Abstract
In response to environmental fluctuations or stresses, bacteria can activate transcriptional and phenotypic programs to coordinate an adaptive response. The intracellular pathogen Legionella pneumophila converts from a noninfectious replicative form to an infectious transmissive form when the bacterium encounters alterations in either amino acid concentrations or fatty acid biosynthesis. Here, we report that L. pneumophila differentiation is also triggered by nicotinic acid, a precursor of the central metabolite NAD+. In particular, when replicative L. pneumophila are treated with 5 mM nicotinic acid, the bacteria induce numerous transmissive-phase phenotypes, including motility, cytotoxicity toward macrophages, sodium sensitivity, and lysosome avoidance. Transcriptional profile analysis determined that nicotinic acid induces the expression of a panel of genes characteristic of transmissive-phase L. pneumophila. Moreover, an additional 213 genes specific to nicotinic acid treatment were altered. Although nearly 25% of these genes lack an assigned function, the gene most highly induced by nicotinic acid treatment encodes a putative major facilitator superfamily transporter, Lpg0273. Indeed, lpg0273 protects L. pneumophila from toxic concentrations of nicotinic acid as judged by analyzing the growth of the corresponding mutant. The broad utility of the nicotinic acid pathway to couple central metabolism and cell fate is underscored by this small metabolite's modulation of gene expression by diverse microbes, including Candida glabrata, Bordetella pertussis, Escherichia coli, and L. pneumophila.
INTRODUCTION
Normally found in fresh water systems as a parasite of protozoa, Legionella pneumophila can also infect alveolar macrophages to cause the life-threatening pneumonia Legionnaires' disease (1). Moreover, in aquatic environments, L. pneumophila can either exist as a planktonic cell or persist within a sessile biofilm community (2). To survive within these diverse ecological niches, L. pneumophila has evolved methods to swiftly adapt to changing conditions by modifying its cellular physiology and morphology in a process known as differentiation (3). Under nutrient-rich conditions, the posttranscriptional regulator CsrA suppresses transmissive traits and activates regulatory pathways that enable robust replication (4, 5). When conditions deteriorate, L. pneumophila synthesizes the second messenger ppGpp to halt proliferation and induce the expression of transmission genes (6–9). Concomittantly, the LetA/LetS two-component system induces the expression of the regulatory RNAs RsmY and RsmZ, which then relieve CsrA repression of transmissive traits (5, 10–12). As a result, L. pneumophila responds to metabolic stress by inducing traits that promote transmission and survival in the harsh environment, including motility, cytotoxicity toward macrophages, resistance to heat and osmotic shock, and the ability to avoid lysosomes (10, 13–17).
To acclimate to local conditions, L. pneumophila must monitor both the external milieu and its own physiological state and then translate a perceived stimulus into a coordinated response. Indeed, when amino acids are depleted, the stringent response enzyme RelA senses the accumulation of uncharged tRNAs at the ribosome and produces the alarmone ppGpp (6, 8, 9). Additionally, L. pneumophila can monitor flux in fatty acid biosynthesis through an interaction between a second stringent response enzyme, SpoT, and a central component of fatty acid metabolism, acyl carrier protein (6, 7, 18, 19). Thus, the RelA and SpoT enzymes equip L. pneumophila to assess its metabolic state and, when necessary, initiate transmission to a new niche. Since L. pneumophila persists in a variety of aquatic and soil environments, it is conceivable that metabolic cues other than amino acids and fatty acids also induce its differentiation.
One common way that microbes respond to external stimuli is via two-component signal transduction systems (20). For many two-component systems, the cues that initiate autophosphorylation and the subsequent phosphorelay are unknown. However, it is predicted that numerous environmental stimuli or conditions can activate these systems (20). At the core of L. pneumophila's differentiation circuitry is LetA/LetS, a two-component system that regulates all known transmission traits (10, 17, 21–23). Although the precise signal that activates the LetA/LetS system has not been identified, genetic studies indicate that multiple cues operate via the phosphorelay to trigger L. pneumophila phase differentiation.
The pyridine derivative nicotinic acid can modulate the activity of microbial two-component systems and, consequently, regulate the genes and phenotypes that are governed by these regulatory proteins. Notably, studies in Bordetella pertussis have deduced that nicotinic acid represses a spectrum of gene expression states and virulence factors, such as pertussis toxin, adenylate cyclase toxin, and filamentous hemagglutinin (24–27). Moreover, the Bordetella two-component system BvgA/BvgS, which controls most known virulence and colonization factors, is inactive when nicotinic acid is present in the medium (28). Likewise, nicotinic acid also regulates the Escherichia coli EvgA/EvgS system, which confers multidrug resistance and acid tolerance (29–34). Both the B. pertussis BvgA/BvgS and the E. coli EvgA/EvgS systems belong to a family of proteins that employ a multistep phosphorelay to activate their response pathways, but the mechanism by which nicotinic acid modulates these two-component systems is not understood.
Since the LetA/LetS system belongs to the same family of signal-transducing proteins as BvgA/BvgS and EvgA/EvgS (22), we postulated that nicotinic acid might similarly modulate the expression of L. pneumophila transmission genes and phenotypes. To test this hypothesis, we performed phenotypic and transcriptional profile analyses of L. pneumophila treated with this small metabolite, which together identified a putative membrane transporter that enhances the pathogen's tolerance of excess nicotinic acid.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and reagents.
E. coli DH5α, HB101, and derivatives were cultured using standard laboratory conditions. L. pneumophila strain Lp02 (thyA hsdR rpsL; MB110), a virulent thymine auxotroph derived from the serogroup 1 clinical isolate Philadelphia 1 (35), was the parental strain for all mutants constructed (Table 1). MB355 contains the pflaG plasmid that encodes thymidylate synthetase as a selectable marker and a transcriptional fusion of the flaA promoter to the green fluorescent protein (GFP) gene (8, 10). MB414 contains letA-22, and MB417 encodes letS-36, mariner insertion alleles of lpg2646 and lpg1912, respectively, that confer resistance to kanamycin (10). Both MB414 and MB417 contain the pflaG reporter plasmid. For phenotypic assays, bacteria were cultured at 37°C in 5 ml of N-(2-acetamido)-2-aminoethanesulfonic acid (ACES; Sigma)-buffered yeast extract (AYE) broth. When necessary, cultures were supplemented with 100 μg/ml thymidine. In all experiments, replicative- or exponential (E)-phase cultures were defined as having an optical density at 600 nm (OD600) of 0.5 to 0.85. Transmissive- or post-exponential (PE)-phase cultures were defined as having an OD600 of 3.4 to 4.5. To obtain CFU, L. pneumophila bacteria were plated on ACES-buffered charcoal-yeast extract agar supplemented with 100 μg/ml thymidine (CYET) and incubated for 4 to 5 days at 37°C. Chloramphenicol (5 and 25 μg/ml), gentamicin (10 μg/ml), ampicillin (100 μg/ml), streptomycin (0.5 and 1 mg/ml), and metronidazole (10 μg/ml) were used for selection of L. pneumophila and E. coli, respectively.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype/phenotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− endA1 hsdR17 (r− m+) supE44 thi-l recA1 gryA (Nal1) relA1 Δ(lacZYA-argF−)U169 ϕ80dlacZΔM15 λpirRK6 | Laboratory Collection |
| DY330 | W3110 ΔlacU169 gal490 λcI857 Δ(cro-bioA) | 77 |
| MB739 | HB101 endA::frt | 36 |
| MB741 | DH5α pMMBGent | 36 |
| MB776 | HB101 λpir endA::frt pR6KcatrdxArpsL | 36 |
| MB870 | DH5α pGEMlpg0272-3 | This work |
| MB871 | DH5α pGEMlpg0272::FRT-cat-FRT | This work |
| MB872 | HB101 endA::frt pGEMlpg0273::cat-rdxA-rpsL | This work |
| MB873 | DH5α pMMBGent-lpg0273 | This work |
| L. pneumophila strains | Wild type; thyA hsdR rpsL | 35 |
| MB110 | ||
| MB355 | MB110 pflaG | 8 |
| MB414 | letA::kan pflaG | 10 |
| MB417 | letS::kan pflaG | 10 |
| MB860 | Lp02 pMMBGent | This work |
| MB861 | Lp02 pMMBGent-lpg0273 | This work |
| MB862 | Lp02 Δlpg0273 pMMBGent | This work |
| MB863 | Lp02 Δlpg0273 pMMBGent-lpg0273 | This work |
| MB864 | Lp02 Δlpg0272::FRT-cat-FRT | This work |
| MB865 | Lp02 Δlpg0272::FRT | This work |
| MB866 | Lp02 lpg0273::cat-rdxA-rpsL | This work |
| MB867 | Lp02 Δlpg0273 | This work |
| Plasmids | ||
| pGEM-T easy | Multiple cloning site within coding region of β-lactamase α fragment linearized with single-T overhangs, 3 kb; Ampr | Promega |
| pflaG | 150-bp flaA promoter fragment fused to GFP gene, encodes thymidylate synthetase, 10.5 kb; Ampr | 8 |
| pKD3 | FRT-cat-FRT allele | 78 |
| pMMBGent | Broad-host-range plasmid pMMB67EH encoding gentamicin resistance | 8 |
Generation of lpg0272 and lpg0273 mutants.
A shuttle vector for IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible expression of lpg0273 was constructed by standard methods using pMMBGent (pMB741) (8). An isogenic mutant of lpg0272 in strain Lp02 was generated by constructing recombinant alleles in E. coli after cloning into pGEM-T easy (Promega) and replacing the desired sequence with a resistance cassette by recombineering (36). An in-frame unmarked deletion in lpg0273 was made by first replacing the first 1,200 bp of the coding sequence with a cat-rdxA-rpsL cassette by recombineering and verifying the phenotype in HB101 λpir endA::frt (MB740) (36, 37). The recombinant selectable/double counterselectable allele was then transferred to Lp02 by natural transformation (38), yielding lpg0273::cat-rdxA-rpsL (MB866). To remove the cassette, an allele with a precise 1,200-bp deletion was constructed by overlap extension PCR, natural transformation of MB866, and selection on metronidazole- and streptomycin-containing agar. The resulting in-frame deletion was confirmed by antibiotic resistance phenotype, PCR, and sequencing.
Fluorimetry.
To monitor expression of the flagellin promoter, L. pneumophila strain MB355, which contains the pflaG reporter plasmid, was cultured in AYE medium without thymidine at 37°C on a rotating wheel. When cultures reached an OD600 of 0.50 to 0.85 (t = 0) they were supplemented with either 5 mM nicotinic acid or water, and, at the times indicated in Fig. 1 and 3 the cell density of each culture was measured at OD600. To analyze similar bacterial concentrations, aliquots were collected by centrifugation, and the cell densities were normalized to an OD600 of 0.01 in phosphate-buffered saline (PBS). An aliquot of each sample (200 μl) was transferred to black 96-well plates (Costar), and the relative fluorescence intensity was measured using a Synergy HT microplate reader (485 nm excitation, 530 nm emission, and sensitivity of 50).
Fig 1.
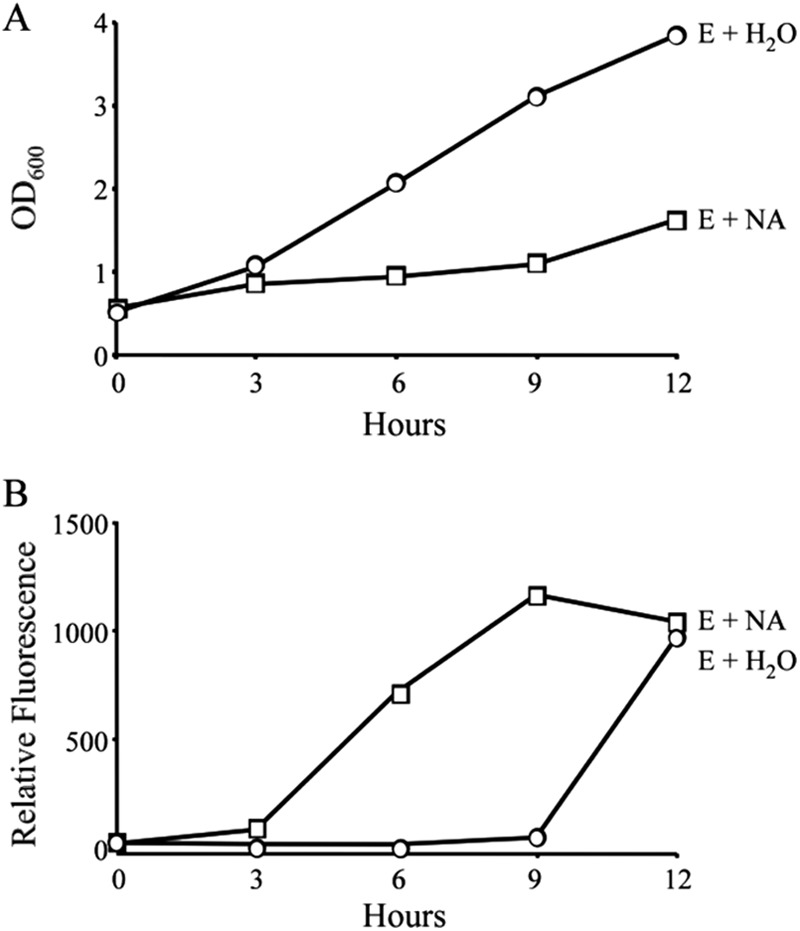
Nicotinic acid treatment triggers growth inhibition and premature motility. E-phase cultures of WT L. pneumophila carrying the flaA-GFP gene reporter were supplemented with water (H2O) or 5 mM nicotinic acid (NA). At the times indicated, samples of each culture were collected to quantify bacterial growth as determined by optical density (A) and expression of the transmission gene flaA by relative fluorescence (B). Shown are representative graphs from three or more independent experiments. At 6 and 9 h posttreatment, the optical densities and fluorescence levels were statistically different between E-phase cultures treated with water and E-phase bacteria treated with 5 mM NA (P < 0.05).
Fig 3.
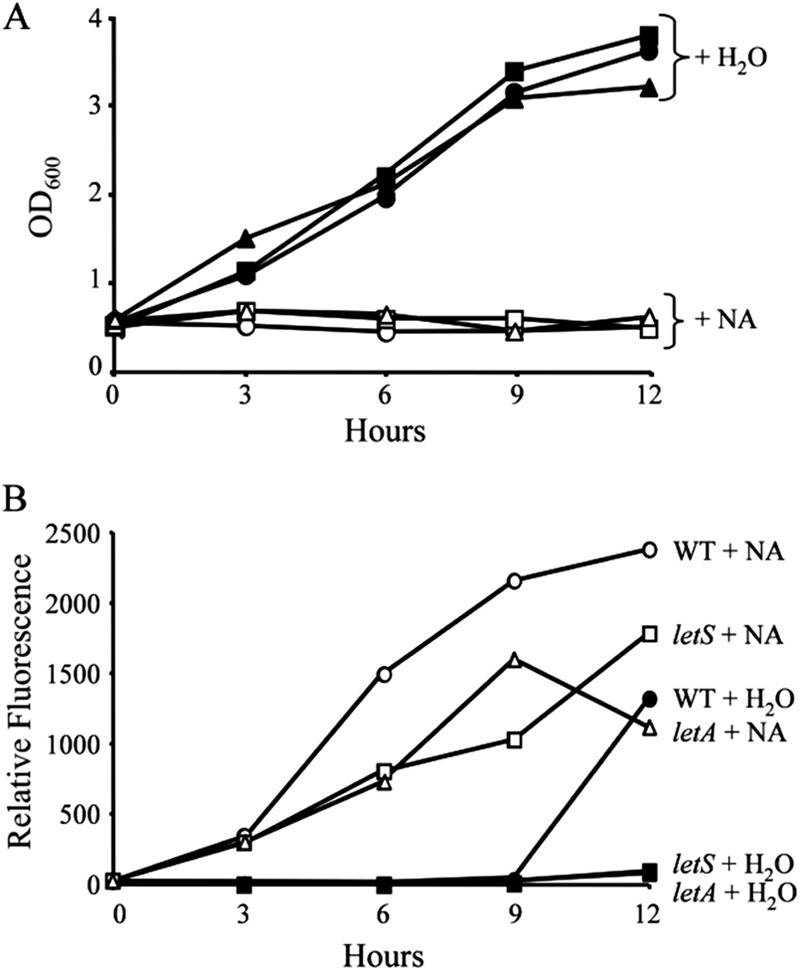
LetA/LetS is dispensable for growth inhibition but required for full induction of motility in response to nicotinic acid treatment. WT (circles), letA (triangles) or letS (squares) L. pneumophila cells containing the flaA-GFP gene reporter were cultured to E phase and then supplemented with water (closed symbols) or 10 mM nicotinic acid (NA; open symbols). At the times indicated, samples were collected and growth was analyzed as OD600 (A) and flaA expression as relative fluorescence (B). Similar fluorescence patterns were observed in three independent experiments using two different fluorometers. At time points after 0 h, the OD600 values for strains treated with NA were significantly different than those for strains treated with water (P < 0.05).
Macrophage cultures.
Macrophages derived from bone marrow obtained from femurs of female A/J mice (Jackson Laboratory) were cultured in RPMI 1640 containing 10% heat-inactivated fetal bovine serum (RPMI-FBS; Gibco BRL) as previously described (39). Following a 7-day incubation in L-cell supernatant-conditioned medium, macrophages were plated at either 2.5 × 105 or 5 × 104 per well for lysosomal degradation and cytotoxicity assays, respectively.
Lysosomal degradation.
The percentage of intracellular L. pneumophila cells that remained intact following a 2-h macrophage infection was quantified by fluorescence microscopy. Briefly, 2.5 × 105 macrophages were plated onto each coverslip in 24-well plates. Then, E-phase microbes treated for 3 h with either water or 5 mM nicotinic acid or PE-phase bacteria were added to macrophage monolayers at a multiplicity of infection (MOI) of ∼1. The cells were centrifuged at 400 × g for 10 min at 4°C and then incubated for 2 h at 37°C. Extracellular bacteria were removed by washing the monolayers with RPMI-FBS three times, and the macrophages were fixed, permeabilized, and stained for L. pneumophila as described previously (40). For each sample, at least 100 macrophages on duplicate coverslips were scored for intact rods versus degraded particles in three independent experiments (5, 13).
Cytotoxicity.
To determine the contact-dependent cytotoxicity of L. pneumophila, E-phase cultures supplemented for 3 h with water or 5 mM nicotinic acid or PE-phase bacteria were added to macrophage monolayers at the MOIs indicated in Fig. 2B. After centrifugation at 400 × g for 10 min at 4°C (40), the cells were incubated at 37°C for 1 h. To quantify macrophage viability, RPMI-FBS containing 10% alamarBlue (Trek Diagnostic Systems) was added to the monolayers for 6 to 12 h, and the reduction of the colorimetric dye was measured spectrophotometrically as described previously (41). Each sample was analyzed in triplicate wells in three or more independent experiments.
Fig 2.
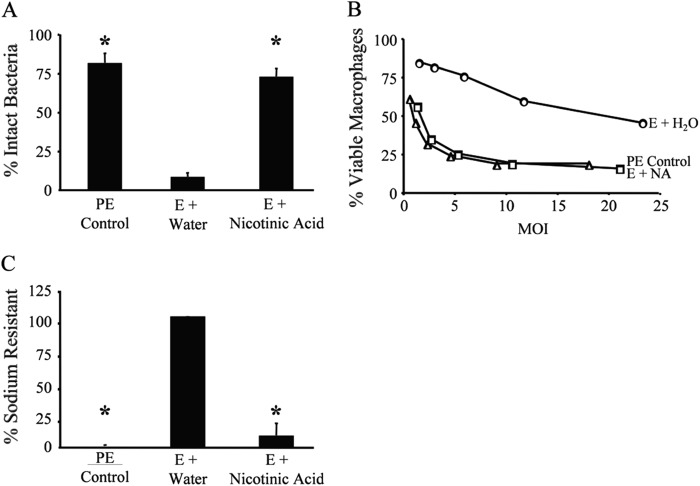
Nicotinic acid treatment induces WT L. pneumophila to express multiple transmissive-phase traits. (A) The extent of lysosome evasion after a 2-h infection of macrophages with L. pneumophila at an MOI of 1 was quantified as the percentage of intracellular bacteria that were intact, observed by fluorescence microscopy. Displayed are the mean results from duplicate samples in three independent experiments. Error bars indicate standard deviations (SD), and asterisks designate significant differences (P < 0.01) compared to the results for the E-phase water control. (B) Macrophage viability after 1-h infection of macrophages at the multiplicities of infection (MOI) shown was quantified with PE-phase (triangles) or E-phase cultures supplemented with water (H2O) or nicotinic acid (NA) using the colorimetric dye alamarBlue. The values plotted represent the means ± SD for triplicate samples determined in one of three similar experiments. At similar MOIs, E-phase cultures supplemented with NA were significantly different than E-phase cultures supplemented with water (P < 0.05). E-phase cultures treated with NA were not statistically different than PE-phase bacteria (P > 0.05). (C) The percentage of sodium-resistant bacteria was measured by plating cultures on medium with or without 100 mM NaCl and then calculating CFU as described previously (41). Values shown = mean [(E + NA or PE control)/(E + H2O) × 100]. Error bars represent SD from duplicate samples in three independent experiments; for E-phase + water results, the error bar is too small to be visible. Asterisks denote statistically significant differences (P < 0.01) compared to the results for the E-phase water control.
Sodium sensitivity.
To calculate the percentage of L. pneumophila cells that are sensitive to sodium, E-phase cultures supplemented for 3 h with either water or 5 mM nicotinic acid or PE-phase bacteria were plated onto CYET and CYET containing 100 mM NaCl. After a 6-day incubation at 37°C, CFU were enumerated and the percentage of sodium-sensitive microbes calculated as described previously (41).
Statistical analyses for phenotypic assays.
To calculate P values for lysosomal degradation and sodium sensitivity assays, one-way analysis of variance (ANOVA) was used for at least 3 independent samples. To calculate P values for cytotoxicity, optical density, and fluorescence assays, t tests were performed on 3 independent samples at similar MOIs or time points.
RNA isolation, RNA labeling, and microarray hybridization.
Wild-type (WT) L. pneumophila cells were cultured on an orbital shaker at 37°C to an OD600 of 0.65 in 150 ml AYE containing 100 μg/ml thymidine. Upon reaching the appropriate optical density, cultures were supplemented with either water or 5 mM nicotinic acid and then incubated for an additional 3 h at 37°C on an orbital shaker. Next, 10-ml aliquots were centrifuged at 6,000 × g for 2 min at 4°C. The culture supernatants were discarded, and the pellets stored at −80°C. Total RNA was extracted using TRIzol (Invitrogen) as described previously (42). The RNA was reverse transcribed and labeled with Cy3 or Cy5 according to the manufacturer's instructions (Amersham Biosciences). The transcription profile was analyzed using microarrays designed to contain gene-specific 70-mer oligonucleotides to represent all predicted genes within the genome of L. pneumophila strain Paris (CR628336) and its plasmid (CR628338) as described previously (43). Hybridizations were performed following the manufacturer's recommendations (Corning) using 250 pmol of Cy3- and Cy5-labeled cDNA. Slides were scanned on a GenePix 4000A scanner (Axon Instruments), and the laser power and photomultiplier tubes (PMTs) were adjusted to balance the two channels. The resulting files were analyzed using Genepix Pro 5.0 software. Spots were excluded from analysis if they contained high background fluorescence, slide abnormalities, or weak intensity. To obtain statistical data for the gene expression profiles, all microarrays were performed in duplicate with a dye swap.
Data and statistical analysis for microarrays.
Data normalization and differential analysis were conducted using R software (http://www.R-project.org). No background subtraction was performed, but a careful graphical examination of all the slides was performed to ensure a homogeneous, low-level background in both channels. A loess normalization (44) was performed on a slide-by-slide basis (BioConductor package marray; http://bioconductor.org/packages/2.2/bioc/html/marray.html). Differential analysis was carried out separately for each comparison between the two time points, using the VarMixt package (45) together with the P value adjustment method pioneered by Benjamini and Yekutieli (46). Only genes with a 1.5-fold or greater change in two or more independent experiments were considered further. Empty and flagged spots were excluded from the data set, and only genes without missing values for the comparison of interest were analyzed. Coordinates for the unannotated genes in L. pneumophila strain Philadelphia 1 and their corresponding gene designations in strain Paris are listed in Table S1 in the supplemental material.
Growth curves.
Bacterial strains were cultured to mid-exponential phase in AYET. Cultures were diluted, and their cell densities normalized in triplicate. A Bioscreen growth curve analyzer, set at continuous shaking at medium amplitude, measured the optical density at 600 nm every hour over a 48-h time period.
RESULTS
Nicotinic acid inhibits L. pneumophila growth and induces motility.
To discern whether nicotinic acid triggers L. pneumophila differentiation, WT bacteria that contained the flaA-GFP gene reporter construct were cultured to the exponential (E) phase and supplemented with either water or 5 mM nicotinic acid. When E-phase cultures were treated with nicotinic acid, replication stopped (Fig. 1A, squares) and the flaA promoter was activated (Fig. 1B, squares). In contrast, control cultures supplemented with water did not induce the flaA promoter until after 9 h of treatment, the time when L. pneumophila normally transitions to the post-exponential (PE) phase (Fig. 1B, circles). In support of the fluorimetry data, direct microscopic examination of cells revealed that nicotinic acid also triggered motility of the majority of cells (data not shown). Thus, in response to a signal generated by exogenous nicotinic acid, replicating L. pneumophila cells initiate a phenotypic switch.
Nicotinic acid supplementation stimulates L. pneumophila differentiation.
To determine whether nicotinic acid induces transmission traits in addition to growth inhibition and motility, we tested nicotinic acid-treated L. pneumophila for other phenotypes characteristic of PE-phase cells, including avoidance of lysosomal degradation, cytotoxicity to macrophages, and sodium sensitivity (41). Indeed, when E-phase L. pneumophila cells were treated with 5 mM nicotinic acid, the majority of the bacteria acquired the capacity to evade lysosomes, as judged by immunofluorescence microscopy. While less than 15% of E-phase Legionella cells avoided degradation, more than 70% of E-phase bacteria exposed to 5 mM nicotinic acid remained intact (P < 0.01) (Fig. 2A). Furthermore, after nicotinic acid treatment, E-phase L. pneumophila cells became as cytotoxic to bone marrow-derived macrophages as PE-phase control cultures (P > 0.05) (Fig. 2B). Finally, 5 mM nicotinic acid also triggered sodium sensitivity of E-phase microbes (P < 0.01) (Fig. 2C). Taken together, our phenotypic data indicate that 5 mM nicotinic acid triggers differentiation of replicative L. pneumophila cells to the transmissive phenotype.
The LetA/LetS two-component system enhances the L. pneumophila response to nicotinic acid.
In B. pertussis and E. coli, nicotinic acid elicits transcriptional and phenotypic responses through a two-component system (30, 34). To analyze whether the response of L. pneumophila to nicotinic acid depends on the homologous LetA/LetS signal transduction system, we cultured letA and letS mutants containing the flaA-GFP gene reporter construct to the E phase, supplemented the cultures with nicotinic acid, and then monitored both culture density and flaA expression over time. When treated with nicotinic acid, the letA and letS mutants resembled WT L. pneumophila by restricting their growth (Fig. 3A). However, L. pneumophila required the LetA/LetS system for full induction of flagellin expression in response to nicotinic acid, since the corresponding mutants expressed the flaA-GFP gene at intermediate levels (Fig. 3B). Furthermore, microscopic examination demonstrated that L. pneumophila requires LetA/LetS to trigger motility in response to nicotinic acid (data not shown). Thus, following nicotinic acid exposure, L. pneumophila activates the flaA promoter and induces motility via processes that are augmented by the LetA/LetS two-component system. On the other hand, nicotinic acid inhibits the motility of B. pertussis and E. coli by signal transduction pathways initiated by BvgA/BvgS and EvgA/EvgS, respectively.
Transcriptome analysis indicates that profound changes in gene expression distinguish the replicative and transmissive phases.
To recognize genes that respond specifically to nicotinic acid, we first compared the transcriptional profiles of L. pneumophila cells in the replicative and the transmissive phase by employing multiple-genome (47, 48) DNA microarrays to analyze E- and PE-phase bacteria. Overall, 840 genes were more highly expressed in the PE phase (or, repressed in the E phase; see Table S2 in the supplemental material) and 714 L. pneumophila genes were repressed in the PE phase (or, elevated in the E phase; see Table S2 in the supplemental material). Importantly, our microarray analysis of strain Lp02 confirms transcriptional and regulatory analysis of other laboratory strains that indicate that L. pneumophila displays distinct gene expression patterns and phenotypes in the replicative and transmissive phases (41, 43, 49).
Several categories of genes were significantly induced in the PE phase compared to their levels of induction in the E phase (see Table S2 in the supplemental material). As expected, these include genes required for flagellum biosynthesis and motility (43, 50). In addition, numerous regulatory genes were activated in the PE phase, such as the sigma factors rpoS and rpoE, the transmission trait enhancer letE, and the stringent response enzyme relA (6, 9, 13–15, 51). Other regulatory genes that were preferentially expressed in the PE phase include several two-component systems and members of the GGDEF/EAL family of proteins. As expected, many genes of the Dot/Icm type IV secretion system had elevated levels of expression in the PE phase, notably, ralF, icmBCGOPRSTVX, dotBV, sidACDFGHH′, sdeBCD, and sdbABC (52). Finally, transcriptional analysis showed that numerous pilus genes, as well as genes encoding transporters and efflux pumps, were more highly expressed in PE than in E phase. Thus, our microarray analysis of the virulent Philadelphia-1 derivative Lp02 validates the model of the reciprocal phases of the L. pneumophila life cycle (3, 41), corroborating the demonstration by Brüggemann et al. and Weissenmayer et al. that the virulent Paris and Philadelphia-1 strains of L. pneumophila undergo a dramatic change in their gene expression profiles both in liquid cultures and during amoeboid infections (3, 41, 43, 53).
Microarrays determine that nicotinic acid regulates a panel of genes similar to PE L. pneumophila.
To learn how L. pneumophila adapts to nicotinic acid treatment, we employed microarrays to compare E-phase L. pneumophila treated with 5 mM nicotinic acid to similar cultures supplemented with water. Since nicotinic acid supplementation triggers L. pneumophila differentiation and transcriptional microarrays indicate that vastly different expression profiles are displayed between the replicative and transmissive phases, we predicted that the transcriptional program induced by nicotinic acid supplementation would also be extensive. Indeed, when E-phase bacteria were treated with nicotinic acid, the expression levels of 678 genes were significantly elevated and 429 genes were repressed compared to their expression in the water control (Fig. 4; also see Table S3 in the supplemental material). Similar to PE-phase L. pneumophila, in E-phase bacteria exposed to nicotinic acid, 17 genes of the flagellar apparatus were induced, including flaA, fliADKS, flgDFIJKLM, fleN, flhB, motA, and motB1B2 (Table 2; also see Table S3) (43, 50). Moreover, the expression of numerous genes that enhance virulence was activated by nicotinic acid supplementation, such as components of the Dot/Icm type IV secretion system (icmBDGJOX), substrates of the secretion system (ralF, lidA, sdcA, sdhB1, sdbAB, sdeBCD, and sidACDFGH), and enhanced entry proteins enhABC (Table 2; also see Table S3) (52). By comparing the microarray data from PE-phase bacteria to the transcriptional data from E-phase L. pneumophila treated with nicotinic acid, it was apparent that the profiles were comparable; bacteria from the two sample sets had in common 595 induced and 299 repressed genes (Fig. 4; also see Table S4). Accordingly, the phenotypic switch triggered by nicotinic acid supplementation is due to the induction of a transcriptional profile that is similar to that of PE-phase L. pneumophila.
Fig 4.
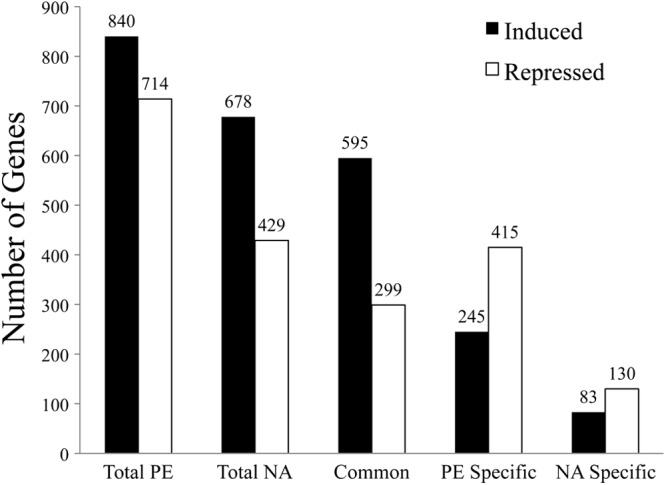
Comparison of genes regulated in the PE-phase and nicotinic acid-treated L. pneumophila. Genes with higher levels of transcriptional activation (black bars) and repression (white bars) than those of E-phase control cells were identified by microarray analysis for the following conditions: Total, genes that responded to either the PE phase or 3-h nicotinic acid treatment (NA); Common, genes that responded to both the PE phase and NA; PE specific, genes that responded to PE but not NA; NA specific, genes that responded to NA but not PE.
Table 2.
Subset of genes upregulated by nicotinic acid
| Functional categorya | Family or system | Locus tag | Name | Fold change |
|---|---|---|---|---|
| Virulence | lpg2157 | sdeA | 70.1 | |
| lpg0910 | enhA2 | 56.0 | ||
| lpg1496 | lem10 | 35.0 | ||
| lpg2509 | sdeD | 25.0 | ||
| lpg2511 | sidC | 20.2 | ||
| lpg2465 | sidD | 16.1 | ||
| lpg2153 | sdeC | 16.0 | ||
| lpg1950 | ralF | 12.3 | ||
| lpg1336 | enhA3 | 12.3 | ||
| lpg2862 | legC8 | 8.3 | ||
| lpg2510 | sdcA | 7.5 | ||
| lpg2482 | sdbB | 7.3 | ||
| lpg1116 | chiA | 7.0 | ||
| lpg2639 | enhC | 6.2 | ||
| lpg2640 | enhB | 5.4 | ||
| lpg0940 | lidA | 5.0 | ||
| lpg0436 | legA11 | 4.6 | ||
| lpg1355 | sidG | 4.6 | ||
| lpg1386 | 4.6 | |||
| lpg2641 | enhA4 | 4.0 | ||
| lpg1948 | legLC4 | 4.0 | ||
| lpg2793 | lepA | 3.3 | ||
| lpg0645 | rtxA-1 | 3.1 | ||
| lpg2689 | icmX | 2.9 | ||
| lpg2300 | ankH | 2.4 | ||
| lpg0525 | lvgA | 2.4 | ||
| lpg2829 | sidH | 2.3 | ||
| lpg2156 | sdeB | 2.3 | ||
| lpg0171 | legU1 | 2.2 | ||
| lpg0454 | icmD/dotP | 2.1 | ||
| lpg0446 | icmO/dotL | 2.0 | ||
| lpg0621 | sidA | 2.0 | ||
| lpg2584 | sidF | 1.8 | ||
| lpg0275 | sdbA | 1.8 | ||
| lpg2190 | 1.7 | |||
| lpg0455 | icmJ/dotN | 1.7 | ||
| lpg0136 | sdhB1 | 1.7 | ||
| lpg0456 | icmB/dotO | 1.6 | ||
| lpg0452 | icmG/dotF | 1.5 | ||
| Pilus | lpg0630 | pilW | 7.4 | |
| lpg1915 | pilE2 | 5.0 | ||
| lpg1522 | pilB | 4.6 | ||
| lpg1523 | pilC | 2.9 | ||
| lpg0632 | 2.4 | |||
| lpg0629 | pilX | 2.1 | ||
| lpg0930 | pilP | 1.9 | ||
| lpg0628 | 1.9 | |||
| lpg0627 | pilE3 | 1.6 | ||
| lpg1914 | pilE1 | 1.5 | ||
| Regulation | GGDEF/EAL | lpg0156 | 10.8 | |
| lpg0277 | 6.3 | |||
| lpg1168 | 5.3 | |||
| lpg0157 | 4.4 | |||
| lpg2642 | 4.4 | |||
| lpg1025 | 4.1 | |||
| lpg1057 | 2.6 | |||
| lpg0744 | 2.4 | |||
| lpg1518 | 2.3 | |||
| lpg0073 | 2.2 | |||
| lpg0029 | 2.1 | |||
| lpg2132 | 1.7 | |||
| lpg1114 | 1.6 | |||
| LysR | lpg1796 | 5.8 | ||
| lpg2383 | 5.0 | |||
| lpg2402 | 4.1 | |||
| lpg2138 | 2.6 | |||
| lpg2376 | 1.9 | |||
| lpg0274 | 1.5 | |||
| σ-Factors | lpg0907 | flgM | 4.1 | |
| lpg0476 | 2.6 | |||
| lpg0845 | 2.4 | |||
| lpg0477 | rpoN | 1.6 | ||
| lpg2667 | rpoH | 1.5 | ||
| lpg1782 | fliA | 4.7 | ||
| Two-component systems | lpg2181 | 31.8 | ||
| lpg0278 | 4.1 | |||
| lpg2145 | 3.7 | |||
| lpg2458 | shkA | 3.5 | ||
| lpg2457 | 2.5 | |||
| lpg2146 | stuC | 2.4 | ||
| lpg1292 | 2.2 | |||
| lpg0098 | 2.1 | |||
| lpg2180 | 1.9 | |||
| lpg1174 | pilR | 1.7 | ||
| Others | lpg1135 | 5.2 | ||
| lpg2524 | 3.8 | |||
| lpg1898 | 3.0 | |||
| lpg1003 | 2.9 | |||
| lpg0433 | 1.8 | |||
| lpg1212 | 1.8 | |||
| Stress response | lpg2493 | 74.5 | ||
| lpg0689 | 5.0 | |||
| lpg2192 | 4.4 | |||
| lpg1540 | uspA | 3.6 | ||
| lpg2191 | gspA | 3.5 | ||
| lpg0192 | hsp20 | 2.6 | ||
| lpg2850 | cspA | 1.9 | ||
| lpg0386 | htpX | 1.9 | ||
| Transport | lpg0273 | 35.0 | ||
| lpg0088 | 22.9 | |||
| lpg0037 | 11.6 | |||
| lpg1454 | 6.3 | |||
| lpg1095 | 6.1 | |||
| lpg1024 | copA1 | 5.3 | ||
| lpg0214 | 4.1 | |||
| lpg1555 | 4.0 | |||
| lpg1125 | 3.9 | |||
| lpg1030 | cecA1 | 3.4 | ||
| lpg2152 | 2.5 | |||
| lpg2516 | smlA | 2.2 | ||
| lpg1029 | cecA2 | 2.2 | ||
| lpg0267 | 2.2 | |||
| lpg2189 | 2.1 | |||
| lpg1123 | 2.1 | |||
| lpg0540 | 2.0 | |||
| lpg0842 | 2.0 | |||
| lpg1159 | 1.8 | |||
| lpg2739 | 1.8 | |||
| lpg1378 | 1.8 | |||
| lpg2126 | 1.8 | |||
| lpg2198 | 1.7 | |||
| lpg0841 | 1.6 | |||
| lpg1835 | 1.5 | |||
| Motility | lpg1340 | flaA | 144.1 | |
| lpg1338 | fliD | 13.4 | ||
| lpg1337 | fliS | 11.9 | ||
| lpg1226 | flgL | 11.8 | ||
| lpg1220 | flgF | 4.3 | ||
| lpg1225 | flgK | 2.8 | ||
| lpg2583 | flhB | 2.7 | ||
| lpg1224 | flgJ | 2.6 | ||
| lpg1218 | flgD | 2.4 | ||
| lpg2318 | motA2 | 2.3 | ||
| lpg1783 | fleN | 2.3 | ||
| lpg2319 | motB2 | 2.0 | ||
| lpg1688 | fliK | 2.0 | ||
| lpg1223 | flgI | 1.8 | ||
| lpg1780 | motB1 | 1.5 |
Functional categories were assigned based on NCBI (National Center for Biotechnology Information) or LegioList (Institut Pasteur) gene annotations or as determined from previously published reports.
Transcriptional analysis identifies genes unique for nicotinic acid modulation.
Nicotinic acid modulates the transcriptional and phenotypic profiles of B. pertussis and E. coli, but the mechanism by which nicotinic acid elicits a response is not understood. To identify the L. pneumophila genes and pathways that respond to nicotinic acid supplementation, we compared the transcriptional profiles of E-phase bacteria treated for 3 h with 5 mM nicotinic acid with that of similar cultures supplemented with water. Using the microarray data sets from replicative- and transmissive-phase L. pneumophila microbes as references, we identified 660 genes that responded specifically to the growth phase (Fig. 4, PE specific; also see Table S5 in the supplemental material), whereas 213 genes were specifically responsive to nicotinic acid supplementation (Fig. 4; also see Table S6). Of the 83 genes that were highly induced specifically following nicotinic acid treatment, nearly 40% have no predicted function. Several regulatory genes had elevated expression levels, including the two-component regulator lpg0098 and two transcriptional regulators from the MarR and LysR families, lpg1212 and lpg0274, respectively. Also, the induction of two virulence genes, icmD/dotP and icmJ/dotN (52), was particular to nicotinic acid addition.
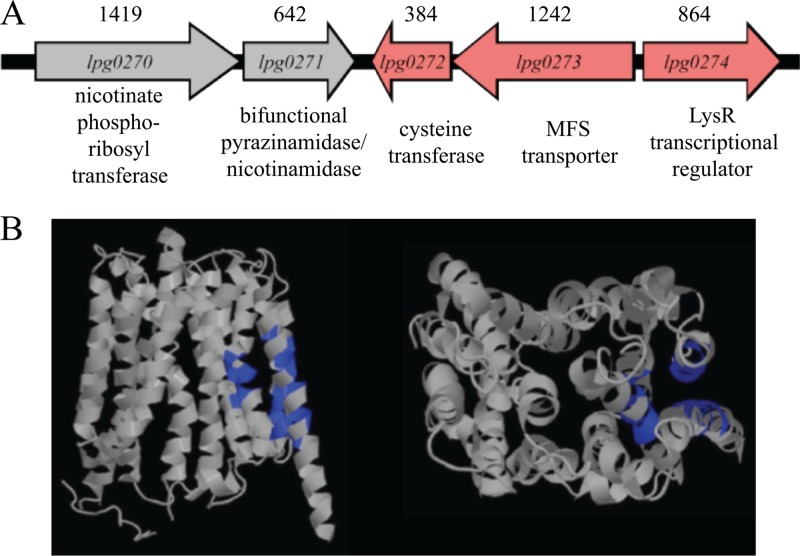
One striking observation was that the two genes with the highest levels of transcriptional activation in response to nicotinic acid lie in an operon; lpg0272 and lpg0273 were 9-fold and 35-fold elevated, respectively (Fig. 5; also see Table S6 in the supplemental material). The lpg0272 locus is annotated as a 14.6-kDa cysteine transferase, and lpg0273 is predicted to encode a 44.9-kDa membrane protein belonging to the major facilitator superfamily (MFS) of transporters (Fig. 5). Directly 3′ of the lpg0272-3 operon but positioned in the opposite orientation is the LysR transcriptional regulator (lpg0274), whose expression is also elevated in response to nicotinic acid (Fig. 5). Positioned 5′ of the lpg0272-3 operon but oriented in the opposite direction are two genes involved in nicotinate and nicotinamide metabolism, lpg0270-1 (Fig. 5). According to our microarray data, following a 3-h treatment with nicotinic acid, lpg0270-1 are neither induced nor repressed. Whether nicotinic acid affects lpg0270-1 expression at times other than 3 h has not been assessed. Together, our transcriptional data indicate that nicotinic acid regulates a distinct set of L. pneumophila genes. Here, we focused on the putative MFS transporter to determine whether it contributes to the bacterial response.
Fig 5.
Region of the L. pneumophila chromosome that encodes lpg0273 and proposed structure of Lpg0273. (A) Chromosomal locus of lpg0273. Arrows indicate orientation of the genes. Sizes (bp) of the genes are indicated above, and the predicted function of each gene is listed below. Red indicates genes with significantly elevated expression levels when treated with 5 mM nicotinic acid. Gray indicates genes that are not affected by a 3-h nicotinic acid treatment. (B) Structure of Lpg0273 and ligand binding site (blue) predicted using 3DLigandSite prediction software (71), showing helices typical of MFS transporter proteins.
lpg0273 enhances the response of L. pneumophila to nicotinic acid.
Protein alignment (54) revealed the highest degree of homology of Lpg0273 to a putative MFS from Coxiella burnetii (ZP_02219825.1), a species phylogenetically close to L. pneumophila (55). Structural prediction (56) yields significant similarity to the solved structure of the MFS multidrug transporter EmrD from E. coli (57). To test more directly whether Lpg0273 or Lpg0272 equips L. pneumophila to respond to nicotinic acid, isogenic mutants were constructed and their growth characterized after exposure to a range of concentrations of nicotinic acid. Bacteria containing mutations in lpg0272 resembled the wild type (data not shown). However, a mutant with an in-frame deletion of the lpg0273 transporter gene was consistently more sensitive to nicotinic acid than wild-type L. pneumophila, as assessed by the OD600 (Fig. 6). The growth defect correlated with the nicotinic acid dose and was complemented by lpg0273 in trans (Fig. 6). Furthermore, plasmid-borne lpg0273 conferred a slight growth advantage to either mutant or wild-type L. pneumophila, even in the absence of nicotinic acid (Fig. 6). Thus, to tolerate excessive nicotinic acid, L. pneumophila induces the expression of an MFS transporter gene encoded in a locus predicted to be dedicated to nicotinate and nicotinamide metabolism.
Fig 6.
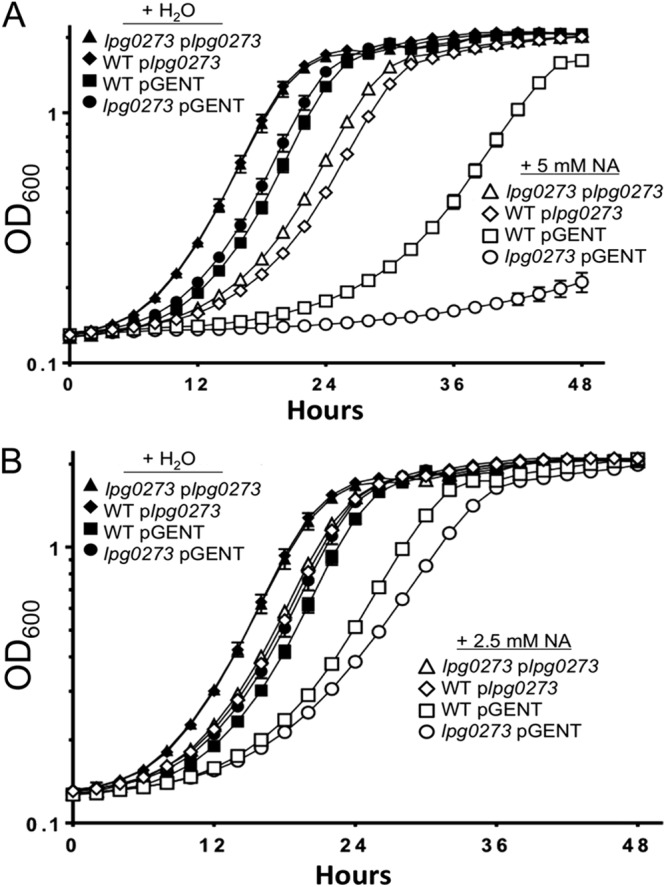
The lpg0273 MFS gene confers resistance to growth inhibition by nicotinic acid. WT or lpg0273 mutant L. pneumophila carrying plasmid-born lpg0273 (plpg0273) or the parent vector (pGent) were cultured in AYET plus 200 μM IPTG and supplemented with H2O or 5 mM (A) or 2.5 mM (B) nicotinic acid (NA), and the optical density was recorded by a Bioscreen growth curve analyzer at the times shown. Displayed are means ± SD calculated from triplicate wells in a single experiment that is representative of at least three independent experiments.
DISCUSSION
A variety of microbes modulate their transcriptional and phenotypic profiles in response to nicotinic acid. Among bacteria, the respiratory pathogen B. pertussis alters the expression of hundreds of genes and virulence traits according to the concentration of nicotinic acid that is present in the culture medium (24–27). In E. coli, nicotinic acid alters the expression of motility, the outer membrane porin protein OmpC, and the alcohol dehydrogenase protein AdhE (34, 58, 59). Here, we demonstrate that exogenous nicotinic acid triggers replicating L. pneumophila cells to differentiate to its transmissive phenotype. In particular, when treated with 5 mM nicotinic acid, L. pneumophila cells stopped dividing, activated the flagellin promoter, and became motile (Fig. 1 and data not shown), cytotoxic to macrophages, sensitive to sodium, and competent to evade lysosomal degradation (Fig. 2). In addition, L. pneumophila cells appear competent to export nicotinic acid, as deduced from the altered sensitivity of cells that either lack or carry extra copies of the putative MFS transporter gene lpg0273. Thus, L. pneumophila provides a tractable experimental system to investigate how bacteria sense and respond to nicotinic acid, a component of central metabolism and energy production pathways.
One of the best-understood mechanisms of microbial gene regulation by nicotinic acid operates in the urinary tract fungal pathogen Candida glabrata. In response to nicotinic acid limitation, its telomeric adhesion genes are derepressed by a class of proteins known as sirtuins (60). These highly conserved NAD+-dependent deacetylase enzymes regulate a variety of processes by affecting the acetylation and deacetylation states of their substrates (61). Sirtuin activity in vivo is thought to be controlled by the metabolite nicotinamide, the amide of nicotinic acid (62). In the case of C. glabrata, the histone deacetylase sirtuin protein Sir2p activates virulence genes encoded in the normally silenced telomeres. Because this pathogenic yeast lacks the enzymes needed to synthesize nicotinic acid, the exogenous supply of this metabolite controls where and when Candida glabrata adheres to host tissues (60). In organisms that are competent to generate nicotinic acid from endogenous or salvaged precursors, sirtuins typically link cellular processes to energy metabolism (63).
Some bacteria encode homologues of the sirtuin proteins (61, 64, 65). For Salmonella enterica, the sirtuin CobB controls the activity of acetyl-coenzyme A (CoA) synthetase (ACS), as well as the conversion of acetate to acetyl-coA (65, 66). In L. pneumophila, alterations in acetate concentrations can perturb the fatty acid biosynthetic pathway and trigger the expression of transmission traits (7). Since sirtuin proteins are conserved not only in eukaryotes but also in prokaryotes and archaea (61), we predict that, similar to S. enterica, L. pneumophila encodes a sirtuin-like protein that regulates acetate levels. By analogy to the C. glabrata pathway, excess nicotinic acid would activate an L. pneumophila sirtuin, which in turn would deacetylate a regulator(s) of fatty acid biosynthesis and microbial differentiation. While in silico analysis of the L. pneumophila genome has not revealed obvious sirtuin candidates, our transcriptional data identified numerous genes of unknown function that are induced following nicotinic acid treatment (see Table S6 in the supplemental material). Perhaps one or more of these genes encodes a novel protein with sirtuin-like activity.
To respond to nicotinic acid, B. pertussis, E. coli, and L. pneumophila each utilize a two-component system, BvgA/BvgS, EvgA/EvgS, and LetA/LetS, respectively. Although many two-component signal transduction systems are comprised of a two-step phosphorelay to activate gene expression, the BvgA/BvgS, EvgA/EvgS, and LetA/LetS systems each likely rely on a four-step phosphorelay to customize their expression profiles (22, 67). In particular, to induce robust motility in response to nicotinic acid, L. pneumophila requires both LetA and LetS (Fig. 3). Whether the residual response by single letA or letS mutants indicates that LetA and LetS interact with another sensor kinase or response regulator protein is an intriguing possibility that remains to be investigated. Numerous other microbes encode a member of this family of four-step signal transduction systems, including GacA/GacS of Acinetobacter baumannii, PigQ/PigW of Serratia marcescens, VarA/VarS of Vibrio cholerae, SirA/BarA of Salmonella, GacA/GacS of Pseudomonas, and the UvrY/BarA and TorR/TorS systems of E. coli (68–70). Whether other bacterial species that encode four-step two-component systems also gauge nicotinic acid concentrations to regulate their genotypic and phenotypic expression profiles warrants testing.
When present at 5 mM, nicotinic acid also acted independently of LetA/LetS to inhibit L. pneumophila growth (Fig. 3). At concentrations of >5 mM, nicotinic acid stopped replication but failed to trigger L. pneumophila differentiation, as judged by lack of lysosome avoidance, cytotoxicity, and sensitivity to sodium (data not shown). Whether higher concentrations of nicotinic acid directly repress genes that govern cellular differentiation or instead reduce viability by nonspecific pathways remains to be determined. Data from a variety of bacterial species clearly indicate that nicotinic acid alters microbial gene transcription and phenotypes, but how this metabolite exerts its effects has not been elucidated. For example, whether nicotinic acid activates four-step two-component regulatory proteins directly or indirectly is not yet known. Furthermore, although we identified an operon that suggests that L. pneumophila is equipped to activate transmission traits as a specific response to perturbations in NAD+ homeostasis (Fig. 5 and 6), whether the concomitant diminished growth rate also indirectly triggers bacterial differentiation remains to be investigated.
By analyzing the gene expression profiles of L. pneumophila cultures treated with nicotinic acid, we identified a putative MFS transport protein that equips L. pneumophila to adapt to excess exogenous nicotinic acid. First, among the 213 genes that responded specifically to nicotinic acid supplementation, the tandem genes lpg0272-3 were the most strongly induced (see Table S6 in the supplemental material). Second, gene annotation and protein prediction identified Lpg0273 as a member of the major facilitator superfamily of proteins (48, 54, 71). In particular, Lpg0273 resembles the multidrug efflux pump EmrD (Fig. 5) (56, 57). Third, the plasmid-born lpg0273 enhances resistance to nicotinic acid, whereas loss of the lpg0273 locus increases sensitivity to nicotinic acid (Fig. 6). Fourth, Saccharomyces cerevisiae encodes transport proteins that balance the import and export of precursors of NAD+, including nicotinic acid (72, 73). Likewise, genetic analysis of B. pertussis also identified a putative efflux pump for nicotinic acid (Scott Stibitz, personal communication). Accordingly, we propose to designate lpg0273 as mnrA, “MFS nicotinic acid responder.” Whether L. pneumophila naturally encounters exogenous nicotinic acid while residing in a vacuole of macrophages or amoebae is an open question. Indeed, since wild-type cells induced to express lpg0273 display a mild growth advantage even in the absence of supplemental nicotinic acid, we postulate that either the putative Lpg0273 transporter promotes NAD+ homeostasis or it contributes to other aspects of L. pneumophila metabolism, such as efflux of toxic compounds.
Additional clues to how L. pneumophila senses and utilizes nicotinic acid were obtained by considering the genes adjacent to the lpg0273 locus. In particular, next to lpg0272-3 is a putative transcriptional regulator gene, lpg0274, whose expression is also elevated 3 h after nicotinic acid treatment (Fig. 5; also see Table S6 in the supplemental material). Although the significance of their genetic linkage has not been assessed experimentally, we postulate that the putative transcriptional regulator lpg0274 contributes to the transcriptional and phenotypic changes elicited by nicotinic acid. Furthermore, two genes that lie upstream from the lpg0272-3 locus, lpg0270-1, are predicted to encode nicotinate phosphoribosyltransferase and nicotinamidase enzymes, components of NAD+ salvage pathways (Fig. 5) (74). Although the gene expression levels of lpg0270-1 were not altered under the conditions tested, their predicted function in the same metabolic pathways as nicotinic acid lends further support to the hypothesis that lpg0272-3 contribute to nicotinic acid metabolism, homeostasis, and signaling.
Further insight into the L. pneumophila response to nicotinic acid was obtained by comparing the complete gene expression profiles of PE-phase bacteria to E-phase cultures that were supplemented with the phenotypic modulator. Microarray analysis indicated that nicotinic acid regulates the expression of 1,107 genes (Fig. 4; also see Table S3 in the supplemental material), a large number of which are also regulated by growth phase (a total of 894 genes are shared between the two conditions tested) (Fig. 4; also see Table S4). Similar to the results for PE-phase bacteria, nicotinic acid supplementation activated the expression of numerous components of the flagellar apparatus (see Tables S2 and S3) (43, 50). This corroborates our phenotypic data indicating that E-phase L. pneumophila cells activate motility 3 h after nicotinic acid treatment (data not shown). Previous data established that both cytotoxicity and lysosome avoidance are promoted by motility (43), and our transcriptional data determined that most of the flagellar genes are induced by nicotinic acid treatment. Therefore, it was not surprising that treatment with 5 mM nicotinic acid stimulated E-phase L. pneumophila cells to become cytotoxic toward macrophages and to avoid the lysosomal compartment more efficiently (Fig. 2 and data not shown). In addition, microarray analysis determined that nicotinic acid activates the expression of several Dot/Icm type IV secretion genes (see Table S3). This secretion system is thought to form a large pore through which sodium enters the bacterial cell (41, 75, 76). Indeed, the expression of the apparatus correlated with the sodium-sensitivity phenotype displayed by PE-phase bacteria and E-phase L. pneumophila cells treated with nicotinic acid (Fig. 2C). Based on our data, we infer that the differentiation that is triggered by nicotinic acid is largely dependent upon a shift in the transcriptional profile of the bacterium toward that of PE-phase L. pneumophila. Finally, although our microarray studies demonstrate that nicotinic acid induces an expression program largely similar to that of PE-phase bacteria, a distinct group of genes is regulated by nicotinic acid. We predict that this class of nicotinic acid-responsive genes is a valuable resource for experimentalists to delineate the biochemical pathways that likely equip L. pneumophila and other microbes to maintain NAD+ homeostasis and to couple energy metabolism to gene expression programs that dictate cell fate.
Supplementary Material
ACKNOWLEDGMENTS
Research in the Swanson lab was supported by a Rackham Graduate Student Research Award and NIH grant RO1 AI044212. Research in the Buchrieser lab was supported by the Institut Pasteur, the Centre National de la Recherche Scientifique (CNRS), and the Fondation de la Recherche Medicale (FRM).
Footnotes
Published ahead of print 14 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00999-12.
REFERENCES
- 1. Carratala J, Garcia-Vidal C. 2010. An update on Legionella. Curr. Opin. Infect. Dis. 23: 152–157 [DOI] [PubMed] [Google Scholar]
- 2. Declerck P. 2010. Biofilms: the environmental playground of Legionella pneumophila. Environ. Microbiol. 12: 557–566 [DOI] [PubMed] [Google Scholar]
- 3. Molofsky AB, Swanson MS. 2004. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53: 29–40 [DOI] [PubMed] [Google Scholar]
- 4. Fettes PS, Forsbach-Birk V, Lynch D, Marre R. 2001. Overexpresssion of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation and pigmentation. Int. J. Med. Microbiol. 291: 353–360 [DOI] [PubMed] [Google Scholar]
- 5. Molofsky AB, Swanson MS. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50: 445–461 [DOI] [PubMed] [Google Scholar]
- 6. Dalebroux ZD, Edwards RL, Swanson MS. 2009. SpoT governs Legionella pneumophila differentiation in host macrophages. Mol. Microbiol. 71: 640–658 [DOI] [PubMed] [Google Scholar]
- 7. Edwards RL, Dalebroux ZD, Swanson MS. 2009. Legionella pneumophila couples fatty acid flux to microbial differentiation and virulence. Mol. Microbiol. 71: 1190–1204 [DOI] [PubMed] [Google Scholar]
- 8. Hammer BK, Swanson MS. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33: 721–731 [DOI] [PubMed] [Google Scholar]
- 9. Zusman T, Gal-Mor O, Segal G. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184: 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hammer BK, Tateda ES, Swanson MS. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44: 107–118 [DOI] [PubMed] [Google Scholar]
- 11. Rasis M, Segal G. 2009. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol. Microbiol. 72: 995–1010 [DOI] [PubMed] [Google Scholar]
- 12. Sahr T, Brüggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C. 2009. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 72: 741–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bachman MA, Swanson MS. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40: 1201–1214 [DOI] [PubMed] [Google Scholar]
- 14. Bachman MA, Swanson MS. 2004. Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect. Immun. 72: 2468–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bachman MA, Swanson MS. 2004. The LetE protein enhances expression of multiple LetA/LetS-dependent transmission traits by Legionella pneumophila. Infect. Immun. 72: 3284–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacobi S, Schade R, Heuner K. 2004. Characterization of the alternative sigma factor sigma54 and the transcriptional regulator FleQ of Legionella pneumophila, which are both involved in the regulation cascade of flagellar gene expression. J. Bacteriol. 186: 2540–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lynch D, Fieser N, Gloggler K, Forsbach-Birk V, Marre R. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol. Lett. 219: 241–248 [DOI] [PubMed] [Google Scholar]
- 18. Battesti A, Bouveret E. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62: 1048–1063 [DOI] [PubMed] [Google Scholar]
- 19. Seyfzadeh M, Keener J, Nomura M. 1993. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 90: 11004–11008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calva E, Oropeza R. 2006. Two-component signal transduction systems, environmental signals, and virulence. Microb. Ecol. 51: 166–176 [DOI] [PubMed] [Google Scholar]
- 21. Broich M, Rydzewski K, McNealy TL, Marre R, Flieger A. 2006. The global regulatory proteins LetA and RpoS control phospholipase A, lysophospholipase A, acyltransferase, and other hydrolytic activities of Legionella pneumophila JR32. J. Bacteriol. 188: 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Edwards RL, Jules M, Sahr T, Buchrieser C, Swanson MS. 2010. The Legionella pneumophila LetA/LetS two-component system exhibits rheostat-like behavior. Infect. Immun. 78: 2571–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shi C, Forsbach-Birk V, Marre R, McNealy TL. 2006. The Legionella pneumophila global regulatory protein LetA affects DotA and Mip. Int. J. Med. Microbiol. 296: 15–24 [DOI] [PubMed] [Google Scholar]
- 24. Cotter PA, DiRita VJ. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54: 519–565 [DOI] [PubMed] [Google Scholar]
- 25. Cummings CA, Bootsma HJ, Relman DA, Miller JF. 2006. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 188: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McPheat WL, Wardlaw AC, Novotny P. 1983. Modulation of Bordetella pertussis by nicotinic acid. Infect. Immun. 41: 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schneider DR, Parker CD. 1982. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect. Immun. 38: 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miller JF, Roy CR, Falkow S. 1989. Analysis of Bordetella pertussis virulence gene regulation by use of transcriptional fusions in Escherichia coli. J. Bacteriol. 171: 6345–6348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eguchi Y, Oshima T, Mori H, Aono R, Yamamoto K, Ishihama A, Utsumi R. 2003. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149: 2819–2828 [DOI] [PubMed] [Google Scholar]
- 30. Masuda N, Church GM. 2002. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 184: 6225–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Masuda N, Church GM. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48: 699–712 [DOI] [PubMed] [Google Scholar]
- 32. Nishino K, Inazumi Y, Yamaguchi A. 2003. Global analysis of genes regulated by EvgA of the two-component regulatory system in Escherichia coli. J. Bacteriol. 185: 2667–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nishino K, Yamaguchi A. 2001. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183: 1455–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Utsumi R, Katayama S, Taniguchi M, Horie T, Ikeda M, Igaki S, Nakagawa H, Miwa A, Tanabe H, Noda M. 1994. Newly identified genes involved in the signal transduction of Escherichia coli K-12. Gene 140: 73–77 [DOI] [PubMed] [Google Scholar]
- 35. Berger KH, Isberg RR. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7: 7–19 [DOI] [PubMed] [Google Scholar]
- 36. Bryan A, Harada K, Swanson MS. 2011. Efficient generation of unmarked deletions in Legionella pneumophila. Appl. Environ. Microbiol. 77: 2545–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bryan A, Swanson MS. 2011. Oligonucleotides stimulate genomic alterations of Legionella pneumophila. Mol. Microbiol. 80: 231–247 [DOI] [PubMed] [Google Scholar]
- 38. Sexton JA, Vogel JP. 2004. Regulation of hypercompetence in Legionella pneumophila. J. Bacteriol. 186: 3814–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swanson MS, Isberg RR. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63: 3609–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Molofsky AB, Shetron-Rama LM, Swanson MS. 2005. Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect. Immun. 73: 5720–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Byrne B, Swanson MS. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66: 3029–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Milohanic E, Glaser P, Coppee JY, Frangeul L, Vega Y, Vazquez-Boland JA, Kunst F, Cossart P, Buchrieser C. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47: 1613–1625 [DOI] [PubMed] [Google Scholar]
- 43. Brüggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, Kunst F, Steinert M, Heuner K, Coppee JY, Buchrieser C. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell. Microbiol. 8: 1228–1240 [DOI] [PubMed] [Google Scholar]
- 44. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15 doi:10.1093/nar/30.4.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Delmar P, Robin S, Daudin JJ. 2005. VarMixt: efficient variance modelling for the differential analysis of replicated gene expression data. Bioinformatics 21: 502–508 [DOI] [PubMed] [Google Scholar]
- 46. Reiner A, Yekutieli D, Benjamini Y. 2003. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375 [DOI] [PubMed] [Google Scholar]
- 47. Cazalet C, Rusniok C, Brüggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, Kunst F, Etienne J, Glaser P, Buchrieser C. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36: 1165–1173 [DOI] [PubMed] [Google Scholar]
- 48. Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, Rineer J, Greenberg JJ, Steshenko V, Park SH, Zhao B, Teplitskaya E, Edwards JR, Pampou S, Georghiou A, Chou IC, Iannuccilli W, Ulz ME, Kim DH, Geringer-Sameth A, Goldsberry C, Morozov P, Fischer SG, Segal G, Qu X, Rzhetsky A, Zhang P, Cayanis E, De Jong PJ, Ju J, Kalachikov S, Shuman HA, Russo JJ. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305: 1966–1968 [DOI] [PubMed] [Google Scholar]
- 49. Edwards RL, Swanson MS. 2006. Regulation of the L. pneumophila life cycle, p 95–111 In Hoffman P, Klein T, Friedman H. (ed), Legionella pneumophila: pathogenesis and Immunity. Springer Publishing Corp., New York, NY [Google Scholar]
- 50. Heuner K, Steinert M. 2003. The flagellum of Legionella pneumophila and its link to the expression of the virulent phenotype. Int. J. Med. Microbiol. 293: 133–143 [DOI] [PubMed] [Google Scholar]
- 51. Hales LM, Shuman HA. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181: 4879–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Segal G, Feldman M, Zusman T. 2005. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 29: 65–81 [DOI] [PubMed] [Google Scholar]
- 53. Weissenmayer BA, Prendergast JG, Lohan AJ, Loftus BJ. 2011. Sequencing illustrates the transcriptional response of Legionella pneumophila during infection and identifies seventy novel small non-coding RNAs. PLoS One 6: e17570 doi:10.1371/journal.pone.0017570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weisburg WG, Dobson ME, Samuel JE, Dasch GA, Mallavia LP, Baca O, Mandelco L, Sechrest JE, Weiss E, Woese CR. 1989. Phylogenetic diversity of the Rickettsiae. J. Bacteriol. 171: 4202–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4: 363–371 [DOI] [PubMed] [Google Scholar]
- 57. Yin Y, He X, Szewczyk P, Nguyen T, Chang G. 2006. Structure of the multidrug transporter EmrD from Escherichia coli. Science 312: 741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Han YW, Uhl MA, Han SJ, Shi W. 1999. Expression of bvgAS of Bordetella pertussis represses flagellar biosynthesis of Escherichia coli. Arch. Microbiol. 171: 127–130 [DOI] [PubMed] [Google Scholar]
- 59. Leonardo MR, Dailly Y, Clark DP. 1996. Role of NAD in regulating the adhE gene of Escherichia coli. J. Bacteriol. 178: 6013–6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Domergue R, Castano I, De Las Penas A, Zupancic M, Lockatell V, Hebel JR, Johnson D, Cormack BP. 2005. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308: 866–870 [DOI] [PubMed] [Google Scholar]
- 61. Blander G, Guarente L. 2004. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73: 417–435 [DOI] [PubMed] [Google Scholar]
- 62. Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. 2004. A link between transcription and intermediary metabolism: a role for Sir2 in the control of acetyl-coenzyme A synthetase. Curr. Opin. Microbiol. 7: 115–119 [DOI] [PubMed] [Google Scholar]
- 63. Canto C, Auwerx J. 2011. NAD+ as a signaling molecule modulating metabolism. Cold Spring Harb. Symp. Quant. Biol. 76: 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9: 2888–2902 [DOI] [PubMed] [Google Scholar]
- 65. Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298: 2390–2392 [DOI] [PubMed] [Google Scholar]
- 66. Wolfe AJ. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69: 12–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cotter PA, Miller JF. 1997. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol. Microbiol. 24: 671–685 [DOI] [PubMed] [Google Scholar]
- 68. Heeb S, Haas D. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant Microbe Interact. 14: 1351–1363 [DOI] [PubMed] [Google Scholar]
- 69. Lapouge K, Schubert M, Allain Haas FH-TD. 2008. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behavior. Mol. Microbiol. 67: 241–253 [DOI] [PubMed] [Google Scholar]
- 70. Perraud A, Weiss V, Gross R. 1999. Signalling pathways in two-component phosphorelay systems. Trends Microbiol. 7: 115–120 [DOI] [PubMed] [Google Scholar]
- 71. Wass MN, Kelley LA, Sternberg MJ. 2010. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 38: W469–W473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Belenky P, Stebbins R, Bogan KL, Evans CR, Brenner C. 2011. Nrt1 and Tna1-independent export of NAD+ precursor vitamins promotes NAD+ homeostasis and allows engineering of vitamin production. PLoS One 6: e19710 doi:10.1371/journal.pone.0019710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lu SP, Kato M, Lin SJ. 2009. Assimilation of endogenous nicotinamide riboside is essential for calorie restriction-mediated life span extension in Saccharomyces cerevisiae. J. Biol. Chem. 284: 17110–17119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Belenky P, Bogan KL, Brenner C. 2007. NAD+ metabolism in health and disease. Trends Biochem. Sci. 32: 12–19 [DOI] [PubMed] [Google Scholar]
- 75. Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61: 5361–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vogel JP, Roy C, Isberg RR. 1996. Use of salt to isolate Legionella pneumophila mutants unable to replicate in macrophages. Ann. N. Y. Acad. Sci. 797: 271–272 [DOI] [PubMed] [Google Scholar]
- 77. Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97: 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.