Background: miR1 is a muscle-specific microRNA, however, its role in muscle atrophy is unclear.
Results: Excess dexamethasone and myostatin induced miR1 via glucocorticoid receptor, resulting in reduced HSP70 and development of muscle atrophy.
Conclusion: miR1 mediates muscle atrophy by regulating the levels of HSP70.
Significance: Given that miR1 has a role in muscle atrophy, its antagonism may be beneficial during atrophy.
Keywords: Akt, Glucocorticoid Receptor, Heat Shock Protein, MicroRNA, Muscle Atrophy, Dexamethasone, HSP70, Myostatin, miR1
Abstract
High doses of dexamethasone (Dex) or myostatin (Mstn) induce severe atrophy of skeletal muscle. Here we show a novel microRNA1 (miR1)-mediated mechanism through which Dex promotes skeletal muscle atrophy. Using both C2C12 myotubes and mouse models of Dex-induced atrophy we show that Dex induces miR1 expression through glucocorticoid receptor (GR). We further show that Mstn treatment facilitates GR nuclear translocation and thereby induces miR1 expression. Inhibition of miR1 in C2C12 myotubes attenuated the Dex-induced increase in atrophy-related proteins confirming a role for miR1 in atrophy. Analysis of miR1 targets revealed that HSP70 is regulated by miR1 during atrophy. Our results demonstrate that increased miR1 during atrophy reduced HSP70 levels, which resulted in decreased phosphorylation of AKT, as HSP70 binds to and protects phosphorylation of AKT. We further show that loss of pAKT leads to decreased phosphorylation, and thus, enhanced activation of FOXO3, up-regulation of MuRF1 and Atrogin-1, and progression of skeletal muscle atrophy. Based on these results, we propose a model whereby Dex- and Mstn-mediated atrophic signals are integrated through miR1, which then either directly or indirectly, inhibits the proteins involved in providing protection against atrophy.
Introduction
Glucocorticoids are used as therapeutic agents due to their potent anti-inflammatory and immunosuppressive functions (1). However, high doses and sustained usage of glucocorticoids result in muscle atrophy (2). The molecular mechanisms of glucocorticoid-induced muscle atrophy have been well studied and two muscle-specific E3 ubiquitin ligases, namely muscle ring finger protein 1 (MuRF1)3 and Atrogin-1/MAFbx (muscle atrophy F-box) have been found to play a key role in glucocorticoid-induced atrophy (3–8). MuRF1 and Atrogin-1 have been shown to ubiquitinate sarcomeric proteins, such as myosin heavy chain, which are then targeted for degradation through the ubiquitin proteosome pathway (9, 10). MuRF1 and Atrogin-1 are transcriptionally activated by FOXO1 and FOXO3, members of the FOXO family of forkhead transcription factors, in skeletal muscle (9, 11). Mechanistically, dephosphorylation and thus activation of FOXO1 and FOXO3 results in their translocation into the nucleus where they transcriptionally activate MuRF1 and Atrogin-1 to induce atrophy. Activation of the insulin-like growth factor 1/PI3K/AKT pathway rescues dexamethasone (Dex)-mediated atrophy through phosphorylating and inactivating FOXO1/FOXO3 (5, 7, 12, 13) implicating inhibition of this pathway as one of the main mechanisms behind Dex-induced atrophy.
Mstn, a member of the transforming growth factor β (TGF-β) superfamily, has also been shown to block insulin-like growth factor 1/PI3K/AKT signaling, resulting in enhanced MuRF1 and Atrogin-1 expression and skeletal muscle atrophy (14). Furthermore, Dex-mediated atrophy results in a dose-dependent increase in Mstn mRNA and protein expression (15) and deletion of the Myostatin gene can prevent Dex-induced muscle wasting (16), suggesting that Dex-mediated muscle atrophy occurs at least in part through the action of Mstn.
Recently, muscle-specific microRNAs (myomiRs) have been shown to participate in a multitude of regulatory pathways in skeletal muscle. Individual microRNAs (miRs) are known to target many mRNAs that control numerous cellular processes (17, 18). A cluster of myomiRs including miR1, miR133, and miR206 plays a role in myogenesis and muscle regeneration (19, 20). Both miR1 and miR133 were found to be down-regulated in skeletal muscle undergoing hypertrophy and it was proposed that Igf-1 mRNA may be negatively regulated by miR1 in skeletal muscle (21) and cardiac muscle (22). Although it appears that miR1 has a role in regulating muscle hypertrophy through IGF-1, little is known about the role of miR1 in skeletal muscle atrophy.
In this study we present evidence to support that miR1 expression is up-regulated during Dex- and Mstn-induced muscle atrophy through a mechanism mediated via glucocorticoid receptor (GR). Through in vitro and in vivo experiments we further show that Dex-induced up-regulation of miR1 resulted in the targeted loss of heat shock protein 70 (HSP70) in muscle. Furthermore, our results demonstrate that during Dex-mediated atrophy, levels of both HSP70 and phosphorylated AKT in association with each other were reduced, indicative of the increased dephosphorylation of AKT observed in response to Dex treatment. Reduced phosphorylation of AKT, in turn, resulted in dephosphorylation of FOXO3, induction of MuRF1 and Atrogin-1, and skeletal muscle atrophy. Thus, taken together these data reveal that both Dex and Mstn can regulate the expression and function of genes associated with skeletal muscle atrophy via miR1.
EXPERIMENTAL PROCEDURES
Animals
Eight-week-old C57Bl/6J male wild type (WT) mice were obtained from National University of Singapore Center for Animal Resources, Singapore. Mstn knock-out (Mstn−/−) mice were originally a gift from Prof. S. J. Lee, Johns Hopkins University, Baltimore, MD, and were maintained at the Nanyang Technological University (NTU) animal house, Singapore. All animals had ad libitum access to chow diet and water. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC), Singapore. Both Mstn−/− and WT mice received subcutaneous injections of either normal saline (0.9% NaCl) or dexamethasone (ilium, Troy Laboratories Pty. Ltd., Australia) daily, until ∼18% body weight loss was seen in the WT mice. Dex was used at a concentration of 5 μg/g body weight. Hind limb muscles were removed, weighed, and stored at −80 °C for further analysis.
Reagents and Proteins
Conditioned medium containing Mstn (conditioned Mstn medium; CMM) was obtained from CHO cells designed to produce and secrete Mstn protein (23). Conditioned medium was also collected from wild type CHO cells (conditioned CHO medium) to act as a control for Mstn treatment experiments. The concentration of Mstn in the CMM was determined by EIA (Immundiagnostik AG, Bensheim, Germany). The activity of the media has been validated previously (24, 25). Dex and RU486 were from Sigma and the HSP70 inhibitor (VER155008) (26) from Tocris Biosciences. The recombinant Mstn inhibitor, Ant1, was expressed and purified as described previously (27).
Cell Culture
The C2C12 murine myoblast cell line (28), obtained from ATCC (American Type Culture Collection, Manassas, VA), was used in the present study and maintained as described previously (24). C2C12 cells were passaged in proliferation medium (DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin) and differentiated in C2C12 differentiation medium containing DMEM, 2% horse serum (Invitrogen) and 1% penicillin and streptomycin. The C2C12 myotubes were differentiated for 72 h and then treated with CMM or conditioned CHO medium, Dex (100 μm), or 100% ethanol (as a control for Dex treatment experiments), RU486 (500 nm), Ant1 (7 μg/ml), or VER155008 (10 and 20 μm), or a combination thereof for 6 and 24 h. To assess the ability of Dex and VER155008 (HSP70 inhibitor) to induce myotubular atrophy, C2C12 myoblasts were plated on Thermanox coverslips (Nunc, Roskilde, Denmark) at a density of 25,000 cells/cm2 and induced to differentiate under low serum conditions (DMEM, 2% horse serum). Following 72 h of differentiation, the myotubes were treated for a further 24 h in differentiation media with ethanol or Dex and DMSO or VER155008. C2C12 myotubes were then fixed with ethanol:formaldehyde:glacial acetic acid (20:2:1) and stained with Gill's hematoxylin and 1% eosin. The myotube area was assessed microscopically using the Image-Pro Plus analysis software package (MediaCybernetics, Bethesda, MD), with individual myotube area assessed for all myotubes present in 10 random images per coverslip, using 3 coverslips per treatment.
Promoter Sequence Analysis and miR1 Target Prediction
The miR1 promoter sequence was obtained from the NCBI genome database. For prediction of the transcription factor binding sites in the miR1 promoter region, the Transcription Element Search System (TESS) program was employed. RNAhybrid version 2.0 was used for checking complementarity between the miR1 sequence and 3′ untranslated region (UTR) of the mouse Hsp-70 gene.
Reverse Transcriptase-Quantitative Polymerase Chain Reaction (RT-qPCR)
RNA was isolated from differentiated C2C12 myotubes and biceps femoris (BF), quadriceps, and tibialis anterior muscles of mice using TRIzol (Invitrogen). For detection of primary miR1, cDNA was synthesized using the iScriptTM cDNA synthesis kit, according to the manufacturer's (Bio-Rad) instructions. For detecting mature miR1, the miScript RT(II) kit (Qiagen, Hamburg, Germany) was used for cDNA synthesis. Quantitative analysis of gene expression by real-time PCR was carried out using the CFX96 (Bio-Rad) system. Each real-time PCR (10 μl) contained 2 μl of cDNA, 5 μl of SsoFast Evagreen (Bio-Rad), and forward and reverse primers at a final concentration of 2.5 μm. For detection of primary miR1 (miR1–1) transcript expression the following primers were used: forward, 5′-AAA CAT ACT TCT TTA TAT GCC CA-3′ and reverse, 5′-TAC ATA CTT CTT TAC ATT CCA TAG C-3′. Mature miR1 expression was detected using miScript Primer Assay 218300 (Qiagen, catalog number MS00005873) and a universal reverse primer. All reactions were performed using the following thermal cycler conditions: 98 °C for 3 min, followed by 45 cycles of a three-step reaction, denaturation at 98 °C for 3 s, annealing at 59 °C for 10 s and extension at 72 °C for 10 s, and final extension at 95 °C for 10 s. The reaction was followed by a melting curve from 65 to 95 °C in 5-s increments of 0.5 °C to ensure amplification specificity. Transcript levels were normalized to the U6 transcript levels. Relative fold-change in expression was calculated using the ΔΔCt method.
Northern Blotting
Northern blotting was performed essentially as described previously (29). miR1 expression was detected using a DNA oligo complementary to the miR1 mature sequence 5′-TAC ATA CTT CTT TAC ATT CCA-3′.
Protein Isolation
C2C12 myotubes, following the various treatments, were harvested in protein lysis buffer (50 mm Tris, pH 7.6, 250 mm NaCl, 5 mm EDTA, 0.1% Igepal-Nonidet P-40 (Fluka, St. Louis, MO)), and Complete protease inhibitor mixture (Roche Diagnostics) to generate protein lysates, as previously described (14). BF muscles from WT and Mstn−/− mice injected with Dex or saline were homogenized in protein lysis buffer and the clear supernatants were collected. These protein lysates were used for Western blotting analysis. Cytoplasmic and nuclear fractions from C2C12 myotubes following Dex and Mstn treatments were isolated as previously described (30).
Western Blot Analysis
Total protein (15–40 μg) was separated on 4–12% BisTris polyacrylamide gels (Invitrogen) and transferred to nitrocellulose membranes by electroblotting. The membranes were blocked for 1 h at room temperature in 5% nonfat milk in 1× TBS-T, then incubated overnight at 4 °C with the respective primary antibodies. The list of antibodies used are: anti-GR (sc-8992), anti-pFOXO3 (sc-101683), anti-FOXO3 (sc-11351), anti-HSP70 (sc-66048), anti-pAKT1/2/3 (Ser473) (SC-7985-R), anti-AKT1/2/3 (sc-8312), and anti-MuRF1 (SC-32920) and all were from Santa Cruz Biotechnology (CA); anti-Mstn (AF788) from R&D Systems, and anti-Atrogin1 (PAB15627) from Abnova (Taipei, Taiwan). Following washes and incubation with the respective secondary antibodies for 1 h at room temperature, the HRP activity was detected using Western Lightning Plus Chemiluminescent reagent (PerkinElmer Life Sciences). The developed films were scanned using the GS-800 Calibrated Densitometer (Bio-Rad), and band density was calculated by Quantity One software.
Immunoprecipitation
Five hundred μg of total protein from BF muscle of WT mice injected with saline or Dex was incubated with 2 μg of anti-pAKT antibody (SC-7985-R) overnight at 4 °C. Protein A-agarose beads (Invitrogen) were then added and the mixture was incubated for a further 2 h at 4 °C. The beads were then washed 4 times with cold 1× PBS and the complexes were eluted. The proteins were denatured by boiling for 5 min in SDS buffer. Samples were then subjected to SDS-polyacrylamide gel electrophoresis and Western blotting as described above.
Electrophoretic Mobility Shift Assay (EMSA)
The biotinylated and cold oligonucleotides (Sigma) containing the glucocorticoid response element (GRE) of the mouse miR1 promoter (5′-CCT GCT GAC ACG GAA GAA CTG AAT GTT CTT TCA GGA GCT-3′) were hybridized to their respective complementary strands. EMSA was performed using the Lightshift Chemiluminescent EMSA kit (Thermo Scientific-Pierce Biotechnology) as per the manufacturer's protocol and as described previously (24). Membranes were probed with stabilized streptavidin-horseradish peroxidase conjugates and developed using the Lightshift Chemiluminescent EMSA kit (Thermo Scientific-Pierce Biotechnology). To confirm the presence of GR in the complex, nuclear extracts from C2C12 myotubes were preincubated for 20 min at room temperature with 1 μl of anti-GR antibody (H300) (sc-8992X) before incubation with the labeled probe for another 20 min at room temperature. To confirm binding of the nuclear protein to the labeled GRE probes, unlabeled primers were added to the reaction mixture to outcompete the labeled GRE oligos.
ChIP Analysis
C2C12 myoblasts were allowed to differentiate for 72 h and then treated with 100 μm Dex or ethanol for a further 24 h. The myotubes were fixed with 1% formaldehyde and collected for the ChIP experiment. ChIP analysis was done using GR (H300) antibody from Santa Cruz (SC-8992X) as previously described (31). The following primers were used for PCR: GR forward primer, 5′-TGTTTCCACACAGGGACAGGGCT-3′; GR reverse primer, 5′-GGCAGTCCCAGCTCCTGAAAGAA-3′.
Transfections and Plasmids
Two miR1–1 promoter fragments, encompassing 1730 and 1518 base pairs (bp), respectively, upstream from the transcription start site were PCR amplified and subsequently directionally cloned into the pGL3B vector (Promega) using NheI and HindIII. The plasmid containing the 1730-bp fragment of the miR1–1 promoter (with the GRE sequence) was designated PC1 and the plasmid containing the truncated 1518-bp fragment of the miR1–1 promoter (without GRE) was designated PC2.
The GR and miR1 overexpression vectors used in the study were a kind gift from Dr. S Stoney Simons (NIDDK, National Institutes of Health) and Prof. T. Akimoto (CDBIM, University of Tokyo, Japan), respectively. AntagomiR1 (5′-AAA UAC AUA CUU CUU UAC AUU CCA UAG C-3′) and nonspecific miR (5′-UCA CAA CCU CCU AGA AAG AGU AGA-3′) sequences were RNA sequences with 2′-O-methyl modifications on all bases and were synthesized by Sigma. The mouse Hsp70 gene 3′ UTR sequence showing the miR1 sequence complementarity was cloned into the pMIR reporter vector (Invitrogen). Both wild type (3′ UTR Hsp70 WT) and mutated forms of the HSP70 3′ UTR sequence (3′ UTR Hsp70 MUT) were cloned, as mentioned above.
C2C12 myoblasts were transfected with either miR1 expression vector or empty vector using Lipofectamine 2000 (Invitrogen) and allowed to differentiate for 72 h prior to collection in protein lysis buffer (50 mm Tris, pH 7.6, 250 mm NaCl, 5 mm EDTA, 0.1% Igepal-Nonidet P-40 (Fluka)).
C2C12 myoblasts were transfected with PC1 and PC2 using Lipofectamine 2000 (Invitrogen). C2C12 myoblasts were then differentiated for 72 h, and protein lysates were collected in 1× passive lysis buffer for luciferase assay. Co-transfection of either PC1 or PC2 with the GR expression vector was also done as mentioned above. Luciferase activity was determined using the GloMax Multidetection system (Promega).
HSP70 overexpression vector (Addgene plasmid 15215, Cambridge, MA) (32) or empty vector was transfected in C2C12 myoblasts using Lipofectamine 2000 (Invitrogen). The myoblasts were differentiated for 72 h and then treated with 100 μm Dex or ethanol for 24 h prior to collection of cells for protein or RNA.
AntagomiR1 Transfections
C2C12 myoblasts were plated on Thermanox coverslips (Nunc, Roskilde, Denmark) for myotube size quantification and in 6-well plates for protein and RNA analysis, at a density of 25,000 cells/cm2 and induced to differentiate under low serum conditions (DMEM + 2% horse serum). Following 72 h of differentiation, the myotubes were transfected with 200 nm AntagomiR1 (5′-AAA UAC AUA CUU CUU UAC AUU CCA UAG C-3′) and nonspecific miR (5′-UCA CAA CCU CCU AGA AAG AGU AGA-3′) oligos with 2′-O-methyl modifications on all bases and synthesized by Sigma. Transfections were done using Lipofectamine 2000 according to the manufacturer's protocol, and 6 h after the transfection, medium was replaced with differentiation medium. The cells were treated with either 100 μm Dex or ethanol for 24 h and collected for RNA or protein. For myotube size quantification, C2C12 myotubes were fixed with ethanol:formaldehyde:glacial acetic acid (20:2:1) after 24 h of Dex treatment of the transfected myotubes. The cells were stained with Gill's hematoxylin and 1% eosin. Myotube area was assessed for all myotubes present in 10 random images in triplicate by the Image-Pro Plus analysis software package (MediaCybernetics).
Statistical Analysis
Experimental replicates are mentioned in relevant figure legends. Transfections were performed at least three times, each in triplicate. Statistical analysis was performed using one-way analysis of variance and Student's t test. p values of p < 0.05 (*) were considered as significant. Error bars are expressed as mean ± S.E.
RESULTS
miR1 Expression Is Increased during in Vitro and in Vivo Skeletal Muscle Atrophy
miR1 is encoded by two different genes, miR1–1 and miR1–2, on different mouse chromosomes. We have concentrated our studies on miR1–1, which is located on chromosome 2 in a cluster with miR133a2 and contributes to the levels of mature miR1. In this article, we will be using the terms “miR1–1” and “miR1” to refer to primary and mature miR1 forms, respectively. Although the expression level of miR1 was found to be altered during load-induced hypertrophy (21), its role in muscular atrophy is currently not known. Therefore we queried if miR1 expression is altered in differentiated C2C12 myotubes undergoing skeletal muscle atrophy induced through treatment with Dex and Mstn. Subsequent real-time PCR and Northern blot revealed an increase in the expression of the primary (Fig. 1A) and mature forms (Fig. 1B) of miR1, respectively, at 6- and 24-h time points of Dex treatment. Consistent with the in vitro results, the expression of miR1–1 was also increased in hind limb muscles, BF, quadriceps, and tibialis anterior (Fig. 1C), collected from mice injected with Dex, when compared with saline-injected controls.
FIGURE 1.
Dex up-regulates miR1 in C2C12 myotubes and mouse skeletal muscle. A, real time qPCR analysis of miR1–1 primary transcript expression in control (Ethanol) and Dex-treated C2C12 myotubes at 6- and 24-h time points. Relative gene expression was normalized to U6 RNA expression. The graphs depict fold-differences relative to controls at each time point. Values are mean ± S.E. (n = 3); **, p < 0.01. B, Northern blot analysis of miR1 in control (Ethanol) and Dex-treated C2C12 myotubes at 6- and 24-h time points. The expression of U6 was measured as a loading control. C, real time qPCR analysis of miR1–1 primary transcript expression in BF, quadriceps, and tibialis anterior muscle from saline- and Dex-injected wild type mice. Relative gene expression analysis was performed using the ΔΔCt method and was normalized to U6 RNA expression. Values are mean ± S.E. (n = 3) and are expressed as the fold-difference relative to saline control; **, p < 0.01.
miR1–1 Expression Is Regulated by Mstn and GR during Dex-mediated Atrophy
After observing an increase in miR1–1 expression during atrophy, we next wanted to investigate how miR1–1 expression may be regulated during atrophy. Unpublished data4 from our lab indicated that Mstn levels were up-regulated in C2C12 myotubes undergoing Dex-mediated atrophy. Therefore, we asked if Mstn plays a role in the regulation of miR1 during Dex-induced atrophy. Mstn levels were analyzed by Western blotting of protein lysates from C2C12 myotubes treated with Dex. The results indicated a significant increase in Mstn levels in response to Dex treatment (Fig. 2A). Next, C2C12 myotubes were treated with CMM (24, 25) and the expression of miR1 was determined by real time PCR. Our results revealed that miR1 expression was up-regulated in C2C12 myotubes treated with CMM (Fig. 2B), supporting a role of Mstn in miR1 regulation. To confirm whether or not Mstn was involved in Dex-mediated up-regulation of miR1, we determined miR1 expression in BF muscle isolated from wild type and Mstn−/− mice treated with Dex. Subsequent Northern blot analysis showed that the increase in miR1 expression was significantly ablated in Mstn−/− muscle treated with Dex (Fig. 2, C and D). Furthermore, when Mstn signaling was inhibited in Dex-treated C2C12 myotubes, through addition of the Mstn-specific antagonist Ant1 (27), we also observed a significant reduction in miR1–1 expression, when compared with myotubes treated with Dex alone (Fig. 2E). These data suggest that Dex may signal via Mstn to induce miR1 expression during skeletal muscle atrophy.
FIGURE 2.
miR1–1 is transcriptionally regulated by Mstn and GR during Dex-mediated muscle atrophy. A, Western blot analysis of Mstn protein levels in control (Ethanol) and Dex-treated C2C12 myotubes. The level of GAPDH was measured as a loading control. B, real time qPCR analysis of miR1–1 primary transcript expression in conditioned CHO media (CCM) and conditioned Mstn media (CMM)-treated C2C12 myotubes at 6- and 24-h time points. Relative gene expression analysis was performed using the ΔΔCT method and was normalized to U6 RNA expression. The graphs depict fold-differences relative to controls at each time point. Values are mean ± S.E. (n = 3); **, p < 0.01. C, Northern blot analysis of miR1 expression in BF muscle from WT (lanes 1 and 2) and Mstn−/− mice (lanes 3 and 4) following either saline or Dex injection. The expression of U6 was measured as the loading control. D, corresponding densitometric analysis of miR1 levels in WT and Mstn−/− muscle following injection of either saline or Dex. Error bars represent mean ± S.E. (n = 3, **, p < 0.01). E, representative graph showing real time qPCR analysis of miR1–1 primary transcript expression in C2C12 myotubes treated with ethanol, Dex, or Dex + Ant1. Values are mean ± S.E. (n = 3) and are expressed as fold-differences relative to ethanol-treated controls; **, p < 0.01. F, Western blot analysis of GR levels in nuclear extracts from control (Ethanol) and Dex-treated C2C12 myotubes. Ponceau S staining was performed to ensure equal loading. G, schematic representation of the miR1–1 promoter showing the location and sequence of the GRE in the miR1–1 upstream enhancer sequence (top). The middle and bottom schematics represent the miR1–1/pGL3b promoter-reporter constructs with and without GRE, respectively. H, assessment of miR1–1 promoter-reporter activity during differentiation of C2C12 myoblasts into myotubes transfected with PC1 or empty vector control (pGL3b). miR1–1 promoter-reporter activity was normalized to Renilla luciferase and expressed as relative luminescence units (RLU). Each bar represents the mean ± S.E. (n = 3); **, p < 0.01. I, assessment of miR1–1 promoter-reporter activity in 72-h differentiated C2C12 myoblasts co-transfected with GR expression vector or empty vector and PC1, PC2, or empty vector control (pGL3b). miR1–1 promoter-reporter activity was normalized to Renilla luciferase and expressed as relative luminescence. Each bar represents the mean ± S.E. (n = 3); ***, p < 0.001. J, electrophoretic mobility shift assay (EMSA) with nuclear extracts from C2C12 myotubes treated with Dex for 6 h using oligonucleotides specific for the GRE within the mouse miR1–1 promoter as a probe. A band shift was noted after the addition of the nuclear extract and was enhanced in the presence of Dex. Lane 1 is the free oligo control and lanes 2 and 3 reveal the shifted band in control (Ethanol) and Dex-treated myotubes, respectively. K, EMSA with nuclear extracts from C2C12 myotubes treated with Dex for 6 h using oligonucleotides specific for the GRE within the mouse miR1–1 promoter as a probe together with competition oligos (unlabeled primers). A band shift was noted after the addition of the nuclear extract and was decreased in a dose-dependent manner upon addition of competition oligos (lanes 2–4). L, EMSA with nuclear extracts from C2C12 myotubes treated with Dex for 6 h using oligonucleotides specific for the GRE within the mouse miR1–1 promoter as a probe together with anti-GR antibody. The supershift upon the addition of anti-GR antibody confirmed the interaction of GR with the GRE in the mouse miR1–1 promoter upon Dex treatment (lanes 2 and 3). M, image of agarose gel depicting the interaction of endogenous GR with endogenous miR1–1 promoter in C2C12 myotubes when treated with 100 μm Dex as analyzed through ChIP. N, real time qPCR analysis of miR1–1 primary transcript expression in ethanol, Dex, and Dex + RU486-treated C2C12 myotubes. The graph displays the fold-change in miR1–1 expression relative to ethanol treated controls. Values are mean ± S.E. (n = 3); **, p < 0.01. O, Western blot analysis of GR levels in nuclear extracts from conditioned CHO media (CCM)- and CMM-treated C2C12 myotubes.
Because Dex is a synthetic glucocorticoid it signals by binding to the GR, which then translocates into the nucleus to regulate the expression of target genes. Our results show that nuclear translocation of GR increased in C2C12 myotubes after 6 h of Dex treatment (Fig. 2F), corresponding with the increased miR1–1 observed at the same time point (Fig. 1A). To further understand the miR1–1 transcriptional regulation, we analyzed the upstream promoter sequence of miR1–1 and identified a putative GRE in the miR1 upstream regulatory sequence (Fig. 2G). To ascertain whether or not the GRE was important for miR1–1 regulation during muscle atrophy we generated a miR1–1 promoter reporter construct containing the GRE binding element (PC1) and a truncated construct where the GRE was removed (PC2) (Fig. 2G). It has been previously shown that miR1 is up-regulated during differentiation (19). Therefore, activity of the miR1–1 promoter construct (PC1) was tested by transfecting it in C2C12 myoblasts and analyzing the luciferase activity of the promoter at different time points during differentiation. The luciferase activity of PC1 gradually increased at 6-, 24-, 48-, and 72-h time points during differentiation (Fig. 2H) as compared with pGL3b.
To determine GR-mediated regulation of miR1–1 promoter, C2C12 cells were transfected with PC1 or PC2 and GR expression vector and the respective control plasmids. A dramatic increase in promoter-luciferase activity was observed in 72-h differentiated myoblasts that were co-transfected with the PC1 and GR expression vector, when compared with 72-h differentiated PC2- and pGL3b-transfected myoblasts (Fig. 2I). These data indicate that the GRE within the miR1–1 upstream region is required for Dex- and GR-mediated regulation of miR1–1 expression. To support these findings we performed EMSA to assess for binding of GR to the mouse miR1–1 promoter. Electrophoretic mobility shift assay (EMSA) was performed with nuclear extracts from Dex-treated C2C12 myotubes using oligonucleotides specific for the GRE within the mouse miR1–1 promoter as a probe. A band shift was observed upon incubation of the probe with the nuclear extract, which was further enhanced upon Dex treatment (Fig. 2J). In addition, the band shift disappeared when unlabeled oligos were added (Fig. 2K) and a supershift was observed upon addition of anti-GR antibody (Fig. 2L), showing the specificity of interaction of the miR1–1 GRE with GR present in the Dex-treated nuclear extracts.
To establish that GR binds to GRE on the miR1–1 promoter in vivo we performed ChIP analysis. C2C12 myotubes (differentiated for 72 h) were treated with Dex and collected for ChIP analysis. Chromatin immunoprecipitation was performed using GR antibody, and DNA was isolated and purified. PCR amplification of immunoprecipitated DNA indicates the binding of endogenous GR on GRE specific to the miR1–1 promoter after Dex treatment (Fig. 2M).
To further confirm that GR was involved in Dex-mediated up-regulation of miR1–1, we treated myotubes with the GR-specific antagonist RU486 and assessed for miR1–1 expression in response to treatment with Dex. Consistent with the above findings, the increase in miR1–1 expression observed after Dex treatment was reversed in myotubes treated with RU486 (Fig. 2N).
To determine whether exogenous Mstn treatment resulted in enhanced GR nuclear translocation, we monitored for GR nuclear localization in the C2C12 myotubes treated with CMM. Subsequent Western blot analysis revealed higher levels of GR in nuclear extracts from CMM-treated C2C12 myotubes (Fig. 2O), indicating that addition of exogenous Mstn protein induced GR nuclear translocation. These results indicate that miR1 expression is mediated by both Mstn and GR during Dex-mediated atrophy.
Inhibition of miR1 Reduces the Levels of Atrophy-related Proteins
To antagonize miR1 we transfected C2C12 myotubes with AntagomiR1, which specifically targets and degrades miR1 thereby blocking miR1 function.
To validate efficacy of AntagomiR1 to inhibit miR1 expression, analysis of miR1 expression after AntagomiR1 transfection was conducted, which confirmed an ∼50% decrease in miR1 expression in AntagomiR1-transfected myotubes when compared with NS miR-transfected myotubes (Fig. 3F). However, after Dex treatment there was a 2-fold increase in miR1 expression in NS miR-transfected C2C12 myotubes but no significant increase was observed in AntagomiR1-transfected myotubes compared with the controls.
FIGURE 3.
miR1 targets HSP70 during Dex-mediated muscle atrophy. A, representative images of C2C12 myoblast differentiated for 72 h and transfected with AntagomiR1 or NS miR followed by 100 μm Dex or ethanol treatments for 24 h. Myotube cultures were fixed and stained with Gill's hematoxylin and eosin. Scale bar represents 100 μm. B, the graph showing frequency distribution of myotube area (μm2) for AntagomiR1 and NS miR transfected C2C12 myotubes from ethanol-treated controls (n = 3). C and D, the graphs show frequency distribution of myotube area (μm2) of ethanol- or Dex-treated NS miR and AntagomiR1-transfected C2C12 myotubes (n = 3). E, the graph represents the average myotube area (μm2) of NS miR or AntagomiR1 transfected and Dex- or ethanol-treated C2C12 myotubes quantified from 10 random images in triplicate per treatment. Values represent mean ± S.E. (n = 3); **, p < 0.01. F, real time quantitative PCR analysis of miR1 expression in mRNA extracted from NS miR- and AntagomiR1-transfected C2C12 myotubes. Values are mean ± S.E. (n = 3) expressed as fold-difference relative to NS miR-transfected controls; **, p < 0.01. G, Western blot analysis of HSP70, pFOXO3, FOXO3, pAKT, AKT, MuRF1, and Atrogin-1 protein levels in NS miR- and AntagomiR1-transfected C2C12 myotubes treated with ethanol or Dex. The level of GAPDH was measured as a loading control. H and I, corresponding densitometric analysis of the levels of phosphorylated FOXO3 and AKT, expressed as a percentage of total FOXO3 and AKT, respectively, in NS miR- and AntagomiR1-transfected C2C12 myotubes following treatment with ethanol or Dex. J, corresponding densitometric analysis of the levels of HSP70, MuRF1, and Atrogin-1 in NS miR- and AntagomiR1-transfected C2C12 myotubes, following treatment with ethanol or Dex. Values represent mean ± S.E. (n = 3); *, p < 0.05. K, bioinformatics analysis, using RNAhybrid version 2.0, showing the complementary miR1 sequence in the murine Hsp70 gene. The matched base pairs are in bold and connected by a vertical line, and the G:U/U:G wobble is indicated by bold letters connected by dots. L, Western blot analysis of HSP70 levels in C2C12 myotubes treated for 24 h with Dex. GAPDH was used as the loading control. M, densitometric analysis of HSP70 protein levels in Dex-treated C2C12 myotubes; **, p < 0.01. N, Western blot analysis of HSP70 levels in C2C12 myotubes transfected with miR1 expression vector, empty vector, and miR1 expression vector with AntagomiR1. GAPDH was used as the loading control. O, densitometric analysis of HSP70 protein levels in 72-h differentiated myoblasts transfected with miR1 expression vector, empty vector, and miR1 expression vector with AntagomiR1; **, p < 0.01. P, assessment of pMIR reporter activity in C2C12 myotubes following overexpression of miR1. C2C12 myoblasts were co-transfected with a miR1 expression vector or empty vector and the pMIR reporter construct containing either a wild type (WT) or mutated (MT) miR1 seed sequence from the murine Hsp70 gene. pMIR reporter activity was normalized to Renilla luciferase and expressed as relative luminescence units (RLU). Each bar represents the mean ± S.E. (n = 3); *, p < 0.05.
Interestingly, AntagomiR1-transfected myotubes appeared to be hypertrophied when compared with control NS miR-transfected myotubes (Fig. 3A). In addition, Dex treatment resulted in severe myotube atrophy in NS miR-transfected myotubes but had a nonsignificant effect on AntagomiR1-transfected myotubes (Fig. 3A). Quantification of the myotube area and frequency distribution analysis of the data shows a larger amount of larger myotubes upon AntagomiR1 transfection as compared with NS miR-transfected myotubes (Fig. 3B). Frequency distribution analysis of the myotube area of NS miR-transfected and Dex-treated myotubes shows a higher number of smaller myotubes (<2500 μm2) and a lower number of larger myotubes (>4000 μm2) after Dex treatment (Fig. 3C) when compared with the controls. However, Dex treatment of AntagomiR1-transfected myotubes did not show such an effect (Fig. 3D). Dex treatment of NS miR and AntagomiR1-transfected myotubes shows a respective ∼34 and ∼14% decrease in myotube area (Fig. 3E) The observed decrease in the myotube area was significant (p < 0.005) in NS miR-transfected myotubes indicating atrophy of these myotubes as compared with the AntagomiR1-transfected myotubes (Fig. 3E). Consistently, quantification data shows an ∼60% increase in myotube area in AntagomiR1-transfected myotubes as compared with NS miR-transfected myotubes (Fig. 3E) confirming hypertrophy.
To confirm the role of miR1 in atrophy we analyzed the levels of phosphorylated AKT and FOXO3 in AntagomiR1-transfected Dex-treated C2C12 myotubes. Subsequent Western blot analysis revealed a significant decrease in the phosphorylation level of both AKT and FOXO3 (pAKT/AKT and pFOXO3/FOXO3) in the NS miR-transfected cells following Dex treatment, however, a significant decrease in phosphorylation of AKT and FOXO3 was not observed upon Dex treatment of AntagomiR1-transfected (miR1 inhibited) C2C12 myotubes (Fig. 3, G–I), confirming a role for miR1 in Dex-mediated muscle atrophy. Because FOXO1/3 induces the expression of muscle-specific E3 ligases, we performed Western blot analysis and as expected, increased MuRF1 and Atrogin-1 levels were observed after Dex treatment (Fig. 3, G and J). Consistent with the above results; the increase in MuRF1 and Atrogin-1 observed in response to Dex treatment was attenuated upon Antagomir1-mediated inhibition of miR1 (Fig. 3, G and J).
miR1 Targets HSP70 during Dex-induced Muscle Atrophy
As the data presented above supports a role for miR1 in muscle atrophy we next wanted to identify potential gene targets of miR1 during atrophy. Subsequent in silico analysis revealed that the 3′ UTR of the mouse HSP70 gene contained a miR1 complementary sequence (Fig. 3K). HSP70 is a 70-kDa heat shock protein that has been shown to confer protection during disuse and Dex-induced skeletal muscle atrophy (33, 34). To investigate if HSP70 was targeted by miR1, first, the level of the HSP70 protein in Dex-treated C2C12 myotubes was initially determined, and as shown in Fig. 3, L and M, HSP70 levels were decreased with Dex treatment, indicating that HSP70 may be targeted by miR1 during Dex-mediated muscle atrophy. Second, the miR1 expression vector was transfected in C2C12 myoblasts, then the cells were allowed to differentiate for 72 h and HSP70 levels were estimated. HSP70 levels were found to be decreased (Fig. 3, N and O) in miR1-transfected cells as compared with empty vector-transfected cells. However, this decrease in HSP70 was rescued with addition of AntagomiR1 confirming that miR1 was responsible for down-regulation of HSP70 (Fig. 3, N and O). To further demonstrate that HSP70 was targeted by miR1 we assessed the levels of HSP70 in NS miR or AntagomiR1-transfected C2C12 myotubes treated with Dex. Subsequent Western blot analysis revealed that Dex-mediated reduction of HSP70 levels was blocked in C2C12 myotubes transfected with AntagomiR1 (Fig. 3, G and J), thus confirming a direct role for miR1 in loss of HSP70 during Dex-induced muscle atrophy.
In addition, we further cloned a wild type and mutated form of the miR1 seed sequence, present in the 3′ UTR of the mouse Hsp70 gene, into the pMIR reporter vector and co-transfected together with a miR1 expression vector into C2C12 myoblasts. A decrease in pMIR reporter activity was observed in 72-h differentiated C2C12 myoblasts co-transfected with the wild type reporter and miR1 expression vector; however, this decrease was not observed in 72-h differentiated myoblasts co-transfected with the mutant pMIR reporter and miR1 expression vector (Fig. 3P). These data confirm that miR1 targets HSP70 in differentiated myoblasts.
Inhibition of HSP70 Resulted in Increased GR Nuclear Translocation, Up-regulated miR1, and Enhanced Levels of Atrophy-related Proteins
The above results confirm that increased miR1 levels lead to reduced HSP70, hypophosphorylation of AKT and FOXO3, and increased MuRF1 and Atrogin-1. To confirm whether or not reduced HSP70, independent of miR1, would also result in similar changes, we inhibited HSP70 with VER155008, a previously characterized inhibitor of HSP70 (26), in C2C12 myotubes. Inhibition of HSP70 resulted in a decrease in abundance of phosphorylated FOXO3 and AKT (Fig. 4, A–C) with an increase of VER155008 concentration. In addition, the levels of MuRF1 and Atrogin-1 were elevated in response to VER155008-mediated inhibition of HSP70 (Fig. 4, D–F), suggesting that loss of HSP70 leads to up-regulation of atrophy-related markers. In agreement with this, we noted an observable myotube atrophy (thinning of myotubes) following inhibition of HSP70 in C2C12 myotubes (Fig. 4G). Quantification of the myotube area reveals a decrease in myotube area after HSP70 inhibition (Fig. 4, H and I) signifying atrophy. Fig. 4, J–L, also shows a similar atrophy when C2C12 myotubes were treated with Dex. Because miR1 reduces HSP70 levels, we further hypothesized that this decrease in HSP70 may lead to increased GR translocation to the nucleus, which would further up-regulate miR1. To prove this hypothesis, we analyzed the nuclear translocation of GR in VER155008-treated C2C12 myotubes. As can be seen in Fig. 4, M and N, inhibition of HSP70 resulted in a dose-dependent increase in nuclear translocation of GR. Moreover, miR1 expression was also increased in VER155008-mediated inhibition of HSP70 (Fig. 4O). Based on these results we conclude that there is positive feedforward inhibition of HSP70 by miR1, because inhibition of HSP70 resulted in enhanced nuclear translocation of GR, which would result in induction of miR1 expression and further inhibition of HSP70.
FIGURE 4.
Inhibition of HSP70 results in decreased AKT and FOXO3 phosphorylation and enhanced myotube atrophy. A, Western blot analysis of pFOXO3, FOXO3, pAKT, and AKT protein levels in C2C12 myotubes treated without (dimethyl sulfoxide; DMSO) or with increasing concentrations (10 and 20 μm) of HSP70 antagonist (VER155008). The level of GAPDH was measured as a loading control. B and C, corresponding densitometric analysis of the levels of phosphorylated FOXO3 and AKT, expressed as a percentage of total FOXO3 and AKT, respectively, in control (DMSO) and VER155008-treated C2C12 myotubes. Values represent mean ± S.E. (n = 3); *, p < 0.05, and **, p < 0.01. D, Western blot analysis of MuRF1 and Atrogin-1 protein levels in control (DMSO) or VER155008-treated C2C12 myotubes. The level of GAPDH was measured as a loading control. E and F, corresponding densitometric analysis of the levels of Atrogin-1 and MuRF1 in control (DMSO) or VER155008-treated C2C12 myotubes. Values represent mean ± S.E. (n = 3); *, p < 0.05 with respect to DMSO-treated controls. G, representative images of C2C12 myotubes treated with DMSO or 10 μm VER155008, fixed, and stained with Gill's hematoxylin and eosin. Scale bar represents 100 μm. H and I, the graphs show frequency distribution of the myotube area (μm2) (H) and average myotube area (μm2) (I) of DMSO- or VER155008-treated C2C12 myotubes quantified from 10 random images in triplicate per treatment (n = 3). Values represent mean ± S.E. (n = 3); **, p < 0.01. J, representative images of C2C12 myotubes treated with ethanol or Dex, fixed, and stained with Gill's hematoxylin and eosin. The scale bar represents 100 μm. K, the graph shows frequency distribution of myotube area (μm2) of C2C12 myotubes treated with ethanol or Dex. L, the graph represents average myotube area (μm2) of ethanol- or Dex-treated C2C12 myotubes quantified from 10 random images in triplicate per treatment. Values represent mean ± S.E. (n = 3); **, p < 0.01. M, Western blot analysis of GR protein levels in nuclear extracts from C2C12 myotubes treated without (DMSO) or with increasing concentrations (10 and 20 μm) of VER155008. N, corresponding densitometric analysis of the levels of GR in C2C12 myotubes treated without (DMSO) or with increasing concentrations (10 and 20 μm) of VER155008. Values represent mean ± S.E. (n = 3); *, p < 0.05 and **, p < 0.01 when compared with DMSO control. O, real time quantitative PCR analysis of miR1 expression in control (DMSO) and VER155008-treated (10 and 20 μm) C2C12 myotubes. Values are fold-change mean ± S.E. (n = 3) when compared with DMSO-treated controls, **, p < 0.01. P, pAKT is associated with HSP70 in skeletal muscle. Western blot analysis of HSP70 (top) and pAKT (middle) co-immunoprecipitated with pAKT in protein lysates from BF muscle of saline and Dex-injected wild type mice. The level of α-tubulin was measured in input fraction (bottom) to ensure an equal amount of protein was used for co-immunoprecipitation. Q, graph showing densitometric analysis of HSP70 protein co-immunoprecipitated with pAKT antibody in BF muscle of saline- and Dex-treated WT mice. HSP70 densitometry values were normalized to α-tubulin and pAKT values and are depicted as a percent decrease after Dex treatment. Data represent mean ± S.E. (n = 3); *, p < 0.05.
pAKT Association with HSP70 Is Blocked during Dex-induced Muscle Atrophy
Recently, Kayama et al. (42) reported that association of HSP70 with pAKT prevents pAKT dephosphorylation in the retina. Because the maintenance of AKT phosphorylation is critical in preventing muscle atrophy, we predicted that miR1-mediated reduction of HSP70 would result in hypophosphorylation of AKT and myotube atrophy. To prove this, we analyzed pAKT by co-immunoprecipitation, using an anti-pAKT-specific antibody, with HSP70. Co-immunoprecipitation confirmed that there was a direct interaction between pAKT and HSP70 in skeletal muscle (Fig. 4P). Furthermore, we noted that after Dex treatment the amount of both HSP70 and AKT in association with each other decreases as compared with the saline-treated controls (Fig. 4, P and Q). These results are consistent with the increased miR1 that can target and degrade HSP70 leading to a decrease in HSP70 after Dex treatment (Fig. 6, A and D). The reduced amount of HSP70 might bind to a lesser amount of pAKT thereby causing the dephosphorylation of the unassociated pAKT, and reducing the phosphorylation of AKT (Fig. 6, A and C).
FIGURE 6.
Mstn regulates miR1 during Dex-mediated muscle atrophy. A, Western blot analysis of pFOXO3, FOXO3, HSP70, pAKT, AKT, MuRF1, and Atrogin-1 levels in whole cell lysates from C2C12 myotubes treated with Dex (lane 2) and Dex + Ant1 (lane 3), as compared with ethanol-treated controls (lane 1). The level of GAPDH was measured as a loading control. B and C, corresponding densitometric analysis of the levels of phosphorylated FOXO3 and AKT, expressed as a percentage of total FOXO3 and AKT, respectively. Error bars represent mean ± S.E. (n = 3); **, p < 0.01, and *, p < 0.05. D, representative graph showing densitometric analysis of HSP70, MuRF1, and Atrogin-1 protein levels (normalized to GAPDH) in ethanol-, Dex-, and Dex + Ant1-treated C2C12 myotubes. Error bars represent mean ± S.E. (n = 3); **, p < 0.01. E, Western blot analysis of pFOXO3, FOXO3, HSP70, pAKT, and AKT protein levels in BF muscle collected from saline- or Dex-injected WT (lanes 1 and 2) and Mstn−/− (lanes 3 and 4) mice. The level of GAPDH was measured as a loading control. F and G, corresponding densitometric analysis of the levels of phosphorylated FOXO3 and AKT, expressed as a percentage of total FOXO3 and AKT, respectively, in WT and Mstn−/− mice after injection with saline or Dex. H, densitometric analysis of HSP70 protein levels in WT and Mstn−/− muscle from saline or Dex-injected mice; *, p < 0.05 and **, p < 0.01. Error bars represent mean ± S.E. (n = 3).
Overexpression of HSP70 during Dex-induced Atrophy Resulted in Rescue of Enhanced Levels of Atrophy-related Proteins
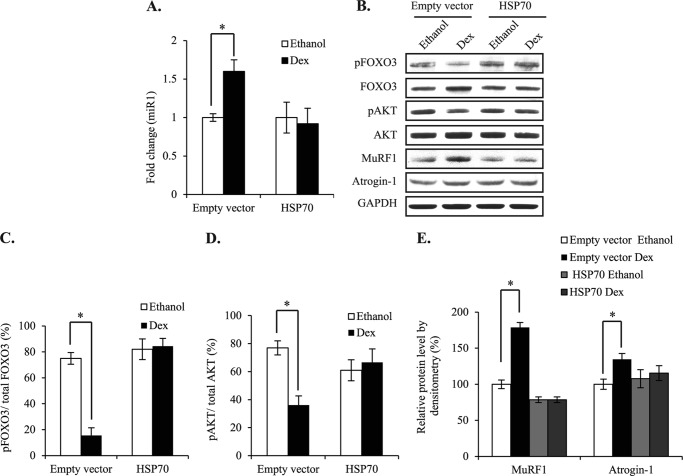
The above results show that inhibition of HSP70 causes atrophy similar to Dex-mediated atrophy. Here we confirm the importance of HSP70 in Dex-mediated atrophy by overexpressing HSP70 in C2C12 myotubes treated with Dex. Empty vector or HSP70 expression plasmids were transfected in C2C12 myoblasts and were allowed to differentiate for 72 h followed by 24 h of ethanol or Dex treatments and collection of RNA and protein. We observed that there was a significant increase in the level of miR1 after Dex treatment in empty vector-transfected myotubes, however, no such increase was observed after Dex treatment in myotubes transfected with HSP70 expression plasmid (Fig. 5A). Furthermore, Western blot results revealed a decrease in phosphorylation of both FOXO3 and AKT (Fig. 5, B–D) and an increase in MURF1 and Atrogin-1 (Fig. 5, B and E) in empty vector-transfected Dex-treated C2C12 myotubes as compared with ethanol-treated controls. There was no significant decrease in the phosphorylation of both FOXO3 and AKT (Fig. 5, B–D) in HSP70 overexpressing Dex-treated C2C12 myotubes as compared with ethanol-treated controls. Also, the increase in the level of MURF1 and Atrogin-1 after Dex treatment was rescued in HSP70 overexpressing C2C12 myotubes (Fig. 5, B and E). These results confirm the role of HSP70 in Dex-mediated atrophy.
FIGURE 5.
Overexpression of HSP70 during Dex-induced atrophy resulted in rescue of enhanced levels of atrophy-related proteins. A, real time qPCR analysis of miR1 expression in mRNA extracted from empty vector or HSP70 expression plasmid-transfected C2C12 myotubes after ethanol or Dex treatments. Values are mean ± S.E. (n = 3) expressed as fold-difference relative to empty vector-transfected controls, *, p < 0.05. B, Western blot analysis of pFOXO3, FOXO3, pAKT, AKT, MuRF1, and Atrogin-1 protein levels in empty vector and HSP70 expression plasmid-transfected C2C12 myotubes treated with ethanol or Dex. The level of GAPDH was measured as a loading control. C and D, corresponding densitometric analysis of the levels of phosphorylated FOXO3 and AKT, expressed as a percentage of total FOXO3 and AKT, respectively, in empty vector or HSP70 expression plasmid-transfected C2C12 myotubes following treatment with ethanol or Dex. E, corresponding densitometric analysis of the levels of MuRF1 and Atrogin-1 in empty vector or HSP70 expression plasmid-transfected C2C12 myotubes, following treatment with ethanol or Dex. Values represent mean ± S.E. (n = 3); *, p < 0.05.
Mstn Regulates Atrophy-related Genes through miR1 during Dex-mediated Muscle Atrophy
Consistent with previous publications (9, 11, 12), treatment with Dex resulted in reduced phosphorylation of AKT (pAKT) and FOXO3 (pFOXO3) (Fig. 6, A–C) and increased MuRF1 and Atrogin-1 expression (Fig. 6, A and D). However, blockade of Mstn through Ant1 treatment rescued pFOXO3 and pAKT levels and blocked the increase in MuRF1 and Atrogin-1 levels observed in Dex-treated C2C12 myotubes (Fig. 6, A–D). Inhibition of Mstn, during Dex treatment, also attenuated the induction of miR1 expression, confirming the involvement of Mstn signaling during Dex-mediated atrophy (Fig. 2E). To confirm these findings, we also checked the phosphorylation status of AKT and FOXO3 in BF muscle from wild type and Mstn−/− mice following Dex injections. As expected, we observed a decrease in the phosphorylation of AKT and FOXO3 in wild type mice following Dex injection; however, no such decrease was observed in Mstn−/− mice upon injection with Dex (Fig. 6, E–G).
Next, we queried whether or not blockade of Mstn with Ant1 would restore HSP70 levels in Dex-treated C2C12 myotubes. Subsequent Western blot analysis revealed that Ant1-mediated inhibition of Mstn resulted in an increase in HSP70 abundance, back to levels comparable with control treated myotubes (Fig. 6, A and D), further demonstrating an involvement of Mstn in Dex signaling during atrophy. As we are proposing that Dex-mediated muscle atrophy also occurs in part through a mechanism involving Mstn regulation of miR1 and HSP70, we determined the levels of HSP70 in Mstn−/− mice after Dex treatment. As shown in Fig. 6, E and H, higher levels of HSP70 were detected in Mstn−/− muscle, as compared with WT muscle. Moreover, Dex treatment failed to inhibit the levels of HSP70 to the same degree in Mstn−/− muscle (32%), when compared with wild type muscle (47%); in fact, the level of HSP70 in Dex-treated Mstn−/− muscle was comparable with that observed in saline-injected wild type muscle (Fig. 6, E and H).
DISCUSSION
MyomiRs have added a new tier of control to muscle gene expression regulation. In particular, the MyomiR, miR1, has been previously shown to regulate myoblast differentiation, apoptosis, and load-induced skeletal muscle hypertrophy (19, 21, 35). Herein, we have elucidated a novel function for miR1 in promoting the skeletal muscle atrophy associated with Dex and Mstn treatment (Fig. 7).
FIGURE 7.
Proposed mechanism behind Dex-induced miR1-mediated skeletal muscle atrophy. Dex activates GR, which translocates to the nucleus and up-regulates miR1. Dex also up-regulates Mstn, which increases nuclear translocation of GR and miR1 expression. miR-1-mediated loss of HSP70 and enhanced activation of GR leads to further up-regulation of miR1 expression. Thus, increased miR1 expression may feedforward to further enhance its own expression. The reduced levels of HSP70 observed following Dex and Mstn treatment would further exacerbate skeletal muscle atrophy by decreasing phosphorylation of AKT, which results in activation of downstream proteosomal signaling components, such as FOXO3, MuRF1, and Atrogin-1. Arrows represent stimulation and blunt-ended lines represent inhibition.
MyomiRs, miR133 and miR206, belong to the miR1 family and like miR1, miR206 has been shown to induce differentiation in C2C12 myoblasts (36). Conversely, miR133 was shown to promote proliferation (19). In spite of their opposing myogenic roles, both miR1 and miR133a were shown to be down-regulated during skeletal muscle hypertrophy (21). In line with these observations, our results reveal that levels of miR133a and miR206 were significantly elevated after Dex treatment of C2C12 myotubes as compared with the controls (data not shown). Thus, it appears that these myomiRs are regulated in a different manner in nonphysiological versus physiological conditions. In this study we focused on miR1, in particular miR1–1, although unpublished results4 also revealed up-regulated miR1–2 expression in Dex-treated C2C12 myotubes suggesting that both miR1–1 and miR1–2 contribute to total mature miR1 levels. Several lines of evidence presented here indicate that Dex directly induces the expression of miR1. In particular, along with miR1–1, we also analyzed the miR1–2 promoter and identified a GRE, 1748 bp upstream of the miR1–2 coding sequences. The presence of a GRE in upstream sequences of both miR1–1 and miR1–2 is indicative of a role for GR-mediated regulation of miR1 in Dex induced atrophy. In addition, we also observed blockade of the GR-mediated increase in miR1–1 promoter-reporter activity in 72-h differentiated myoblasts co-transfected with the GRE-truncated miR1 promoter construct (where the GRE has been removed) and GR expression vector (Fig. 2I). Furthermore, EMSA and ChIP analyses confirmed that GR was able to directly bind to the GRE within the miR1–1 promoter region in response to Dex treatment (Fig. 2, J–M).
Consistent with the above hypothesis, inhibition of miR1 resulted in observable myotube hypertrophy (Fig. 3, A and B), which is in agreement with previous reports demonstrating a reduction in miR1 expression during cardiac and skeletal muscle hypertrophy (21, 22). MicroRNAs act by base pairing to complementary sequences within the 3′ UTR of target mRNAs and either inhibit translation or promote mRNA degradation. Previously published reports have identified Igf-1 mRNA as one of the targets of miR1 in cardiac and skeletal muscle (21, 22). Given the role of IGF-1 in hypertrophy of skeletal (12) and cardiac muscle (37) and inhibition of atrophy through enhancing AKT signaling (12), Igf-1 mRNA may be targeted by miR1 in response to Dex and Mstn treatments. However, in silico analysis of potential targets of miR1 revealed that HSP70 may be targeted by miR1 in response to Dex treatment. In support, reduced levels of HSP70 were observed in Dex-treated myotubes and skeletal muscle, concomitant with elevated miR1 expression. This result was corroborated by Western blot data showing a decrease in HSP70 in miR1-transfected C2C12 myotubes when compared with control myotubes and through the fact that HSP70 levels in the myotubes were restored by addition of AntagomiR1 (Fig. 3N). Furthermore, a rescue of pMIR reporter activity was observed in 72-h differentiated myoblasts co-transfected with the mutated Hsp70 gene miR1 seed sequence and miR1 expression vector, when compared with the 72-h differentiated myoblasts co-transfected with the WT Hsp70 gene miR1 seed sequence and miR1 expression vector (Fig. 3P). Taken together, these results show that HSP70 is a bona fide target of miR1 and thus it was speculated that miR1 targets and represses HSP70 during Dex and Mstn induced atrophy.
HSP70 is one of the most important members of heat shock protein family. HSP70 is a constitutively active, cytoprotective protein, which acts like a molecular chaperone and is elevated during cellular stress in skeletal muscle. In fact, previously published reports revealed that overexpression of HSP70 in mouse skeletal muscle protects against muscle damage and age-related muscle dysfunction (38, 39). In contrast, decreased HSP70 levels have been shown to contribute to the pathogenicity of multiple models of skeletal muscle atrophy (40). Mechanistically, HSP70 has been shown to directly regulate FOXO3 during disuse muscle atrophy and overexpression of HSP70 was shown to reduce MuRF1 and Atrogin-1 promoter activity (34, 41). In the present study we observed down-regulation of HSP70 in Dex-treated samples, concomitant with a decrease in the levels of phosphorylated AKT and FOXO3, indicating that the reduced levels of HSP70 detected following Dex treatment may be responsible for hypophosphorylation of both AKT and FOXO3. Consistent with this, VER155008-mediated inhibition of HSP70 resulted in reduced phosphorylation of AKT and FOXO3 and myotubular atrophy (Fig. 4, A–C and G–I). A recent publication by Senf et al. (41) revealed that HSP70 regulates FOXO3 through both AKT-dependent and -independent mechanisms. Here, we observed changes in both pAKT and pFOXO3 in response to HSP70 inhibition. Furthermore, we noted that in BF muscle after Dex treatment the amount of HSP70 associated with pAKT decreased, when compared with the saline-treated controls (Fig. 4, P and Q), suggesting that miR1-mediated inhibition of HSP70 may result in the dephosphorylation of AKT observed following Dex treatment. In support, a recent study by Kayama et al. (42) revealed that HSP70 binds to pAKT in mouse retina and prevents AKT dephosphorylation. Thus, we propose that one of the mechanisms by which Dex-induced skeletal muscle atrophy occurs is through miR1-mediated down-regulation of HSP70. Dephosphorylation of FOXO3 results in FOXO3 activation/nuclear translocation, up-regulation of FOXO3 downstream target genes, including MuRF1 and Atrogin-1, and the development of skeletal muscle atrophy (11). Blockade of HSP70 also resulted in dephosphorylation of FOXO3 and increased levels of MuRF1 and Atrogin-1, suggesting a role for HSP70 in regulation of Atrogin-1 and MuRF1 levels. Role of HSP70 in Dex-mediated atrophy was further confirmed in HSP70 overexpressing C2C12 myotubes treated with Dex. Reduced phosphorylation of AKT and FOXO3; and increased MURF1 and Atrogin-1 in Dex-treated C2C12 myotubes were rescued in HSP70 overexpressing myotubes (Fig. 5, B–E). Indeed, inhibition of miR1 through AntagomiR1 treatment also rescued the levels of HSP70, pAKT, and pFOXO3 and reduced the levels of both MuRF1 and Atrogin-1 during Dex-induced muscle atrophy. It is important to mention that although previous reports have shown that both Dex and Mstn treatments resulted in reduced AKT phosphorylation (12, 14), we have shown here for the first time the involvement of miR1 and HSP70 in Dex- and Mstn-mediated inhibition of AKT phosphorylation
Another mechanism by which HSP70 can offer resistance to atrophy is through associating directly with GR in the cytoplasm (43) thereby preventing nuclear translocation of GR. Therefore, if miR1 was responsible for reducing the levels of HSP70 during Dex-induced atrophy then increased nuclear localization of GR would be expected. Our results confirm that with inhibition of HSP70 there was a dose-dependent increase in nuclear translocation of GR and concomitant up-regulation of miR1, which we propose would in turn further reduce HSP70 levels. With respect to this we propose a feedforward mechanism during Dex-mediated atrophy, whereby increased miR1 expression and enhanced nuclear localization of GR would result in further up-regulation of miR1 and reduction of HSP70.
Dex is also known to up-regulate Mstn, at the transcription level, through putative GREs located within the Mstn promoter region (15, 44) and post-transcriptionally through miR-27 (45). Results presented here show up-regulation of Mstn in C2C12 myotubes upon Dex treatment (Fig. 2A). Importantly Ant1-mediated blockade of Mstn rescued the levels of phosphorylated AKT and FOXO3 following Dex treatment (Fig. 6, A–C). These data are in agreement with previously published results from Gilson et al. (16), which demonstrated that deletion of the myostatin gene prevents glucocorticoid-induced muscle atrophy. Interestingly, we noted that inhibition of Mstn during Dex treatment of C2C12 myotubes not only blunted the expression of atrophy-related genes but also miR1, thereby supporting a role for Mstn in regulation of miR1 during Dex-induced atrophy.
In addition to cell culture data, we also confirmed the above findings in vivo, by injecting WT and Mstn−/− mice with Dex and assessing the expression of miR1 through Northern blot analysis. Our results confirmed elevated expression of miR1 in WT mice, following Dex injection. We did not observe a significant difference in the expression of miR1 between saline-treated WT or saline-treated Mstn−/− muscle and even after Dex treatment miR1 levels were comparable with saline-injected controls in Mstn−/− mice (Fig. 2, C and D). A previous study by Rachagani et al. (46) has shown that myomiRs, miR1, miR133, and miR206, were up-regulated in the pectoralis muscle of Mstn−/− mice (46). It is plausible that in our study only nonsignificant differences were observed in mature miR1 expression in Mstn−/− mice muscle, when compared with WT muscle. This may be due to the fact that we used Northern blotting to detect mature miR1, whereas Rachagani et al. (46) used real-time PCR, which might be more sensitive than Northern blotting to detect the differences in miR1 expression between Mstn−/− and WT muscle. Our results also show that Mstn−/− muscle had higher levels of HSP70 as compared with WT muscle. However, upon Dex treatment in Mstn−/− mice the levels of HSP70 decreased to untreated WT levels (Fig. 6, E and H). It can be speculated that due to genetic deletion of the Mstn gene, Mstn independent mechanisms may be in place that modify the levels of HSP70 in Mstn−/− mice during Dex treatment.
Taken together these findings prove that Mstn also plays a role in Dex-mediated up-regulation of miR1. As Dex-mediated induction of miR1 expression occurred through transcriptional activation of GR, we thus reasoned that Mstn may also promote up-regulation of miR1 by enhancing nuclear localization of GR. Indeed our results showed for the first time that Mstn treatment resulted in increased nuclear localization of GR.
In conclusion, here we report a novel miR1-mediated mechanism through which Dex and Mstn induce skeletal muscle atrophy. We demonstrate that both excess Dex and Mstn led to activation of GR and increased miR1 expression. We also suggest that increased miR1 expression may feedforward to further enhance its own expression. Most certainly, increased miR1 was shown to promote loss of HSP70, which normally blocks GR activation via associating with GR in the cytoplasm. Subsequent miR-1-mediated loss of HSP70 and enhanced activation of GR would thus lead to further up-regulation of miR1 expression. In addition, the reduced levels of HSP70 observed following Dex and Mstn treatments would further exacerbate skeletal muscle atrophy by decreasing phosphorylation of AKT, which results in activation of downstream proteosomal signaling components, such as FOXO3, MuRF1, and Atrogin-1.
Acknowledgments
We thank Prof. S. J. Lee for Mstn+/− heterozygous mice, Dr. S. Stoney Simons for GR expression plasmid, Dr. Evan Eisenberg for HSP70 expression vector (Addgene) and Prof. T. Akimoto for miR1 expression vector. We also acknowledge Ildasolha Bte Jamari for help. We thank Dr. Esther Latres (Regeneron Pharmaceuticals, Tarrytown, NY) for gifting the MuRF1 antibodies utilized in this present study.
This work was supported by the BioMedical Research Council (BMRC) and National Research Foundation (NRF), Republic of Singapore.
H. Kukreti, K. Amuthavalli, A. Harikumar, S. Sathiyamoorthy, P. Z. Feng, R. Anantharaj, S. L. K. Tan, S. Lokireddy, S. Bonala, S. Sriram, C. McFarlane, R. Kambadur, and M. Sharma, unpublished data.
- MuRF1
- muscle ring finger protein 1
- miR1
- microRNA1
- GR
- glucocorticoid receptor
- Dex
- dexamethasone
- myomiRs
- muscle-specific microRNAs
- BF
- biceps femoris
- GRE
- glucocorticoid response element
- Mstn
- myostatin
- NS
- nonspecific
- CMM
- conditioned Mstn medium
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- qPCR
- quantitative PCR.
REFERENCES
- 1. Rhen T., Cidlowski J. A. (2005) Anti-inflammatory action of glucocorticoids. New mechanisms for old drugs. N. Engl. J. Med. 353, 1711–1723 [DOI] [PubMed] [Google Scholar]
- 2. Hasselgren P. O. (1999) Glucocorticoids and muscle catabolism. Curr. Opin. Clin. Nutr. Metab. Care 2, 201–205 [DOI] [PubMed] [Google Scholar]
- 3. Wing S. S., Goldberg A. L. (1993) Glucocorticoids activate the ATP-ubiquitin-dependent proteolytic system in skeletal muscle during fasting. Am. J. Physiol. 264, E668–676 [DOI] [PubMed] [Google Scholar]
- 4. Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., Pan Z. Q., Valenzuela D. M., DeChiara T. M., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708 [DOI] [PubMed] [Google Scholar]
- 5. Latres E., Amini A. R., Amini A. A., Griffiths J., Martin F. J., Wei Y., Lin H. C., Yancopoulos G. D., Glass D. J. (2005) Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J. Biol. Chem. 280, 2737–2744 [DOI] [PubMed] [Google Scholar]
- 6. Cohen S., Brault J. J., Gygi S. P., Glass D. J., Valenzuela D. M., Gartner C., Latres E., Goldberg A. L. (2009) During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J. Cell Biol. 185, 1083–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schakman O., Gilson H., Kalista S., Thissen J. P. (2009) Mechanisms of muscle atrophy induced by glucocorticoids. Horm. Res. 72, 36–41 [DOI] [PubMed] [Google Scholar]
- 8. Zhao W., Qin W., Pan J., Wu Y., Bauman W. A., Cardozo C. (2009) Dependence of dexamethasone-induced Akt/FOXO1 signaling, up-regulation of MAFbx, and protein catabolism upon the glucocorticoid receptor. Biochem. Biophys. Res. Commun. 378, 668–672 [DOI] [PubMed] [Google Scholar]
- 9. Clarke B. A., Drujan D., Willis M. S., Murphy L. O., Corpina R. A., Burova E., Rakhilin S. V., Stitt T. N., Patterson C., Latres E., Glass D. J. (2007) The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 6, 376–385 [DOI] [PubMed] [Google Scholar]
- 10. Lokireddy S., McFarlane C., Ge X., Zhang H., Sze S. K., Sharma M., Kambadur R. (2011) Myostatin induces degradation of sarcomeric proteins through a Smad3 signaling mechanism during skeletal muscle wasting. Mol. Endocrinol. 25, 1936–1949 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004) Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stitt T. N., Drujan D., Clarke B. A., Panaro F., Timofeyva Y., Kline W. O., Gonzalez M., Yancopoulos G. D., Glass D. J. (2004) The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 14, 395–403 [DOI] [PubMed] [Google Scholar]
- 13. Li B. G., Hasselgren P. O., Fang C. H., Warden G. D. (2004) Insulin-like growth factor-I blocks dexamethasone-induced protein degradation in cultured myotubes by inhibiting multiple proteolytic pathways. 2002 ABA paper. J. Burn Care Rehabil. 25, 112–118 [DOI] [PubMed] [Google Scholar]
- 14. McFarlane C., Plummer E., Thomas M., Hennebry A., Ashby M., Ling N., Smith H., Sharma M., Kambadur R. (2006) Myostatin induces cachexia by activating the ubiquitin proteolytic system through an NF-κB-independent, FoxO1-dependent mechanism. J. Cell Physiol. 209, 501–514 [DOI] [PubMed] [Google Scholar]
- 15. Ma K., Mallidis C., Bhasin S., Mahabadi V., Artaza J., Gonzalez-Cadavid N., Arias J., Salehian B. (2003) Glucocorticoid-induced skeletal muscle atrophy is associated with up-regulation of myostatin gene expression. Am. J. Physiol. Endocrinol. Metab. 285, E363–371 [DOI] [PubMed] [Google Scholar]
- 16. Gilson H., Schakman O., Combaret L., Lause P., Grobet L., Attaix D., Ketelslegers J. M., Thissen J. P. (2007) Myostatin gene deletion prevents glucocorticoid-induced muscle atrophy. Endocrinology 148, 452–460 [DOI] [PubMed] [Google Scholar]
- 17. Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G. (2002) miRNPs. A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16, 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stefani G., Slack F. J. (2008) Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9, 219–230 [DOI] [PubMed] [Google Scholar]
- 19. Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakasa T., Ishikawa M., Shi M., Shibuya H., Adachi N., Ochi M. (2010) Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J. Cell Mol. Med. 14, 2495–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McCarthy J. J., Esser K. A. (2007) MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J. Appl. Physiol. 102, 306–313 [DOI] [PubMed] [Google Scholar]
- 22. Elia L., Contu R., Quintavalle M., Varrone F., Chimenti C., Russo M. A., Cimino V., De Marinis L., Frustaci A., Catalucci D., Condorelli G. (2009) Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation 120, 2377–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zimmers T. A., Davies M. V., Koniaris L. G., Haynes P., Esquela A. F., Tomkinson K. N., McPherron A. C., Wolfman N. M., Lee S. J. (2002) Induction of cachexia in mice by systemically administered myostatin. Science 296, 1486–1488 [DOI] [PubMed] [Google Scholar]
- 24. Sriram S., Subramanian S., Sathiakumar D., Venkatesh R., Salerno M. S., McFarlane C. D., Kambadur R., Sharma M. (2011) Modulation of reactive oxygen species in skeletal muscle by myostatin is mediated through NF-κB. Aging Cell 10, 931–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McFarlane C., Hui G. Z., Amanda W. Z., Lau H. Y., Lokireddy S., Xiaojia G., Mouly V., Butler-Browne G., Gluckman P. D., Sharma M., Kambadur R. (2011) Human myostatin negatively regulates human myoblast growth and differentiation. Am. J. Physiol. Cell Physiol. 301, C195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Massey A. J., Williamson D. S., Browne H., Murray J. B., Dokurno P., Shaw T., Macias A. T., Daniels Z., Geoffroy S., Dopson M., Lavan P., Matassova N., Francis G. L., Graham C. J., Parsons R., Wang Y., Padfield A., Comer M., Drysdale M. J., Wood M. (2010) A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother. Pharmacol. 66, 535–545 [DOI] [PubMed] [Google Scholar]
- 27. Siriett V., Salerno M. S., Berry C., Nicholas G., Bower R., Kambadur R., Sharma M. (2007) Antagonism of myostatin enhances muscle regeneration during sarcopenia. Mol. Ther. 15, 1463–1470 [DOI] [PubMed] [Google Scholar]
- 28. Yaffe D., Saxel O. (1977) Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270, 725–727 [DOI] [PubMed] [Google Scholar]
- 29. Kambadur R., Sharma M., Smith T. P., Bass J. J. (1997) Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 7, 910–916 [DOI] [PubMed] [Google Scholar]
- 30. Ye S., Eriksson P., Hamsten A., Kurkinen M., Humphries S. E., Henney A. M. (1996) Progression of coronary atherosclerosis is associated with a common genetic variant of the human stromelysin-1 promoter which results in reduced gene expression. J. Biol. Chem. 271, 13055–13060 [DOI] [PubMed] [Google Scholar]
- 31. Bonala S., Lokireddy S., Arigela H., Teng S., Wahli W., Sharma M., McFarlane C., Kambadur R. (2012) Peroxisome proliferator-activated receptor β/δ induces myogenesis by modulating myostatin activity. J. Biol. Chem. 287, 12935–12951 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Zeng X. C., Bhasin S., Wu X., Lee J. G., Maffi S., Nichols C. J., Lee K. J., Taylor J. P., Greene L. E., Eisenberg E. (2004) Hsp70 dynamics in vivo. Effect of heat shock and protein aggregation. J. Cell Sci. 117, 4991–5000 [DOI] [PubMed] [Google Scholar]
- 33. Luo G., Sun X., Hungness E., Hasselgren P. O. (2001) Heat shock protects L6 myotubes from catabolic effects of dexamethasone and prevents down-regulation of NF-κB. Am. J. Physiol. Regul. Integr. Comp. Physiol. 281, R1193–1200 [DOI] [PubMed] [Google Scholar]
- 34. Senf S. M., Dodd S. L., McClung J. M., Judge A. R. (2008) Hsp70 overexpression inhibits NF-κB and Foxo3a transcriptional activities and prevents skeletal muscle atrophy. FASEB J. 22, 3836–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shan Z. X., Lin Q. X., Deng C. Y., Zhu J. N., Mai L. P., Liu J. L., Fu Y. H., Liu X. Y., Li Y. X., Zhang Y. Y., Lin S. G., Yu X. Y. (2010) miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes. FEBS Lett. 584, 3592–3600 [DOI] [PubMed] [Google Scholar]
- 36. Kim H. K., Lee Y. S., Sivaprasad U., Malhotra A., Dutta A. (2006) Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 174, 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Welch S., Plank D., Witt S., Glascock B., Schaefer E., Chimenti S., Andreoli A. M., Limana F., Leri A., Kajstura J., Anversa P., Sussman M. A. (2002) Cardiac-specific IGF-1 expression attenuates dilated cardiomyopathy in tropomodulin-overexpressing transgenic mice. Circ. Res. 90, 641–648 [DOI] [PubMed] [Google Scholar]
- 38. McArdle A., Dillmann W. H., Mestril R., Faulkner J. A., Jackson M. J. (2004) Overexpression of HSP70 in mouse skeletal muscle protects against muscle damage and age-related muscle dysfunction. FASEB J. 18, 355–357 [DOI] [PubMed] [Google Scholar]
- 39. Dodd S., Hain B., Judge A. (2009) Hsp70 prevents disuse muscle atrophy in senescent rats. Biogerontology 10, 605–611 [DOI] [PubMed] [Google Scholar]
- 40. Lecker S. H., Jagoe R. T., Gilbert A., Gomes M., Baracos V., Bailey J., Price S. R., Mitch W. E., Goldberg A. L. (2004) Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 18, 39–51 [DOI] [PubMed] [Google Scholar]
- 41. Senf S. M., Dodd S. L., Judge A. R. (2010) FOXO signaling is required for disuse muscle atrophy and is directly regulated by Hsp70. Am. J. Physiol. Cell Physiol. 298, C38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kayama M., Nakazawa T., Thanos A., Morizane Y., Murakami Y., Theodoropoulou S., Abe T., Vavvas D., Miller J. W. (2011) Heat shock protein 70 (HSP70) is critical for the photoreceptor stress response after retinal detachment via modulating anti-apoptotic Akt kinase. Am. J. Pathol. 178, 1080–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dittmar K. D., Pratt W. B. (1997) Folding of the glucocorticoid receptor by the reconstituted Hsp90-based chaperone machinery. The initial hsp90·p60·hsp70-dependent step is sufficient for creating the steroid binding conformation. J. Biol. Chem. 272, 13047–13054 [DOI] [PubMed] [Google Scholar]
- 44. Ma K., Mallidis C., Artaza J., Taylor W., Gonzalez-Cadavid N., Bhasin S. (2001) Characterization of 5′-regulatory region of human myostatin gene. Regulation by dexamethasone in vitro. Am. J. Physiol. Endocrinol. Metab. 281, E1128–1136 [DOI] [PubMed] [Google Scholar]
- 45. Allen D. L., Loh A. S. (2011) Posttranscriptional mechanisms involving microRNA-27a and b contribute to fast-specific and glucocorticoid-mediated myostatin expression in skeletal muscle. Am. J. Physiol. Cell Physiol. 300, C124–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rachagani S., Cheng Y., Reecy J. M. (2010) Myostatin genotype regulates muscle-specific miRNA expression in mouse pectoralis muscle. BMC Res. Notes 3, 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]