Abstract
Myopia, or nearsightedness, is the most common eye disorder, resulting primarily from excess elongation of the eye. The etiology of myopia, although known to be complex, is poorly understood. Here we report the largest ever genome-wide association study (45,771 participants) on myopia in Europeans. We performed a survival analysis on age of myopia onset and identified 22 significant associations (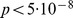 ), two of which are replications of earlier associations with refractive error. Ten of the 20 novel associations identified replicate in a separate cohort of 8,323 participants who reported if they had developed myopia before age 10. These 22 associations in total explain 2.9% of the variance in myopia age of onset and point toward a number of different mechanisms behind the development of myopia. One association is in the gene PRSS56, which has previously been linked to abnormally small eyes; one is in a gene that forms part of the extracellular matrix (LAMA2); two are in or near genes involved in the regeneration of 11-cis-retinal (RGR and RDH5); two are near genes known to be involved in the growth and guidance of retinal ganglion cells (ZIC2, SFRP1); and five are in or near genes involved in neuronal signaling or development. These novel findings point toward multiple genetic factors involved in the development of myopia and suggest that complex interactions between extracellular matrix remodeling, neuronal development, and visual signals from the retina may underlie the development of myopia in humans.
), two of which are replications of earlier associations with refractive error. Ten of the 20 novel associations identified replicate in a separate cohort of 8,323 participants who reported if they had developed myopia before age 10. These 22 associations in total explain 2.9% of the variance in myopia age of onset and point toward a number of different mechanisms behind the development of myopia. One association is in the gene PRSS56, which has previously been linked to abnormally small eyes; one is in a gene that forms part of the extracellular matrix (LAMA2); two are in or near genes involved in the regeneration of 11-cis-retinal (RGR and RDH5); two are near genes known to be involved in the growth and guidance of retinal ganglion cells (ZIC2, SFRP1); and five are in or near genes involved in neuronal signaling or development. These novel findings point toward multiple genetic factors involved in the development of myopia and suggest that complex interactions between extracellular matrix remodeling, neuronal development, and visual signals from the retina may underlie the development of myopia in humans.
Author Summary
The genetic basis of myopia, or nearsightedness, is believed to be complex and affected by multiple genes. Two genetic association studies have each identified a single genetic region associated with myopia in European populations. Here we report the results of the largest ever genetic association study on myopia in over 45,000 people of European ancestry. We identified 22 genetic regions significantly associated with myopia age of onset. Two are replications of the previously identified associations, and 20 are novel. Ten of the novel associations replicate in a small separate cohort. Sixteen of the novel associations are in or near genes implicated in eye development, neuronal development and signaling, the visual cycle of the retina, and general morphology: BMP3, BMP4, DLG2, DLX1, KCNMA1, KCNQ5, LAMA2, LRRC4C, PRSS56, RBFOX1, RDH5, RGR, SFRP1, TJP2, ZBTB38, and ZIC2. These findings point to numerous biological pathways involved in the development of myopia and, in particular, suggest that early eye and neuronal development may lead to the eventual development of myopia in humans.
Introduction
Myopia, or nearsightedness, is the most common eye disorder worldwide. In the United States, an estimated 30–40% of the adult population has clinically relevant myopia (more severe than −1 diopter), and the prevalence has increased markedly in the last 30 years [1], [2]. Myopia is a refractive error that results primarily from increased axial length of the eye [3]. The increased physical length of the eye relative to optical length causes images to be focused in front of the retina, resulting in blurred distance vision.
The etiology of myopia is multifactorial [3]. Briefly, postnatal eye growth is directed by visual stimuli that evoke a signaling cascade within the eye. This cascade is initiated in the retina and passes through the retinal pigment epithelium (RPE) and choroid to guide remodeling of the sclera (the white outer wall of the globe) (cf. [4], [5]). Animal models implicate these visually-guided alterations of the scleral extracellular matrix in the eventual development of myopia [4], [6].
The human eye grows from an average of 17 mm at birth to 21–22 mm in adulthood [7]. By ages 5–6 only about 2% of children are myopic [7]. Although the eye grows only 0.5 mm through puberty [8], the incidence of myopia increases sevenfold during this time [7], peaking between the ages 9–14 [9]. Myopia developed during childhood or early adolescence generally worsens throughout adolescence and then stabilizes by age 20. Compared to myopia that develops in childhood or adolescence, adult onset myopia tends to be less severe [10]–[12]. The majority of myopia cases are primary and nonsyndromic [3]; however, myopia can arise as a complication of other conditions, such as severe prematurity, cataracts, and keratoconus [13], [14], and is sometimes associated with certain connective tissue disorders, such as Stickler syndrome [15].
Although epidemiological studies have implicated numerous environmental factors in the development of myopia, most notably education, outdoor exposure, reading, and near work [3], it is well established that genetics plays a substantial role. Twin and sibling studies have provided heritability estimates that range from 50% to over 90% [16]–[20]. Children of myopic parents tend to have longer eyes and are at higher than average risk of developing myopia in childhood [21]. Segregation analyses suggest that multiple genes are involved in the development of myopia [22], [23]. To date, there have been seven genome-wide association studies (GWAS) on myopia or related phenotypes (pathological myopia, refractive error, and ocular axial length): two in Europeans [24], [25] and five in Asian populations [26]–[30]. Each of these publications has identified a different single association with myopia. In addition there have been several linkage studies (see [3], [31] for reviews) and an exome sequencing study of severe myopia [32].
In contrast to the previous GWAS that used degree of refractive error as a quantitative dependent measure, we analyzed data for 45,771 individuals from the 23andMe database who reported whether they had been diagnosed with nearsightedness, and if so, at what age. We performed a genome-wide survival analysis on age of onset of myopia, discovering 22 genome-wide significant associations with myopia age of onset, 20 of which are novel. Ten of the novel and one of the previously identified associations replicate in a separate (smaller and more coarsely phenotyped) cohort of 8,323 individuals.
Results/Discussion
Participants reported via web-based questionnaires whether they had been diagnosed with nearsightedness, and if so, at what age. Only those participants who reported onset between five and 30 years of age were included to limit cases of secondary myopia (e.g., myopia due to premature birth or cataracts). Further filtering was performed to limit errors in reporting (see Methods).
All participants were customers of 23andMe and of primarily European ancestry; no pair was related at the level of first cousins or closer. We performed a genome-wide survival analysis using a Cox proportional hazards model on data for 45,771 individuals (“discovery set”). The Cox model assumes that there is an (unknown) baseline probability of developing myopia at every year of age. The model then tests whether each single nucleotide polymorphism (SNP) is associated with a significantly higher or lower probability of developing myopia compared to baseline. The Cox model can be thought of as a generalization of an analysis of myopia age of onset. In contrast to an analysis of age of onset, the Cox model allows for the inclusion of non-myopic controls, resulting in increased statistical power. Analyses controlled for sex and five principal components of genetic ancestry. An additional, non-overlapping set of 8,323 participants who reported on their use of corrective eyewear for nearsightedness before the age of ten were used as a replication set. See Table 1 for characteristics of the two cohorts.
Table 1. Cohort statistics.
| Number | % female | Age (SE) | Age of onset (SE) | |
| Discovery, myopic | 25,999 | 45.9 | 47.7 (15.5) | 13.6 (5.8) |
| Discovery, not myopic | 19,772 | 40.3 | 49.6 (17.1) | — |
| Replication, myopic at 10 | 1,488 | 45.3 | 46.7 (14.9) |
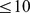
|
| Replication, not myopic at 10 | 6,835 | 45.2 | 53.7 (15.1) | — |
Sex, current age, and age of onset for discovery and replication cohorts.
Table 2 shows the top SNPs for all 35 genetic regions associated with myopia with a 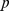 -value smaller than
-value smaller than 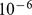 . All
. All 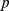 -values from the GWAS have been corrected for the inflation factor of GC = 1.167. A total of 22 of the SNPs cross our threshold for genome-wide significance (
-values from the GWAS have been corrected for the inflation factor of GC = 1.167. A total of 22 of the SNPs cross our threshold for genome-wide significance (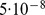 , see Figure S1). These 22 include two SNPs previously associated with refractive error in GWAS of European populations: rs524952 near GJD2 and ACTC1 and rs28412916 near RASGRF1
[24], [25], [33].
, see Figure S1). These 22 include two SNPs previously associated with refractive error in GWAS of European populations: rs524952 near GJD2 and ACTC1 and rs28412916 near RASGRF1
[24], [25], [33]. 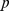 -values genome-wide are shown in Figure 1; Figure S2 shows the quantile-quantile plot for the analysis. Table S3 shows all SNPs with
-values genome-wide are shown in Figure 1; Figure S2 shows the quantile-quantile plot for the analysis. Table S3 shows all SNPs with 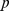 -values under
-values under 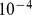 .
.
Table 2. Index SNPs for regions with 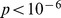 .
.
| rsid | chr | Position | Genes | MAF |
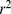
|
allele | HR (CI) |
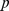 -value -value |
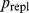
|
| rs12193446 | 6 | 129820038 | LAMA2 | 0.094 | 0.991 | A/G | 0.788 (0.763–0.813) |
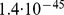
|
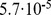
|
| rs1381566 | 11 | 40149607 | LRRC4C | 0.181 | 0.873 | T/G | 1.149 (1.122–1.176) |
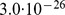
|
0.0038 |
| rs17648524 | 16 | 7459683 | RBFOX1 | 0.365 | 0.974 | G/C | 1.102 (1.082–1.122) |
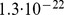
|
0.36 |
| rs7744813 | 6 | 73643289 | KCNQ5 | 0.405 | 0.958 | A/C | 0.909 (0.893–0.926) |
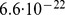
|
0.15 |
| rs3138142 | 12 | 56115585 | RDH5 | 0.218 | 0.831 | C/T | 0.890 (0.870–0.911) |
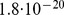
|
0.011 |
| chr8:60178580 | 8 | 60178580 | TOX/CA8 | 0.358 | 0.971 | C/G | 0.914 (0.897–0.931) |
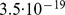
|
0.0061 |
| rs524952 | 15 | 35005886 | GOLGA8B/GJD2 | 0.469 | 0.982 | T/A | 1.089 (1.070–1.108) |
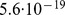
|
0.040 |
| rs2137277 | 8 | 40734662 | SFRP1 | 0.189 | 0.922 | A/G | 0.901 (0.880–0.923) |
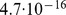
|
0.46 |
| rs1550094 | 2 | 233385396 | PRSS56 | 0.305 | 0.965 | A/G | 1.087 (1.067–1.107) |
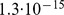
|
0.031 |
| rs2908972 | 17 | 11407259 | SHISA6 | 0.397 | 0.969 | T/A | 1.074 (1.055–1.093) |
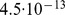
|
0.034 |
| rs17412774 | 2 | 146773948 | PABPCP2 | 0.450 | 0.976 | A/C | 0.933 (0.917–0.950) |
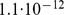
|
0.067 |
| rs11145746 | 9 | 71834380 | TJP2 | 0.198 | 0.887 | G/A | 1.087 (1.063–1.112) |
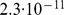
|
0.87 |
| rs28412916 | 15 | 79378167 | RASGRF1 | 0.401 | 0.989 | A/C | 1.067 (1.048–1.086) |
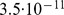
|
0.08 |
| rs5022942 | 4 | 81959966 | BMP3 | 0.229 | 0.991 | G/A | 1.076 (1.054–1.098) |
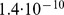
|
0.0093 |
| rs745480 | 10 | 85986554 | RGR | 0.473 | 0.975 | C/G | 1.063 (1.044–1.081) |
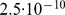
|
0.095 |
| rs2155413 | 11 | 84634790 | DLG2 | 0.466 | 0.997 | C/A | 1.061 (1.043–1.080) |
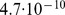
|
0.023 |
| rs13091182 | 3 | 141133960 | ZBTB38 | 0.333 | 0.994 | G/A | 0.940 (0.923–0.958) |
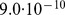
|
0.31 |
| rs17400325 | 2 | 178565913 | PDE11A | 0.050 | 0.933 | T/C | 1.144 (1.099–1.190) |
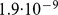
|
0.027 |
| rs17428076 | 2 | 172851936 | DLX1 | 0.237 | 0.985 | C/G | 0.935 (0.916–0.955) |
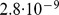
|
0.53 |
| rs6480859 | 10 | 79081948 | KCNMA1 | 0.363 | 0.987 | C/T | 1.058 (1.039–1.077) |
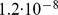
|
0.40 |
| chr14:54413001 | 14 | 54413001 | BMP4 | 0.489 | 0.933 | G/C | 0.946 (0.929–0.963) |
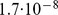
|
0.21 |
| rs4291789 | 13 | 100672921 | ZIC2 | 0.326 | 0.724 | C/G | 1.069 (1.046–1.092) |
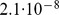
|
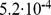
|
| rs10963578 | 9 | 18338649 | SH3GL2/ADAMTSL1 | 0.200 | 0.958 | G/A | 0.936 (0.915–0.957) |
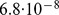
|
0.15 |
| rs11939401 | 4 | 80818417 | ANTXR2 | 0.203 | 0.999 | C/T | 0.939 (0.919–0.959) |
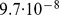
|
0.13 |
| rs1843303 | 3 | 4185124 | SETMAR | 0.303 | 0.981 | T/C | 1.055 (1.036–1.075) |
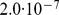
|
0.042 |
| chr11:65348347 | 11 | 65348347 | EHBP1L1 | 0.017 | 0.558 | G/A | 0.770 (0.700–0.846) |
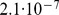
|
0.54 |
| rs4367880 | 10 | 114795256 | TCF7L2 | 0.199 | 0.959 | G/C | 1.063 (1.040–1.087) |
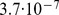
|
0.57 |
| rs61988414 | 14 | 42313443 | LRFN5 | 0.168 | 0.878 | A/G | 1.071 (1.045–1.097) |
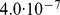
|
0.84 |
| rs9365619 | 6 | 164251746 | QKI | 0.457 | 0.999 | C/A | 1.048 (1.031–1.067) |
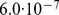
|
0.047 |
| rs4245599 | 10 | 60365755 | BICC1 | 0.466 | 0.952 | G/A | 1.049 (1.031–1.068) |
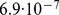
|
0.44 |
| rs10512441 | 17 | 31239645 | MYO1D/TMEM98 | 0.203 | 0.919 | C/T | 1.062 (1.039–1.085) |
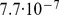
|
0.39 |
| rs9902755 | 17 | 47220726 | B4GALNT2 | 0.470 | 0.668 | C/T | 1.059 (1.037–1.081) |
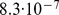
|
0.91 |
| rs6702767 | 1 | 200844547 | GPR25 | 0.485 | 0.982 | G/A | 1.048 (1.030–1.066) |
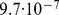
|
0.40 |
| chr17:79585492 | 17 | 79585492 | NPLOC4 | 0.393 | 0.604 | G/A | 1.063 (1.039–1.087) |
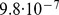
|
0.032 |
| rs6487748 | 12 | 9435768 | PZP | 0.491 | 0.981 | A/G | 1.048 (1.030–1.066) |
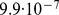
|
0.42 |
Index SNPs for regions with (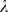 -corrected)
-corrected) 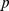 -values under
-values under 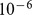 . Positions and alleles are given relative to the positive strand of build 37 of the human genome; alleles are listed as major/minor. The listed genes are the postulated candidate gene in each region.
. Positions and alleles are given relative to the positive strand of build 37 of the human genome; alleles are listed as major/minor. The listed genes are the postulated candidate gene in each region. 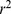 is the estimated imputation accuracy; HR is the hazard ratio per copy of the minor allele;
is the estimated imputation accuracy; HR is the hazard ratio per copy of the minor allele; 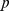 -value is the
-value is the 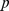 -value in the discovery cohort;
-value in the discovery cohort; 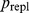 is the
is the 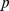 -value in the replication cohort. Significant replication
-value in the replication cohort. Significant replication 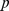 -values are bolded.
-values are bolded.
Figure 1. Negative 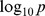 -values genome wide for myopia.
-values genome wide for myopia.
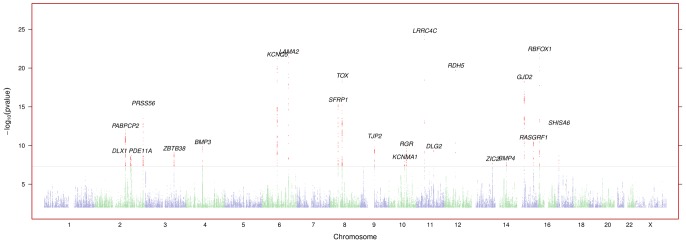
Regions are named with their postulated candidate gene or genes. 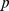 -values under
-values under 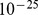 have been cut off (only the LAMA2 and LRRC4C regions are affected). See Figure S1 for plots in each region with a significant association.
have been cut off (only the LAMA2 and LRRC4C regions are affected). See Figure S1 for plots in each region with a significant association.
Of the 22 SNPs significant in the discovery set, 11 were also significant in the replication set (Table 2). Of the 11 SNPs that did not replicate, only two showed different signs between the discovery and replication sets ( ). Given these results, and considering that the replication set was much smaller than the discovery set and measured age of onset less exactly, we suspect that much of the lack of replication is due to lack of power.
). Given these results, and considering that the replication set was much smaller than the discovery set and measured age of onset less exactly, we suspect that much of the lack of replication is due to lack of power.
We defined a genetic myopia propensity score as the number of copies of the risk alleles across all 22 SNPs identified via the discovery set. The propensity score showed a strong association with early onset myopia (less than 10 years old) in our replication cohort (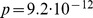 , odds ratio 1.075 per risk allele). The top decile of genetic propensity had 1.97 greater odds of developing myopia before the age of 10 than the bottom decile. In a Cox model fit to the discovery set, the propensity score explains 2.9% of the total variance. Note that this estimate may be inflated, as it is calculated on the discovery population. In this model, someone in the 90th percentile of risk (a score of 21.95) is nearly twice as likely to develop myopia by the age of 25 as someone in the 10th percentile of risk (score of 15.01), Figure 2.
, odds ratio 1.075 per risk allele). The top decile of genetic propensity had 1.97 greater odds of developing myopia before the age of 10 than the bottom decile. In a Cox model fit to the discovery set, the propensity score explains 2.9% of the total variance. Note that this estimate may be inflated, as it is calculated on the discovery population. In this model, someone in the 90th percentile of risk (a score of 21.95) is nearly twice as likely to develop myopia by the age of 25 as someone in the 10th percentile of risk (score of 15.01), Figure 2.
Figure 2. Estimated survival curves by genetic propensity score.
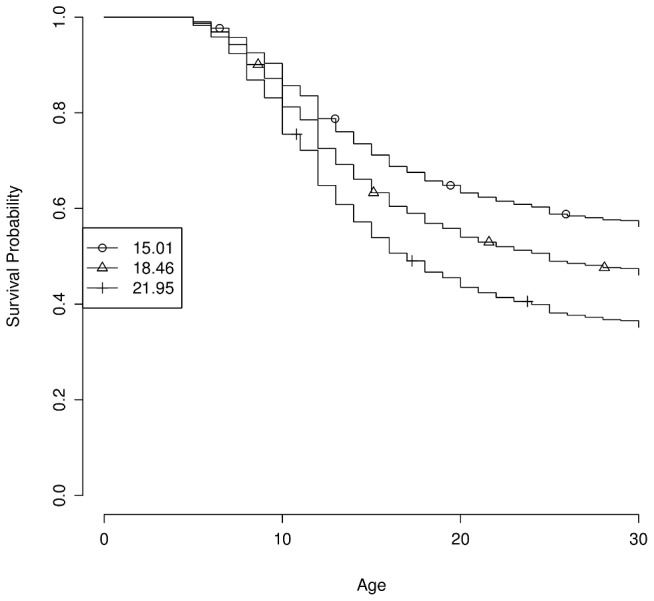
The genetic propensity score is computed as the number of risk alleles across the 22 genome-wide significant SNPs. Curves show estimated survival probability (i.e., the probability of not having developed myopia) by age under the fitted Cox model for the 10th, 50th, and 90th percentiles of scores (15.01, 18.46, and 21.95, respectively).
Of the 20 novel associations, many lie in or near genes with direct links to processes related to myopia development. Two of them lie in regions associated with myopia in linkage studies: rs1550094 in PRSS56 (MIM: 609995) [34] and chr14:54413001 near BMP4 (MIM: 255500) [35]. Two suggestive associations also are in such regions: rs4245599 in BICC1 (MIM: 612717) [36] and rs9902755 in B4GALNT2 (MIM: 608474) [37]. Below, we briefly sketch out possible connections between these associations and extracellular matrix (ECM) remodeling, the visual cycle, eye and body growth, retinal neuron development, and general neuronal development or signaling.
Extracellular Matrix Remodeling
The strongest association is a SNP in an intron of LAMA2 (laminin, alpha 2 subunit, rs12193446, 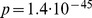 , hazard ratio (HR) = 0.79). Laminins are extracellular structural proteins that are integral parts of the ECM. Changes in the composition of the ECM of the sclera have been shown to alter the axial length of the eye [5]. Laminins play a role in the development and maintenance of different eye structures [38], [39]. The laminin alpha 2 chain in particular is found in the extraocular muscles during development [38], and may act as an adhesive substrate and possibly a guidance cue for retinal ganglion cell growth cones [40]. We also found a suggestive association related to laminin (rs11939401,
, hazard ratio (HR) = 0.79). Laminins are extracellular structural proteins that are integral parts of the ECM. Changes in the composition of the ECM of the sclera have been shown to alter the axial length of the eye [5]. Laminins play a role in the development and maintenance of different eye structures [38], [39]. The laminin alpha 2 chain in particular is found in the extraocular muscles during development [38], and may act as an adhesive substrate and possibly a guidance cue for retinal ganglion cell growth cones [40]. We also found a suggestive association related to laminin (rs11939401, 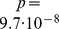 , HR = 0.939) approximately 17 kb upstream of ANTXR2 (anthrax toxin receptor 2). ANTXR2 binds laminin and possibly collagen type IV [41] and thus may also be involved in extracellular matrix remodeling.
, HR = 0.939) approximately 17 kb upstream of ANTXR2 (anthrax toxin receptor 2). ANTXR2 binds laminin and possibly collagen type IV [41] and thus may also be involved in extracellular matrix remodeling.
The Visual Cycle
Two of the novel associations are in or near genes involved in the regeneration of 11-cis-retinal, the light sensitive component of photoreceptors, a process commonly referred to as the visual cycle of the retina. These associations are with rs3138142, 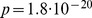 , HR = 0.89, in RDH5 (retinol dehydrogenase 5 (11-cis/9-cis)) and rs745480 (
, HR = 0.89, in RDH5 (retinol dehydrogenase 5 (11-cis/9-cis)) and rs745480 (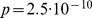 , HR = 1.06), a SNP 18 kb upstream of RGR, which encodes the retinal G protein-coupled receptor. The SNP rs3138142 is a synonymous change in RDH5. It has been linked to RDH5 expression [42], [43], and it is part of an Nr2f2 (nuclear receptor subfamily 2, group F, member 2) transcription factor binding motif in mouse [44], [45]. Both RDH5 and RGR play crucial roles in the regeneration of 11-cis retinal in the RPE [46]. Mutations in RDH5 have been linked with fundus albipunctatus, a rare form of congenital stationary night blindness (for a recent review, see [47]) and progressive cone dystrophy [48], and mutations in RGR have been linked with autosomal recessive and autosomal dominant retinitis pigmentosa [49], [50].
, HR = 1.06), a SNP 18 kb upstream of RGR, which encodes the retinal G protein-coupled receptor. The SNP rs3138142 is a synonymous change in RDH5. It has been linked to RDH5 expression [42], [43], and it is part of an Nr2f2 (nuclear receptor subfamily 2, group F, member 2) transcription factor binding motif in mouse [44], [45]. Both RDH5 and RGR play crucial roles in the regeneration of 11-cis retinal in the RPE [46]. Mutations in RDH5 have been linked with fundus albipunctatus, a rare form of congenital stationary night blindness (for a recent review, see [47]) and progressive cone dystrophy [48], and mutations in RGR have been linked with autosomal recessive and autosomal dominant retinitis pigmentosa [49], [50].
We also identified an association within another gene that functions in the RPE: rs7744813 (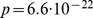 , HR = 0.91), a SNP in KCNQ5 (potassium voltage-gated channel, KQT-like subfamily, member 5). KCNQ5 encodes a potassium channel found in the RPE and neural retina. These channels are believed to contribute to ion flow across the RPE [51], [52] and to affect the function of cone and rod photoreceptors [52].
, HR = 0.91), a SNP in KCNQ5 (potassium voltage-gated channel, KQT-like subfamily, member 5). KCNQ5 encodes a potassium channel found in the RPE and neural retina. These channels are believed to contribute to ion flow across the RPE [51], [52] and to affect the function of cone and rod photoreceptors [52].
Eye and Body Growth
Five of our associations show possible links to eye or overall morphology. The first is a missense mutation in PRSS56 (A224T, rs1550094, 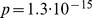 , HR = 1.09). Other mutations in PRSS56 have been shown to cause strikingly small eyes with severe decreases in axial length [53]–[55]. Two other associated SNPs are near genes that encode bone morphogenetic proteins: chr14:54413001 (
, HR = 1.09). Other mutations in PRSS56 have been shown to cause strikingly small eyes with severe decreases in axial length [53]–[55]. Two other associated SNPs are near genes that encode bone morphogenetic proteins: chr14:54413001 (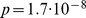 , HR = 0.95) near BMP4 (bone morphogenic protein-4), and rs5022942 (
, HR = 0.95) near BMP4 (bone morphogenic protein-4), and rs5022942 (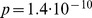 , HR = 1.08) in BMP3 (bone morphogenic protein-3). Inherited BMP4 mutations have been associated with syndromic microphthalmia and various eye, brain and digital malformations [56], [57]. Although BMP3 is primarily known for its role in bone development (e.g., it is linked to skeletal defects in humans and skull shapes in dogs [58], [59]), it was found to be uniquely expressed in keratocytes, specialized mesenchymal cells that are important for development of the cornea by producing and maintaining the extracellular matrix of the corneal stroma [60]. One associated SNP, rs13091182 (
, HR = 1.08) in BMP3 (bone morphogenic protein-3). Inherited BMP4 mutations have been associated with syndromic microphthalmia and various eye, brain and digital malformations [56], [57]. Although BMP3 is primarily known for its role in bone development (e.g., it is linked to skeletal defects in humans and skull shapes in dogs [58], [59]), it was found to be uniquely expressed in keratocytes, specialized mesenchymal cells that are important for development of the cornea by producing and maintaining the extracellular matrix of the corneal stroma [60]. One associated SNP, rs13091182 (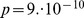 , HR = 0.94), in ZBTB38 (zinc finger and BTB domain-containing protein 38), is in linkage disequilibrium (LD) with a SNP previously associated with height (rs6763931;
, HR = 0.94), in ZBTB38 (zinc finger and BTB domain-containing protein 38), is in linkage disequilibrium (LD) with a SNP previously associated with height (rs6763931; 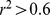 ) [61]. The final SNP with a link to morphology is rs17428076 (
) [61]. The final SNP with a link to morphology is rs17428076 (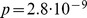 , HR = 0.94), near DLX1 (homo sapiens distal-less homeobox 1). Disruption of DLX1 has been shown to result in poor optic cup regeneration in planarians and small eyes in mice [62], [63].
, HR = 0.94), near DLX1 (homo sapiens distal-less homeobox 1). Disruption of DLX1 has been shown to result in poor optic cup regeneration in planarians and small eyes in mice [62], [63].
Retinal Ganglion Cell Projections
Two of the novel associations are near genes that affect the outgrowth of retinal ganglion neurons during development. The first is rs4291789 (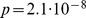 , HR = 1.07), which lies 34 kb downstream of ZIC2 (Zic family member 2). ZIC2 regulates two independent parts of ipsilateral retinal ganglion cell development: axon repulsion at the optic chiasm midline [64], [65], and organization of the axonal projections at their final targets in the brain [66].
, HR = 1.07), which lies 34 kb downstream of ZIC2 (Zic family member 2). ZIC2 regulates two independent parts of ipsilateral retinal ganglion cell development: axon repulsion at the optic chiasm midline [64], [65], and organization of the axonal projections at their final targets in the brain [66].
The second, rs2137277 (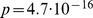 , HR = 0.90), is a variant in ZMAT4 (zinc finger, matrin-type 4). ZMAT4 has no known link to vision, but this variant also lies 385 kb downstream of SFRP1 (secreted frizzled-related protein 1). SFRP1 is involved in the differentiation of the optic cup from the neural retina [67], retinal neurogenesis [68], the development and function of photoreceptor cells [69], [70], and the growth of retinal ganglion cells [71].
, HR = 0.90), is a variant in ZMAT4 (zinc finger, matrin-type 4). ZMAT4 has no known link to vision, but this variant also lies 385 kb downstream of SFRP1 (secreted frizzled-related protein 1). SFRP1 is involved in the differentiation of the optic cup from the neural retina [67], retinal neurogenesis [68], the development and function of photoreceptor cells [69], [70], and the growth of retinal ganglion cells [71].
Neuronal Signaling and Development
Finally, we found five associations with SNPs in genes involved in neuronal development and signaling, but without a known role in vision development or the vision cycle: in KCNMA1 (potassium large conductance calcium-activated channel, subfamily M, alpha member 1; rs6480859, 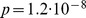 , HR = 1.06); in RBFOX1 (RNA binding protein, fox-1 homolog; rs17648524,
, HR = 1.06); in RBFOX1 (RNA binding protein, fox-1 homolog; rs17648524, 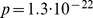 , HR = 1.10); in LRRC4C, leucine rich repeating region containing 4C, also known as NGL-1 (rs1381566,
, HR = 1.10); in LRRC4C, leucine rich repeating region containing 4C, also known as NGL-1 (rs1381566, 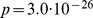 , HR = 1.15); in DLG2 (discs, large homolog 2; rs2155413,
, HR = 1.15); in DLG2 (discs, large homolog 2; rs2155413, 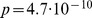 , HR = 1.06); and in TJP2 (tight junction protein 2; rs11145746,
, HR = 1.06); and in TJP2 (tight junction protein 2; rs11145746, 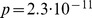 , HR = 1.09).
, HR = 1.09).
KCNMA1 encodes the pore-forming alpha subunit of a MaxiK channel, a family of large conductance, voltage and calcium-sensitive potassium channels involved in the control of smooth muscle and neuronal excitation. RBFOX1 belongs to a family of RNA binding proteins that regulates the alternative splicing of several neuronal transcripts implicated in neuronal development and maturation [72]. LRRC4C encodes a binding partner for netrin G1 and promotes the outgrowth of thalamocortical axons [73]. DLG2 plays a critical role in the formation and regulation of protein scaffolding at postsynaptic sites [74]. TJP2 has been linked with hearing loss: its duplication and subsequent overexpression are found in adult-onset progressive nonsyndromic hearing loss [75].
Conclusion
This study represents the largest GWAS on myopia in Europeans to date. This cohort of 45,771 individuals led to the discovery of 20 novel associations as well as replication of the two previously reported associations in Europeans. Ten of these novel associations replicate in our much smaller replication set of 8,323 individuals. In contrast to the earlier studies that used refractive error as a quantitative outcome, we used a Cox proportional hazards model with age of onset of myopia as our major endpoint. This model yielded greater statistical power than a simple case-control study of myopia. Of the 22 significant SNPs found using this model, all but two had smaller 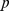 -values when a hazards model was employed, and only 20 would be genome-wide significant using a case-control analysis on the same dataset (Table S1).
-values when a hazards model was employed, and only 20 would be genome-wide significant using a case-control analysis on the same dataset (Table S1).
The proportional hazards model assumes that the effect of each SNP on myopia risk does not vary by age. When we tested the validity of this assumption for the 22 significant SNPs, only the one in LAMA2 (rs12193446) showed evidence of different effects at different ages (Table S2). While this violation should not lead to overly small 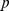 -values for this SNP in the GWAS, it does make risk prediction based on these results less straightforward. For example, rs12193446 shows a large effect on myopia hazard at an early age, peaking around 11 years, and then a null or even negative effect on hazard at older ages (Figure S3). This age dependent hazard suggests that different biological processes may affect the development of myopia at different ages.
-values for this SNP in the GWAS, it does make risk prediction based on these results less straightforward. For example, rs12193446 shows a large effect on myopia hazard at an early age, peaking around 11 years, and then a null or even negative effect on hazard at older ages (Figure S3). This age dependent hazard suggests that different biological processes may affect the development of myopia at different ages.
Our findings further suggest that there may be somewhat different genetic factors underlying myopia age of onset and refractive error. Because adult onset myopia tends to be less severe than myopia developed in childhood or adolescence [10]–[12], age of onset is likely correlated with refractive error, but it is not known how strongly. Many of the associations with myopia age of onset that we found are stronger than the two previously detected associations with refractive error (near GJD2 and near RASGRF1). Notably, the latter association, near RASGRF1, also failed to replicate in a recent meta-analysis [33]. The fact that many of our associations with strong effects on age of onset have not shown up in previous refractive error GWAS implies that some genetic factors may affect the age of onset independent of eventual severity, and that the strength of different genetic associations with myopia may depend on the specific phenotype under study.
We also note that our phenotype was based on participants' reports rather than clinical assessments. Although in theory errors in recall could have affected our results, we expect that the vast majority of people are able to recall when they first wore glasses with at most a few years of error. In fact, a subset of those eligible to be part of our discovery cohort provided age of myopia diagnosis in two independent places (see Methods for details). Out of 1,463 people who reported age of diagnosis in both surveys and met our inclusion criteria (European ancestry, age at diagnosis between five and 30 and less than current age), 96.0% reported ages that differed by at most three years and 97.8% by at most five years.
The five associations previously reported in pathological myopia or refractive error GWAS in Asian populations [26]–[30] show no overlap with the significant or suggestive regions found here. Nor did we find an association with the ZNF644 locus that was identified as the site of high-penetrance, autosomal dominant mutations in Han Chinese families with apparent monogenic inheritance of high-grade myopia [32]. This lack of overlap could result from different genetic factors being involved in myopia across populations. It has been suggested that pathological myopia, which represents less than 2% of cases in the United States [1], has different underlying genetic factors than non-pathological myopia [31].
Our identification of 20 novel genetic associations suggests several novel genetic pathways in the development of human myopia. These findings augment existing research on the development of myopia, which to date has been studied primarily in animal models of artificially induced myopia. Some of the associations are consistent with the current view, based largely on animal models, that a visually-triggered signaling cascade from the retina ultimately guides the scleral remodeling that leads to eye growth, and that the RPE plays a key role in this process [4]. A number of the novel associations point to the potential importance of early neuronal development in the eventual development of myopia, particularly the growth and topographical organization of retinal ganglion cells. These associations suggest that early neuronal development may also contribute to future refractive errors. We expect that these findings will drive new research into the complex etiology of myopia.
Methods
Human Subjects
All participants were drawn from the customer base of 23andMe, Inc., a consumer genetics company. This cohort has been described in detail previously [76], [77]. Participants provided informed consent and participated in the research online, under a protocol approved by the external AAHRPP-accredited IRB, Ethical & Independent Review Services (E&I Review).
Phenotype Data
Participants in the discovery cohort were asked online as part of a medical history questionnaire or an eyesight questionnaire: “Have you ever been diagnosed by a doctor with any of the following vision conditions?: Nearsightedness (near objects are clear, far objects are blurry) (Yes/No/I don't know)”. If they answered “yes”, they were asked, “At what age were you first diagnosed with nearsightedness (near objects are clear, far objects are blurry)? Your best guess is fine.” Those reporting an age of onset either greater than their current age or outside of the range 5–30 were removed from analysis. All participants also reported current age. A total of 4,758 participants reporting age of onset outside of 5–30 and 87 reporting age of onset in the future were removed.
To limit errors in reporting, we excluded from the discovery cohort those who provided discordant answers in the medical history and eyesight questionnaires. We defined discordance as a disagreement in diagnosis or a difference in more than 5 years in age of onset. A total of 92 people with discordant age of onset and 276 with discordant diagnosis were removed. Many of these people would have been filtered out by our other restrictions: only 32 of the 92 with discordant ages of onset would not have been removed for other reasons (mostly because their stated age of onset was not between 5–30), and only 139 of the 276 with discordant diagnoses. These 32 and 139 are out of 1,463 and 2,845 eligible people, respectively, leading us to estimates of 97.8% and 95.1% concordance in age of onset and myopia diagnosis (after the filters mentioned above were applied).
The replication cohort consisted of 8,323 23andMe customers who were not part of the discovery cohort and were not closely related (at 700 cM or greater IBD) to each other or to anyone in the discovery cohort. They provided information on myopia age of onset in one of two ways. 5,265 answered a single question: “Did you wear glasses or other corrective eyewear for nearsightedness before the age of 10? (Yes/No/I'm not sure).” The other 3,058 provided age of onset in the same manner as the discovery cohort. Note that these 3,058 were people that would have been eligible for the discovery cohort, however, they provided data in the time in between our analysis of the discovery and replication cohorts. Their data was converted to the same binary scale as the first group.
Genotyping and Imputation
Participants were genotyped and additional SNP genotypes were imputed against the August 2010 release of the 1000 genomes data as described previously [78]. Briefly, they were genotyped on at least one of three genotyping platforms, two based on the Illumina HumanHap550+ BeadChip, the third based on the Illumina Human OmniExpress+ BeadChip. The platforms included assays for 586,916, 584,942, and 1,008,948 SNPs respectively. Genotypes for a total of 11,914,767 SNPs were imputed in batches of roughly 10,000 individuals, grouped by genotyping platform. Of these, 7,087,609 met our criteria of 0.005 minor allele frequency, average 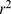 across batches of at least 0.5, and minimum
across batches of at least 0.5, and minimum 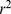 across batches of at least 0.3. (The minimum
across batches of at least 0.3. (The minimum 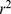 requirement was added to filter out SNPs that imputed poorly in the batches consisting of the less dense platform.)
requirement was added to filter out SNPs that imputed poorly in the batches consisting of the less dense platform.)
Statistical Analysis
In order to minimize population substructure while maximizing statistical power, the study was limited to individuals with European ancestry. Ancestry was inferred from the genome-wide genotype data, and principal component analysis was performed as in [76], [79]. The combined discovery and replication populations were filtered by relatedness to remove participants at a first cousin or closer relationship. More precisely, no two participants shared more than 700 cM of DNA identical by descent (IBD, approximately the lower end of sharing between a pair of first cousins). IBD was calculated using the methods described in [80].
For the survival analysis, let the hazard function 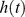 be the rate of developing myopia at time
be the rate of developing myopia at time  . Then the Cox proportional hazards model is
. Then the Cox proportional hazards model is
for an arbitrary baseline hazard function 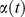 and covariates
and covariates 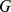 (genotype),
(genotype), 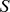 (sex),
(sex), 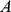 (age), and
(age), and  (projections onto principal components).
(projections onto principal components). 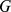 was coded as a dosage from 0–2 as the estimated number of minor alleles present.
was coded as a dosage from 0–2 as the estimated number of minor alleles present.
For each SNP, we fit a Cox proportional hazards model using R [81] and computed a p-value using a likelihood ratio test for the genotype term. All SNPs with 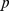 -values under
-values under 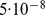 after genomic control correction were considered genome-wide significant. The hazard ratio (HR) reported throughout can be interpreted as the multiplicative change in the rate of onset of myopia per copy of the minor allele (e.g.,
after genomic control correction were considered genome-wide significant. The hazard ratio (HR) reported throughout can be interpreted as the multiplicative change in the rate of onset of myopia per copy of the minor allele (e.g., 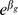 ). To test the proportional hazards assumption, we tested for independence of the scaled Schoenfeld residuals for each significant SNP and time using cox.zph (Table S2). Replication
). To test the proportional hazards assumption, we tested for independence of the scaled Schoenfeld residuals for each significant SNP and time using cox.zph (Table S2). Replication 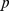 -values in Table 2 are one-sided
-values in Table 2 are one-sided 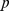 -values from a likelihood ratio test for a logistic regression model controlling for age, sex, and five principal components.
-values from a likelihood ratio test for a logistic regression model controlling for age, sex, and five principal components.
For Figure 2, we computed a myopia propensity score for each individual as the (estimated) number of risk alleles among the 22 genome-wide significant SNPs. We then fit a Cox model including that score, sex, and five principal components. To estimate proportion variance explained for this model, we used a pseudo-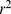 using likelihoods (similar to the Nagelkerke pseudo
using likelihoods (similar to the Nagelkerke pseudo 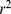 for logistic regression). That is, we calculated the variance explained as
for logistic regression). That is, we calculated the variance explained as
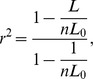 |
where 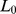 is the null likelihood and
is the null likelihood and 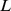 the likelihood for the full model. This is one of several methods used to compute variance explained for Cox proportional hazards models [82].
the likelihood for the full model. This is one of several methods used to compute variance explained for Cox proportional hazards models [82].
Supporting Information
Region plots for genome-wide significant associations Colors depict the squared correlation (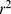 ) of each SNP with the most associated SNP (shown in purple). Gray indicates SNPs for which
) of each SNP with the most associated SNP (shown in purple). Gray indicates SNPs for which 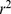 information was missing.
information was missing.
(PDF)
Quantile-quantile plot for myopia survival analysis Actual (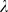 -corrected)
-corrected) 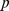 -values versus the null.
-values versus the null.
(PDF)
Smoothed log-hazard ratios as a function of age for four SNPs In each plot, the straight line shows the estimated log-hazard ratio (beta) for each SNP in the proportional hazards model. The solid curve is a spline fit to beta estimated at different ages; the dotted curves are approximate 95% confidence intervals. The 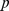 -value in each caption is the result of a test of the proportional hazards assumption. The sign of all coefficients has been made positive for ease of comparison (so (a), (c), and (d) are flipped relative to the main text). Note that among the examples here, only (a) shows evidence of deviation from the proportional hazards assumption after correction for 22 tests.
-value in each caption is the result of a test of the proportional hazards assumption. The sign of all coefficients has been made positive for ease of comparison (so (a), (c), and (d) are flipped relative to the main text). Note that among the examples here, only (a) shows evidence of deviation from the proportional hazards assumption after correction for 22 tests.
(PDF)
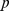 -values for survival and case-control analyses.
-values for survival and case-control analyses. 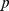 -values for SNPs in the survival analysis used in the paper as well as in a case-control logistic regression on the same set of individuals. The survival analysis gives a smaller
-values for SNPs in the survival analysis used in the paper as well as in a case-control logistic regression on the same set of individuals. The survival analysis gives a smaller 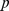 -value for 30 of 35 SNPs and has 22 genome-wide significant (
-value for 30 of 35 SNPs and has 22 genome-wide significant (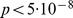 ) as compared to 20 for the case-control.
) as compared to 20 for the case-control. 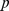 -values in both cases are adjusted for the genomic control inflation factor of 1.16.
-values in both cases are adjusted for the genomic control inflation factor of 1.16.
(PDF)
Tests of deviation from the proportional hazards assumption. 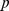 -values for significant SNPs for deviation from the proportional hazards assumption in the Cox model. For each SNP, we fit a Cox proportional hazards model including the SNP, sex, and five principal components as predictors, and then tested for independence of the scaled Schoenfeld residuals with time. Only one SNP deviates significantly from this assumption after correction for 22 tests. Plots for four example SNPs are shown in Figure S3.
-values for significant SNPs for deviation from the proportional hazards assumption in the Cox model. For each SNP, we fit a Cox proportional hazards model including the SNP, sex, and five principal components as predictors, and then tested for independence of the scaled Schoenfeld residuals with time. Only one SNP deviates significantly from this assumption after correction for 22 tests. Plots for four example SNPs are shown in Figure S3.
(PDF)
Statistics for all SNPs with 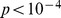 . All 6,141 SNPs with (
. All 6,141 SNPs with (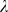 -corrected)
-corrected) 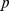 -values under
-values under 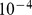 in the discovery cohort. Positions and alleles are given relative to the positive strand of build 37 of the human genome; alleles are listed as major/minor. The gene column shows the position of the SNP in context of the nearest genes. The SNP position is within the brackets, and the number of dashes gives approximate
in the discovery cohort. Positions and alleles are given relative to the positive strand of build 37 of the human genome; alleles are listed as major/minor. The gene column shows the position of the SNP in context of the nearest genes. The SNP position is within the brackets, and the number of dashes gives approximate 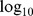 distances. The MAF is the minor allele frequency in Europeans,
distances. The MAF is the minor allele frequency in Europeans, 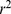 is the estimated imputation accuracy, HR is the hazard ratio per copy of the minor allele, and
is the estimated imputation accuracy, HR is the hazard ratio per copy of the minor allele, and 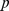 -value is the
-value is the 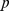 -value in the discovery cohort.
-value in the discovery cohort.
(XLS)
Acknowledgments
We thank the customers of 23andMe for participating in this research and all the employees of 23andMe for their contributions to this work.
Funding Statement
This study was funded by the participants and by 23andMe. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vitale S, Sperduto RD, Ferris FL (2009) Increased prevalence of myopia in the United States between 1971–1972 and 1999–2004. Arch Ophthalmol 127: 1632–1639. [DOI] [PubMed] [Google Scholar]
- 2. Kempen JH, Mitchell P, Lee KE, Tielsch JM, Broman AT, et al. (2004) The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol 122: 495–505. [DOI] [PubMed] [Google Scholar]
- 3. Wojciechowski R (2011) Nature and nurture: the complex genetics of myopia and refractive error. Clin Genet 79: 301–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rymer J, Wildsoet CF (2005) The role of the retinal pigment epithelium in eye growth regulation and myopia: a review. Vis Neurosci 22: 251–261. [DOI] [PubMed] [Google Scholar]
- 5. Rada JA, Shelton S, Norton TT (2006) The sclera and myopia. Exp Eye Res 82: 185–200. [DOI] [PubMed] [Google Scholar]
- 6. Wildsoet C, Wallman J (1995) Choroidal and scleral mechanisms of compensation for spectacle lenses in chicks. Vision Res 35: 1175–1194. [DOI] [PubMed] [Google Scholar]
- 7. Mutti DO, Zadnik K, Adams AJ (1996) Myopia. The nature versus nurture debate goes on. Invest Ophthalmol Vis Sci 37: 952–957. [PubMed] [Google Scholar]
- 8. Fledelius HC (1982) Ophthalmic changes from age of 10 to 18 years. A longitudinal study of sequels to low birth weight. IV. Ultrasound oculometry of vitreous and axial length. Acta Ophthalmol (Copenh) 60: 403–411. [DOI] [PubMed] [Google Scholar]
- 9. Fan DS, Lam DS, Lam RF, Lau JT, Chong KS, et al. (2004) Prevalence, incidence, and progression of myopia of school children in Hong Kong. Invest Ophthalmol Vis Sci 45: 1071–1075. [DOI] [PubMed] [Google Scholar]
- 10. Fledelius HC (1995) Myopia of adult onset. Can analyses be based on patient memory? Acta Ophthalmol Scand 73: 394–396. [DOI] [PubMed] [Google Scholar]
- 11. Iribarren R, Cortinez MF, Chiappe JP (2009) Age of first distance prescription and final myopic refractive error. Ophthalmic Epidemiol 16: 84–89. [DOI] [PubMed] [Google Scholar]
- 12. Iribarren R, Cerrella MR, Armesto A, Iribarren G, Fornaciari A (2004) Age of lens use onset in a myopic sample of office-workers. Curr Eye Res 28: 175–180. [DOI] [PubMed] [Google Scholar]
- 13. Isaza G, Arora S (2012) Incidence and severity of retinopathy of prematurity in extremely premature infants. Can J Ophthalmol 47: 296–300. [DOI] [PubMed] [Google Scholar]
- 14. Kennedy RH, Bourne WM, Dyer JA (1986) A 48-year clinical and epidemiologic study of keratoconus. Am J Ophthalmol 101: 267–273. [DOI] [PubMed] [Google Scholar]
- 15. Richards AJ, McNinch A, Martin H, Oakhill K, Rai H, et al. (2010) Stickler syndrome and the vitreous phenotype: mutations in COL2A1 and COL11A1. Hum Mutat 31: E1461–1471. [DOI] [PubMed] [Google Scholar]
- 16. Hammond CJ, Snieder H, Gilbert CE, Spector TD (2001) Genes and environment in refractive error: the twin eye study. Invest Ophthalmol Vis Sci 42: 1232–1236. [PubMed] [Google Scholar]
- 17. Lyhne N, Sjolie AK, Kyvik KO, Green A (2001) The importance of genes and environment for ocular refraction and its determiners: a population based study among 20–45 year old twins. Br J Ophthalmol 85: 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teikari JM, O'Donnell J, Kaprio J, Koskenvuo M (1991) Impact of heredity in myopia. Hum Hered 41: 151–156. [DOI] [PubMed] [Google Scholar]
- 19. Wojciechowski R, Congdon N, Bowie H, Munoz B, Gilbert D, et al. (2005) Heritability of refractive error and familial aggregation of myopia in an elderly American population. Invest Ophthalmol Vis Sci 46: 1588–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peet JA, Cotch MF, Wojciechowski R, Bailey-Wilson JE, Stambolian D (2007) Heritability and familial aggregation of refractive error in the Old Order Amish. Invest Ophthalmol Vis Sci 48: 4002–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goss DA, Jackson TW (1996) Clinical findings before the onset of myopia in youth: 4. Parental history of myopia. Optom Vis Sci 73: 279–282. [DOI] [PubMed] [Google Scholar]
- 22. Klein AP, Duggal P, Lee KE, Klein R, Bailey-Wilson JE, et al. (2005) Support for polygenic influences on ocular refractive error. Invest Ophthalmol Vis Sci 46: 442–446. [DOI] [PubMed] [Google Scholar]
- 23. Ashton GC (1985) Segregation analysis of ocular refraction and myopia. Hum Hered 35: 232–239. [DOI] [PubMed] [Google Scholar]
- 24. Solouki AM, Verhoeven VJ, van Duijn CM, Verkerk AJ, Ikram MK, et al. (2010) A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet 42: 897–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hysi PG, Young TL, Mackey DA, Andrew T, Fernandez-Medarde A, et al. (2010) A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet 42: 902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li YJ, Goh L, Khor CC, Fan Q, Yu M, et al. (2011) Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology 118: 368–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakanishi H, Yamada R, Gotoh N, Hayashi H, Yamashiro K, et al. (2009) A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet 5: e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Z, Qu J, Xu X, Zhou X, Zou H, et al. (2011) A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum Mol Genet 20: 2861–2868. [DOI] [PubMed] [Google Scholar]
- 29. Shi Y, Qu J, Zhang D, Zhao P, Zhang Q, et al. (2011) Genetic variants at 13q12.12 are associated with high myopia in the Han Chinese population. Am J Hum Genet 88: 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fan Q, Barathi VA, Cheng CY, Zhou X, Meguro A, et al. (2012) Genetic variants on chromosome 1q41 influence ocular axial length and high myopia. PLoS Genet 8: e1002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Young TL, Metlapally R, Shay AE (2007) Complex trait genetics of refractive error. Arch Ophthalmol 125: 38–48. [DOI] [PubMed] [Google Scholar]
- 32. Shi Y, Li Y, Zhang D, Zhang H, Li Y, et al. (2011) Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet 7: e1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Verhoeven VJ, Hysi PG, Saw SM, Vitart V, Mirshahi A, et al. (2012) Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Hum Genet 131: 1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paluru PC, Nallasamy S, Devoto M, Rappaport EF, Young TL (2005) Identification of a novel locus on 2q for autosomal dominant high-grade myopia. Invest Ophthalmol Vis Sci 46: 2300–2307. [DOI] [PubMed] [Google Scholar]
- 35. Yang Z, Xiao X, Li S, Zhang Q (2009) Clinical and linkage study on a consanguineous Chinese family with autosomal recessive high myopia. Mol Vis 15: 312–318. [PMC free article] [PubMed] [Google Scholar]
- 36. Nallasamy S, Paluru PC, Devoto M, Wasserman NF, Zhou J, et al. (2007) Genetic linkage study of high-grade myopia in a Hutterite population from South Dakota. Mol Vis 13: 229–236. [PMC free article] [PubMed] [Google Scholar]
- 37. Paluru P, Ronan SM, Heon E, Devoto M, Wildenberg SC, et al. (2003) New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci 44: 1830–1836. [DOI] [PubMed] [Google Scholar]
- 38. Bystrom B, Virtanen I, Rousselle P, Gullberg D, Pedrosa-Domellof F (2006) Distribution of laminins in the developing human eye. Invest Ophthalmol Vis Sci 47: 777–785. [DOI] [PubMed] [Google Scholar]
- 39. Peterson PE, Pow CS, Wilson DB, Hendrickx AG (1995) Localisation of glycoproteins and glycosaminoglycans during early eye development in the macaque. J Anat 186 (Pt 1) 31–42. [PMC free article] [PubMed] [Google Scholar]
- 40. Morissette N, Carbonetto S (1995) Laminin alpha 2 chain (M chain) is found within the pathway of avian and murine retinal projections. J Neurosci 15: 8067–8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bell SE, Mavila A, Salazar R, Bayless KJ, Kanagala S, et al. (2001) Differential gene expression during capillary morphogenesis in 3D collagen matrices: regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. J Cell Sci 114: 2755–2773. [DOI] [PubMed] [Google Scholar]
- 42. Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, et al. (2008) High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet 4: e1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, et al. (2007) Population genomics of human gene expression. Nat Genet 39: 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, et al. (2009) Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boyle A, Hong E, Hariharan M, Cheng Y, Shaub M, et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22: 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Strauss O (2005) The retinal pigment epithelium in visual function. Physiol Rev 85: 845–881. [DOI] [PubMed] [Google Scholar]
- 47. Wang NK, Chuang LH, Lai CC, Chou CL, Chu HY, et al. (2012) Multimodal fundus imaging in fundus albipunctatus with RDH5 mutation: a newly identified compound heterozygous mutation and review of the literature. Doc Ophthalmol 125: 51–62. [DOI] [PubMed] [Google Scholar]
- 48. Nakamura M, Hotta Y, Tanikawa A, Terasaki H, Miyake Y (2000) A high association with cone dystrophy in Fundus albipunctatus caused by mutations of the RDH5 gene. Invest Ophthalmol Vis Sci 41: 3925–3932. [PubMed] [Google Scholar]
- 49. Wang Q, Chen Q, Zhao K, Wang L, Wang L, et al. (2001) Update on the molecular genetics of retinitis pigmentosa. Ophthalmic Genet 22: 133–154. [DOI] [PubMed] [Google Scholar]
- 50. Bernal S, Calaf M, Garcia-Hoyos M, Garcia-Sandoval B, Rosell J, et al. (2003) Study of the involvement of the RGR, CRPB1, and CRB1 genes in the pathogenesis of autosomal recessive retinitis pigmentosa. J Med Genet 40: e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pattnaik BR, Hughes BA (2012) Effects of KCNQ channel modulators on the M-type potassium current in primate retinal pigment epithelium. Am J Physiol, Cell Physiol 302: C821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang X, Yang D, Hughes BA (2011) KCNQ5/K(v)7.5 potassium channel expression and subcellular localization in primate retinal pigment epithelium and neural retina. Am J Physiol, Cell Physiol 301: C1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nair KS, Hmani-Aifa M, Ali Z, Kearney AL, Ben Salem S, et al. (2011) Alteration of the serine protease PRSS56 causes angle-closure glaucoma in mice and posterior microphthalmia in humans and mice. Nat Genet 43: 579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Orr A, Dube MP, Zenteno JC, Jiang H, Asselin G, et al. (2011) Mutations in a novel serine protease PRSS56 in families with nanophthalmos. Mol Vis 17: 1850–1861. [PMC free article] [PubMed] [Google Scholar]
- 55. Gal A, Rau I, El Matri L, Kreienkamp HJ, Fehr S, et al. (2011) Autosomal-recessive posterior microphthalmos is caused by mutations in PRSS56, a gene encoding a trypsin-like serine protease. Am J Hum Genet 88: 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reis LM, Tyler RC, Schilter KF, Abdul-Rahman O, Innis JW, et al. (2011) BMP4 loss-of-function mutations in developmental eye disorders including SHORT syndrome. Hum Genet 130: 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bakrania P, Efthymiou M, Klein JC, Salt A, Bunyan DJ, et al. (2008) Mutations in BMP4 cause eye, brain, and digit developmental anomalies: overlap between the BMP4 and hedgehog signaling pathways. Am J Hum Genet 82: 304–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lipska BS, Brzeskwiniewicz M, Wierzba J, Morzuchi L, Piotrowski A, et al. (2011) 8.6 Mb interstitial deletion of chromosome 4q13.3q21.23 in a boy with cognitive impairment, short stature, hearing loss, skeletal abnormalities and facial dysmorphism. Genet Couns 22: 353–363. [PubMed] [Google Scholar]
- 59. Schoenebeck JJ, Hutchinson SA, Byers A, Beale HC, Carrington B, et al. (2012) Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet 8: e1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Scott SG, Jun AS, Chakravarti S (2011) Sphere formation from corneal keratocytes and phenotype specific markers. Exp Eye Res 93: 898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, et al. (2008) Many sequence variants affecting diversity of adult human height. Nat Genet 40: 609–615. [DOI] [PubMed] [Google Scholar]
- 62. Lapan SW, Reddien PW (2011) dlx and sp6-9 Control optic cup regeneration in a prototypic eye. PLoS Genet 7: e1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stoykova A, Fritsch R, Walther C, Gruss P (1996) Forebrain patterning defects in Small eye mutant mice. Development 122: 3453–3465. [DOI] [PubMed] [Google Scholar]
- 64. Garcia-Frigola C, Carreres MI, Vegar C, Mason C, Herrera E (2008) Zic2 promotes axonal divergence at the optic chiasm midline by EphB1-dependent and -independent mechanisms. Development 135: 1833–1841. [DOI] [PubMed] [Google Scholar]
- 65. Herrera E, Brown L, Aruga J, Rachel RA, Dolen G, et al. (2003) Zic2 patterns binocular vision by specifying the uncrossed retinal projection. Cell 114: 545–557. [DOI] [PubMed] [Google Scholar]
- 66. Garcia-Frigola C, Herrera E (2010) Zic2 regulates the expression of Sert to modulate eye-specific refinement at the visual targets. EMBO J 29: 3170–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Esteve P, Sandonis A, Ibanez C, Shimono A, Guerrero I, et al. (2011) Secreted frizzled-related proteins are required for Wnt/-catenin signalling activation in the vertebrate optic cup. Development 138: 4179–4184. [DOI] [PubMed] [Google Scholar]
- 68. Esteve P, Sandonis A, Cardozo M, Malapeira J, Ibanez C, et al. (2011) SFRPs act as negative modulators of ADAM10 to regulate retinal neurogenesis. Nat Neurosci 14: 562–569. [DOI] [PubMed] [Google Scholar]
- 69. Garcia-Hoyos M, Cantalapiedra D, Arroyo C, Esteve P, Rodriguez J, et al. (2004) Evaluation of SFRP1 as a candidate for human retinal dystrophies. Mol Vis 10: 426–431. [PubMed] [Google Scholar]
- 70. Esteve P, Trousse F, Rodriguez J, Bovolenta P (2003) SFRP1 modulates retina cell differentiation through a beta-catenin-independent mechanism. J Cell Sci 116: 2471–2481. [DOI] [PubMed] [Google Scholar]
- 71. Rodriguez J, Esteve P, Weinl C, Ruiz JM, Fermin Y, et al. (2005) SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat Neurosci 8: 1301–1309. [DOI] [PubMed] [Google Scholar]
- 72. Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, et al. (2012) RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet 21: 4171–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lin JC, Ho WH, Gurney A, Rosenthal A (2003) The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat Neurosci 6: 1270–1276. [DOI] [PubMed] [Google Scholar]
- 74. Oliva C, Escobedo P, Astorga C, Molina C, Sierralta J (2012) Role of the MAGUK protein family in synapse formation and function. Dev Neurobiol 72: 57–72. [DOI] [PubMed] [Google Scholar]
- 75. Walsh T, Pierce SB, Lenz DR, Brownstein Z, Dagan-Rosenfeld O, et al. (2010) Genomic duplication and overexpression of TJP2/ZO-2 leads to altered expression of apoptosis genes in progressive nonsyndromic hearing loss DFNA51. Am J Hum Genet 87: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Eriksson N, Macpherson JM, Tung JY, Hon LS, Naughton B, et al. (2010) Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet 6: e1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tung JY, Do CB, Hinds DA, Kiefer AK, Macpherson JM, et al. (2011) Efficient Replication of over 180 Genetic Associations with Self-Reported Medical Data. PLoS ONE 6: e23473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Eriksson N, Benton GM, Do CB, Kiefer AK, Mountain JL, et al. (2012) Genetic variants associated with breast size also influence breast cancer risk. BMC Med Genet 13: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Eriksson N, Tung JY, Kiefer AK, Hinds DA, Francke U, et al. (2012) Novel associations for hypothyroidism include known autoimmune risk loci. PLoS ONE 7: e34442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Henn B, Hon L, Macpherson JM, Eriksson N, Saxonov S, et al. (2012) Cryptic distant relatives are common in both isolated and cosmopolitan genetic samples. PLoS ONE 7: e34267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Therneau T (2012) A Package for Survival Analysis in S. R package version 2.36-14.
- 82. Schemper M, Stare J (1996) Explained variation in survival analysis. Stat Med 15: 1999–2012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Region plots for genome-wide significant associations Colors depict the squared correlation (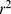 ) of each SNP with the most associated SNP (shown in purple). Gray indicates SNPs for which
) of each SNP with the most associated SNP (shown in purple). Gray indicates SNPs for which 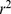 information was missing.
information was missing.
(PDF)
Quantile-quantile plot for myopia survival analysis Actual (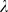 -corrected)
-corrected) 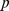 -values versus the null.
-values versus the null.
(PDF)
Smoothed log-hazard ratios as a function of age for four SNPs In each plot, the straight line shows the estimated log-hazard ratio (beta) for each SNP in the proportional hazards model. The solid curve is a spline fit to beta estimated at different ages; the dotted curves are approximate 95% confidence intervals. The 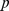 -value in each caption is the result of a test of the proportional hazards assumption. The sign of all coefficients has been made positive for ease of comparison (so (a), (c), and (d) are flipped relative to the main text). Note that among the examples here, only (a) shows evidence of deviation from the proportional hazards assumption after correction for 22 tests.
-value in each caption is the result of a test of the proportional hazards assumption. The sign of all coefficients has been made positive for ease of comparison (so (a), (c), and (d) are flipped relative to the main text). Note that among the examples here, only (a) shows evidence of deviation from the proportional hazards assumption after correction for 22 tests.
(PDF)
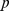 -values for survival and case-control analyses.
-values for survival and case-control analyses. 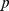 -values for SNPs in the survival analysis used in the paper as well as in a case-control logistic regression on the same set of individuals. The survival analysis gives a smaller
-values for SNPs in the survival analysis used in the paper as well as in a case-control logistic regression on the same set of individuals. The survival analysis gives a smaller 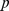 -value for 30 of 35 SNPs and has 22 genome-wide significant (
-value for 30 of 35 SNPs and has 22 genome-wide significant (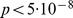 ) as compared to 20 for the case-control.
) as compared to 20 for the case-control. 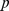 -values in both cases are adjusted for the genomic control inflation factor of 1.16.
-values in both cases are adjusted for the genomic control inflation factor of 1.16.
(PDF)
Tests of deviation from the proportional hazards assumption. 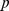 -values for significant SNPs for deviation from the proportional hazards assumption in the Cox model. For each SNP, we fit a Cox proportional hazards model including the SNP, sex, and five principal components as predictors, and then tested for independence of the scaled Schoenfeld residuals with time. Only one SNP deviates significantly from this assumption after correction for 22 tests. Plots for four example SNPs are shown in Figure S3.
-values for significant SNPs for deviation from the proportional hazards assumption in the Cox model. For each SNP, we fit a Cox proportional hazards model including the SNP, sex, and five principal components as predictors, and then tested for independence of the scaled Schoenfeld residuals with time. Only one SNP deviates significantly from this assumption after correction for 22 tests. Plots for four example SNPs are shown in Figure S3.
(PDF)
Statistics for all SNPs with 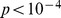 . All 6,141 SNPs with (
. All 6,141 SNPs with (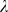 -corrected)
-corrected) 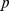 -values under
-values under 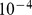 in the discovery cohort. Positions and alleles are given relative to the positive strand of build 37 of the human genome; alleles are listed as major/minor. The gene column shows the position of the SNP in context of the nearest genes. The SNP position is within the brackets, and the number of dashes gives approximate
in the discovery cohort. Positions and alleles are given relative to the positive strand of build 37 of the human genome; alleles are listed as major/minor. The gene column shows the position of the SNP in context of the nearest genes. The SNP position is within the brackets, and the number of dashes gives approximate 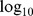 distances. The MAF is the minor allele frequency in Europeans,
distances. The MAF is the minor allele frequency in Europeans, 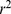 is the estimated imputation accuracy, HR is the hazard ratio per copy of the minor allele, and
is the estimated imputation accuracy, HR is the hazard ratio per copy of the minor allele, and 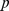 -value is the
-value is the 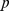 -value in the discovery cohort.
-value in the discovery cohort.
(XLS)


