of biofilm formation. We demonstrate here that prolonged (>4 h) exposure of S. Typhimurium to Sal4 at 37°C (but not at ambient temperature [25°C]) results in measurable exopolysaccharide (EPS) accumulation and biofilm formation on both borosilicate glass surfaces and polystyrene microtiter plates. The polysaccharide produced by S. Typhimurium in response to Sal4 contains cellulose, in addition to O-Ag capsule and colanic acid. EPS production was dependent on YeaJ, a proposed inner membrane-localized diguanylate cyclase (DGC) and a known regulator of cellulose biosynthesis. An S. Typhimurium ΔyeaJ strain was unable to produce cellulose or form a biofilm in response to Sal4. Conversely, the overexpression of yeaJ in S. Typhimurium enhanced Sal4-induced biofilm formation and resulted in increased intracellular levels of cyclic dimeric guanosine monophosphate (c-di-GMP) compared to that of a wild-type control; this strongly suggests that YeaJ is indeed a functional DGC. Based on these data, we speculate that Sal4, by virtue of its ability to associate with the O-Ag and to induce OM stress, renders S. Typhimurium avirulent by triggering a c-di-GMP-dependent signaling pathway via YeaJ that leads to the suppression of bacterial motility while simultaneously stimulating EPS production.
INTRODUCTION
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a Gram-negative facultative intracellular bacterium that causes acute gastroenteritis in humans and typhoid-like fever in mice (1). A pivotal step in infection is the invasion of Peyer's patch M cells and villous enterocytes, a highly complex event mediated by the Salmonella pathogenicity island 1 (SPI-1) type 3 secretion system (T3SS) (2–8). However, infection of the intestinal epithelium occurs only after a variety of host-associated environmental signals have primed S. Typhimurium to express virulence and colonization factors necessary for attachment and invasion (9–13). For example, iron limitation encountered in the intestinal lumen promotes the expression of thin aggregative fimbriae (Tafi), which are involved in epithelial attachment (13), while oxygen limitation activates the expression of the SPI-1 genes involved in invasion (14). S. Typhimurium uses a panoply of global regulatory circuits, such as two-component systems, like CpxR-CpxA and PhoP-PhoQ, as a means to adapt to its local environment and to coordinate virulence factor expression levels (15, 16). The transition from a motile and virulent state to a nonmotile and biofilm state, in particular, is regulated by members of the cyclic dimeric guanosine monophosphate (c-di-GMP) regulatory pathways (17–21). Thus, various environmental signals influence the virulence of S. Typhimurium and alter the ability of the bacterium to infect a host.
Secretory IgA (SIgA) is the most abundant class of antibodies in intestinal secretions, and it serves as the first line of defense against enteroinvasive pathogens, like S. Typhimurium (22). In experimental animal models, IgA antibodies against the O antigen (O-Ag) are highly effective at preventing Salmonella infection (23, 24). In fact, O-Ag-specific monoclonal IgA antibodies (MAbs) are more effective than flagellum-specific IgA MAbs at providing protective immunity (25). One of the best examples of this is Sal4, a monoclonal dimeric IgA specific for the S. Typhimurium O5 serotype. Using the so-called backpack tumor model, it was shown that, when secreted into the intestinal lumen as SIgA, Sal4 blocks S. Typhimurium entry into the intestinal mucosa (23). Invasion of an isogenic O5 mutant was unaffected by Sal4, thereby demonstrating the specificity of the antibody's activity. When examined in vitro, Sal4 prevented SPI-1 T3SS-mediated entry into polarized epithelial cell monolayers, but it did not affect bacterial attachment (24, 26).
Although it is now well established that Sal4 prevents S. Typhimurium from invading intestinal epithelial cells, the mechanism by which this occurs is unknown. Sal4 does not possess complement-fixing or bactericidal activity (23, 26). Moreover, immune exclusion, a process involving bacterial agglutination, entrapment in mucus, and clearance from the intestinal lumen, cannot fully account for the activity of Sal4 (27). For these reasons, we have focused on the possibility that the association of Sal4 with the O-Ag may directly affect bacterial virulence. In support of this hypothesis, we demonstrated in a series of recent studies that the association of Sal4 with the O-Ag of S. Typhimurium has three immediate effects on bacterial physiology and virulence. First, Sal4 (5 to 15 μg/ml) treatment results in a rapid (<15 min) and dose-dependent arrest of S. Typhimurium flagellum-based motility in both liquid and semisolid media (26). Second, the binding of Sal4 to the bacterial outer membrane (OM) causes a significant reduction in SPI-1 T3SS activity, as evidenced by a decrease in SPI-1-dependent pore formation and the diminished delivery of SPI-1 effector proteins into host cells (28). Finally, the exposure of S. Typhimurium to Sal4 results in rapid (<15 min) alterations in membrane integrity, including enhanced OM permeability and increased shedding of lipopolysaccharides (LPS), as well as a transient reduction in the proton motive force (PMF) across the inner membrane (28).
In addition to these effects on virulence and physiology, we also observed by cryo-electron microscopy (cryo-EM) that Sal4-treated bacteria were surrounded by an electron-dense material that we have simply referred to as a fuzzy coat (22, 26). The electron-dense material was evident as early as 15 min post-antibody exposure and became more pronounced with time. In fact, within an hour after Sal4 treatment, cells were surrounded by a mucoid capsular material that is remarkably similar in appearance to the exopolysaccharides (EPS) secreted by S. Typhimurium during the initial stages of biofilm formation in response to environmental perturbations (22, 28, 29–31).
Based on these data, we postulate that Sal4 treatment generates a form of OM stress and that the early onset of EPS production by S. Typhimurium may represent an adaptive response to this sublethal stimulus. In this paper, we sought to characterize in more detail the effects of Sal4 on EPS production by S. Typhimurium. We demonstrate here that the prolonged exposure of S. Typhimurium to Sal4 results in EPS accumulation and biofilm formation on both glass and plastic surfaces. Moreover, we determined that this response is dependent on a putative diguanylate cyclase (DGC) known as YeaJ. In Escherichia coli, YeaJ is involved in regulating the bacterial transition from motility to biofilm formation (32). Based on these data, we propose that the association of Sal4 with the O-Ag induces OM stress, triggering a c-di-GMP signaling pathway in S. Typhimurium that effectively promotes the transition of the bacterium from a motile and invasive state to a nonmotile noninvasive biofilm state, thereby rendering the bacterium unable to invade intestinal epithelial cells.
MATERIALS AND METHODS
Bacterial strains, mammalian cell lines, and growth media.
The bacterial strains and plasmids used in this study are shown in Table 1. Cultures were grown in Luria-Bertani (LB) or M9 medium with 0.4% glucose at 37°C, or at room temperature (25°C); however, the strains carrying a temperature-sensitive pKD46, a plasmid that encodes λ Red recombinase to enhance homologous recombination, were maintained at 30°C. Cell growth was monitored and measured by the optical density at a wavelength of 600 nm (OD600). Media were supplemented with ampicillin (100 μg/ml) or l-arabinose (0.2%) as needed. HeLa cells were purchased from ATCC (CCL-2) and were maintained in Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2-95% air atmosphere.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source |
|---|---|---|
| Strains | ||
| Salmonella enterica serovar Typhimurium | ||
| ATCC 14028 | WT S. Typhimurium | ATCC |
| AMD212 | ATCC 14028-ΔthyA; WT strain with a deletion in thyA gene | 34 |
| SJF12 | oafA126::Tn10d-Tc | 26 |
| SJF11 | fkpA-lacZ oafA126::Tn10d-Tc; Kanr, Tcr | Lab collection |
| JSG1919 | bscE::kan; kanamycin cassette insertion mutation in bscE gene; Kanr | J. S. Gunn |
| JSG1546 | wcaA::luc; a mutant of wcaA constructed using luciferase-reporter suicide vector pGPL01; Ampr | 59 |
| JSG2897 | ΔyihO; λ Red deletion of yihO | 12 |
| STM14_1306 | ΔcsgD; deletion mutant of csgD; Kanr | M. McClelland |
| JA001 | ΔoafA; scar-free deletion mutant of oafA | 34 |
| JA002 | ΔyeaJ; scar-free mutant of yeaJ | This study |
| JA002+pYeaJ | ΔyeaJ(pYeaJ); JA002 strain complemented with pYeaJ plasmid; Ampr | This study |
| WT+pYeaJ | WT(pYeaJ); overexpression of YeaJ in WT background with pYeaJ plasmid; Ampr | This study |
| WT+pBAD24 | WT(pBAD24); WT strain carrying pBAD24 plasmid alone; Ampr | This study |
| JSG1919+pYeaJ) | bscE::kan(pYeaJ); overexpression of YeaJ in JSG1919 background with pYeaJ plasmid; Kanr Ampr | This study |
| Escherichia coli DH5α | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Invitrogen |
| Plasmids | ||
| pBAD24 | Arabinose-inducible, pBR322 ori; Ampr | 35 |
| pYeaJ | pBAD24 with yeaJ insertion at XbaI and HindIII sites | This study |
| P17 | pGEM-T-thyA; pGEM-T with thyA insertion; Ampr | 34 |
| pWSK29 | Ampr | 52 |
| pWYeaJ | pWSK29 with yeaJ insertion at SmaI and HindIII sites; Ampr | This study |
Antibiotic resistance phenotypes: Kanr, kanamycin; Tcr, tetracycline; Ampr, ampicillin.
Antibodies.
The murine hybridoma secreting Sal4, the monoclonal polymeric IgA antibody, was obtained from Marian Neutra (Children's Hospital, Boston, MA) (23). Hybridomas secreting the IgA antibodies Sal4 and 23D7 (an isotype control used throughout this study) (33) were maintained as described previously (26). Rabbit polyclonal antiserum against Salmonella O antigens (group B factors 1, 4, 5, and 12) was purchased from BD Difco (Franklin Lakes, NJ).
Construction of ΔyeaJ, ΔyeaJ(pYeaJ), bscE::kan(pYeaJ), and YeaJ overexpression strains.
The yeaJ gene of S. Typhimurium was deleted using a Flexible Recombineering Using Integration of thyA (FRUIT)-based method described previously (34). The primer sequences used to construct the mutant strains used in this study are listed in Table S1 in the supplemental material. Briefly, to construct the ΔyeaJ scar-free mutant, the thyA cassette was amplified using a primer pair (no. 319 to 320) with a ∼40-nucleotide (nt) 5′ sequence that matched the desired site of recombination from the plasmid pGEM-T-thyA. PCR products were purified using a MinElute PCR purification kit (Qiagen), electroporated into strain ATCC 14028 containing the pKD46 plasmid but lacking thyA (WT14028, ΔthyA), and grown on LB agar plates containing ampicillin, thymine (100 μg/ml), and arabinose (0.2%) at 37°C to induce expression of the λ Red genes. After 1 h, recovered cells were plated onto M9 minimal medium (lacking thymine) containing ampicillin at 30°C. Recombinants were verified using colony PCR for thyA insertion at the correct genomic location (yeaJ::thyA intermediate strain). Utilizing splicing by overlap extension (SOEing) PCR, an overlapping PCR product of the desired site of scar-free mutagenesis was constructed and electroporated into the yeaJ::thyA intermediate strain containing pKD46 and then grown on LB agar containing ampicillin and arabinose (0.2%) to induce expression of the λ Red genes. Recovered cells were plated at 30°C onto M9 minimal medium containing thymine (100 μg/ml), trimethoprim (20 μg/ml), and ampicillin. Recombinants were verified using colony PCR with primers flanking the expected site of mutagenesis and were further confirmed via DNA sequencing; then, thyA was reintroduced at its native locus.
The overexpression of yeaJ in a wild-type (WT) background (WT+pYeaJ) and bscE::kan background (bscE::kan+pYeaJ) and complementation of the ΔyeaJ mutant were accomplished as follows. The coding sequence of yeaJ from the S. Typhimurium ATCC 14028 genome was PCR amplified with yeaJ-specific primers containing built-in restriction sites as indicated in Table S1 and cloned into the XbaI and HindIII (New England BioLabs, Ipswich, MA) sites of a pBAD24 vector (35). E. coli DH5α competent cells were transformed with the resulting yeaJ construct (pYeaJ). Transformants were selected on LB agar plates containing ampicillin, and the sequence of the selected clone was verified by nucleotide sequencing. To construct the WT(pYeaJ) strain, the plasmid (pYeaJ) was isolated using QIAprep spin miniprep columns (Qiagen) and was transformed into S. Typhimurium strain ATCC 14028 by electroporation. Transformants were selected on LB agar containing ampicillin and were confirmed using PCR primers (pBAD-F and yeaJ-R-XbaI; see Table S1) and nucleic acid sequencing. To induce YeaJ expression, the selected transformants were grown in LB broth containing ampicillin and l-arabinose (0.2%), and the culture was shaken at 37°C overnight. Complementation of ΔyeaJ and overexpression of YeaJ in the bscE::kan background were achieved by transforming pYeaJ plasmid into the ΔyeaJ and bscE::kan mutant strains, respectively. Transformants were selected on LB agar containing ampicillin and were confirmed by PCR using primers (pBAD-F and yeaJ-R-XbaI; see Table S1) and by nucleic acid sequencing. To induce the expression of pYeaJ, the selected transformants were grown in LB broth containing ampicillin and l-arabinose (0.2%) with shaking at 37°C overnight.
Crystal violet-based biofilm assays.
Sal4-induced biofilms were measured using a previously described crystal violet (CV) staining method (36). Overnight cultures of each test strain were initially standardized in fresh LB broth containing either Sal4 (15 μg/ml), 23D7 (15 μg/ml), an IgA isotype control of irrelevant specificity, or chemically defined medium (CDM) with no antibody, to an OD600 of 0.05 (approximately 3 × 107 initial CFU/ml). Biofilm assays were performed either in borosilicate glass tubes or on 96-well polystyrene microtiter plates (Nunc, Denmark). Cultures in borosilicate glass tubes (1.3-ml final volume) were incubated upright on a rotary shaker set at 150 rpm at 37°C. After 24 h, pellicle formation was assessed at the air-liquid interface by visual observation and was digitally photographed. To assess biofilm formation, culture supernatants were decanted and the tubes were washed with phosphate-buffered saline (PBS) (pH 7.4) to remove unbound bacteria; then they were air-dried for 15 min. The remaining cells and cell-associated materials were fixed with 2-volume equivalents of methanol (Macron Chemicals, PA) for 15 min, air-dried for 30 min, and then stained with 0.1% CV (Sigma-Aldrich Co.) for 5 min. After this, the unbound dye was decanted and the tubes were washed with distilled water and digitally photographed.
For polystyrene microtiter plate-based assays, standardized cultures (200 μl per well) were incubated (after being covered and sealed with Parafilm) for 1 to 7 days at 25°C or 37°C. The media were not replaced over the course of these experiments. The OD595s of the bacterial cultures were monitored spectrophotometrically using a VersaMax microplate reader (Molecular Devices, Sunnyvale, CA). After optical density measurements were taken, culture supernatants were decanted and unbound bacteria were removed by washing with PBS (pH 7.4). The remaining cells and cell-associated materials were fixed (250 μl methanol per well), stained with CV, and solubilized with ethanol; absorbance was quantitated at 570 nm as indicated above.
Calcofluor and Congo red binding assays.
Cellulose production and the rdar (red, dry, and rough) phenotype were assessed essentially as described previously (37). The Congo red (CR) binding assay is often used to detect rdar colony morphology that produces cellulose and curli fimbriae (also known as Tafi) in Salmonella (38–41), while the calcofluor (CF) binding assay is used to detect the cellulose production of the colonies. Briefly, 5 μl of overnight cultures from WT and yeaJ mutant strains was spotted on agar plates containing CF (fluorescence brightener 28; 200 μg/ml) (Sigma) or CR (40 μg/ml) (Sigma) and Coomassie brilliant blue (20 μg/ml) (Sigma), and then was overlaid with 5 μl Sal4 (47 μg/ml) or CDM. Plates were incubated for 1 to 7 days at 25°C or 37°C. The development of the colony morphology and the dye binding activity were analyzed over time.
Isolation of EPS.
Isolation of EPS was performed as described previously (42, 43), but with minor modifications. Briefly, bacterial cells were grown in LB broth at 37°C overnight and subcultured into 5-ml fresh LB broth (OD600, 0.05) with Sal4 (15 μg/ml) or 23D7 (15 μg/ml), or simply an equal volume of CDM; then they were grown at 37°C for 4 h without shaking before the removal of 4 ml of bacterial supernatants. The cells in the remaining 1 ml were then collected by centrifugation, washed twice with PBS, and resuspended in sterile PBS to a final OD600 of 0.5. One milliliter of each culture was collected by centrifugation, resuspended in 0.5 ml of 50 mM sodium acetate buffer (pH 5.8)-100 mM NaCl, and then transferred to a 1.5-ml microcentrifuge tube. An equal volume containing 10 mM Tris-HCl (pH 8.0), 5 mM EDTA, and 0.5% SDS was added, and the tube was vortexed briefly and incubated at 100°C for 5 min. After centrifugation, the resulting pellet containing the crude exopolysaccharide was suspended in 0.5 ml of a solution containing 25 mM Tris-HCl (pH 8.0), 5 mM β-mercaptoethanol, 0.5% SDS, and 0.5 ml of 2× SDS-PAGE sample buffer; it was incubated at 100°C for 10 min. EPS samples (20 μl each) were separated by 12% SDS-PAGE with an extra-long (10-mm) stacking gel and a 35-mm-long resolving gel. Silver staining and Western blotting were used to determine relative EPS expression levels.
Western blotting.
Samples separated by SDS-PAGE were transferred to a pure nitrocellulose membrane (0.45 μM) (Bio-Rad). The membrane was blocked overnight with 3% bovine serum albumin in Tris-buffered saline containing 0.05% Tween 20 (TBST), washed three times with TBST, and incubated with rabbit anti-Salmonella O-antigen antiserum (group B factors 1, 4, 5, and 12) (BD Difco, Franklin Lakes, NJ) diluted (1:500) in blocking buffer. After washing with TBST, the membrane was incubated with secondary antibody (goat anti-rabbit horseradish peroxidase [HRP]-conjugated IgG) (SouthernBiotech) diluted 1:1,000 in blocking buffer. Antibody bound to the target antigen was detected by enhanced chemiluminescent (ECL) Western blotting substrate (Pierce Chemicals, Rockford, IL).
c-di-GMP quantification.
To determine the levels of c-di-GMP produced by YeaJ, the amount of c-di-GMP produced by specific strains of S. Typhimurium was quantified by using ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS-MS) as described previously (44, 45). Briefly, standardized cultures of WT, ΔyeaJ, WT(pYeaJ), and WT(pBAD24) strains were grown at 37°C with shaking at 150 rpm for 3 h in M9 minimal medium containing 0.4% glucose until the cultures reached a density of ∼2.8 × 108 CFU/ml (equivalent to an OD600 of 0.4). The final CFU/ml for each strain was determined by plating serial dilutions of the cultures onto nonselective LB medium. Bacteria were collected by centrifugation and nucleotides were extracted by resuspending the pellet in extraction buffer (MeOH to acetonitrile to distilled water [dH2O] [40:40:20] plus 0.1 N formic acid) as described previously (45). Ten microliters of each sample was analyzed by UPLC-MS-MS on a Quattro Premier XE mass spectrometer (Waters Corporation) coupled with an Acquity UPLC system (Waters Corporation). c-di-GMP was detected with electrospray ionization using multiple reaction monitoring in negative ion mode at m/z 689.16 to 344.31. Chemically synthesized c-di-GMP (Axxora) was used to generate a standard curve for calculating the c-di-GMP concentration in each extract.
HeLa cell invasion assay.
HeLa cell invasion and gentamicin protection assays were done as described previously (24, 26, 28). S. Typhimurium ATCC 14028, SJF11 (ΔoafA), ΔyeaJ, and WT(pYeaJ) strains were grown to mid-log phase in LB with aeration (∼2.8 × 108 CFU/ml), diluted 1:10, and then mixed at equal proportions. The exact ratio of the two strains was determined by replica plating. The bacteria were then incubated with Sal4 (5 μg/ml) for 45 min before being applied to HeLa cells in 12-well microtiter plates. The microtiter plates were subjected to low-speed centrifugation (10 min at 1,000 × g) at 4°C to promote bacterium-epithelial cell contact and then were incubated at 37°C for 1 h, followed by gentamicin treatment and cell lysis, as described previously (24). The competitive index (CI) was calculated as CI = ([% strain A recovered/% strain B recovered]/[% strain A inoculated/% strain B inoculated]) (46). Statistical significances were determined by one-way analysis of variance (ANOVA).
RESULTS
Sal4 induces biofilm formation in S. Typhimurium.
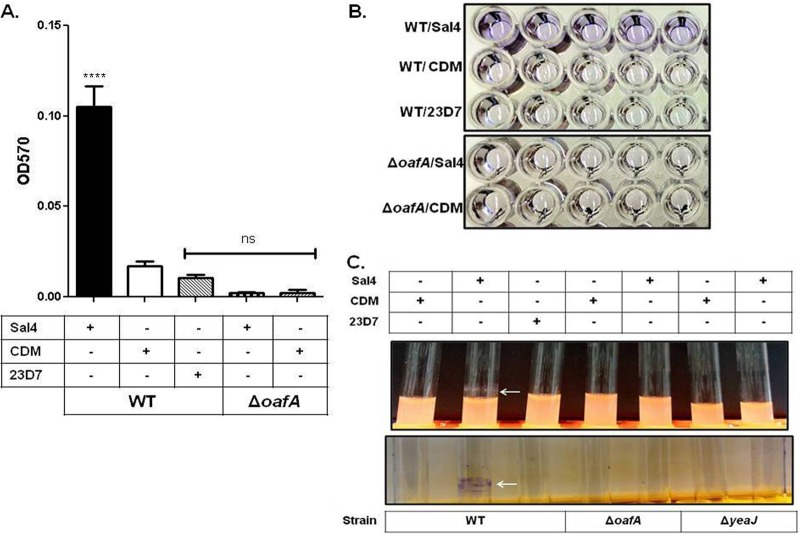
To determine whether Sal4 treatment induces EPS production and biofilm formation, we performed quantitative biofilm assays using uncoated polystyrene microtiter plates (Fig. 1A and B) or borosilicate glass tubes (Fig. 1C). After incubation at 37°C for 24 h, there was a significant increase in CV staining on both the polystyrene microtiter plates and glass tubes in the presence of Sal4 (15 μg/ml) compared to that on 23D7, an IgA isotype control MAb (15 μg/ml), or CDM alone (Fig. 1). As an additional control, we used an S. Typhimurium oafA deletion (ΔoafA) mutant (strain JA001), which lacks the Sal4 epitope due to the loss of O-acetyltransferase activity (47). The ΔoafA mutant did not exhibit biofilm formation on polystyrene microtiter plates or glass tubes in response to Sal4 (Fig. 1), demonstrating that the effects of Sal4 are specific and related to its ability to associate with the bacterial O-Ag.
Fig 1.
S. Typhimurium biofilm formation in response to Sal4. WT and JA001 (ΔoafA) strains of S. Typhimurium in LB broth were incubated on microtiter plates or in borosilicate glass culture tubes at 37°C for 24 h in the presence of Sal4 (15 μg/ml), 23D7 (15 μg/ml), or chemically defined medium (CDM). The cultures were then assayed for EPS production using CV, as described in Materials and Methods. (A) Quantification of biofilm mass, as measured by absorbance (optical density) at 570 nm (OD570). Statistical significance was determined by a one-way ANOVA, followed by Bonferroni's correction, compared to the WT strain treated with CDM. ****, P < 0.0001; ns, nonsignificant. Each bar represents the mean ± the standard error of the mean (SEM). (B) Representative images of CV-stained microtiter plates highlighting the effect of Sal4 on WT S. Typhimurium (top panel, top row). In contrast, there was no EPS production by strain JA001 (ΔoafA) in response to Sal4 (bottom panel), demonstrating the specificity of the antibody response. (C) Representative images demonstrating pellicle formation (top panel) and EPS production (bottom panel) on borosilicate culture tubes in response to Sal4 (arrows). The data in each panel represent the averages of three independent assays. In each assay, experimental samples were performed in five-replicate (quintuplicate) wells or culture tubes.
A modified SDS-PAGE and silver-staining protocol (see Materials and Methods) confirmed that there were elevated amounts of EPS associated with Sal4-treated WT cells compared to that with 23D7-treated cells or the CDM-treated control (Fig. 2A). As expected, EPS production in response to the Sal4 antibody was not observed with either heat-killed bacteria or the ΔoafA mutant (Fig. 2A). Furthermore, Western blot analysis revealed that the EPS material secreted by S. Typhimurium in response to Sal4 was reactive with O-Ag polyclonal antiserum, suggesting that, in part, the EPS may be composed of O-Ag capsule (Fig. 2B) (31). Sal4-treated colonies also bound to CF, a specific and sensitive probe for β-1,4-linked d-glucopyranosyl residues of cellulose (Fig. 2C) (48). Together, these data demonstrate that EPS material is produced in response to the Sal4 antibody.
Fig 2.
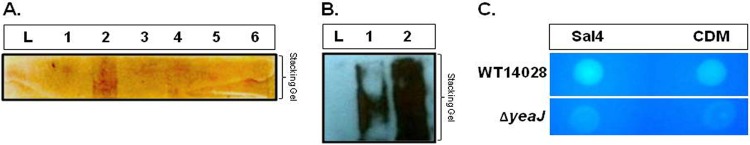
Expression of an EPS containing O-Ag capsule-like material by S. Typhimurium in response to Sal4. (A and B) EPS preparations from WT strain 14028 or strain SJF12 (ΔoafA) treated with Sal4 (10 μg/ml), 23D7 (10 μg/ml), or CDM for 4 h at 37°C were separated by SDS-PAGE on 12% gels with an extra-long (35-mm) stacking gel. (A) Silver stain. Lane L, molecular weight ladder; lane 1, WT treated with CDM; lane 2, WT treated with Sal4; lane 3, WT treated with 23D7; lane 4, heat-killed WT treated with Sal4; lane 5, SJF12 treated with CDM; and lane 6, SJF12 treated with Sal4. (B) Following SDS-PAGE, EPS preparations were transferred onto nitrocellulose membranes and subjected to Western blotting using Salmonella-specific O-Ag polyclonal antisera, as described in Materials and Methods. Lane L, ladder; lane 1, WT treated with CDM; lane 2, WT treated with Sal4. (C) Cellulose production in response to Sal4. WT and ΔyeaJ strains were spotted on CF agar plates and then overlaid with 5 μl Sal4 (47 μg/ml) or CDM. Plates were then incubated at 37°C for 24 h and photographed on a UV light box.
To examine the complexity of the EPS induced by Sal4, we assessed the Sal4-induced EPS production by strains carrying mutations in genes known to be important in S. Typhimurium exopolysaccharide synthesis, including ΔyihO (O-Ag capsule), wcaA::luc (colanic acid), and bscE::kan (cellulose). The bcsE::kan mutant was completely defective in Sal4-induced biofilm formation, while the ΔyihO and wcaA::luc strains each demonstrated reduced biofilms compared to that shown by the WT strain (see Fig. S1 in the supplemental material). These data further support the notion that the EPS elicited in response to Sal4 consists primarily of cellulose, and likely also O-Ag capsule and colanic acid.
YeaJ is important in Sal4 biofilm formation at 37°C.
In an effort to identify the specific genes and proteins of S. Typhimurium involved in biofilm formation in response to Sal4, we screened a library of chromosomal deletion mutants for those that were incapable of forming a biofilm in the presence of Sal4 (J. Amarasinghe, K. Levinson, and N. Mantis, unpublished data). One of the mutants we identified carried a chromosomal deletion in yeaJ (STM1283), a gene that encodes a putative inner membrane-localized DGC (49). DGCs synthesize c-di-GMP to regulate cellulose production, biofilm formation, and virulence in S. Typhimurium (50). In E. coli, yeaJ regulates the transition between motility and biofilm formation states (32). In S. Typhimurium, yeaJ as expressed on a plasmid was capable of complementing the biofilm deficiency of a strain lacking gcpA, a known DGC (51). Therefore, we hypothesized that yeaJ is involved in Sal4-induced biofilm formation in S. Typhimurium.
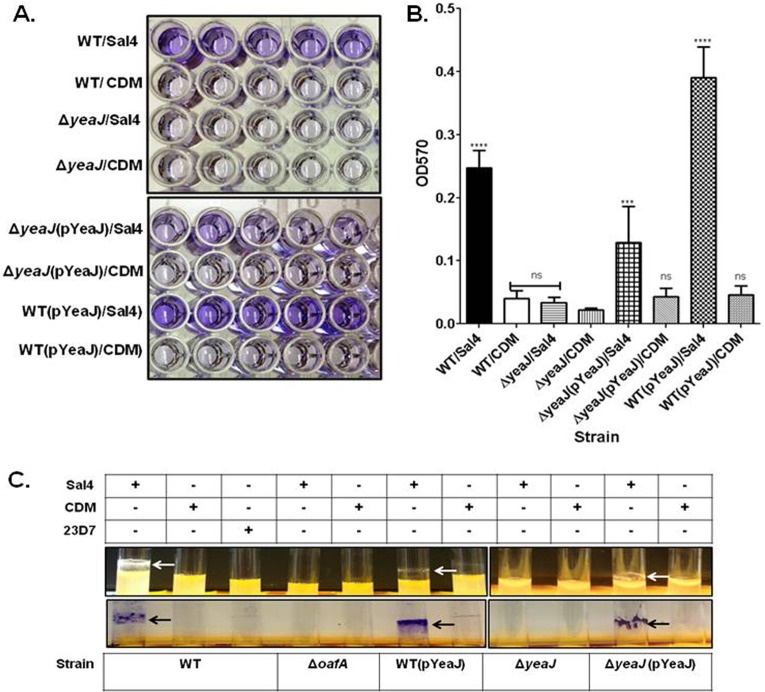
To determine the role of YeaJ in the induction of biofilm formation by Sal4, we constructed a nonpolar chromosomal yeaJ deletion (ΔyeaJ) in an otherwise WT background. To investigate the effect of this mutation on Sal4-induced biofilm formation, ΔyeaJ was tested using a quantitative microtiter biofilm assay in the presence or absence of Sal4. In this assay, the ΔyeaJ strain was unable to form a biofilm in response to Sal4 (Fig. 3A and B), or to produce cellulose in response to Sal4 (Fig. 2C). In an effort to complement this mutation, we cloned the wild-type allele of yeaJ, under the control of its own promoter, into a low-copy-number plasmid (pWSK29) (52), and introduced the resulting plasmid (pYeaJ) into the ΔyeaJ strain. Although we confirmed by nucleotide sequencing that yeaJ was properly cloned into this vector, the resulting low-copy-number plasmid, pWYeaJ, did not restore the Sal4-induced biofilm phenotype or cellulose production. We speculated that the failure of this plasmid to complement the ΔyeaJ mutant might be due to the insufficient (or improper) expression of YeaJ possibly because of the absence of cis or trans-acting elements involved in the regulation of yeaJ. We therefore cloned the native yeaJ gene into a commonly used medium-copy-number plasmid (pBAD24), in which the expression of yeaJ was now under the control of the arabinose-inducible araC promoter. The introduction of yeaJ as expressed from this pBAD-based plasmid resulted in the full restoration of cellulose production, as indicated by CF (Fig. 4A) and CR binding (Fig. 4A), as well as in the partial restoration of biofilm formation in response to Sal4 (Fig. 3A and B).
Fig 3.
S. Typhimurium biofilm formation in response to Sal4 is dependent on YeaJ. WT, JA002 (ΔyeaJ), JA002 complemented with pYeaJ, and WT (pYeaJ) strains of S. Typhimurium in LB broth were incubated on microtiter plates (A and B) or in borosilicate glass (C) culture tubes at 37°C for 24 h in the presence of Sal4 (15 μg/ml), 23D7 (15 μg/ml), or CDM. The cultures were then assayed for biofilm formation using CV, as described in Materials and Methods. (A) Representative images of CV-stained microtiter plate demonstrating that Sal4-induced biofilm formation is dependent on YeaJ. (B) Quantification of biofilm mass, as measured by absorbance (optical density) at 570 nm (OD570). Statistical significance was determined by a one-way ANOVA, followed by Bonferroni's correction, compared to the WT strain treated with CDM. ****, P < 0.0001; ***, P < 0.001; ns, nonsignificant. Each bar represents the mean ± the standard error of the mean (SEM). (C) Representative images demonstrating that pellicle formation (top panel) and biofilm formation (bottom panel) on borosilicate culture tubes in response to Sal4 (arrows) are dependent on YeaJ. The data in each panel represent the average of three independent assays. In each assay, experimental samples were performed in quintuplicate wells and duplicate culture tubes.
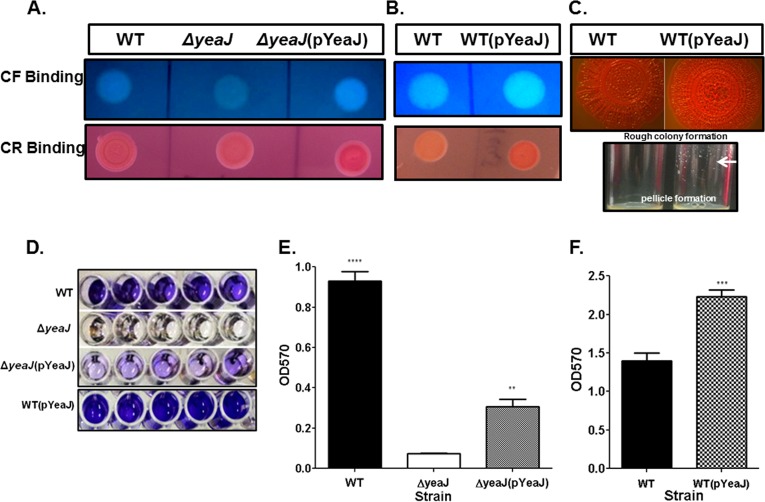
Fig 4.
Cellulose production and biofilm formation at 25°C by S. Typhimurium are dependent on YeaJ. (A) WT, JA002 (ΔyeaJ), and JA002+pYeaJ [ΔyeaJ(pYeaJ)] strains were spotted on CF and CR agar plates and incubated at 25°C for 72 h to assess cellulose production and rough colony formation. Under these conditions, ΔyeaJ was deficient in cellulose production and failed to form rough colonies compared to the WT strain. See the text for details. (B) Phenotype of WT and WT(pYeaJ) on CR (top panel) and CF (bottom panel) agar plates following incubation at 25°C for 24 h. (C) WT and WT(pYeaJ) rough colony formation on CR agar after 7 days at 25°C (top panel) and pellicle formation (arrow) after 3 h of incubation in LB at 37°C (bottom panel). (D to F) WT, JA002 (ΔyeaJ), JA002 + pYeaJ [ΔyeaJ(pYeaJ)], and WT(pYeaJ) strains incubated in microtiter plates for 7 days at 25°C. The cultures were then assayed for biofilm formation using CV, as described in Materials and Methods. (D) Representative images of CV-stained microtiter plates highlighting the role of YeaJ in biofilm formation at 25°C; (E and F) quantification of biofilm mass, as measured by absorbance (optical density) at 570 nm (OD570). The data in each panel represent the average of three independent assays. In each assay, experimental samples were performed in quintuplicate wells and duplicate culture tubes. In panel E, statistical significance was determined by a one-way ANOVA, followed by Bonferroni’s correction, compared to the WT strain treated with CDM. ⁎⁎⁎⁎, P < 0.0001; ⁎⁎, P < 0.01. In panel F, significance was determined by paired Student’s t test, compared to the control (WT). ⁎⁎⁎, P = 0.0003. In panels E and F, each bar represents the mean with the SEM.
If yeaJ encodes a bona fide DGC, then we postulated that overexpression of the yeaJ gene via an inducible promoter in an otherwise WT background will enhance baseline (as well as Sal4-induced) biofilm formation and EPS production. To test this, we introduced the plasmid carrying the full-length yeaJ (pYeaJ) cloned under the control of the arabinose-inducible araC promoter into wild-type S. Typhimurium. As expected, the ectopic expression of YeaJ resulted in enhanced Sal4-induced biofilm formation at 37°C on borosilicate glass and microtiter plates (Fig. 3A and B). Ectopic expression of YeaJ in the absence of Sal4, however, did not significantly induce biofilm formation. This result suggests that the DGC activity of YeaJ is dependent on Sal4-mediated changes to S. Typhimurium, arguing that Sal4 and YeaJ are components of a common signaling pathway. Based on these data, we concluded that YeaJ is involved in Sal4-induced biofilm formation at 37°C, likely functioning as a DGC enzyme to synthesize c-di-GMP in the presence of Sal4.
YeaJ is also necessary for biofilm formation at 25°C.
Our above-mentioned data indicate that the activity of YeaJ is Sal4-dependent at 37°C. However, we noticed that Sal4 does not induce biofilm formation at 25°C (data not shown), a temperature that has been shown to induce robust biofilm formation by S. Typhimurium due to the optimal production of cellulose and fimbriae under this condition (53, 54). Therefore, we wondered whether the activity of YeaJ can transpire at 25°C in the absence of Sal4. We first tested whether the disruption of yeaJ impacts cellulose production at room temperature, as assessed using the CF binding assay. In this assay, the WT strain produced bright colonies (by 3 days), an indicator of cellulose production (Fig. 4A, top). In contrast, the ΔyeaJ mutant colonies appeared dull under a UV light source, even when they had been incubated at room temperature for as long as 3 days (Fig. 4A, top). Cellulose production was restored to WT levels when the ΔyeaJ strain was transformed with a plasmid carrying a functional copy of yeaJ (Fig. 4A, top). Furthermore, the overexpression of YeaJ in an otherwise WT background accelerated the appearance of CF-positive colonies (Fig. 4B, top) and biofilm formation (Fig. 4D and F).
In S. Typhimurium, the rdar (red, dry, and rough) colony morphotype is linked to biofilm formation at low ambient temperatures (≤28°C). To examine the effect of the yeaJ mutation on the rdar morphotype, the yeaJ mutant and the WT control were plated on CR agar and incubated at room temperature for 7 days. The WT strain displayed CR-positive colonies within 48 h, and it assumed the rdar morphotype within 3 days (Fig. 4A, bottom). In contrast, the ΔyeaJ mutant was defective in CR binding and in the formation of rdar colonies, as evidenced by smooth moist colonies, not rough and dry colonies, at day 3 (Fig. 4A, bottom). Complementation of the ΔyeaJ mutant partially restored the rdar phenotype in that the colonies remained mildly rough and not dry at day 3 (Fig. 4A, bottom) or even after prolonged incubation (>14 days) (data not shown). On the other hand, the overexpression of YeaJ in a WT background (WT+pYeaJ) accelerated and accentuated the rdar morphotype, as CR-bound colonies appeared within 24 h (Fig. 4B, bottom) and colony roughness was notably pronounced at 7 days compared to that of the WT control at the same time point (Fig. 4C, top). The WT strain with pBAD24 vector alone exhibited similar levels of CR and CF binding as the WT strain, indicating no evidence of a deleterious effect or the metabolic burden of pBAD24 plasmid (see Fig. S4 in the supplemental material). From these data, we conclude that YeaJ is required for the production of cellulose, and possibly fimbriae, in S. Typhimurium at low ambient temperatures.
Next, we evaluated the capacity of the ΔyeaJ mutant to produce a biofilm at room temperature over a 1-week period. The WT S. Typhimurium strain 14028 formed a robust biofilm at 25°C within 7 days, as evidenced by CV staining in untreated polystyrene 96-well plates (Fig. 4D). In contrast, at the same time point, wells inoculated with the ΔyeaJ mutant were CV-negative (Fig. 4D), suggesting that the mutant is defective in biofilm production under these conditions. The failure of the ΔyeaJ mutant to produce a biofilm was not due to a growth defect, because it achieved the same optical density as the WT strain (data not shown). Complementation of the ΔyeaJ strain resulted in partial restoration of the biofilm phenotype (Fig. 4D), while overexpression of YeaJ in a WT background resulted in pronounced pellicle formation at the air-liquid interface within 3 h of growth in LB broth and in significantly greater biofilm formation (Fig. 4C, D, and F). These data demonstrate that YeaJ, a putative DGC, is critical in S. Typhimurium biofilm formation and is possibly a major regulator of cellulose production at 25°C and 37°C in response to Sal4.
YeaJ is an active DGC in S. Typhimurium.
To determine whether YeaJ is indeed an active DGC responsible for c-di-GMP production at 37°C in S. Typhimurium, bacterial cell lysates from the WT, ΔyeaJ, WT(pYeaJ), or WT(pBAD24) strains grown at 37°C were subjected to UPLC-MS-MS. c-di-GMP levels in lysates from WT(pYeaJ) cells were significantly higher (8.3-fold) than WT levels (Fig. 5; P < 0.001). In contrast, c-di-GMP levels in the ΔyeaJ strain of S. Typhimurium were reduced ∼0.6-fold compared to the WT levels, although this difference was not statistically significant (Fig. 5). Finally, c-di-GMP levels were identical between the wild-type strains with and without the pBAD24 vector, confirming that the plasmid itself did not influence c-di-GMP production (see Fig. S3 in the supplemental material). These data demonstrate that YeaJ contributes to the total c-di-GMP pool at 37°C in S. Typhimurium, likely by functioning as an active DGC.
Fig 5.
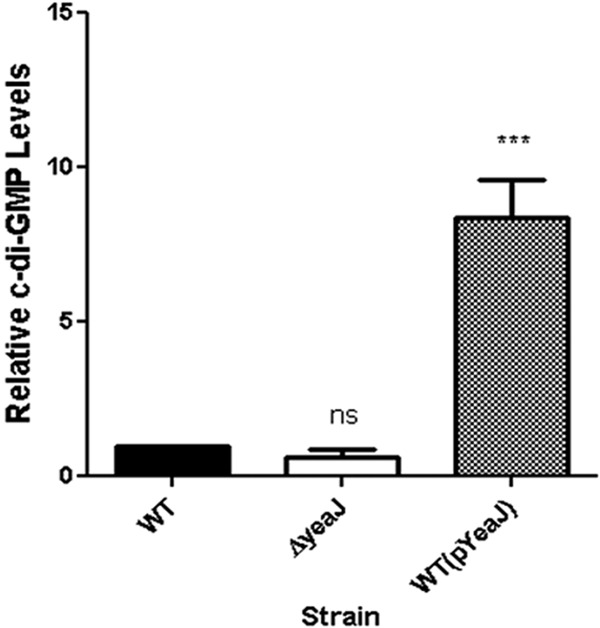
YeaJ possesses DGC activity shown by c-di-GMP levels in S. Typhimurium. WT, JA002 (ΔyeaJ), and WT+pYeaJ strains of S. Typhimurium were grown on M9 medium supplemented with 0.4% glucose to mid-log phase and were collected by centrifugation. Nucleotides were extracted from the cell pellets as described in Materials and Methods and then were subjected to UPLC-MS-MS. Levels of c-di-GMP were normalized relative to the WT strain (set to a value of 1). Statistical significance was determined by a one-way ANOVA, followed by Bonferroni's correction, compared to the control (WT): ***, P < 0.001; ns, not significant. Each bar represents the mean with the SEM.
A role for YeaJ in S. Typhimurium invasion of epithelial cells in vitro.
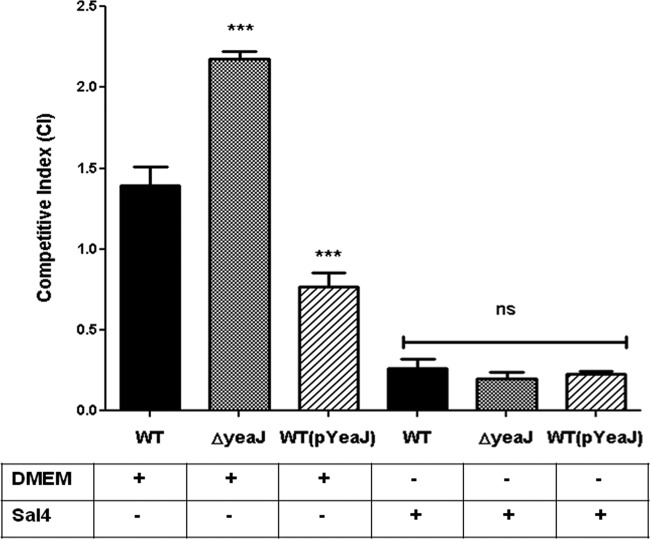
A number of DGCs, including STM1283, have been implicated in the regulation of the capacity of S. Typhimurium to invade intestinal epithelial cells (19, 21). In order to assess the specific role of YeaJ in this process, as well as to evaluate the impact of Sal4, we performed competitive uptake assays in HeLa cells. We expected that if YeaJ does indeed contribute to the overall pool of c-di-GMP in the cell, then a ΔyeaJ mutant would be expected to be more invasive than the WT S. Typhimurium, while a strain overexpressing YeaJ would be expected to be less invasive. To test this, WT strain 14028, the ΔyeaJ mutant, or the WT strain overexpressing YeaJ (WT+pYeaJ) was mixed one-to-one with SJF11 (ΔoafA) and incubated with or without Sal4 (5 μg/ml) for 45 min before being applied to HeLa cell cultures. As predicted, the ΔyeaJ mutant was slightly more invasive than the wild-type strain, whereas the YeaJ-overexpressing strain was slightly less invasive (Fig. 6). In the presence of Sal4, the invasion of all three strains was reduced by >80%, demonstrating that neither deletion nor overexpression of YeaJ bypasses the effects of Sal4 on bacterial invasion.
Fig 6.
YeaJ plays a role in invasion of epithelial cells. The WT, ΔyeaJ, and WT+pYeaJ strains of serovar Typhimurium were mixed with the oafA mutant (SJF11) at a 1:1 ratio, incubated without antibody (DMEM alone) or with Sal4 (5 μg/ml) for 45 min, applied to HeLa cells seeded in 24-well plates, and subjected to centrifugation (10 min at 4°C) to promote bacterium-epithelial cell adherence, followed by incubation for 1 h at 37°C. The number of invasive bacteria was determined via a gentamicin protection assay (24). The competitive index (CI) reflects the invasion of WT, ΔyeaJ, or WT+pYeaJ strain cells to HeLa cells relative to the invasion of oafA mutant cells (data not shown). Statistical significance was determined by a one-way ANOVA, followed by Bonferroni's correction, compared to the control (WT/ΔoafA+DMEM): ***, P < 0.001. Each bar represents the mean with the SEM.
DISCUSSION
Sal4 was described more than 20 years ago as an IgA MAb against the O5 epitope of LPS of S. Typhimurium that was protective against intestinal Salmonella infection, although the exact mechanism for this protection remains unclear (23, 24). Sal4 is neither bactericidal nor bacteriostatic (23, 55, 56). Sal4 does not activate complement nor does it promote Fc (fragment, crystallizable)-mediated clearance of opsonized bacteria (23, 57). Furthermore, although Sal4 is effective at promoting bacterial agglutination and entrapment in mucus, a phenomenon known as immune exclusion, these properties alone cannot fully account for the protective capacity of Sal4 (22, 26). We recently demonstrated that Sal4 treatment leads to an immediate inhibition in motility and decreased T3S activity in S. Typhimurium (28). Sal4 also triggers changes in OM integrity, accompanied by the appearance of a mucoid capsular material (28). In this study, we have demonstrated that prolonged exposure (>4 h) of S. Typhimurium to Sal4 results in detectable levels of EPS production that ultimately result in biofilm formation, at least in vitro. Sal4-induced EPS production and biofilm formation were found to be dependent on YeaJ, a member of the c-di-GMP network in S. Typhimurium. YeaJ has been demonstrated to regulate the bacterial transition from a motile and invasive state to a biofilm and noninvasive state in E. coli (32). Based on these data, we speculate that Sal4, by virtue of its ability to associate with the O-Ag and to induce OM stress, renders S. Typhimurium avirulent by triggering the c-di-GMP-dependent signaling pathway via YeaJ that leads to the suppression of bacterial motility and T3S, while simultaneously stimulating EPS production and biofilm formation.
In S. Typhimurium, EPS production is proposed to facilitate the ability of the bacterium to colonize inert and host-associated surfaces and to respond to various environmental stress conditions (58–61). For example, S. Typhimurium produces biofilms on abiotic and biotic surfaces, including glass, plastic (62), gallstones (12, 59), epithelial cells, and chicken intestinal tissues (63). In addition, S. Typhimurium has been shown to form biofilms as a defense against phagocytosis by host immune cells (64). Gunn and colleagues have demonstrated that S. Typhimurium is capable of producing biofilms in vitro in response to prolonged exposure to bile (e.g., 14 days), revealing that host factors can stimulate S. Typhimurium to initiate biofilm formation (30). In S. Typhimurium, EPS is primarily composed of three major polysaccharides: cellulose, O-Ag capsule, and colanic acid (29, 31, 40, 41, 65). In addition, fimbriae and the surface protein BapA are known to be critical components of the extracellular matrix (13, 65) (for review, see 20). The actual composition of the EPS produced by S. Typhimurium, however, varies based on environmental conditions and the nature of the colonized surfaces. For example, O-Ag capsule, but not cellulose or colanic acid, is a vital part of the biofilm formed on gallstones in response to bile (12). On the other hand, cellulose and colanic acid (as well as fimbriae), but not O-Ag capsule, are critical factors in biofilm formation on epithelial surfaces (12, 66).
EPS production in response to Sal4 is unique in at least two aspects. First, the onset of EPS production is virtually instantaneous and appears to persist for hours. The appearance of Sal4-triggered EPS production was initially observed by cryo-EM as being like a fuzzy coat within 15 min of antibody exposure (28). Scanning and transmission electron microscopy revealed that by 1 h, Sal4-treated cells were surrounded by a mucoid layer and formed a meshwork that is reminiscent of the earliest stages of biofilm formation (28). We speculate that the appearance of CV-positive staining at the air-liquid interface of S. Typhimurium grown for 24 h in the presence of Sal4 on borosilicate glass tubes and polystyrene microtiter plates represents a continuation of EPS production. Furthermore, our data suggest that Sal4-induced EPS consists of cellulose, O-Ag capsule, and colanic acid, because mutations in any one of these polysaccharide biosynthesis pathways resulted in reduced EPS production based on Western blot analysis of the EPS preparations and indicator agar plates. However, we cannot exclude the possibility that Sal4 triggers the production of an additional unidentified EPS. Therefore, studies are under way to determine the structure and exact composition of the EPS secreted by S. Typhimurium in response to Sal4.
Second, EPS production in response to Sal4 occurs at 37°C, not at ambient (25°C) temperature. This is unique because, in general, S. Typhimurium biofilm production is largely suppressed at elevated temperatures (i.e., 37°C), due to the temperature-dependent regulation of biofilm determinant genes by CsgD (53, 54). The fact that Sal4-induced biofilm production requires a functional CsgD (see Fig. S2 in the supplemental material) is consistent with the previous findings that show that CsgD is essential for the c-di-GMP-mediated induction of biofilm formation (67). This finding also suggests that the antibody stimulates a signaling pathway(s) in S. Typhimurium that overrides the temperature-dependent regulation by CsgD. This would not be unprecedented, as the temperature-dependent regulation by CsgD can be circumvented (at least genetically) at 37°C in order to express curli fimbriae and cellulose (13, 68–70). In addition, Römling and colleagues have demonstrated that upon iron starvation, the temperature-independent expression of Tafi occurred at 37°C (13). Thus, Sal4 is a unique environmental signal that induces biogenesis of EPS in S. Typhimurium at host temperatures. To our knowledge, these data represent the first demonstration of a specific host-derived molecule that stimulates a c-di-GMP network in S. Typhimurium.
It has been demonstrated that SIgA can promote the formation of biofilms by commensal bacteria, like E. coli, on intestinal epithelial surfaces (22, 71). Bacteria grown in vitro in the presence of high SIgA concentrations (e.g., 0.5 mg/ml) were capable of colonizing epithelial cell surfaces and of forming a microbial mat, whereas cells grown in the absence of SIgA were not (71). This phenomenon was determined to be the result of SIgA serving as a scaffold for bacterial colonization, not as a response of E. coli to SIgA per se. Specifically, E. coli bound to oligosaccharide side chains on SIgA by virtue of its mannose-specific type-1 pilli. Thus, the interaction between E. coli and IgA is bacterially mediated and is independent of the variable (Fv) regions of IgA. This is in contrast to the interaction between S. Typhimurium and Sal4, which occurs solely through the association of Sal4 with O5-Ag, as a strain of S. Typhimurium that lacks the O5-Ag does not produce EPS or form a biofilm in response to Sal4.
Sal4 promotes the transition of S. Typhimurium from a virulent to an avirulent modality, in which the loss of motility and T3S is accompanied by increased EPS production. This inversely proportional regulation of motility and EPS production is reminiscent of that of the c-di-GMP regulatory pathways in enteric bacteria (32, 50, 72–75). Thus, it is intriguing that we found that EPS production and biofilm formation in response to Sal4 are dependent on YeaJ, a suspected member of the DGC family. The S. Typhimurium YeaJ was annotated as a DGC based on its sequence similarity with known DGCs (49). Garcia and colleagues provided the first functional evidence to support YeaJ as a possible DGC, in that they demonstrated that YeaJ expressed on a plasmid complemented the biofilm deficiency of a ΔgcpA mutant (gcpA is a known member of the c-di-GMP network) (51). Furthermore, recent genetic studies have indicated YeaJ as being involved in rdar colony formation, flagellum-based motility, and cellulose production in S. Typhimurium (21, 76).
We put forth several lines of evidence that demonstrate that the YeaJ protein does indeed possess DGC activity that ultimately contributes to c-di-GMP levels in S. Typhimurium. We found that the ectopic overexpression of YeaJ in S. Typhimurium at room temperature results in pronounced pellicle formation at the air-liquid interface and in significantly greater biofilm formation at 7 days. Overexpression of YeaJ in the WT background also enhanced EPS production and biofilm formation at 37°C in response to Sal4. Another line of evidence for the significant capacity of YeaJ to govern the Sal4-induced biofilm formation at 37°C is its ability to complement the biofilm defect of a bscE::kan mutant, a cellulose mutant. We observed that when a functional copy of yeaJ was introduced in trans to the bscE::kan mutant, the Sal4-induced biofilm defect of this mutant was fully restored (see Fig. S1 in the supplemental material). Finally, we found that the overexpression of YeaJ resulted in an 8-fold increase in baseline c-di-GMP levels in S. Typhimurium compared to that in a WT control (Fig. 5). It should be noted that we quantitated c-di-GMP levels using UPLC-MS-MS, a technique that is considerably more sensitive than standard high-performance liquid chromatography (HPLC), which likely explains why others have not noted YeaJ-induced changes in c-di-GMP levels (21).
Interestingly, although the overexpression of YeaJ in S. Typhimurium resulted in an increase in the intracellular pool of c-di-GMP at 37°C, this itself was not sufficient to promote biofilm formation at this elevated temperature. We think that this finding is significant because it may imply that EPS production and biofilm formation by S. Typhimurium in response to Sal4 require two signals. In addition to elevated intracellular c-di-GMP levels, we propose that a second signal related to Sal4-induced changes to the OM may be required. We recently reported that the association of Sal4 with the O-Ag of S. Typhimurium results in the shedding of LPS, a transient reduction in the PMF, and a corresponding decrease in intracellular ATP levels (28). Any one of these physiologic changes in the OM can serve as the secondary cue to initiate EPS production and biofilm formation by S. Typhimurium.
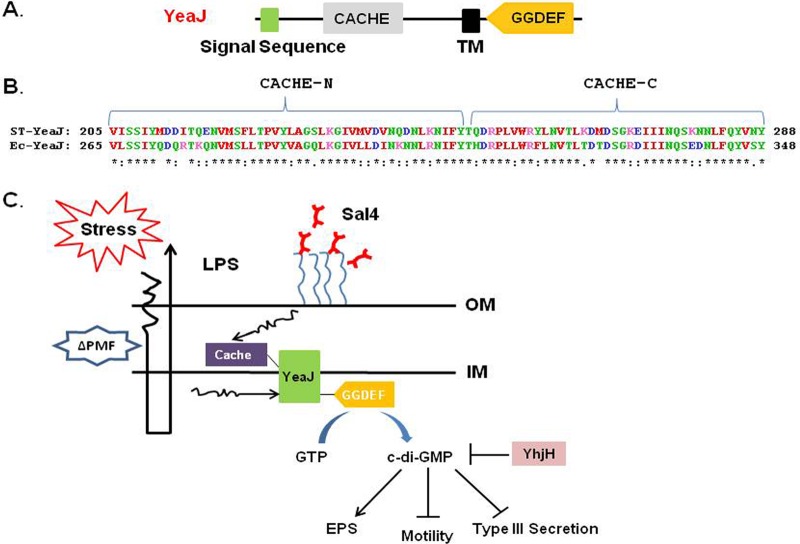
Indeed, it is interesting to speculate that YeaJ may sense Sal4 binding to the bacterial surface, possibly as a consequence of antibody-mediated OM stress (28). In addition to its conserved GGDEF domain that is involved in diguanylate cyclase activity, YeaJ has a putative extracellular sensory Cache (Ca2+ channels, chemotaxis receptors) domain that is virtually identical to that of YeaJ in E. coli (Fig. 7A and B). In general, Cache domains sense stimuli present in the periplasm and transmit signals to an output domain, such as GGDEF, resulting in a specific adaptive response (77–80). It has been proposed that Cache domains sense changes in the periplasm or inner membrane. Current studies are under way to understand the role of the Cache domain of YeaJ in sensing Sal4 antibody and in inducing EPS and biofilm formation.
Fig 7.
Model for Sal4-induced EPS production and biofilm formation. (A) Schematic linear depiction of the structure of YeaJ, highlighting the putative N-terminal signal sequence, periplasmic Cache domain, a transmembrane (TM) domain, and the GGDEF domain; (B) ClustalW-sequence alignment of the Cache domains of YeaJ from S. Typhimurium (ST) and E. coli (Ec). (C) Proposed mechanism for YeaJ-mediated c-di-GMP signal transduction in S. Typhimurium in response to Sal4. Sal4 binding to the O-Ag results in OM stress, including a transient reduction in the PMF. We propose that the Cache domain of YeaJ senses Sal4-induced changes in membrane integrity and, in turn, activates the GGDEF domain of YeaJ to produce c-di-GMP. Elevated levels of c-di-GMP stimulate EPS production and reduce both bacterial motility and T3S. We speculate that the cognate phosphodiesterase (PDE) YhjH degrades the c-di-GMP produced by YeaJ.
In conclusion, we propose that the binding of Sal4 to the O-Ag of S. Typhimurium activates YeaJ, which in turn stimulates intracellular levels of c-di-GMP, thereby leading to a loss in motility, a reduction in T3S activity, and an increase in EPS production (Fig. 7C). Moreover, we speculate that Sal4-induced EPS production represents an adaptive mechanism employed by S. Typhimurium to shed antibody and/or to increase resistance to secondary insults, such as antimicrobial peptides and other factors present in intestinal secretions. This study provides further insight into how SIgA-mediated immunity may occur in the gut, and it underscores how antibodies may exert their effects differently depending on the environmental conditions (27). In systemic compartments, for example, the opsonization of Salmonella by surface antigen-specific IgG leads to complement-mediated killing and Fc-dependent phagocytosis (81). In the gut, we propose that the opsonization of S. Typhimurium by Sal4 (or another IgA antibody of similar specificity) induces a form of outer membrane stress that is sufficient to incapacitate both SPI-1 T3S and flagellum-based motility (28). Moreover, we would now argue that prolonged exposure to Sal4 promotes the transition of S. Typhimurium from a motile and invasive state to a nonmotile and noninvasive state. Once S. Typhimurium has been coated with SIgA and effectively immobilized in the intestinal lumen, it is likely cleared through peristalsis. Additionally, there is evidence that SIgA-coated microbes are actively sampled by intestinal M cells and are transported to underlying gut-associated lymphoid tissues (22, 82). Taken together, our study advances our fundamental understanding of how SIgA may exert its protective effects at mucosal surfaces, without the assistance of Fc-mediated effector functions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joseph Wade and Anne Stringer for their assistance in mutant construction, as well as for their generosity in providing us with bacterial strains and plasmids. We also thank Richelle Reid, Kara Levinson, Stephen Forbes, Benjamin Koestler, Jocelyn Cole, the Wadsworth Center Applied Genomic Technologies Core facility, and the MSU Mass Spectrometry facility for technical assistance. We thank John Gunn (Ohio State University), Michael McClelland (San Diego Institute for Biological Research), and Sidney Kushner (University of Georgia) for providing us with bacterial strains.
J.J.A. is the recipient of a Wadsworth Center Biodefense and Emerging Infectious Diseases training program (T32AI055429; principal investigator [PI], McDonough) postdoctoral fellowship.
This work was supported in part by grants 2-U54-AI-057153 (to C.M.W.) and R01HD061916 (to N.J.M.) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD).
We declare no conflicts of interest.
Footnotes
Published ahead of print 10 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00813-12.
REFERENCES
- 1. Rabsch W, Tschäpe H, Bäumler AJ. 2001. Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 3:237–247 [DOI] [PubMed] [Google Scholar]
- 2. Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. 2010. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. U. S. A. 107:17733–17738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsolis RM, Adams LG, Ficht TA, Bäumler AJ. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67:4879–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang S, Santos RL, Tsolis RM, Stender S, Hardt WD, Bäumler AJ, Adams LG. 2002. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect. Immun. 70:3843–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. 2005. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 73:3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Galan JE, Curtiss R., III 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 86:6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Groisman EA, Ochman H. 1993. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 12:3779–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 93:2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ellermeier JR, Slauch JM. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24–29 [DOI] [PubMed] [Google Scholar]
- 10. Löber S, Jaäckel D, Kaiser N, Hensel M. 2006. Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int. J. Med. Microbiol. 296:435–447 [DOI] [PubMed] [Google Scholar]
- 11. Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759–1773 [DOI] [PubMed] [Google Scholar]
- 12. Crawford RW, Gibson DL, Kay WW, Gunn JS. 2008. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect. Immun. 76:5341–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Römling U, Sierralta WD, Eriksson K, Normark S. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249–264 [DOI] [PubMed] [Google Scholar]
- 14. Bajaj V, Lucas RL, Hwang C, Lee CA. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703–714 [DOI] [PubMed] [Google Scholar]
- 15. García Véscovi E, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174 [DOI] [PubMed] [Google Scholar]
- 16. Humphreys S, Rowley G, Stevenson A, Anjum MF, Woodward MJ, Gilbert S, Kormanec J, Roberts M. 2004. Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect. Immun. 72:4654–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 18. Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385–407 [DOI] [PubMed] [Google Scholar]
- 19. Sondermann H, Shikuma NJ, Yildiz FH. 2012. You've come a long way: c-di-GMP signaling. Curr. Opin. Microbiol. 15:140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Steenackers H, Hermans K, Vanderleyden J, De Keersmaecker SCJ. 2011. Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res. Int. 45:502–531 [Google Scholar]
- 21. Ahmad I, Lamprokostopoulou A, Le Guyon S, Streck E, Barthel M, Peters V, Hardt WD, Romling U. 2011. Complex c-di-GMP signaling networks mediate transition between virulence properties and biofilm formation in Salmonella enterica serovar Typhimurium. PLoS One 6:e28351 doi:10.1371/journal.pone.0028351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mantis NJ, Rol N, Corthésy B. 2011. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 4:603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michetti P, Mahan MJ, Slauch JM, Mekalanos JJ, Neutra MR. 1992. Monoclonal secretory immunoglobulin A protects mice against oral challenge with the invasive pathogen Salmonella typhimurium. Infect. Immun. 60:1786–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Michetti P, Porta N, Mahan MJ, Slauch JM, Mekalanos JJ, Blum AL, Kraehenbuhl JP, Neutra MR. 1994. Monoclonal immunoglobulin A prevents adherence and invasion of polarized epithelial cell monolayers by Salmonella typhimurium. Gastroenterology 107:915–923 [DOI] [PubMed] [Google Scholar]
- 25. Iankov ID, Petrov DP, Mladenov IV, Haralambieva IH, Mitov IG. 2002. Lipopolysaccharide-specific but not anti-flagellar immunoglobulin A monoclonal antibodies prevent Salmonella enterica serotype Enteritidis invasion and replication within HEp-2 cell monolayers. Infect. Immun. 70:1615–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Forbes SJ, Eschmann M, Mantis NJ. 2008. Inhibition of Salmonella enterica serovar Typhimurium motility and entry into epithelial cells by a protective antilipopolysaccharide monoclonal immunoglobulin A antibody. Infect. Immun. 76:4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mantis NJ, Forbes SJ. 2010. Secretory IgA: arresting microbial pathogens at epithelial borders. Immunol. Invest. 39:383–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Forbes SJ, Martinelli D, Hsieh C, Ault JG, Marko M, Mannella CA, Mantis NJ. 2012. Association of a protective monoclonal IgA with the O antigen of Salmonella enterica serovar Typhimurium impacts type 3 secretion and outer membrane integrity. Infect. Immun. 80:2454–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452–1463 [DOI] [PubMed] [Google Scholar]
- 30. Prouty AM, Schwesinger WH, Gunn JS. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70:2640–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gibson DL, White AP, Snyder SD, Martin S, Heiss C, Azadi P, Surette M, Kay WW. 2006. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J. Bacteriol. 188:7722–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanchez-Torres V, Hu H, Wood TK. 2011. GGDEF proteins YeaI, YedQ, and YfiN reduce early biofilm formation and swimming motility in Escherichia coli. Appl. Microbiol. Biotechnol. 90:651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mantis NJ, McGuinness CR, Sonuyi O, Edwards G, Farrant SA. 2006. Immunoglobulin A antibodies against ricin A and B subunits protect epithelial cells from ricin intoxication. Infect. Immun. 74:3455–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stringer AM, Singh N, Yermakova A, Petrone BL, Amarasinghe JJ, Reyes-Diaz L, Mantis NJ, Wade JT. 2012. FRUIT, a scar-free system for targeted chromosomal mutagenesis, epitope tagging, and promoter replacement in Escherichia coli and Salmonella enterica. PLoS One 7:e44841 doi:10.1371/journal.pone.0044841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amarasinghe JJ, Connell TD, Scannapieco FA, Haase EM. 2012. Novel iron-regulated and Fur-regulated small regulatory RNAs in Aggregatibacter actinomycetemcomitans. Mol. Oral Microbiol. 27:327–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Römling U. 2001. Genetic and phenotypic analysis of multicellular behavior in Salmonella typhimurium. Methods Enzymol. 336:48–59 [DOI] [PubMed] [Google Scholar]
- 38. White AP, Surette MG. 2006. Comparative genetics of the rdar morphotype in Salmonella. J. Bacteriol. 188:8395–8406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guard-Petter J. 2001. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 3:421–430 [DOI] [PubMed] [Google Scholar]
- 40. Solano C, Garcia B, Valle J, Berasain C, Ghigo JM, Gamazo C, Lasa I. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793–808 [DOI] [PubMed] [Google Scholar]
- 41. Römling U, Bian Z, Hammar M, Sierralta WD, Normark S. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaplan JB, Velliyagounder K, Ragunath C, Rohde H, Mack D, Knobloch JK, Ramasubbu N. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 186:8213–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Amarasinghe JJ, Scannapieco FA, Haase EM. 2009. Transcriptional and translational analysis of biofilm determinants of Aggregatibacter actinomycetemcomitans in response to environmental perturbation. Infect. Immun. 77:2896–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Waters CM, Lu W, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190:2527–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, Fetherston JD, Mack D, Goldman WE, Gomelsky M, Perry RD. 2011. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 79:533–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ellermeier CD, Ellermeier JR, Slauch JM. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57:691–705 [DOI] [PubMed] [Google Scholar]
- 47. Slauch JM, Lee AA, Mahan MJ, Mekalanos JJ. 1996. Molecular characterization of the oafA locus responsible for acetylation of Salmonella typhimurium O-antigen: oafA is a member of a family of integral membrane trans-acylases. J. Bacteriol. 178:5904–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Perry JL, Miller GR. 1989. Quality control slide for potassium hydroxide and cellufluor fungal preparations. J. Clin. Microbiol. 27:1411–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 50. Römling U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 62:1234–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Garcia B, Latasa C, Solano C, Garcia-del Portillo F, Gamazo C, Lasa I. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54:264–277 [DOI] [PubMed] [Google Scholar]
- 52. Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199 [PubMed] [Google Scholar]
- 53. Arnqvist A, Olsén A, Normark S. 1994. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13:1021–1032 [DOI] [PubMed] [Google Scholar]
- 54. Römling U, Rohde M, Olsén A, Normark S, Reinköster J. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10–23 [DOI] [PubMed] [Google Scholar]
- 55. Apter FM, Michetti P, Winner LS, III, Mack JA, Mekalanos JJ, Neutra MR. 1993. Analysis of the roles of antilipopolysaccharide and anti-cholera toxin immunoglobulin A (IgA) antibodies in protection against Vibrio cholerae and cholera toxin by use of monoclonal IgA antibodies in vivo. Infect. Immun. 61:5279–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wijburg OL, Uren TK, Simpfendorfer K, Johansen FE, Brandtzaeg P, Strugnell RA. 2006. Innate secretory antibodies protect against natural Salmonella typhimurium infection. J. Exp. Med. 203:21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mestecky J, Russell MW. 2009. Specific antibody activity, glycan heterogeneity and polyreactivity contribute to the protective activity of S-IgA at mucosal surfaces. Immunol. Lett. 124:57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Costerton JW. 1995. Overview of microbial biofilms. J. Ind. Microbiol. 15:137–140 [DOI] [PubMed] [Google Scholar]
- 59. Prouty AM, Gunn JS. 2003. Comparative analysis of Salmonella enterica serovar Typhimurium biofilm formation on gallstones and on glass. Infect. Immun. 71:7154–7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mouslim C, Hilbert F, Huang H, Groisman EA. 2002. Conflicting needs for a Salmonella hypervirulence gene in host and non-host environments. Mol. Microbiol. 45:1019–1027 [DOI] [PubMed] [Google Scholar]
- 61. Römling U, Yaron PDS. 2007. Biofilms of Salmonella enterica, p 127–145 In Rhen DMM, Mastroeni P, Threlfall EJ. (ed), Salmonella molecular biology and pathogenesis. Horizon Press, Norfolk, United Kingdom [Google Scholar]
- 62. Stepanović S, Cirković I, Ranin L, Svabić-Vlahović M. 2004. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett. Appl. Microbiol. 38:428–432 [DOI] [PubMed] [Google Scholar]
- 63. Ledeboer NA, Jones BD. 2005. Exopolysaccharide sugars contribute to biofilm formation by Salmonella enterica serovar Typhimurium on HEp-2 cells and chicken intestinal epithelium. J. Bacteriol. 187:3214–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Crull K, Rohde M, Westphal K, Loessner H, Wolf K, Felipe-López A, Hensel M, Weiss S. 2011. Biofilm formation by Salmonella enterica serovar Typhimurium colonizing solid tumours. Cell. Microbiol. 13:1223–1233 [DOI] [PubMed] [Google Scholar]
- 65. Latasa C, Roux A, Toledo-Arana A, Ghigo JM, Gamazo C, Penadés JR, Lasa I. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58:1322–1339 [DOI] [PubMed] [Google Scholar]
- 66. Ledeboer NA, Frye JG, McClelland M, Jones BD. 2006. Salmonella enterica serovar Typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect. Immun. 74:3156–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kader A, Simm R, Gerstel U, Morr M, Römling U. 2006. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60:602–616 [DOI] [PubMed] [Google Scholar]
- 68. Gerstel U, Römling U. 2001. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ. Microbiol. 3:638–648 [DOI] [PubMed] [Google Scholar]
- 69. Olsén A, Wick MJ, Mörgelin M, Björck L. 1998. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect. Immun. 66:944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Römling U, Bokranz W, Rabsch W, Zogaj X, Nimtz M, Tschäpe H. 2003. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol. 293:273–285 [DOI] [PubMed] [Google Scholar]
- 71. Bollinger RR, Everett ML, Palestrant D, Love SD, Lin SS, Parker W. 2003. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 109:580–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Boles BR, McCarter LL. 2002. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 184:5946–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lamprokostopoulou A, Monteiro C, Rhen M, Römling U. 2010. Cyclic di-GMP signalling controls virulence properties of Salmonella enterica serovar Typhimurium at the mucosal lining. Environ. Microbiol. 12:40–53 [DOI] [PubMed] [Google Scholar]
- 74. Simm R, Lusch A, Kader A, Andersson M, Römling U. 2007. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123–1134 [DOI] [PubMed] [Google Scholar]
- 76. Solano C, García B, Latasa C, Toledo-Arana A, Zorraquino V, Valle J, Casals J, Pedroso E, Lasa I. 2009. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 106:7997–8002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Iyer LM, Koonin EV, Aravind L. 2003. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Anantharaman V, Aravind L. 2000. Cache—a signaling domain common to animal Ca(2+)-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 25:535–537 [DOI] [PubMed] [Google Scholar]
- 79. Galperin MY. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188:4169–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Galperin MY, Nikolskaya AN, Koonin EV. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11–21 [DOI] [PubMed] [Google Scholar]
- 81. Goh YS, Grant AJ, Restif O, McKinley TJ, Armour KL, Clark MR, Mastroeni P. 2011. Human IgG isotypes and activating Fcgamma receptors in the interaction of Salmonella enterica serovar Typhimurium with phagocytic cells. Immunology 133:74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Boullier S, Tanguy M, Kadaoui KA, Caubet C, Sansonetti P, Corthésy B, Phalipon A. 2009. Secretory IgA-mediated neutralization of Shigella flexneri prevents intestinal tissue destruction by down-regulating inflammatory circuits. J. Immunol. 183:5879–5885 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.