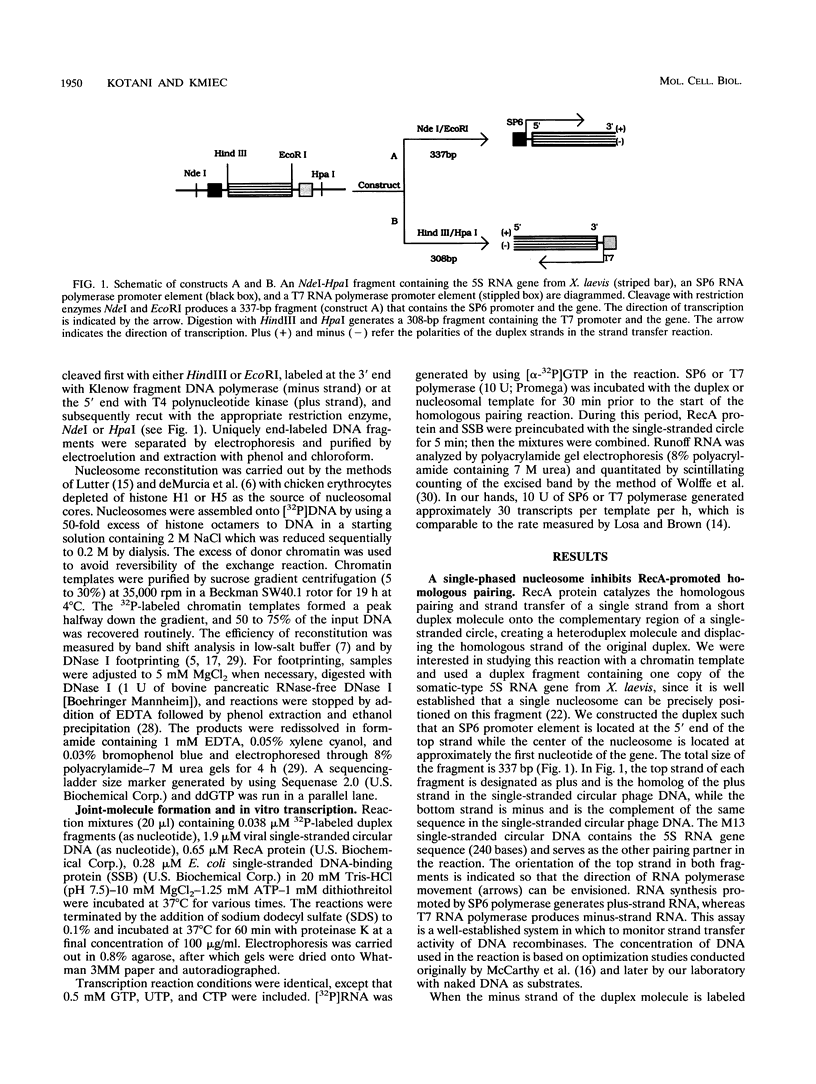
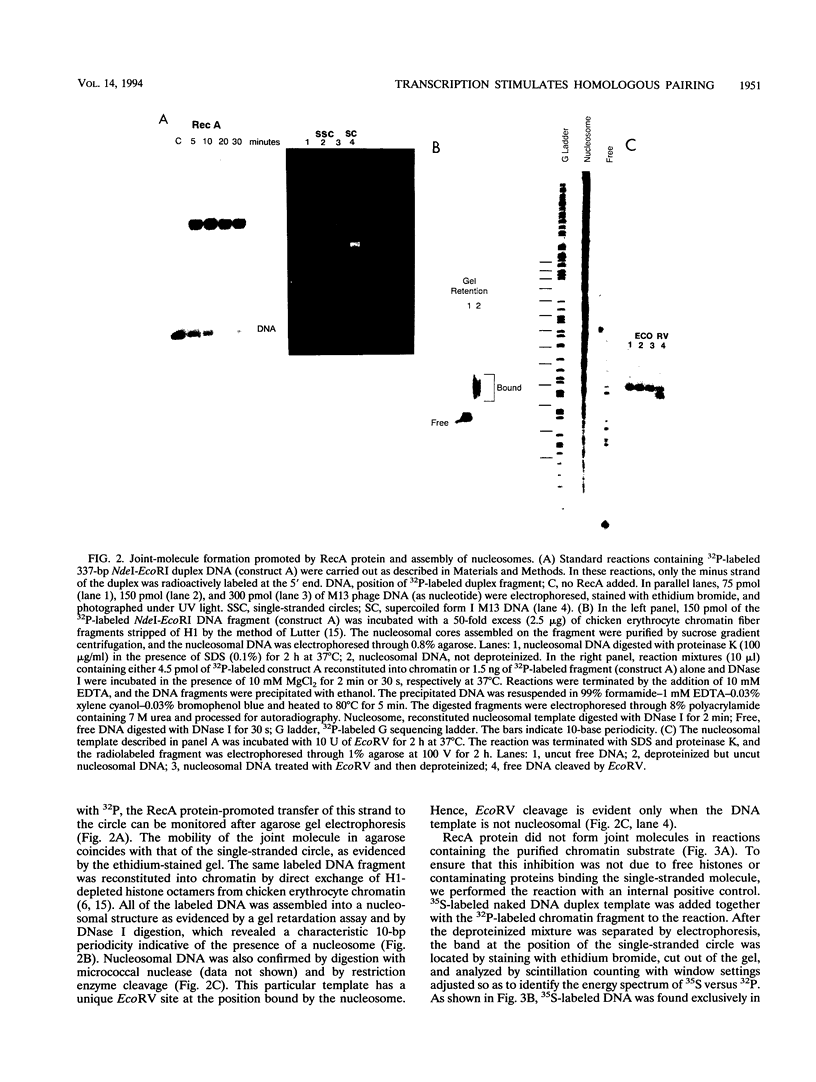
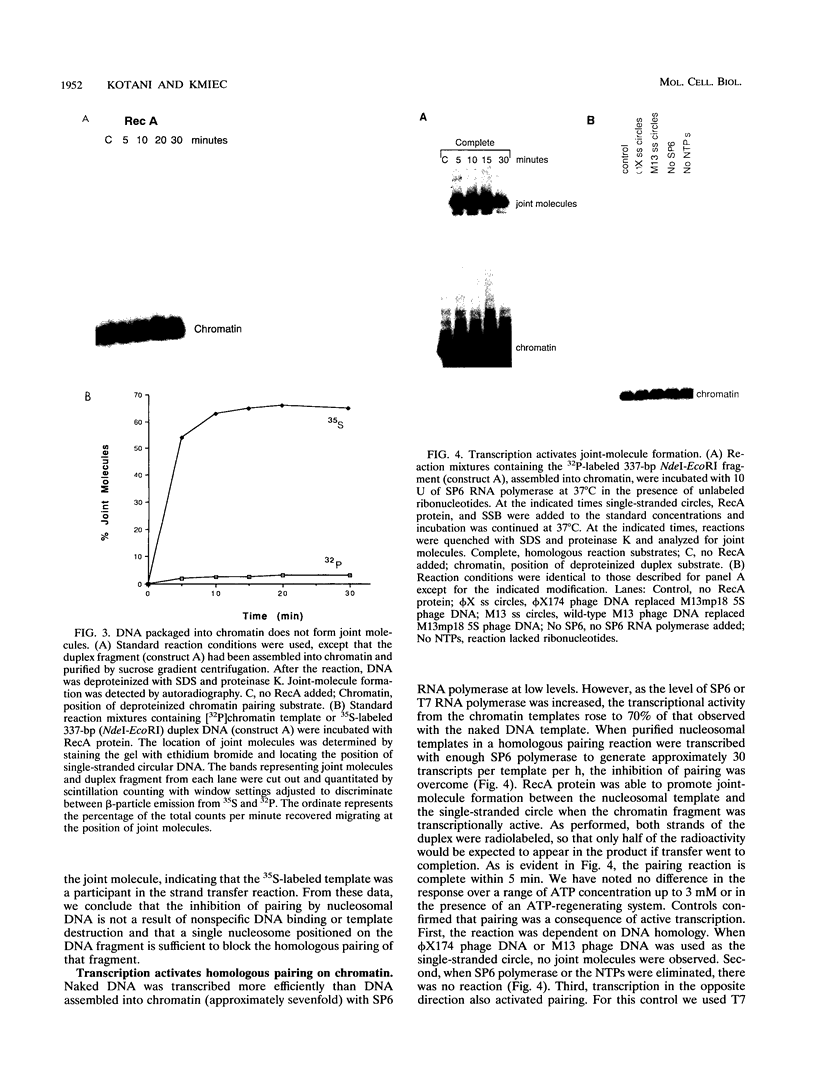
Abstract
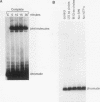
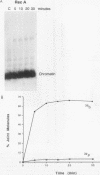
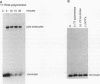
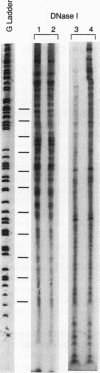
RecA protein catalyzes the homologous pairing of a single-stranded circular DNA and a linear duplex DNA molecule. When the duplex is packaged into chromatin, formation of homologously paired complexes is blocked. We have established a system for studying the RecA-promoted reaction by using a duplex fragment containing a single-phased nucleosome. Under these conditions there is no reaction leading to formation of joint molecule complexes. However, transcription on the chromatin template activates the formation of complexes. Reaction is dependent on RNA synthesis and DNA sequence homology and proceeds regardless of the direction of transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Blackwell T. K., Yancopoulos G. D. Development of the primary antibody repertoire. Science. 1987 Nov 20;238(4830):1079–1087. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- Bonne-Andrea C., Wong M. L., Alberts B. M. In vitro replication through nucleosomes without histone displacement. Nature. 1990 Feb 22;343(6260):719–726. doi: 10.1038/343719a0. [DOI] [PubMed] [Google Scholar]
- Brown D. D. The role of stable complexes that repress and activate eucaryotic genes. Cell. 1984 Jun;37(2):359–365. doi: 10.1016/0092-8674(84)90366-0. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Felsenfeld G. A nucleosome core is transferred out of the path of a transcribing polymerase. Cell. 1992 Oct 2;71(1):11–22. doi: 10.1016/0092-8674(92)90262-b. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Wolffe A. P. Superhelical stress and nucleosome-mediated repression of 5S RNA gene transcription in vitro. EMBO J. 1991 Nov;10(11):3419–3428. doi: 10.1002/j.1460-2075.1991.tb04906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990 Dec;6(12):395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- Hsieh C. L., McCloskey R. P., Lieber M. R. V(D)J recombination on minichromosomes is not affected by transcription. J Biol Chem. 1992 Aug 5;267(22):15613–15619. [PubMed] [Google Scholar]
- Kirov N., Tsaneva I., Einbinder E., Tsanev R. In vitro transcription through nucleosomes by T7 RNA polymerase. EMBO J. 1992 May;11(5):1941–1947. doi: 10.1002/j.1460-2075.1992.tb05247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Losa R., Brown D. D. A bacteriophage RNA polymerase transcribes in vitro through a nucleosome core without displacing it. Cell. 1987 Aug 28;50(5):801–808. doi: 10.1016/0092-8674(87)90338-2. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- McCarthy J. G., Sander M., Lowenhaupt K., Rich A. Sensitive homologous recombination strand-transfer assay: partial purification of a Drosophila melanogaster enzyme and detection of sequence effects on the strand-transfer activity of RecA protein. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5854–5858. doi: 10.1073/pnas.85.16.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Roth S. Y., Simpson R. T. A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol Cell Biol. 1992 Sep;12(9):4015–4025. doi: 10.1128/mcb.12.9.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol Cell Biol. 1992 Dec;12(12):5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persiani D. M., Selsing E. DNase I sensitivity of immunoglobulin light chain genes in Abelson murine leukemia virus transformed pre-B cell lines. Nucleic Acids Res. 1989 Jul 11;17(13):5339–5348. doi: 10.1093/nar/17.13.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdas J., Mythili E., Muniyappa K. Nucleosomes on linear duplex DNA allow homologous pairing but prevent strand exchange promoted by RecA protein. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1344–1348. doi: 10.1073/pnas.88.4.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razvi F., Gargiulo G., Worcel A. A simple procedure for parallel sequence analysis of both strands of 5'-labeled DNA. Gene. 1983 Aug;23(2):175–183. doi: 10.1016/0378-1119(83)90049-5. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985 Dec 16;4(13A):3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherdin U., Rhodes K., Breindl M. Transcriptionally active genome regions are preferred targets for retrovirus integration. J Virol. 1990 Feb;64(2):907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M. S., Corcoran L. M., Baltimore D. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J Exp Med. 1991 Mar 1;173(3):711–720. doi: 10.1084/jem.173.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M., Freisem-Rabien U., Jessberger R., Doerfler W. Transcriptional activities of mammalian genomes at sites of recombination with foreign DNA. J Virol. 1987 Feb;61(2):344–353. doi: 10.1128/jvi.61.2.344-353.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil R. L., Roeder G. S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987 Mar 27;48(6):1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Drew H. R. Initiation of transcription on nucleosomal templates. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9817–9821. doi: 10.1073/pnas.86.24.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P., Jordan E., Brown D. D. A bacteriophage RNA polymerase transcribes through a Xenopus 5S RNA gene transcription complex without disrupting it. Cell. 1986 Feb 14;44(3):381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]
- de Murcia G., Mazen A., Erard M., Pouyet J., Champagne M. Isolation and physical characterization of a stable core particle. Nucleic Acids Res. 1980 Feb 25;8(4):767–779. [PMC free article] [PubMed] [Google Scholar]