Background: SFMBT1 is a poorly characterized chromatin reader.
Results: SFMBT1 is associated with multiple transcriptional corepressor complexes and required for MyoD-mediated transcriptional silencing.
Conclusion: Novel mechanisms of SFMBT1-mediated transcription repression and its regulation of myogenesis are identified.
Significance: Our findings contribute to the understanding of how histone modification language is interpreted to regulate gene expression and biological processes.
Keywords: Differentiation, Epigenetics, Myogenesis, Polycomb, Stem Cells, LSD1, MBT, MyoD, SFMBT1
Abstract
SFMBT1 belongs to the malignant brain tumor domain-containing chromatin reader family that recognizes repressive histone marks and represses transcription. The biological functions and molecular basis underlying SFMBT1-mediated transcriptional repression are poorly elucidated. Here, our proteomic analysis revealed that SFMBT1 is associated with multiple transcriptional corepressor complexes, including CtBP/LSD1/HDAC complexes, polycomb repressive complexes, and malignant brain tumor family proteins, that collectively contribute to SFMBT1 repressor activity. During myogenesis, Sfmbt1 represses myogenic differentiation of cultured and primary myoblasts. Mechanistically, Sfmbt1 interacts with MyoD and mediates epigenetic silencing of MyoD target genes via recruitment of its associated corepressors and subsequent induction of epigenetic modifications and chromatin compaction. Therefore, our study identified novel mechanisms accounting for SFMBT1-mediated transcription repression and revealed an essential role of Sfmbt1 in regulating MyoD-mediated transcriptional silencing that is required for the maintenance of undifferentiated states of myogenic progenitor cells.
Introduction
Chromatin is composed of repetitive units called nucleosomes that consist of DNA (147 base pairs) wrapped around an octamer of histone proteins (two subunits of each of the histones H2A, H2B, H3, and H4) (1, 2). Posttranslational modifications of the N-terminal tails of histones affect chromatin structure, consequently serving as “histone marks” for transcriptional repression or activation (3, 4). One important type of histone mark is histone methylation, which is regulated by specific sets of proteins that carry out the addition, removal, or specific binding of methyl groups (4).
The MBT3 domain-containing proteins bind to low methylated histones, functioning as the chromatin “readers,” and have been linked to development and tumor suppression by genetic analyses (5). The MBT domain was first reported in Drosophila dL3mbt (6, 7). Mutations in the MBT domain of dL3mbt caused malignant overgrowth in the larval brain, implicating a tumor suppressing function of dL3mbt and the importance of the MBT domain (6, 7). In mammals, three different groups of MBT proteins have been identified, with hSCML2 and hSCMH1 containing two MBT domains; L3MBTL1, L3MBTL3, and L3MBTL4 containing three MBT domains; and L3MBTL2, SFMBT1, and SFMBT2 containing four MBT domains (5). The MBT proteins prevalently bind to mono- and dimethylated histone lysines in vitro and repress transcription via interaction with various repressors (5). Different MBT proteins have been identified in various protein complexes (5), suggesting that MBT proteins have distinct functional activities and modes of action in regulating chromatin.
In this study, we focused on a poorly characterized MBT protein, SFMBT1 (Scm-like with four mbt domains 1). Mammalian SFMBT1 contains four MBT repeat domains that are essential for mediating histone H3 N-terminal tail binding and transcriptional repression (8). In Drosophila, dSfmbt contains four MBT repeats and is a polycomb protein because dSfmbt knockout displays a classic polycomb phenotype with strong and widespread derepression of HOX genes (9). dSfmbt is part of the Pho repressive complex, which contains the DNA-binding protein Pho and recruits other polycomb proteins, mediating H3K27me3 for transcriptional silencing at target loci (10). The MBT repeat domain of dSfmbt preferentially binds to mono- or dimethylated lysine containing histone peptides (11). Although SFMBT1 was considered as a mammalian homologue for Drosophila dSfmbt in the literature, mammalian SFMBT1 and dSfmbt are probably not homologues, and SFMBT1 is likely unique to mammals on the basis of the following evidence: 1) SFMBT1 was not found in the mammalian Pho homolog YY1 protein complex in mammalian cells (12); 2) human SFMBT1 binds selectively with the N-terminal tail of histone H3 in a manner that appears to be independent of histone modification (8); and 3) SFMBT1 belongs to a different branch from dSfmbt on the basis of evolutionary relationship and domain organizations (5). Therefore, alternative mechanisms might account for the role of mammalian SFMBT1 in transcriptional regulation, and SFMBT1 might have unique functions. However, the molecular mechanisms underlying SFMBT1 transcriptional repression as well as its biological functions are unknown.
To gain unbiased biochemical insights into how SFMBT1 exerts its transcription repressor function, we performed affinity purification and MS analysis of the SFMBT1 protein complex. Our data revealed a novel biochemical connection of SFMBT1 with CtBP/LSD1/HDAC complexes, polycomb protein complexes (PRC), and other MBT proteins, suggesting functional cooperation of these corepressor proteins in establishing repressive chromatin states. We subsequently utilized a skeletal myogenesis model to investigate the biological functions of Sfmbt1 because epigenetic regulation has critical roles in the highly regulated myogenic process. Through gain of function and loss of function studies in combination with gene expression profiling studies, we found that Sfmbt1 critically regulates the myogenic programs through transcriptional silencing of the master regulator of myogenic process, MyoD.
EXPERIMENTAL PROCEDURES
Plasmids
pLKO.1-based lentiviral shRNA plasmids targeting mouse Sfmbt1 and MyoD genes were purchased from Open Biosystems. Human SFMBT1 truncation mutants (N: 1–473 aa, M: 494–699 aa, and C: 721–866 aa) were cloned to the pGex vector to generate GST fusion proteins. GFP-SFMBT1 was generated with the pEGFP-C3 vector. pQCXIP-FLAG-SFMBT1 was described previously (8). pMyog-luc was kindly provided by Dr. Stephen J. Tapscott (13). pCMV2-FLAG-L3MBTL3 was kindly provided by Dr. Toru Miyazaki (14). HA-tagged, full-length (FL) MyoD and truncated mutants (N: 1–66 aa, ΔN: 84–318 aa, C: 173–318 aa, and ΔC: 1–240 aa) were kindly provided by Drs. Serge A. Leibovitch and Slimane Ait-Si-Ali (15).
Antibodies
The antibodies were obtained from the following commercial sources: LSD1 (Abcam, catalog no. ab17721), anti-FLAG (M2, Sigma, catalog no. F-3165), anti-HA (Covance, catalog no. MMS-101P), GFP (Santa Cruz Biotechnology, catalog no. sc8334), CoREST (Millipore, catalog no. 07-455), BHC80 (Abcam, catalog no. ab41631), HDAC1 (Thermo, catalog no. PA1-860, and Santa Cruz Biotechnology, catalog no. sc7872), HDAC2 (Thermo, catalog no. PA1-861), EZH2 (Millipore, 07-400, and Active Motif, catalog no. 39875), RNF2 (Active Motif, catalog no. 39663), PHC1 (Active Motif, catalog no. 39723), SUZ12 (Millipore, catalog no. 07-379), β-actin (Sigma, catalog no. A5316), H3K4me2 (Millipore, catalog no. 07-030), H3K27me3 (Millipore, catalog no. 07-449), H3Ac (Millipore, catalog no. 06-599), H4Ac (Millipore, catalog no. 06-598), myosin (R & D Systems, catalog no. MF20), Myogenin (BD Biosciences, catalog no. 556358), and MyoD (BD Biosciences, catalog no. 556130).
Cell Culture
293FT and 293T cells were cultured in DMEM with 10% heat-inactivated fetal bovine serum and penicillin/streptomycin. C2C12 cells (ATCC) were cultured in DMEM with 20% heat-inactivated fetal bovine serum and penicillin/streptomycin. Primary myoblasts were isolated and maintained as described previously (16). Briefly, limb muscles from newborn mice were minced and then digested with collagenase/dispase at 37 °C for 30 min. The dissociated cells were collected and cultured in a collagen-coated dish with F-10-based primary myoblast selective growth medium (Ham F-10 nutrient mixture supplied with 20% FBS, 2.5 ng/ml bFGF/ml, 100 units/ml penicillin, and 100 μg/ml streptomycin). To selectively enrich myoblasts, myoblasts were physically dissociated from the culture dish during splitting, whereas the majority of fibroblasts remained attached. To further exclude the easily attaching fibroblasts, the dissociated cells were preplated to a fresh dish for 15 min, and then the unattached myoblasts were collected and cultured in a new dish. The selection process was repeated until a high purity of myoblasts was attained. For the myogenesis assay, myoblasts were grown to 80–90% confluence and induced for differentiation by switching from growth medium to differentiation medium (DMEM containing 2% horse serum).
Nuclear Extract Preparation, Protein Complex Isolation, and Western Blot Analysis
The detailed purification procedure was described previously (17). Briefly, nuclear extracts were prepared from 293T cells stably transduced with FLAG-SFMBT1. FLAG-SFMBT1 protein complexes were then purified using anti-FLAG M2 mAb-conjugated agarose beads (Sigma). After extensive washing, protein complexes were eluted using the elution buffer containing 50 μg/ml FLAG peptides at room temperature for 30 min. The eluted protein samples were separated on 4–12% NuPAGE Bis-tris gels (Invitrogen) and analyzed by LC-MS/MS. The procedures for Western blotting and immunoprecipitation studies were described previously (18).
Viral Production, Titering, and Transduction
Viral production and transduction in cells was described previously (19). Stably transduced cells (polyclonal) were obtained and maintained by puromycin selection. To prepare concentrated viruses, supernatants containing lentiviruses were centrifuged at 25,000 rpm for 2 h at 4 °C using a SW28 rotor (Beckman). The pellets were suspended with PBS, aliquoted, and stored at −80 °C. The viral titers were determined following a procedure provided by Open Biosystems.
Immunofluorescence Staining and Luciferase Assays
Immunofluorescence staining and luciferase reporter assays were performed as described previously (18).
Microarray Analyses
Microarray experiments were performed using RNA isolated from the C2C12 scramble shRNA control and C2C12 Sfmbt1 knockdown. The total RNA was extracted using TRIzol reagents (Invitrogen) and purified using a Qiagen column with in-column DNase digestion. Hybridizations of Affymetrix mouse genome 430 2.0 arrays were performed at the Dana Farber Microarray core facility. The data were preprocessed and analyzed using dChip software. To reduce the false positives, probe sets defined as “Absent” detection calls crossing all samples were removed from the data analysis. Genes with a fold change ≥ 1.5 and p < 0.05 were considered differentially expressed.
GST Pull-down Assays
GST pull-down assays were performed as described previously (18). Briefly, bacterially expressed GST or GST-SFMBT1 fusion proteins were induced and purified with glutathione-Sepharose. Equivalent amounts of GST or GST-SFMBT1 immobilized on glutathione-Sepharose were then incubated with a nuclear extract of 293T transfected with HA-MyoD. After extensive washing, the pull-down HA-MyoD was detected by Western blot analysis.
Real-time RT-PCR
cDNA was generated with 2 μg of total RNA using a GeneAmp RNA PCR kit (Applied Biosystems). Real-time PCR was performed using the StepOneTM real-time PCR system (Applied Biosystems) with the SYBR Green PCR core reagents kit (Applied Biosystems). The sequences for the primers used are listed in supplemental Table S1.
ChIP
ChIP assays were performed as described previously (20). Briefly, cells were fixed with formaldehyde for 10 min at room temperature to cross-link the DNA to chromatin-associated protein complexes. The cross-linking was stopped with glycine, and cells were then collected and sonicated with a MISONIX S-4000 sonicator to shear DNA to lengths between 200 and 800 bp. The DNA-protein complexes were then immunoprecipitated with the indicated antibodies and washed extensively. The ChIP DNA was then purified and eluted with 100 μl of H2O. A total of 2.5 μl of ChIP DNA was used for real-time PCR analysis. The primer sequences are listed in supplemental Table S1.
DNase I Sensitivity Assay
A DNase I sensitivity assay was performed with RQ1 RNase-free DNse (Promega). Nuclei of 5 × 105 cells were treated with DNase at 37 °C for 30 min, and then the reactions were stopped with EDTA at a final concentration of 25 mm. Genomic DNA samples were purified by protease K digestion and phenol chloroform extraction. The purified DNA samples were analyzed by real-time PCR to measure the relative amount of uncut DNA around the promoter region of Myogenin.
Statistical Analyses of Experimental Data
An independent Student's t test was used to analyze data from various experiments.
RESULTS
SFMBT1 Is Associated with Multiple Transcriptional Corepressor Complexes
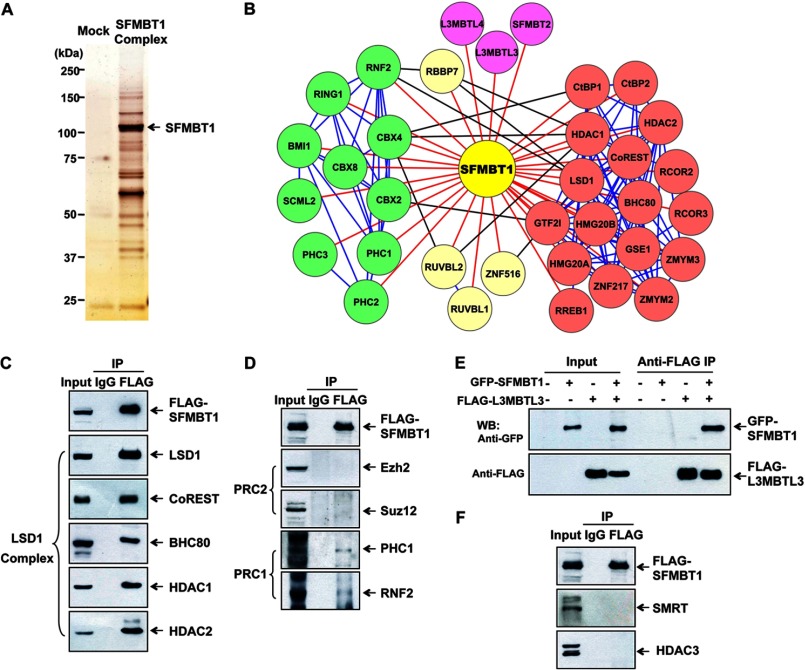
To elucidate the mechanisms underlying transcriptional repression function of SFMBT1, we performed a proteomic analysis to identify SFMBT1-interacting partners in mammalian cells. The SFMBT1 protein complex was immunoprecipitated using anti-FLAG beads from nuclear extracts of FLAG-SFMBT1-expressing 293T cells followed by elution with FLAG peptides. The SFMBT1-associated proteins were next separated by SDS-PAGE (Fig. 1A) and subjected to MS analysis. We found that SFMBT1-associated proteins consist of distinct transcriptional corepressors, including 17 components of the CtBP/LSD1/HDAC protein complex, 11 members of PRC, and four MBT proteins (Fig. 1B and supplemental Fig. S1). We further validated the interaction of SFMBT1 with these corepressors identified from our MS analysis. As shown in Fig. 1C, we performed immunoprecipitation of FLAG-SFMBT1 followed by Western blotting and validated that LSD1 core complex components, including LSD1, CoREST, BHC80, HDAC1, and HDAC2, were present in SFMBT1 immunoprecipitates. Using similar assays, we observed that SFMBT1 interacted with PRC1 proteins PHC1 and RNF2 but not with PRC2 members EZH2 and SUZ12 (Fig. 1D), which is consistent with our MS data (B). Because our MS data also indicated an interaction between SFMBT1 and L3MBTL3, we performed coimmunoprecipitation and detected an interaction between GFP-SFMBT1 and FLAG-L3MBTL3 in mammalian cells (Fig. 1E). Also, consistent with our MS data, we failed to detect SFMBT1 interaction with SMRT/HDAC3 corepressors (Fig. 1F). Therefore, we identified the biochemical components of the SFMBT1 protein complex, which suggest that SFMBT1 mediates transcriptional repression via interactions with multiple transcriptional repression complexes.
FIGURE 1.
SFMBT1 is associated with multiple transcriptional corepressor complexes. A, silver staining of the SFMBT1-associated proteins. IPs with anti-FLAG (M2)-agarose beads were performed using nuclear extracts of transduced 293T cells expressing either vector (Mock) or FLAG-SFMBT1. The protein complex was eluted with FLAG peptides after extensive washing and separated by 4–12% gradient SDS-PAGE. B, a list of potential SFMBT1-associated proteins was identified by mass spectrometric analysis. A cytoscape program was used to show protein interactions. Red lines indicate the interactions between SFMBT1 and its associated partners. Blue lines indicate the interactions within the same transcriptional repressive protein complex. Black lines indicate the interactions among different repressive complexes. C and D, SFMBT1 interacted with the LSD1 protein complex and PRC1 but not PRC2. FLAG-SFMBT1-expressing 293T cells were subjected to IP with anti-IgG or anti-FLAG antibodies, followed by Western blotting using the indicated antibodies. E, SFMBT1 interacted with L3MBTL3. 293T cells were transfected with GFP-SFMBT1 and FLAG-L3MBTL3 individually or in combination, and then IP using anti-FLAG antibodies followed by Western blotting (WB) were performed. F, SFMBT1 failed to coimmunoprecipitate with SMRT and HDAC3. IPs were performed with anti-IgG or anti-FLAG antibodies using nuclear extracts from FLAG-SFMBT1-expressing cells, followed by Western blotting with the indicated antibodies.
Sfmbt1 Expression Is Down-regulated During Myogenic Differentiation
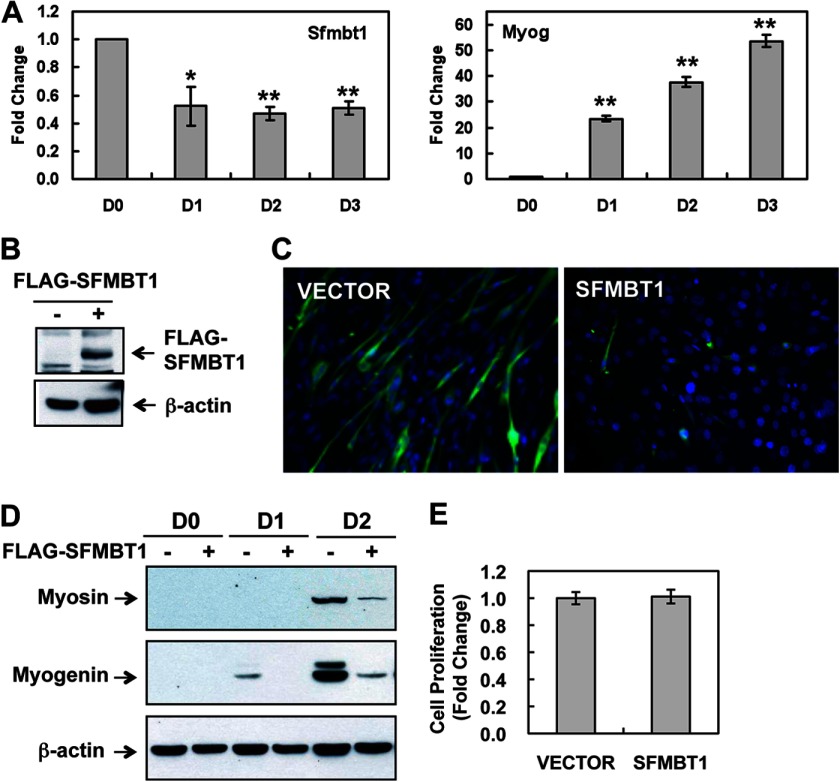
The biological functions of SFMBT1 are unknown. Previous observations that mouse Sfmbt1 is expressed in skeletal muscles (21) and that Drosophila dSfmbt is associated with the promoters of genes involved in muscle development in embryos and larvae (22) suggest a possible role of Sfmbt1 in regulating skeletal myogenesis. Importantly, epigenetic silencing is critical in restricting the expression of muscle genes in muscle progenitors and maintaining proper growth and differentiation (23–25). To investigate the function of Sfmbt1 in myogenesis, we utilized the C2C12 myoblast differentiation model and first investigated the pattern of Sfmbt1 expression during myogenic differentiation. We found that the Sfmbt1 transcript level was highest in undifferentiated myoblasts and reduced during the course of differentiation (Fig. 2A). Thus, these data suggest a possible role for Sfmbt1 in repressing myogenic differentiation.
FIGURE 2.
Sfmbt1 is a negative regulator of myogenic differentiation. A, Sfmbt1 expression was down-regulated during myoblast differentiation. C2C12 cells were induced to differentiate by switching from growth medium to differentiation medium, and RNA samples were collected at different time points. The transcript levels of Sfmbt1 (left panel) and Myogenin (Myog, right panel) were determined by real-time RT-PCR and presented as fold changes (mean ± S.E.). *, p < 0.05; **, p < 0.01. B, FLAG-SFMBT1 expression in transduced C2C12 cells was detected by Western blotting. C, SFMBT1 overexpression blocked myotube formation. FLAG-SFMBT1-expressing and control C2C12 cells were induced to undergo differentiation. Myosin staining (green) was performed 3 days later using an anti-Myosin antibody. Nuclei were counterstained with DAPI. D, SFMBT1 overexpression inhibited expression of muscle-specific differentiation markers. C2C12 cell lysates were collected at various time points after differentiation and detected for expression of Myosin and Myogenin by Western blotting. D1, day 1. E, SFMBT1 overexpression did not affect proliferation of C2C12 myoblasts. Cells were seeded to 10-cm culture plates at 1 × 105 cells/plate, and cell numbers were counted 3 days later. n = 3. Data are represented as fold change with cells transduced with empty vector defined as 1.
SFMBT1 Overexpression Blocks and Sfmbt1 Depletion Enhances Myogenic Differentiation
We next investigated the respective functional consequences of SFMBT1 overexpression and Sfmbt1 depletion on myoblast proliferation and differentiation. First, C2C12 myoblasts were transduced with human FLAG-SFMBT1 retroviruses or empty vector control viruses followed by puromycin selection. SFMBT1 expression was confirmed by Western blot analysis (Fig. 2B). The transduced C2C12 myoblasts were induced to differentiate, and myotube formation and muscle-specific gene expression were then examined various days after differentiation. We found that ectopic SFMBT1 expression resulted in a block in myotube formation (Fig. 2C) and a decrease in expression of muscle differentiation markers (myosin and Myogenin) (D). Ectopic SFMBT1 expression appeared to have little effect on the proliferation of undifferentiated myoblasts in growth medium (Fig. 2E). Thus, SFMBT1 represses myogenic differentiation, likely contributing to the maintenance of undifferentiated states of myoblasts.
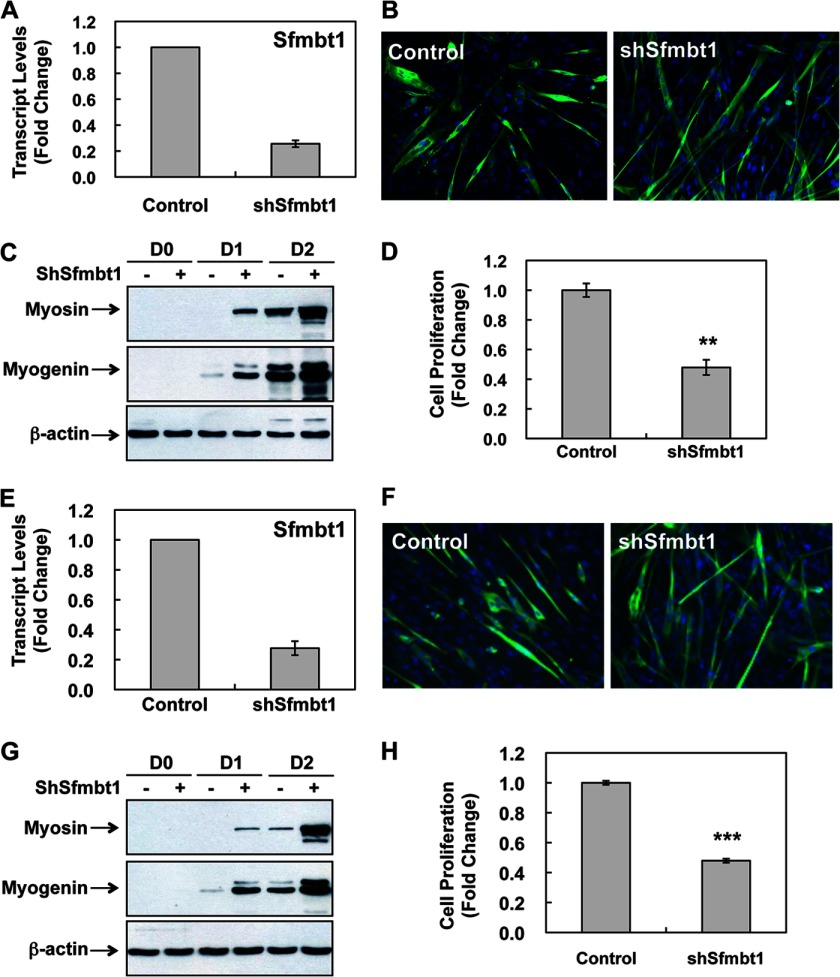
Next, we knocked down Sfmbt1 expression in C2C12 myoblasts using lentiviral-mediated shRNA targeting Sfmbt1 (shSfmbt1) or scrambled shRNA as a control (Fig. 3A). When induced for differentiation, Sfmbt1 knockdown cells displayed a more differentiated phenotype than control cells, as shown by the formation of larger and more myotubes (Fig. 3B and supplemental Fig. S2) and earlier and more robust expression of myogenic transcription factor Myogenin and muscle gene Myosin (Fig. 3C). Moreover, Sfmbt1 knockdown significantly reduced the proliferation rate of C2C12 myoblasts cultured in growth medium (Fig. 3D). The shRNA knockdown effects were specific, as we observed similar effects with an independent shRNA construct targeting a different region of Sfmbt1 (supplemental Fig. S3). Importantly, we found that Sfmbt1 knockdown promoted differentiation and reduced proliferation of primary mouse myoblasts (Fig. 3, E–H, and supplemental Fig. S2). Therefore, our gain of function and loss of function studies (Figs. 2 and 3) strongly indicate that Sfmbt1 is required for proper proliferation of myogenic progenitor cells and prevention of premature differentiation.
FIGURE 3.
Sfmbt1 knockdown promotes myogenic differentiation in cultured and primary myoblasts. A, Sfmbt1 transcript levels in Sfmbt1 knockdown and control C2C12 cells were measured by real-time RT-PCR. Data are presented as mean ± S.E. B, Sfmbt1 knockdown and control C2C12 cells were induced for differentiation for 3 days. Myotubes were stained with anti-Myosin antibodies (green), and the nuclei were counterstained with DAPI (blue). C, whole cell lysates were collected at various days (D) after differentiation for Western blot analysis of Myosin and Myogenin levels. D, Sfmbt1 knockdown significantly decreased C2C12 proliferation. n = 3. **, p < 0.01. Data are presented as fold change with cells transduced with empty vector viruses as 1. E–H, Sfmbt1 knockdown promoted differentiation of primary myoblasts isolated from neonatal mice. Sfmbt1 knockdown in primary myoblasts was determined by real-time RT-PCR (E). Sfmbt1 knockdown cells showed enhanced myogenic differentiation, as indicated by increased myotube formation (F) by immunofluorescence staining and expression of muscle-specific proteins Myosin and Myogenin (G) by Western blotting. H, Sfmbt1 knockdown resulted in reduced cell proliferation in primary myoblasts. n = 3. ***, p < 0.001.
Sfmbt1 Represses Transcription of Muscle Transcription Factors Such as Myogenin, Mef2C, and Muscle Structural Genes
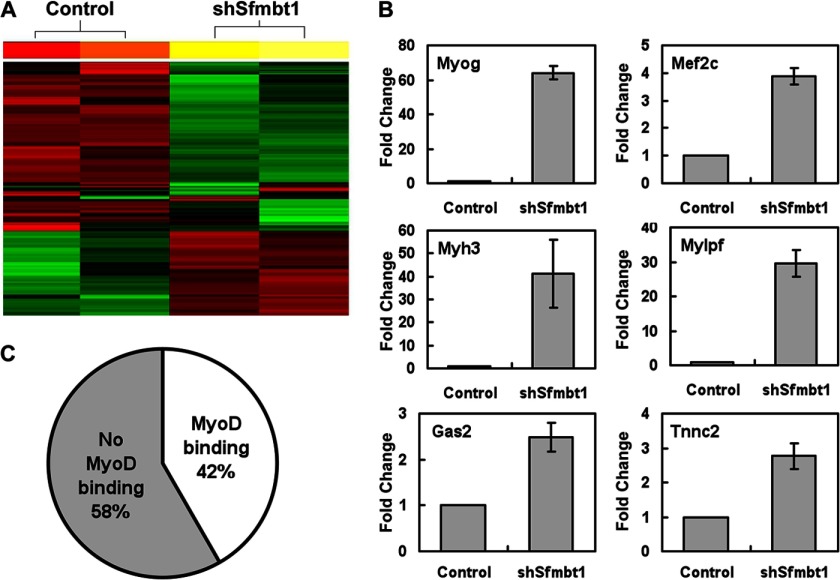
To investigate the molecular mechanisms underlying Sfmbt1 regulation of myogenesis, we examined Sfmbt1-regulated genes in undifferentiated myoblasts. We performed a microarray analysis to compare gene expression between proliferating Sfmbt1 knockdown and control cells and identified a total of 206 up-regulated genes and 307 down-regulated genes with the criteria of fold change ≥ 1.5 and p < 0.05 (Fig. 4A and supplemental Table S2, S3). The up-regulated genes with highest fold changes in Sfmbt1 knockdown cells were numerous muscle differentiation genes whose expression is normally repressed in proliferating, undifferentiated muscle progenitors. These genes included muscle transcription factors, Myogenin and Mef2C, and many myofibrillar genes such as troponin, Tnnc, Tnnt, Tnni, Myosin light and heavy chains, and α-actin. Elevated expression levels of several typical muscle genes such as Myogenin (myog), Mef2c, Myh3, Mylpf, Gas2, and Tnnc2 in Sfmbt1 knockdown cells were confirmed by real-time RT-PCR (Fig. 4B). On the other hand, expression of the muscle stem cell factors Bmi1 (26) and Nestin (27) was decreased when Sfmbt1 was depleted, suggesting a role of Sfmbt1 in the maintenance of the muscle stem cell state. Our findings thus support that Sfmbt1 critically confers transcriptional silencing of muscle genes in myogenic progenitor cells.
FIGURE 4.
Sfmbt1 target genes in C2C12 myoblasts have a significant overlap with direct MyoD target genes. A, a heat map showed changes in Sfmbt1 knockdown and control cell gene expression profiling on the basis of microarray analysis. B, several Sfmbt1 target genes were validated by quantitative RT-PCR. Data are presented as mean ± S.E. C, a significant subset of Sfmbt1 target genes showed MyoD binding on their promoters. The comparisons of the up-regulated gene set because of Sfmbt1 knockdown, and the gene set showing MyoD binding in the promoter (28) reveals that about 42% of the Sfmbt1 target genes showed MyoD binding in the promoters.
Sfmbt1 Interacts with the Muscle Transcription Factor MyoD
The myogenic transcription factor MyoD is associated with transcriptional corepressors to repress MyoD-mediated transcription in undifferentiated myogenic progenitors (25). Because Sfmbt1 depletion resulted in enhanced expression of important MyoD target genes such as Myogenin and Mef2c in myoblasts, we investigated a possible interaction between Sfmbt1 and MyoD. We compared the Sfmbt1 target gene set from our microarray data with the published MyoD ChIP-Sequence gene set (28) and found that about 42% of the up-regulated genes because of Sfmbt1 knockdown showed physical MyoD binding to their promoters (Fig. 4C). These data suggest that MyoD interacts with and recruits Sfmbt1 to MyoD target loci for transcriptional repression in myogenic progenitors.
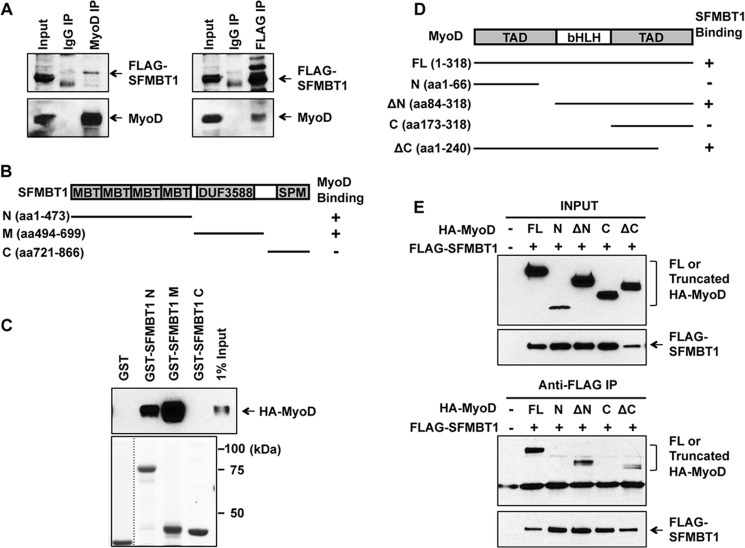
To test this possibility, we determined a possible physical interaction between Sfmbt1 and MyoD. Our reciprocal coimmunoprecipitation assays using FLAG-SFMBT1-expressing C2C12 cells showed that FLAG-SFMBT1 is readily detectable in endogenous MyoD immunoprecipitates and endogenous MyoD in FLAG-SFMBT1 immunoprecipitates (Fig. 5A), indicating an interaction between SFMBT1 and MyoD. Moreover, our mapping studies using a series of SFMBT1 (GST fusion) and MyoD (HA-tagged) truncated mutants revealed that MyoD interacts with the N-terminal (four-MBT repeat domain) and middle region (DUF 3588, domain of unknown function 3588) of SFMBT1 (Fig. 5, B and C) and that the bHLH domain of MyoD is required for its interaction with SFMBT1 (D and E). Because the muscle transcription factor Myogenin is also a bHLH member, we performed coimmunoprecipitation and found that Sfmbt1 failed to interact with Myogenin (supplemental Fig. S4). Therefore, our data indicate that Sfmbt1 specifically interacts with MyoD and represses MyoD-dependent transcription.
FIGURE 5.
Sfmbt1 interacts with myogenic master transcription factor MyoD. A, FLAG-SFMBT1 interacted with endogenous MyoD in C2C12 by reciprocal IP. Nuclear extracts from FLAG-SFMBT1-expressing C2C12 cells were immunoprecipitated with anti-IgG and anti-MyoD antibodies, respectively (left panel), or anti-IgG and anti-FLAG antibodies, respectively (right panel). Western blotting was subsequently performed using the indicated antibodies. B, diagram of full-length and truncated mutants of SFMBT1. C, GST pull-down assays showed that MyoD binds to the N-terminal and central regions of the SFMBT1 protein. Various GST-SFMBT1 fusion proteins immobilized on glutathione-Sepharose were incubated with whole cell lysates of HA-MyoD-transfected 293T cells. The pull-down products were analyzed by Western blotting with anti-HA antibodies. D, the domain structure of MyoD and various truncated mutants are shown. E, the bHLH domain of MyoD interacted specifically with SFMBT1 by coimmunoprecipitation assays. 293T cells were cotransfected with FLAG-SFMBT1, and full-length (FL) or truncated mutants of HA-MyoD and whole cell lysates were harvested 48 h after transfection. SFMBT1 IP was performed with anti-FLAG-agaroses followed by Western blotting with HA antibodies.
Sfmbt1 Recruits Multiple Transcriptional Corepressor Complexes to Mediate Epigenetic Silencing of MyoD Target Genes
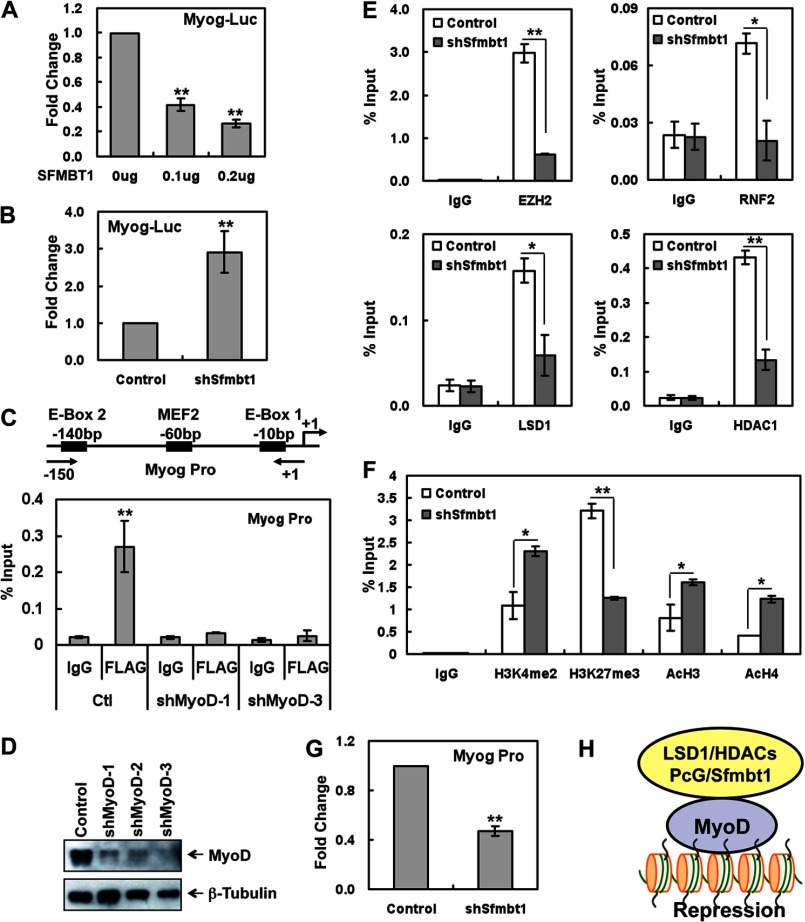
One key MyoD target is the muscle transcription factor Myogenin, which is essential for muscle cell differentiation (29). Therefore, we focused on the Myogenin gene and investigated how Sfmbt1 regulates MyoD-mediated transcriptional repression. First, we confirmed that exogenous SFMBT1 repressed the activity of a Myogenin promoter luciferase reporter in a dose-dependent manner (Fig. 6A). Conversely, depletion of endogenous Sfmbt1 in C2C12 resulted in increased Myogenin promoter activity (Fig. 6B). Consistent with the promoter assays, Sfmbt1 knockdown resulted in enhanced transcription levels of endogenous myogenin (Fig. 4B). Moreover, Sfmbt1-depleted C2C12 cells showed an increase in Myogenin protein 1 day after differentiation by Western blotting (Fig. 3C) and immunofluorescence staining (supplemental Fig. S5). Next, we carried out ChIP assays using FLAG-SFMBT1-expressing C2C12 cells and found that FLAG-SFMBT1 is enriched on the MyoD binding region of the Myogenin promoter, indicating that Sfmbt1 directly regulates MyoD target myogenin gene transcription (Fig. 6C). Moreover, the depletion of MyoD decreased the binding of SFMBT1 to the Myogenin promoter (Fig. 6, C and D), which suggests that SFMBT1 depends on MyoD to localize on the Myogenin promoter and repress muscle genes in the myoblasts. Therefore, our overall data indicate that Sfmbt1 is localized on the MyoD target Myogenin promoter and represses MyoD-dependent transcription, which is required for myogenic differentiation.
FIGURE 6.
Sfmbt1 recruits multiple transcriptional repressor complexes and epigenetically represses MyoD-mediated transcription. A, SFMBT1 overexpression inhibited Myogenin promoter reporter activity. C2C12 cells were transfected with pMyog-Luc, a Renilla luciferase construct, and different amounts of SFMBT1 plasmids. Firefly luciferase activities were analyzed and normalized against Renilla luciferase activities 48 h after transfection. **, p < 0.01. B, Sfmbt1 knockdown resulted in enhanced Myogenin promoter reporter activity. Sfmbt1 knockdown and control cells were transfected with Myog-Luc and Renilla luciferase plasmids for 48 h. **, p < 0.01. C, SFMBT1 depends on MyoD to bind to the Myogenin promoter. ChIP assays with anti-FLAG-agarose or anti-IgG-agarose control were performed using FLAG-SFMBT1-expressing C2C12 cells without or with MyoD depletion, and the purified ChIP DNA was used as templates for quantitative PCR with Myogenin promoter primers. A percentage of the input is presented. D, Western blotting confirmed reduced MyoD expression in MyoD knockdown cells. E, Sfmbt1 depletion resulted in reduced binding of transcriptional repressors, including EZH2, RNF2, LSD1, and HDAC1, to the Myogenin promoter by ChIP assays. *, p < 0.05. **, p < 0.01. F, Sfmbt1 depletion caused enhanced levels of active histone marks and reduced levels of repressive marks on the myogenin promoter region by ChIP assays. *, p < 0.05. **, p < 0.01. G, DNase I sensitivity assays showed that Sfmbt1 knockdown leads to increased chromatin accessibility around the TSS region of the myogenin gene. Intact nuclei isolated from Sfmbt1 knockdown and control C2C12 cells were treated with DNase I at 37 °C for 30 min. Genomic DNA was then isolated and amplified with the primers spanning the TSS site of the Myogenin gene. The relative levels of uncut DNA around the TSS site of the myogenin gene were presented. *, p < 0.05; **, p < 0.01. H, proposed model of Sfmbt1 in regulating MyoD-mediated epigenetic silencing of myogenesis.
SFMBT1 is associated with multiple transcriptionally repressive complexes, including the CtBP/LSD1/HDAC complex, PRC, and MBT family proteins (Fig. 1). In addition, SFMBT1 interacts with MyoD and directly regulates MyoD target gene transcription (Figs. 4–6). Therefore, we further examined the effect of Sfmbt1 depletion on the recruitment of multiple corepressors to the MyoD target Myogenin promoter by ChIP assays. As shown in Fig. 6E, Sfmbt1 knockdown resulted in decreased binding of multiple transcription corepressors such as LSD1, HDAC1, EZH2, and RING2 to the Myogenin promoter region that spans the MyoD binding sites. These data thus indicate that Sfmbt1 is required for the recruitment of multiple transcriptional corepressor complexes to MyoD target loci.
Because the Sfmbt1-associated CtBP/LSD1/HDACs complex and PRC components have enzymatic activities to catalyze histone methylation/demethylation and deacetylation, we further investigated whether Sfmbt1 depletion affects the histone modification status on the MyoD target loci. Our ChIP assays showed that Sfmbt1 knockdown resulted in decreased levels in the transcriptionally repressive histone mark H3K27me3 and increased levels of active histone marks, H3K4me2, AcH3, and AcH4 (Fig. 6F), in the MyoD-binding region of the Myogenin gene promoter.
Moreover, a protein complex analysis showed that SFMBT1 is associated with the MBT family proteins and PRC1 proteins that function in chromatin compaction, suggesting that the Sfmbt1-associated complex might contribute to chromatin structure compaction on target promoters. Therefore, we compared chromatin accessibility flanking the transcription initiation site (TSS) of MyoD target gene Myogenin before and after Sfmbt1 knockdown using a DNase I sensitivity assay. In brief, intact nuclei isolated from Sfmbt1-depleted and control C2C12 cells were treated with DNase I. Genomic DNA was then extracted for real-time RT-PCR analysis to measure the uncut DNA spanning the TSS site of the Myogenin gene. We found that Sfmbt1 knockdown reduces the level of uncut DNA at the Myogenin promoter region, suggesting that the region spanning the TSS site of the Myogenin gene is relatively more accessible to DNase I digestion in Sfmbt1 knockdown cells (Fig. 6G). These data support that the Sfmbt1-mediated chromatin compaction of the target gene promoter contributes to transcriptional repression.
DISCUSSION
The molecular mechanisms and biological functions of SFMBT1 were previously uncharacterized. In this study, through protein complex analysis, gene expression profiling, and myogenic differentiation assays, we identified novel mechanisms accounting for SFMBT1-mediated transcription repression and revealed a critical role of Sfmbt1 in regulating muscle cell growth and differentiation programs that contribute to the maintenance of undifferentiated states of myogenic progenitors.
Our unbiased proteomic analysis of the SFMBT1 protein complex and subsequent biochemical interaction validations strongly indicate that SFMBT1 is associated with multiple transcriptional corepressors, including the CtBP/LSD1/HDAC complex components, the PRC components, and other MBT proteins. Multiple studies have revealed functional interplay among these protein complexes in transcriptional repression. For instance, the CtBP/LSD1/HDAC complex components catalyze histone demethylation/deacetylation (30, 31) and recruit polycomb proteins to target promoters (32, 33). The PRC1 complex maintains transcriptional repression and compacts chromatin at target promoters (10). Moreover, the MBT protein Scm in the PRC1 complex recruits other components of PRC2 and PRC1 to mediate the histone modifications at target loci and repress transcription (34). In addition, the L3MBTL proteins function as chromatin readers and compact the chromatin (35, 36). Because these SFMBT1-associated proteins are critical regulators of histone modifications, polycomb protein recruitment, and chromatin compaction, our study thus identified a novel, central role of SFMBT1 in coordinating transcriptional repressive complexes in transcription repression.
It was shown previously that Drosophila dSfmbt binds to low methylated repressive histone marks in vitro (9, 11), yet human SFMBT1 was able to bind to unmodified H3 (8). In light of different associated protein complexes, it is possible that mammalian SFMBT1 might interact with histone marks differently from dSfmbt in terms of the requirement of posttranslational modifications. Our data support that SFMBT1 might interact with histone tails and function as a repressive chromatin reader and recruit multiple corepressor complexes to regulate the histone modifications and chromatin compaction at target loci, subsequently repressing transcription.
The biological functions of SFMBT1 have not been elucidated. In this study, we investigated the function and mechanism of Sfmbt1-mediated transcriptional regulation in skeletal myogenesis, as current evidence strongly indicates that skeletal myogenesis is under epigenetic control (37). By utilizing the skeletal myogenesis model, we showed that 1) Sfmbt1 expression is down-regulated during myogenic differentiation, 2) overexpression of SFMBT1 in C2C12 myoblasts blocks the myogenic differentiation, and 3) Sfmbt1 knockdown promotes the myogenic differentiation in C2C12 and primary myoblasts and reduces proliferation. Overall, gain of function and loss of function of Sfmbt1 significantly impact growth and differentiation of myogenic cells in vitro. Therefore, our data suggest a critical role of Sfmbt1 in regulating skeletal myogenesis, likely via supporting muscle stem cell proliferation and preventing premature differentiation.
The mechanisms of polycomb and MBT protein recruitment to specific genomic loci and regulation of target gene expression in a mammalian system remains unclear (5, 10, 38). Many studies have revealed that transcription factors are crucial for polycomb and MBT protein recruitment to target promoters (34–36). It has been reported that the bHLH transcription factor MyoD recruits different epigenetic regulators to myogenic target loci and establishes spatiotemporal muscle-specific gene expression patterns in the myogenic developmental process (25). About 42% of the derepressed genes in Sfmbt1 knockdown C2C12 cells are direct MyoD targets (Fig. 4C), strongly suggesting that MyoD might recruit Sfmbt1 to target promoters to mediate the transcription silencing. This is supported by our findings that SFMBT1 interacts with MyoD and represses MyoD target Myogenin transcription (Figs. 5 and 6). Moreover, SFMBT1 directly binds to the MyoD target Myogenin promoter and is required for the recruitment of its associated transcriptional repressors to the Myogenin promoter (Fig. 6). Accordingly, the transcriptional repressive histone mark H3K27me3 decreases and the active marks H3K4me2, AcH3, and AcH4 increase on the MyoD target promoter when Sfmbt1 is depleted. Our chromatin accessibility assay revealed that the chromatin structure in the promoter region of MyoD target Myogenin became more accessible in Sfmbt1 knockdown C2C12 cells, suggesting that Sfmbt1 is required for the chromatin compaction of MyoD binding regions. Therefore, our data indicated that Sfmbt1 is required for the recruitment of multiple repressive complexes to MyoD and maintenance of repressive histone marks and chromatin compaction on MyoD target promoters.
These findings thus allow us to propose a model in which the MBT domain-containing protein Sfmbt1 associates with multiple transcriptional repressive complexes and functions as a negative regulator of myogenic differentiation (Fig. 6H). In undifferentiated myoblasts, Sfmbt1 interacts with MyoD and recruits its associated repressors to MyoD to mediate histone modifications and chromatin compaction on MyoD target promoters, which excludes the binding of coactivators and results in epigenetic silencing of muscle genes and maintenance of the undifferentiated state of myogenic progenitor cells. When Sfmbt1 is depleted, transcriptional corepressors are no longer associated with MyoD, which facilitates transcriptional derepression of MyoD target genes and promotes myogenic differentiation. In summary, our study provides strong evidence that Sfmbt1 links functionally to Polycomb proteins, MBT proteins, and the LSD1 complex and critically regulates target gene transcription and, subsequently, cell growth and differentiation programs.
Supplementary Material
Acknowledgments
We are grateful to Drs. Serge A. Leibovitch and Slimane Ait-Si-Ali for sharing the MyoD plasmids, Dr. Stephen J Tapscott for kindly providing the pMyog-luc plasmid, and Dr. Toru Miyazaki for the pCMV2-FLAG-L3MBTL3 plasmid.
This work was supported by Muscular Dystrophy Association Grant MDA 4315 (to L. W.), by the University of Florida Shands Cancer Center Startup fund, and by the Florida Bankhead-Coley Cancer Research Program.
The microarray data were deposited in the NCBI Gene Expression Omnibus and are accessible through GEO series number GSE 24346.

This article contains supplemental Figs. S1–S5 and Tables S1–S3.
- MBT
- malignant brain tumor
- PRC
- polycomb protein complex
- aa
- amino acids
- bFGF
- basic fibroblast growth factor
- IP
- immunoprecipitation
- bHLH
- basic helix-loop-helix
- TSS
- transcription start site
- CtBP
- C-terminal binding protein
- HDAC
- histone deacetylase.
REFERENCES
- 1. Luger K., Richmond T. J. (1998) DNA binding within the nucleosome core. Curr. Opin. Struct. Biol. 8, 33–40 [DOI] [PubMed] [Google Scholar]
- 2. Luger K., Hansen J. C. (2005) Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 15, 188–196 [DOI] [PubMed] [Google Scholar]
- 3. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 4. Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonasio R., Lecona E., Reinberg D. (2010) MBT domain proteins in development and disease. Semin. Cell Dev. Biol. 21, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gateff E., Löffler T., Wismar J. (1993) A temperature-sensitive brain tumor suppressor mutation of Drosophila melanogaster. Developmental studies and molecular localization of the gene. Mech. Dev. 41, 15–31 [DOI] [PubMed] [Google Scholar]
- 7. Wismar J., Löffler T., Habtemichael N., Vef O., Geissen M., Zirwes R., Altmeyer W., Sass H., Gateff E. (1995) The Drosophila melanogaster tumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech. Dev. 53, 141–154 [DOI] [PubMed] [Google Scholar]
- 8. Wu S., Trievel R. C., Rice J. C. (2007) Human SFMBT is a transcriptional repressor protein that selectively binds the N-terminal tail of histone H3. FEBS Lett. 581, 3289–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klymenko T., Papp B., Fischle W., Köcher T., Schelder M., Fritsch C., Wild B., Wilm M., Müller J. (2006) A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 20, 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simon J. A., Kingston R. E. (2009) Mechanisms of polycomb gene silencing. Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708 [DOI] [PubMed] [Google Scholar]
- 11. Grimm C., Matos R., Ly-Hartig N., Steuerwald U., Lindner D., Rybin V., Müller J., Müller C. W. (2009) Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 28, 1965–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu S., Shi Y., Mulligan P., Gay F., Landry J., Liu H., Lu J., Qi H. H., Wang W., Nickoloff J. A., Wu C., Shi Y. (2007) A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat. Struct. Mol. Biol. 14, 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berkes C. A., Bergstrom D. A., Penn B. H., Seaver K. J., Knoepfler P. S., Tapscott S. J. (2004) Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14, 465–477 [DOI] [PubMed] [Google Scholar]
- 14. Arai S., Miyazaki T. (2005) Impaired maturation of myeloid progenitors in mice lacking novel Polycomb group protein MBT-1. EMBO J. 24, 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Philipot O., Joliot V., Ait-Mohamed O., Pellentz C., Robin P., Fritsch L., Ait-Si-Ali S. (2010) The core binding factor CBF negatively regulates skeletal muscle terminal differentiation. PLoS ONE 5, e9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Springer M. L., Rando T. A., Blau H. M. (2002) Gene delivery to muscle. Curr. Protoc. Hum. Genet. Chapter 13, 13.4.1–13.4.19 [DOI] [PubMed] [Google Scholar]
- 17. Ogawa H., Ishiguro K., Gaubatz S., Livingston D. M., Nakatani Y. (2002) A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296, 1132–1136 [DOI] [PubMed] [Google Scholar]
- 18. Wu L., Aster J. C., Blacklow S. C., Lake R., Artavanis-Tsakonas S., Griffin J. D. (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26, 484–489 [DOI] [PubMed] [Google Scholar]
- 19. Shen H., McElhinny A. S., Cao Y., Gao P., Liu J., Bronson R., Griffin J. D., Wu L. (2006) The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 20, 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu Y., Lin S., Li J. L., Nakagawa H., Chen Z., Jin B., Tian L., Ucar D. A., Shen H., Lu J., Hochwald S. N., Kaye F. J., Wu L. (2012) Altered LKB1/CREB-regulated transcription co-activator (CRTC) signaling axis promotes esophageal cancer cell migration and invasion. Oncogene 31, 469–479 [DOI] [PubMed] [Google Scholar]
- 21. Usui H., Ichikawa T., Kobayashi K., Kumanishi T. (2000) Cloning of a novel murine gene Sfmbt, Scm-related gene containing four mbt domains, structurally belonging to the Polycomb group of genes. Gene 248, 127–135 [DOI] [PubMed] [Google Scholar]
- 22. Oktaba K., Gutiérrez L., Gagneur J., Girardot C., Sengupta A. K., Furlong E. E., Müller J. (2008) Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell 15, 877–889 [DOI] [PubMed] [Google Scholar]
- 23. Palacios D., Mozzetta C., Consalvi S., Caretti G., Saccone V., Proserpio V., Marquez V. E., Valente S., Mai A., Forcales S. V., Sartorelli V., Puri P. L. (2010) TNF/p38α/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 7, 455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caretti G., Di Padova M., Micales B., Lyons G. E., Sartorelli V. (2004) The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 18, 2627–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aziz A., Liu Q. C., Dilworth F. J. (2010) Regulating a master regulator: establishing tissue-specific gene expression in skeletal muscle. Epigenetics 5, 691–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robson L. G., Di Foggia V., Radunovic A., Bird K., Zhang X., Marino S. (2011) Bmi1 is expressed in postnatal myogenic satellite cells, controls their maintenance and plays an essential role in repeated muscle regeneration. PLoS ONE 6, e27116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Day K., Shefer G., Richardson J. B., Enikolopov G., Yablonka-Reuveni Z. (2007) Nestin-GFP reporter expression defines the quiescent state of skeletal muscle satellite cells. Dev. Biol. 304, 246–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G. J., Parker M. H., MacQuarrie K. L., Davison J., Morgan M. T., Ruzzo W. L., Gentleman R. C., Tapscott S. J. (2010) Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell 18, 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berkes C. A., Tapscott S. J. (2005) MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 16, 585–595 [DOI] [PubMed] [Google Scholar]
- 30. Shi Y., Sawada J., Sui G., Affar el B., Whetstine J. R., Lan F., Ogawa H., Luke M. P., Nakatani Y., Shi Y. (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422, 735–738 [DOI] [PubMed] [Google Scholar]
- 31. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 32. Srinivasan L., Atchison M. L. (2004) YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 18, 2596–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Basu A., Atchison M. L. (2010) CtBP levels control intergenic transcripts, PHO/YY1 DNA binding, and PcG recruitment to DNA. J. Cell Biochem. 110, 62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang L., Jahren N., Miller E. L., Ketel C. S., Mallin D. R., Simon J. A. (2010) Comparative analysis of chromatin binding by Sex Comb on Midleg (SCM) and other polycomb group repressors at a Drosophila Hox gene. Mol. Cell Biol. 30, 2584–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trojer P., Li G., Sims R. J., 3rd, Vaquero A., Kalakonda N., Boccuni P., Lee D., Erdjument-Bromage H., Tempst P., Nimer S. D., Wang Y. H., Reinberg D. (2007) L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129, 915–928 [DOI] [PubMed] [Google Scholar]
- 36. Trojer P., Cao A. R., Gao Z., Li Y., Zhang J., Xu X., Li G., Losson R., Erdjument-Bromage H., Tempst P., Farnham P. J., Reinberg D. (2011) L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol. Cell 42, 438–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bharathy N., Taneja R. (2012) Methylation muscles into transcription factor silencing. Transcription 3, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schuettengruber B., Cavalli G. (2009) Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136, 3531–3542 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.