Tomato fruit development is subject to connections between auxin signaling, chloroplastic activity, and sugar metabolism.
Abstract
Successful completion of fruit developmental programs depends on the interplay between multiple phytohormones. However, besides ethylene, the impact of other hormones on fruit quality traits remains elusive. A previous study has shown that down-regulation of SlARF4, a member of the tomato (Solanum lycopersicum) auxin response factor (ARF) gene family, results in a dark-green fruit phenotype with increased chloroplasts (Jones et al., 2002). This study further examines the role of this auxin transcriptional regulator during tomato fruit development at the level of transcripts, enzyme activities, and metabolites. It is noteworthy that the dark-green phenotype of antisense SlARF4-suppressed lines is restricted to fruit, suggesting that SlARF4 controls chlorophyll accumulation specifically in this organ. The SlARF4 underexpressing lines accumulate more starch at early stages of fruit development and display enhanced chlorophyll content and photochemical efficiency, which is consistent with the idea that fruit photosynthetic activity accounts for the elevated starch levels. SlARF4 expression is high in pericarp tissues of immature fruit and then undergoes a dramatic decline at the onset of ripening concomitant with the increase in sugar content. The higher starch content in developing fruits of SlARF4 down-regulated lines correlates with the up-regulation of genes and enzyme activities involved in starch biosynthesis, suggesting their negative regulation by SlARF4. Altogether, the data uncover the involvement of ARFs in the control of sugar content, an essential feature of fruit quality, and provide insight into the link between auxin signaling, chloroplastic activity, and sugar metabolism in developing fruit.
The fruit developmental process is controlled by an intricate interplay between multiple phytohormones that influences the overall fruit quality. However, with the exception of ethylene, which has been shown to control many ripening-associated metabolic pathways such as those leading to pigment and aroma volatile production, the impact of other hormones on fruit quality traits remains poorly known (Pech et al., 2012). Auxin is, however, an important phytohormone for initiation of fleshy fruit development, since it was shown to play a key role in triggering fruit set upon flower fertilization (Pandolfini et al., 2002; Wang et al., 2005; de Jong et al., 2009a, 2009b). Auxin is also essential in determining final fruit size through the control of cell division and cell expansion (Devoghalaere et al., 2012). In support of the potential role of auxin in fruit development is the finding that the highest auxin concentrations in different parts of the plant were found in developing fruit (Müller et al., 2002). Auxin was shown to repress amyloplast development in tobacco (Nicotiana tabacum) cells, and the accumulation of the major enzymes for starch biosynthesis is affected by auxin, including ADP-Glc pyrophosphorylase (AGPase) small subunit genes, granule-bound starch synthase (STS), and starch-branching enzyme (SBE) transcripts (Miyazawa et al., 1999).
It is well established that auxin modulates plant development through transcriptional regulation of target genes (Ulmasov et al., 1999) and that the regulation of auxin-responsive genes is mediated by two gene families, Auxin Response Factor (ARF) and Auxin/Indole-3-Acetic Acid (Aux/IAA; Ulmasov et al., 1999; Guilfoyle and Hagen, 2007, 2012; Audran-Delalande et al., 2012). ARFs can either activate or repress transcription of auxin-responsive genes. Auxin is known to regulate various aspects of plant development, including apical dominance, tropisms, and vascular patterning, and plays a crucial role in cell division and cell expansion during the developmental stages spanning and subsequent to the fruit set (Abel and Theologis, 1996; Inzé and De Veylder, 2006). Even though the direct role of ARFs during fruit ripening remains to be clearly established, experimental evidence supporting such a hypothesis was provided by the down-regulation of DEVELOPMENTALLY REGULATED12 (DR12), a tomato (Solanum lycopersicum) ARF gene now named SlARF4, which results in enhanced fruit firmness and increased chlorophyll content associated with a larger number of chloroplasts, leading to dark-green fruits at preripening stages (Jones et al., 2002). Taken together, these findings suggest the ability of auxin to regulate sugar accumulation during fruit development via SlARF4. A number of studies have demonstrated the role of specific ARFs in early stages of fruit development such as fruit set (Wang et al., 2005; Goetz et al., 2006; de Jong et al., 2009b), but the putative role of these transcriptional regulators in controlling some ripening-related events and the overall quality of the fruit remains largely unknown.
Tomato organoleptic quality is strongly influenced by the increase in total sugar and acidity in mature fruit (Bucheli et al., 1999), while the sugar/organic acid ratio is considered an important indicator of the flavor and nutritional quality of fruits (Davies and Hobson, 1981; Bassi and Selli, 1990; Salles et al., 2003). It is well accepted that fruit growth comprises three main stages (Ho and Hewitt, 1986), with the first stage being characterized by an intense mitotic activity leading to an increase in cell number. During this stage, starch, which represents the major carbon reserve in the fruit, reaches a maximal accumulation (Ho, 1996). The second stage corresponds to cell enlargement associated with the degradation of starch into soluble sugars (Davies and Cocking, 1965; Schaffer and Petreikov, 1997). The last stage corresponds to a slow growth phase comprising the fruit-ripening phase, characterized by intensive metabolic changes that lead to Glc and Fru accumulation (Carrari et al., 2006). All three growth stages are essential for final sugar accumulation in the fruit, and early studies have shown that the level of soluble solids in ripe tomato fruit is related to the starch level in immature and mature green fruit (Davies and Cocking). At the physiological and molecular levels, sugar accumulation in tomato fruit is the consequence of various linked physiological processes that are genetically programmed under multihormonal control (Bouzayen et al., 2010).
To further address the link between auxin signaling and sugar metabolism, this study carries out metabolic and transcriptomic analyses of antisense and cosuppressed transgenic lines for the SlARF4 gene, showing that SlARF4 controls chlorophyll accumulation specifically in the fruit. The data support the hypothesis that fruit photosynthetic activity accounts for the photoassimilate production and therefore for the elevated starch levels in the transgenic fruit.
RESULTS
SlARF4 Genomic Structure and Expression Pattern
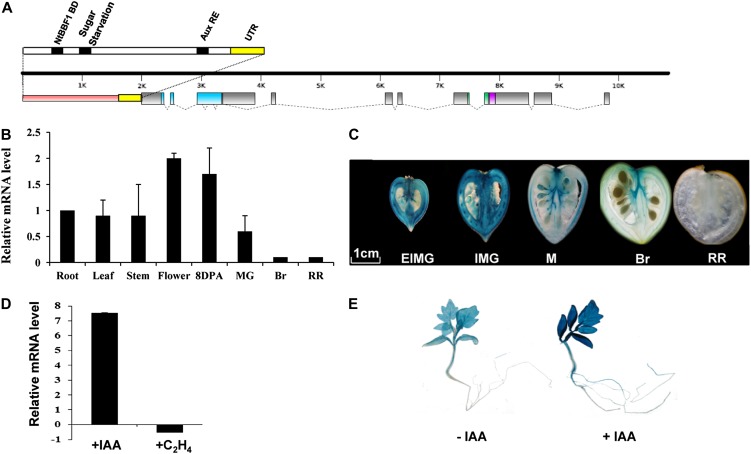
SlARF4, formerly named DR12, is the first ARF gene isolated and characterized in the tomato (Jones et al., 2002). The SlARF4 coding sequence is 2,436 bp long, and the genomic clone is composed of 12 exons and 11 introns (Fig. 1A). The derived protein contains 811 amino acids, sharing the three highly conserved domains (DNA-binding domain and protein/protein domains III and IV) that are typical of the ARF family (Guilfoyle and Hagen, 2007). In silico analysis of the 1.8-kb promoter sequence performed using the PLACE signal scan search tool (http://www.dna.affrc.go.jp/PLACE/signalscan.html) identified several cis-elements, including the canonical auxin response element (AuxRE), TGTCTC, at position –220, a sugar starvation element, TATCCA, at position –960, and an auxin induction element, ACTTTA, at position –977. This latter sequence has been shown to be involved in mediating tissue-specific and auxin-inducible expression of the rolB oncogene (Baumann et al., 1999).
Figure 1.
Structural features and expression patterns of the SlARF4 gene. A, Genomic structure of the SlARF4 gene. The pink portion represents the promoter region, the gray dots represent the introns, the gray boxes represent the exons, the yellow boxes represent the untranslated regions, the blue box represents the DNA-binding domain, the green box represents domain III, and the purple box represents domain IV. The putative cis-acting elements found in the promoter region are indicated by black bars. B, Expression pattern of SlARF4 monitored by qPCR. Expression in the root was taken as reference. C, Expression pattern of SlARF4 revealed by the expression of the GUS reporter gene driven by the SlARF4 promoter during fruit development and maturation. D, Auxin and ethylene regulation of SlARF4 expression. qPCR analysis of SlARF4 transcript levels in RNA samples extracted from 3-week-old light-grown seedlings treated with buffer (control), auxin (20 µm IAA for 2 h), or ethylene (50 µL L–1 for 5 h). E, Auxin-responsiveness of SlARF4 promoter revealed by the expression of the GUS reporter gene driven by the SlARF4 promoter in seedlings treated with auxin (20 µm IAA for 2 h). UTR, Untranslated region; DBD, DNA-binding domain; 8DPA, fruit at 8 DPA; MG, fruit at mature green stage; Br, fruit at breaker; RR, red ripe fruit.
The expression pattern of the SlARF4 gene in tomato ‘Micro-Tom’ was analyzed by real-time PCR to assess its transcript accumulation in roots, leaves, stems, flowers, and fruit 8 DPA and at mature green, breaker, and red ripe stages. The data reveal ubiquitous expression in all tissues tested, with the highest levels of SlARF4 transcript accumulation found in flowers and young fruit 8 DPA. During fruit development, the transcript levels decrease dramatically, showing the lowest levels at the ripening stages (Fig. 1B). The expression pattern of SlARF4 was also assessed in planta using a promoter-GUS fusion construct (proARF4::GUS) stably introduced into tomato lines. GUS staining performed on proARF4::GUS homozygous lines revealed a strong expression in the pericarp and vascular tissues of young fruit 15 and 25 DPA. Thereafter, the SlARF4 expression dramatically decreases throughout ripening, with the GUS staining being no longer detectable at 55 DPA (Fig. 1C). The presence of the canonical AuxRE TGTCTC in the promoter region of SlARF4 (Fig. 1A) prompted the investigation of the auxin responsiveness of the SlARF4 promoter. Quantitative PCR (qPCR) analyses indicated that exogenous auxin treatment induces SlARF4 transcript accumulation up to nearly 8-fold in light-grown seedlings when compared with untreated plants (Fig. 1D). The auxin responsiveness is then confirmed using transgenic lines expressing the GUS reporter gene driven by the SlARF4 promoter (ProARF4::GUS), where 2-h auxin treatment resulted in a strong induction of SlARF4 promoter activity in tomato seedlings (Fig. 1E). In contrast, qPCR analysis revealed no significant change in SlARF4 expression upon ethylene treatment of light-grown seedlings (Fig. 1D).
SlARF4 Acts as a Repressor of Auxin Response
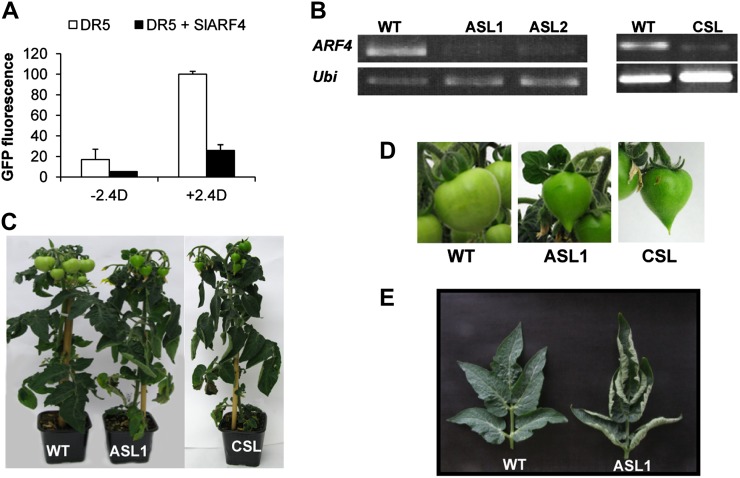
To better characterize the function of the SlARF4-encoded protein, the ability of this protein to regulate the activity of auxin-responsive promoters in a single-cell system was evaluated. A reporter construct, consisting of the synthetic auxin-responsive promoter DR5 fused to a GFP coding sequence (Ottenschläger et al., 2003), was cotransfected into tobacco protoplasts with an effector construct, allowing a constitutive expression of the SlARF4 protein. Transient expression experiments using this dedicated single-cell system revealed that the DR5-driven GFP expression was enhanced by auxin (2,4-D) treatment in the absence of the effector construct providing the SlARF4 protein. However, the presence of the SlARF4 protein strongly inhibited the auxin-induced activity of the DR5 promoter, clearly demonstrating the ability of SlARF4 to act in vivo as a transcriptional repressor of auxin-dependent gene transcription (Fig. 2A).
Figure 2.
Altered phenotypes of SlARF4 down-regulated plants. A, SlARF4 protein represses in vivo the activity of the synthetic promoter DR5. Tobacco protoplasts were transformed either with the reporter construct (DR5::GFP) alone or with both the reporter and effector constructs (35S-SlARF4) and incubated in the presence or absence of 50 μm 2,4-D. GFP fluorescence was measured 16 h after transfection. A mock effector construct lacking SlARF4 was used as a control for the cotransfection experiments. Transformations were performed in triplicate. Mean fluorescence is indicated in arbitrary units ± se. B, Expression of SlARF4 in transgenic lines analyzed by semiquantitative real-time PCR analysis in leaves. In each PCR reaction, the internal reference ubiquitin (Ubi) gene was coamplified with the SlARF4 gene. C, Wild-type and SlARF4 antisense plants at the same stage of development (6-week-old plants). D, Dark-green and heart-shaped phenotype of SlARF4 down-regulated fruit at 35 DPA compared with wild-type fruit at the same stage. E, Upward-curled leaf phenotype of SlARF4 down-regulated fruit. The leaves of SlARF4 down-regulated lines exhibit severe in-rolling along the longitudinal axis of the leaf compared with wild-type plants grown in the same conditions at the same stage. WT, Wild type. [See online article for color version of this figure.]
Down-Regulation of the SlARF4 Gene in Tomato
A previous study has shown that down-regulation of DR12/SlARF4 in tomato results in a dark-green fruit phenotype that is associated with a dramatic increase in chloroplast number (Jones et al., 2002). To gain insight into the physiological significance of the SlARF4-encoded protein, transgenic lines expressing either sense or antisense constructs of the SlARF4 gene were generated using the tomato ‘Micro-Tom’ genotype, and several homozygous lines corresponding to independent transformation events were obtained. Several independent antisense lines (ASLs) and cosuppressed sense lines displayed substantial down-regulation of SlARF4 (Fig. 2B) and reproduced the same phenotypes as those previously described for DR12 ASLs within the genetic background of tomato ‘Kemer’ (Jones et al., 2002). Importantly, the phenotypes of the transgenic lines are very consistent and independent of the genetic background, since they are reproducible between the DR12 ASL lines generated in the Kemer cultivar and those obtained in the Micro-Tom cultivar used in this study. That is, SlARF4 down-regulated lines display severe upward leaf curling along the longitudinal axis of the leaf (Fig. 2E) and dark-green fruits at the preripening stages, with a slightly heart-shaped phenotype (Fig. 2, C and D). Although, overall, more than 10 independent lines showing the above-described phenotypes were generated, three down-regulated lines, ASL1, ASL2, and cosuppressed line1 (CSL1), showing the strongest phenotypes were selected for deep molecular and physiological characterization.
Physiological and Biochemical Characterization of Transgenic Lines
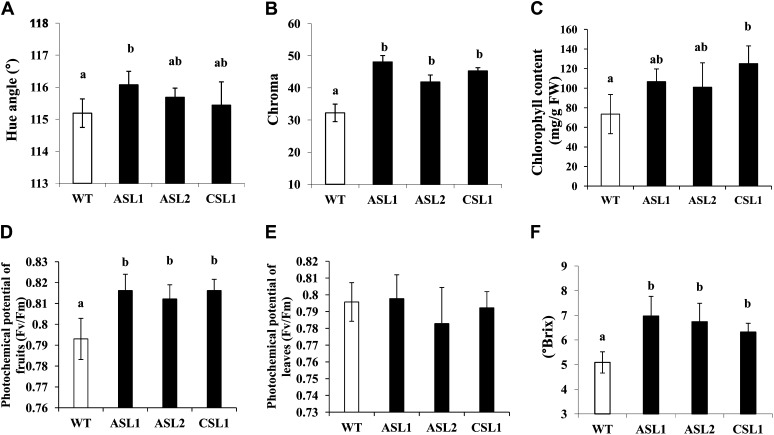
The impact of SlARF4 silencing on fruit and leaf development was investigated at the biochemical and physiological levels in the two selected transgenic lines. The assessment of color parameters in SlARF4 down-regulated fruits at 35 DPA, corresponding to the mature green stage, indicated that the hue angle values, indicative of color saturation, are higher than in the wild type, thus confirming the observed dark-green phenotype (Fig. 3A). Furthermore, measurement of color saturation (chroma, which is indicative of color intensity) provided significantly higher values in SlARF4 down-regulated fruit than in wild-type fruit (Fig. 3B). Chlorophyll quantification in 35-DPA fruits indicated that SlARF4 down-regulated fruits accumulate higher amount than the wild type (Fig. 3C), although no increase in chlorophyll accumulation was found in leaves (data not shown). The dark-green phenotype and the associated elevated chlorophyll content in the fruit tissues may potentially confer higher photosynthetic performance to the transgenic fruit. This hypothesis was assessed by measuring the photochemical potential in wild-type and SlARF4 antisense or cosuppressed leaves and fruits. In fruits, the photochemical potential was more important in SlARF4 down-regulated lines than in the wild type (Fig. 3D), whereas no significant differences were observed for leaves (Fig. 3E). Because sugar is the main product of chloroplast activity, it became relevant to assess whether the enhanced chlorophyll content and higher photochemical potential in SlARF4 down-regulated fruits results in higher sugar accumulation. Indeed, Brix determination in fruits at 55 DPA indicates that total soluble solids showed significantly higher values in SlARF4 down-regulated fruit than in the wild type (Fig. 3F).
Figure 3.
Physiological and biochemical analysis of SlARF4 down-regulated lines. Color parameters measured in wild-type and SlARF4 down-regulated fruits. A, Hue angle. B, Chroma. C, Chlorophyll content in wild-type and SlARF4 down-regulated fruit. D and E, Potential photochemical efficiency of fruits (D) and leaves (E) of wild-type and SlARF4 down-regulated plants. Fruits were analyzed at the same stage of development. F, Total soluble solids content measured in fruit of wild-type and SlARF4 down-regulated plants at 55 DPA. Small letters show significant difference using ANOVA at P < 0.05. WT, Wild type, Fv/Fm, maximum photochemical efficiency of PSII in the dark-adapted state, FW, fresh weight.
SlARF4 Down-Regulation Leads to Enhanced Sugar Accumulation in Fruit
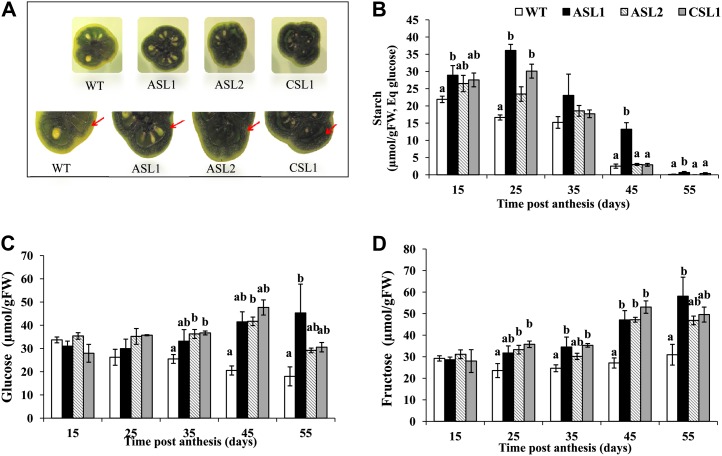
In further characterizing the effects of SlARF4 down-regulation on fruit biology and quality, increased starch levels in green fruit were observed. Performing an iodine-staining experiment to uncover whether starch accumulation localizes to a particular tissue in the fruit revealed that the blue-purple color, indicative of the presence of starch, was mainly found in the pericarp tissue, with more intense staining found in SlARF4 down-regulated fruit than in the wild type (Fig. 4A).
Figure 4.
A, Starch content evaluated by Lugol staining in wild-type and SlARF4 down-regulated 35-d-old fruits. Red arrows show the starch accumulation in the pericarp of the fruit revealed by blue-purple color indicative of starch reaction with iodine. B to D, Starch and soluble sugar contents in wild-type and SlARF4 down-regulated fruit during development and maturation (15, 25, 35, 45, and 55 DPA). For each developmental stage, the samples consist of a mixture of six different fruits, and the data represent the mean ± se of three independent biological repeats. Small letters show significant difference using ANOVA at P < 0.05. FW, Fresh weight. [See online article for color version of this figure.]
The changes in sugar metabolism occurring in the SlARF4 down-regulated lines were assessed by following sugar and starch content at different stages of fruit development and ripening (15, 25, 35, 45, and 55 DPA). Starch content declined steadily throughout fruit development in both wild-type and SlARF4 down-regulated lines when expressed on fresh weight (Fig. 4B). Starch accumulated over the early stages of fruit development and then underwent rapid degradation starting at the preripening stages. However, comparatively, in SlARF4 down-regulated fruit, starch content stayed above the levels found in the wild type, particularly at early stages (15 and 25 DPA) of fruit development (Fig. 4B). Because starch degradation is known to be the main source of soluble sugars, we assessed the impact of underexpressing SlARF4 on Glc and Fru contents. The levels of Glc and Fru became significantly higher in the SlARF4 down-regulated fruit than in the wild type (Fig. 4, C and D) as fruit development advanced toward ripening (stages 35, 45, and 55 DPA). This difference, which represents 30 and 50 µmol g–1 fresh weight in CSL1 and ASL1, respectively, can be explained at least in part by the higher levels of transient starch accumulation in the transgenic lines.
Expression Profiling of Starch Biosynthesis Genes in the Tomato
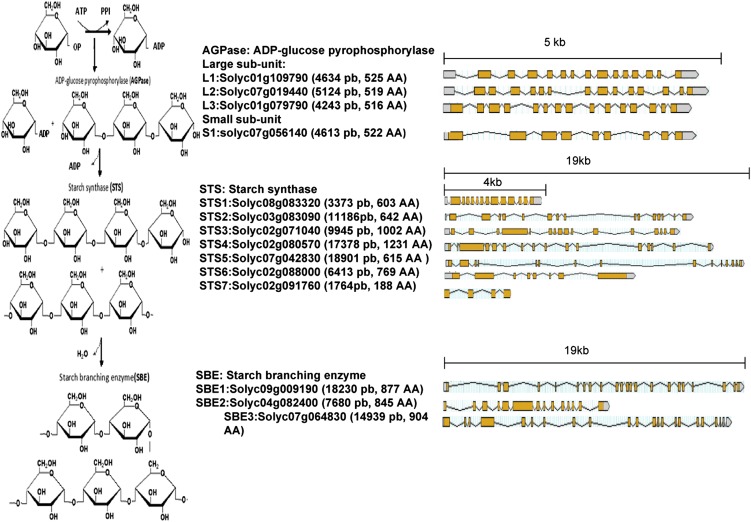
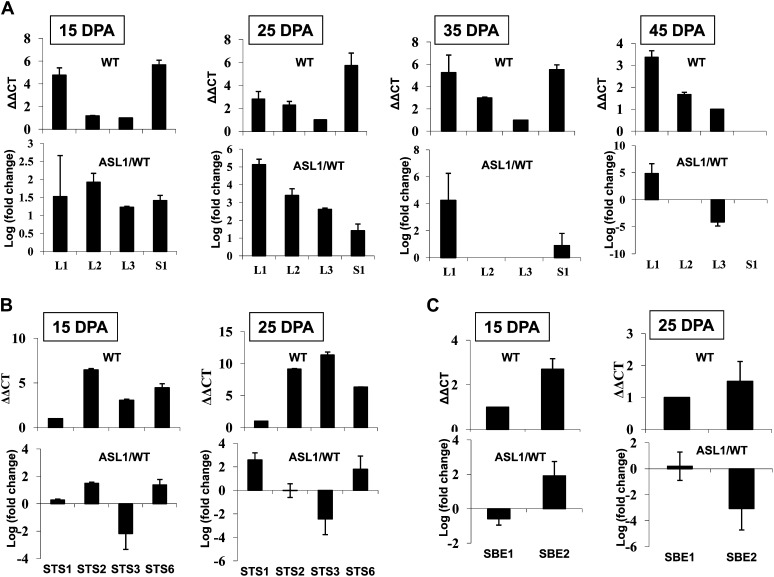
To gain more insight into the mechanism by which sugar metabolism is impacted in SlARF4 down-regulated lines, we investigated the expression pattern of starch biosynthesis genes. Starch biosynthesis is known to involve a series of enzyme-catalyzed processes (Smith, 1999; Liang et al., 2001; James et al., 2003) belonging to three separate enzyme families (Fig. 5), AGPase, STS, and SBE (Yelle et al., 1988; Schaffer and Petreikov, 1997). Building on the annotated tomato genome sequence, genome-wide in silico screening allowed for the identification of all members of the three enzyme families involved in starch synthesis in tomato. The tomato genome contains three genes encoding the large AGPase subunit, SlAGPaseL1 (L1), SlAGPaseL2 (L2), and SlAGPaseL3 (L3), and one gene encoding the small subunit, SlAGPaseS (S1). STS enzymes are encoded by seven genes, SlSTS1 to SlSTS7, and SBE enzymes are encoded by a small gene family made up of three members, SlSBE1, SlSBE2, and SlSBE3 (Fig. 5). However, the lack of a reference expression pattern for starch synthesis genes in the tomato prompted us to establish their expression profile in wild-type tomato fruit. Transcript accumulation was assessed for all members of AGPase (Fig. 6A), STS (Fig. 6B), and SBE (Fig. 6C) gene families by qPCR throughout fruit development (15, 25, 35, and 45 DPA). With respect to the AGPase family, L1 and S1 show the highest level of expression concomitant with the starch accumulation phase (15–35 DPA). However, the expression of S1 dramatically decreases as fruit ripening proceeds (45 DPA), suggesting that the regulation of the AGPase activity may take place primarily at the level of the small subunit. Among the seven SlSTS genes, transcripts were detected only for SlSTS1, SlSTS2, SlSTS3, and SlSTS6 (Fig. 6B), suggesting that SlSTS4, SlSTS5, and SlSTS7 genes may contribute to starch synthesis in nonfruit tissues. SlSBE1 and SlSBE2 display fruit-associated expression at early stages of fruit development (15 and 25 DPA), with no contribution of the SlSBE3 gene at any of the fruit developmental stages tested (Fig. 6C). Moreover, the expression of SlSTS and SlSBE genes was undetectable at late stages of fruit development (35–55 DPA).
Figure 5.
Gene structure of the different enzymes involved in the biosynthesis of starch in tomato fruit. Left, Steps of starch biosynthesis. Center, Different members of the three enzyme families involved in AGPase, STS, and SBE. Genomic and protein lengths are also indicated. Right, Representation of the genomic structure of each gene showing the introns and exons. [See online article for color version of this figure.]
Figure 6.
Expression profile of SlAGPase genes in wild-type and ASL1 tomato fruits. The levels of transcripts were assessed in tomato fruit by qPCR at 15, 25, 35, and 45 DPA for (A) SlAGPaseL1 (L1), SlAGPaseL2 (L2), SlAGPaseL3 (L3), and SlAGPaseS (S1); at 15 and 25 DPA for (B) SlSTS1 (STS1), SlSTS2 (STS2), SlSTS3 (STS3) and SlSTS6 (STS6); and at 15 and 25 DPA for (C) SlSBE1 (SBE1) and SlSBE2 (SBE2). ΔΔCT refers to the fold difference in the expression of SlAGPase, SlSTS, and SlSBE relative to the isoforms L3, STS1, and SBE1, respectively. Levels of STS4, STS5, STS7, and SBE3 were not detectable. Log (fold change) refers to the expression of SlAGPase, SlSTS, and SlSBE isoforms in ASL1 relative to the expression of the same isoform in the wild type. The data represent mean values obtained with three replicates. WT, Wild type.
SlARF4 Down-Regulation Alters the Expression of Starch Biosynthetic Genes and the Corresponding Enzyme Activities
Comparative expression analysis of starch biosynthesis genes assessed in the wild type and ASL1 indicated a significant up-regulation of all AGPase genes (Fig. 6A) at the preripening stages lasting 25 DPA and thereafter; during the ripening phase, transcript accumulation of L1 remained higher in ASL1 than in the wild type. In contrast, the expression of L2 and S1 was similar in wild-type and transgenic lines at late stages of fruit development (35 and 45 DPA). It was noteworthy that the expression of L3 undergoes strong down-regulation at the ripening stage in ASLs (Fig. 6A).
Assessing the transcript levels of SlSTS genes (Fig. 6B) revealed that the expression of STS2 and STS6 was induced at 15 DPA, while that of STS3 was clearly repressed (Fig. 6B). At 25 DPA, the expression of STS1 and STS6 was induced in the SlARF4 down-regulated lines, while that of STS3 remained repressed. At 15 DPA, the expression of SlSBE1 was repressed and that of SlSBE2 induced; while at 25 DPA, SlSBE2 was strongly repressed (Fig. 6C).
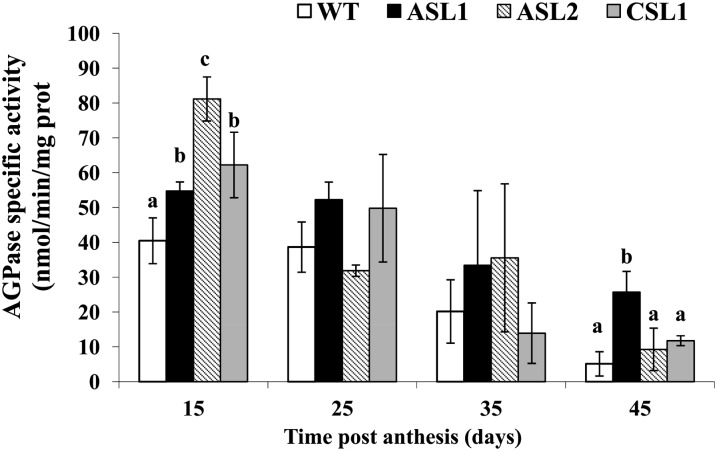
Overall, the data indicate that down-regulation of SlARF4 leads to an increase in transcript levels for SlAGPase genes at the preripening stages of fruit development concomitant with the observed starch accumulation at the same stages. To further unravel the impact of SlARF4 down-regulation on the expression of SlAGPase genes, the corresponding enzyme activity was assessed at different stages of fruit development and ripening (Fig. 7). In line with the increase in transcript accumulation, SlAGPase activity was greater in the SlARF4 down-regulated fruits than in wild-type fruit, especially at the preripening stages, providing a good correlation between transcript levels and enzyme activity. The SlAGPase activity dramatically decreased at the onset of fruit ripening in both wild-type and transgenic lines, though it remained significantly higher in SlARF4 down-regulated fruits (Fig. 7).
Figure 7.
Specific activity of SlAGPase. The AGPase specific activity was quantified in wild-type (WT) and SlARF4 down-regulated fruits (ASL1, ASL2, and CSL1). The data represent the mean ± se of six replicates. Small letters show significant difference using ANOVA at P = 0.05.
Expression Analysis of SlGLK Genes
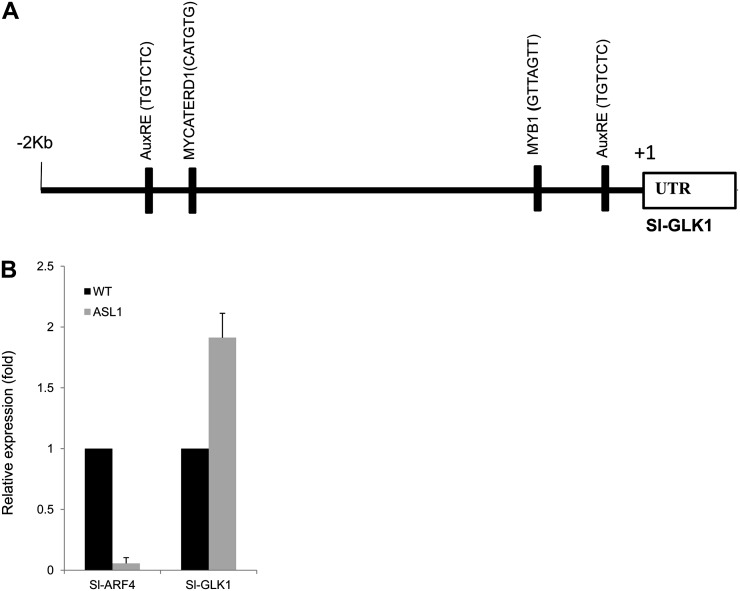
Considering that the chlorophyll and starch phenotypes of SlARF4 ASLs are reminiscent of those described in tomato GOLDEN2-LIKE (SlGLK) overexpressing lines (Powell et al., 2012), we addressed the putative link between the expression of this MYB-type transcription factor and the phenotypes displayed by the SlARF4 transgenic lines. Two GLK genes (SlGLK1 and SlGLK2) are present in the tomato genome, and it was reported that in most domesticated genotypes the SlGLK2 gene bears the uniform ripening (u) mutation that results in light-green fruit phenotype. Transcript accumulation analysis revealed that SlGLK2 expression is up-regulated in transgenic lines underexpressing SlARF4 (data not shown). However, verifying the sequence of the SlGLK2 gene in the Micro-Tom cultivar revealed that this genotype carries the inactive u allele of SlGLK2, which rules out the possibility that the dark-green phenotype of SlARF4 antisense fruit may result from the up-regulation of SlGLK2. We then checked whether ARF4 might regulate the expression of SlGLK1, whose expression has been reported to be low in the fruit tissues. The data presented in Figure 8B show an enhanced accumulation of SlGLK1 transcripts in SlARF4-ASL fruit tissues, suggesting that the down-regulation of ARF4 expression results in the up-regulation of SlGLK1, which in turn may increase chlorophyll accumulation. In support of this hypothesis, we found that the promoter region of the SlGLK1 gene contains two perfectly conserved canonical ARF binding sites, the so-called TGTCTC box (Fig. 8A).
Figure 8.
SlARF4 is a possible repressor of the SlGLK1 gene. A, SlGLK1 promoter sequence analysis. The promoter region of SlGLK1 was analyzed for putative cis-acting elements. The identified sites are represented by black bars: MYB-binding site (GTTAGTT), AuxRE (TGTCTC), and MYC box (CATGTG). B, Expression pattern of SlGLK1 and SlARF4 monitored by qPCR in down-regulated fruit compared with the wild type at 25 DPA. The relative mRNA level for each gene was normalized with respect to the actin housekeeping gene. The results were expressed using the wild type as a reference for each gene (relative mRNA level 1). WT, Wild type.
DISCUSSION
Auxin has long been reported to be involved in fruit development, and exogenous application of auxin was shown to disturb normal fruit ripening in many crop species (Vendrell, 1985; Cohen, 1996). Moreover, the link between auxin biosynthesis or signaling and sugar accumulation in the fruit tissues has been highlighted by a number of studies (Pandolfini et al., 2002; Wang et al., 2009), though the mechanisms by which this hormone impacts sugar metabolism and therefore fruit quality remain poorly understood. Previous work demonstrated that DR12/ARF4, a member of the tomato ARF gene family of transcription factors, is involved in the regulation of fruit development; that is, transgenic tomato plants with decreased SlARF4 mRNA levels produced dark-green fruit at immature stages, with increased chlorophyll content, a larger number of chloroplasts, and unusual cell division at late stages of fruit development, as well as blotchy ripening and enhanced fruit firmness (Jones et al., 2002; Guillon et al., 2008). In further characterizing the role of this auxin transcriptional regulator, the current study addresses more specifically the impact of down-regulation of SlARF4 on sugar metabolism throughout fruit development. Both metabolic and transcriptomic data lead to the conclusion that SlARF4 underexpressing lines accumulate more starch at early stages of fruit development and more sugar at the ripening stages. Overall, the data provide insight into the link between auxin signaling, chloroplastic activity in the fruit tissues, and sugar metabolism.
Several tomato mutants such as dark green, high pigment1, and high pigment2 (Sanders et al., 1975; Jarret et al., 1984) displayed fruit phenotypes similar to those showed by SlARF4 down-regulated lines with regard to high chlorophyll content. However, in contrast to these mutants where the dark-green phenotype can be observed in both leaf and fruit tissues, the enhanced chlorophyll content in SlARF4 underexpressing plants is restricted to immature fruits. This feature suggests that SlARF4 controls chlorophyll accumulation specifically in the fruit. Furthermore, the enhanced chlorophyll content in SlARF4 down-regulated fruits correlates with a higher photochemical efficiency compared with wild-type fruits, supporting the idea that fruit photosynthetic activity may account, at least partially, for photoassimilate production and therefore for the elevated starch levels in the transgenic fruit. Consistent with this idea, cells in developing fruit were shown to contain photosynthetically active chloroplasts (Piechulla et al., 1987), suggesting that photosynthesis may provide a significant contribution to both metabolism and growth of the fruit organ. This hypothesis is further supported by global transcriptomic profiling of transgenic lines altered in auxin response owing to down-regulation of SlIAA9, an Aux/IAA gene, which revealed that the activation of photosynthesis-related genes is a major phenomenon in developing tomato fruit (Schauer et al., 2006; Wang et al., 2009; Matas et al., 2011). In tomato, photosynthesis in developing fruit can contribute up to 20% of the fruit photosynthate, and light-harvesting electron transfer and CO2 fixation proteins are conserved in the active state in fruit tissue (Blanke and Lenz, 1989; Hetherington et al., 1998; Carrara et al., 2001; Matas et al., 2011). Yet, the prevailing idea is that fruit growth and metabolism are predominantly supported by photoassimilate supply from the source (Ruan et al., 2012), and in this regard, our data cannot rule out that the higher sugar content observed in the transgenic lines could also arise from a more efficient import of photoassimilate into fruit. Indeed, altering auxin sensitivity via down-regulation of tomato IAA9 has been reported to promote the development of vascular bundles (Wang et al., 2005), which may enhance sink strength and sugar supply to the fruit.
Starch is the end product of photosynthesis and the predominant carbohydrate reserve in many plants, and in addition to being important for plant development, starch biosynthesis is also a critical factor for fruit quality. The regulation of starch synthesis has received much attention in tomato fruit (Beckles et al., 2001a, 2001b), and it has been reported that the reaction catalyzed by AGPase is the limiting step for starch biosynthesis in potato (Solanum tuberosum) tubers (Tiessen et al., 2002), a Solanaceae species close to tomato. Indeed, modifying AGPase activity and properties has a direct impact on starch levels in plants (Tsai and Nelson, 1966; Smidansky et al., 2002; Berger et al., 2004; Hädrich et al., 2012). Of particular note, the enhanced starch content in SlARF4 down-regulated fruit correlates well with the up-regulated expression of key genes involved in starch biosynthesis, especially genes coding for AGPase. The data revealed a net increase, compared with wild-type fruit, in both transcript accumulation and enzyme activity for SlAGPase at the preripening stages in the SlARF4 down-regulated fruit. The expression of SlAGPase is highly correlated with the accumulation of starch in both wild-type fruits and SlARF4 down-regulated fruits. The presence of three conserved motifs in the promoter region of SlAGPase (Supplemental Table S1) corresponding to putative AuxREs is supportive of a direct regulation of AGPase gene expression by SlARF4. Together, the data strongly suggest that SlARF4 controls starch accumulation in fruit mainly by repressing the expression of the SlAGPase gene. In the same way, previous studies showed a negative effect of auxin on the expression of the SlAGPase gene (Miyazawa et al., 1999). SlARF4 down-regulated fruit displayed higher soluble solids (Brix) at the ripening stages, likely owing to the overaccumulation of starch in green fruit that could be degraded into soluble sugars. This is in agreement with previous work stressing the decisiveness of starch content at green fruit stage in the determination of soluble solid content at the ripening stage (Schaffer et al., 2000; Baxter et al., 2005). In addition to auxin, it was recently reported that malate levels impact starch metabolism (Centeno et al., 2011); however, the putative link between auxin regulation of carbohydrate accumulation and malate metabolism is still to be elucidated.
Expression pattern revealed by the ProARF4::GUS fusion reporter construct uncovered a significant expression of SlARF4 in all tissues analyzed, with the highest level of expression observed in flower and pericarp and in vascular tissues of young fruit. SlARF4 expression reaches a maximum at 25 DPA and then decreases at the end of ripening. In addition, the up-regulation of SlARF4 expression by auxin suggests an auxin control of this gene. These findings are in accordance with previous studies showing that auxin concentration increases at the beginning (10 to 25 DPA) of fruit development (Müller et al., 2002). Using a single-cell approach, we showed that similar to its Arabidopsis (Arabidopsis thaliana) ortholog, SlARF4 is also able to strongly repress in vivo the activity of the synthetic DR5 auxin-responsive promoter. Taken together, these data suggest that SlARF4 is involved in the auxin regulation of young fruit development by repressing the expression of auxin-responsive genes.
It was recently reported that the u mutation of the SlGLK2 gene is responsible for the light-green phenotype in cultivated tomato varieties (Powell et al., 2012), and that overexpression of SlGLK2 and its paralog SlGLK1 leads to dark-green fruit similar to those described in down-regulated SlARF4. However, even though the expression of SlGLK2 was found to be significantly enhanced in SlARF4 down-regulated lines, it cannot account for the dark-green phenotype of the transgenic fruit, since the cv Micro-Tom variety bears the inactive u allele of SlGLK2. Interestingly, transcript accumulation of the SlGLK1 gene was also significantly enhanced in SlARF4 transgenic lines, suggesting that down-regulation of SlARF4 leads to derepression of the SlGLK1 gene in the fruit tissue, which may be responsible for the increase in chlorophyll accumulation. This hypothesis is further supported by the presence of two perfectly conserved canonical ARF binding sites, the so-called TGTCTC box, in the promoter region of the SlGLK1 gene. The data could also suggest that in wild-type tomato, SlARF4 may act through the transcriptional repression of SlGLK1 gene expression in fruits. It is noteworthy that many of the phenotypes displayed by SlGLK overexpressing lines are shared by the antisense SlARF4 plants, including the increased number of green fruit chloroplasts (Jones et al., 2002) and enhanced sugar accumulation. The possible ability of the SlARF4 protein to repress the transcriptional activity of the SlGLK promoter supports the idea that these transcription factors may control the photosynthetic activity in the fruit through a common route. Overall, the current study brings insight into the ability of auxin to control starch accumulation during fruit development and therefore to impact fruit quality. The data also shed some light on the molecular actors involved in auxin action and define SlARF4 as a major player in mediating the auxin control of sugar metabolism in tomato fruit.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum ‘Micro-Tom’) plants were grown under standard greenhouse conditions. Conditions in the culture chamber room were set as follows: 14-h-day/10-h-night cycle, 25°C /20°C day/night temperature, 80% relative humidity, and 250 mol m–2 s–1 intense light. Seeds were sterilized, rinsed in sterile water, and sown in Magenta vessels containing 50 mL of one-half-strength Murashige and Skoog medium added with R3 vitamin (0.5 mg L–1 thiamine, 0.25 mg L–1 nicotinic acid, and 0.5 mg L–1 pyridoxine), 1.5% (w/v) Suc, and 0.8% (w/v) agar, pH 5.9.
Plant Transformation
To generate SlARF4 overexpressing plants, the forward 5′-ATGGAAATTGATCTGAATCATGC-3′ and reverse 5′-TCAAATCCTGATTACAGTTGGAGATG-3′ primers were used to amplify the 2,436 bp of full-length SlARF4 coding sequence. Two SlARF4 antisense constructs were made, one corresponding to the 5′ region (5′ untranslated region and DNA-binding domain) of ARF4 and the other to the 3′ region. The forward 5′-ATGGAAATTGATCTGAATCATGC-3′ and reverse 5′-TGGCTGTCCAGTACTGATGGTG-3′ primers were used to amplify the 1,300-bp 5′ sequence. The forward 5′-CATGTCGATTTCGTTGTACCTTAC-3′ and reverse 5′-CCACATAGTTTTCATCATACAAGC-3′ primers were used to amplify the 1.6-kb nucleotide 3′ sequence. These two fragments were then cloned into the pGA643 binary vector in the antisense orientation under the transcriptional control of the Cauliflower mosaic virus 35S (35SCaMV) promoter and the nopaline synthase terminator. All transgenic plants were generated by Agrobacterium tumefaciens-mediated transformation according to Wang et al. (2005). All experiments were carried out using homozygous lines from F3 or later generations.
Isolation and Cloning of the SlARF4 Promoter
PCR was performed on the genomic DNA of tomato ‘Micro-Tom’ (10 ng µL–1). PCR primers are detailed in Supplemental Table S2. The corresponding amplified fragment was cloned into the pMDC162 vector containing the GUS reporter gene using Gateway technology (Invitrogen). The cloned SlARF promoter was sequenced from both sides using vector primers in order to see whether the end of the promoter is matching with the beginning of the reporter gene. Sequence results were carried out using the Vector NTI (Invitrogen) and ContigExpress software by referring to ARF promoter sequences. Transgenic plants were generated by A. tumefaciens-mediated transformation according to Wang et al. (2005).
Transient Expression Using a Single-Cell System
For cotransfection assays, the coding sequence of SlARF4 was cloned into the pGreen vector and expressed under the control of the 35SCaMV promoter. Protoplasts were transformed either with 10 µg of the reporter vector alone containing the DR5 synthetic AuxRE fused to the GFP reporter gene (Ottenschläger et al., 2003) or in combination with 10 µg of the SlARF4 construct as the effector plasmid, allowing for the constitutive expression of the SlARF4 protein. Protoplasts were obtained from suspension-cultured tobacco (Nicotiana tabacum) Bright Yellow-2 cells and transfected according to the method described previously (Leclercq et al., 2005). After 16 h of incubation in the presence or absence of 2.4-D (50 µm), GFP expression was analyzed and quantified by flow cytometry (FACSCalibur II, BD Biosciences) as indicated in Audran-Delalande et al. (2012). All transient expression assays were repeated at least three times with similar results.
GUS Staining and Analysis
Tissues from transgenic lines transformed with the SlARF4 promoter-GUS fusion construct (ProARF4::GUS) were taken and put in GUS staining solution (100 mm sodium phosphate buffer, pH 7.2, 10 mm EDTA). Vacuum was made twice for 15 min. Tissues were then incubated in GUS staining solution at 37°C overnight. Samples were then decolorated using several washes of graded ethanol series.
Auxin and Ethylene Treatment
For qPCR expression studies, 21-d-old tomato seedlings were harvested and treated with auxin (20 μm IAA for 2 h). The tissues were then immediately frozen in liquid nitrogen and stored at –80°C until RNA extraction. For GUS analysis, 21-d-old tomato ProARF4::GUS-transformed seedlings were incubated for 2 h in one-half-strength Murashige and Skoog buffer with or without 20 μm IAA. Tissues were then immediately incubated in GUS staining buffer. Ethylene treatments were performed for 5 h in sealed glass boxes. Five-day-old etiolated seedlings were treated with 50 μL L–1 ethylene, and control seedlings were exposed to air alone. The tissues were immediately frozen in liquid nitrogen and stored at –80°C until RNA extraction.
RNA Extraction and Quantitative Real-Time PCR
Total RNA from tissues was extracted using a plant RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. Total RNA was then DNase-treated with Ambion (Invitrogen) to remove any contaminating genomic DNA. Complementary DNA synthesis was done by reverse transcription of first strand complementary DNA from 2 μg of total RNA using Omniscript (Qiagen) according to the manufacturer’s instructions. Gene-specific primers were designed by Primer3 software (version 0.4.0). Primers sequences are listed in Supplemental Table S2). The relative transcript abundance was monitored on an ABI PRISM 7900HT sequencer using SYBR Green PCR Master Mix (Applied Biosystems). The relative expression for each gene of interest was calculated using the comparative threshold cycle values and the SlActin (forward 5′-TGTCCCTATCTACGAGGGTTATGC-3′, reverse 5′-AGTTAAATCACGACCAGCAAGAT-3′) as an internal standard, as described previously (Pirrello et al., 2006).
Chlorophyll Fluorescence Parameter Measurements
Chlorophyll fluorescence parameters were measured with a PAM-2000 pulse-amplitude modulation fluorometer (Walz). The measurements were made on fruits at 35 DPA. The chlorophyll fluorescence parameter measurements were done according to the method described in detail by Maury et al. (1996).
Color Measurement
L, a, and b values (International Commission on Illumination) were measured on fruit with a Konica Minolta CR-200 Chroma Meter at 35 DPA. The chromameter was calibrated against a standard white tile. The different color indexes were calculated according to the following equations: Hue = tan–1 (b/a), if a > 0 and 180 + tan–1 (b/a), if a < 0; Chroma = (a2 + b2)0.5.
Starch was colored in situ with Lugol solution (Sigma-Aldrich) by dipping tomato halves for 10 s, then removing the excess stain by gently tipping onto a paper tissue. The starch was then revealed by turning the pale-brown color of the iodine solution to a dark-blue color.
Fruit Brix Measurement
Breaker and breaker-plus-10-d fruit tissue was homogenized in a razor blade and centrifuged for 1 min at 12,000 rpm. The soluble solids (Brix) content of the resulting juice was measured on the MASTER-20T portable refractometer (Atago).
Chemicals and Enzymes
ADP-Glc, AMP, ATP, 6-aminocaproic acid, benzamidine, Bradford reagent, Fru-6-P, Glc-1-P, Glc-1,6-bisP, phenazine ethosulfate, Suc, thiazolyl blue tetrazolium bromide, Tricine, Triton X-100, amyloglucosidase, catalase, and NAD glyceraldehyde-3-P dehydrogenase were purchased from Sigma-Aldrich. Dithiothreitol, leupeptin, NAD+, NADH, NADP+, NADPH, and α-amylase were purchased from Roche.
Extraction and Assay of Enzymes
Samples were powdered under liquid nitrogen and stored at –80°C until use. Aliquots of approximately 20 mg fresh weight were extracted as in Gibon et al. (2009). Assays were prepared in 96-well polystyrene microplates (Sarstedt) using a robotized platform (Hamilton). Absorbances were read at 340 nm in MP96 readers (SAFAS). ADP-Glc pyrophosphorylase and NADP-glyceraldehyde-3-P dehydrogenase were assayed as described in Gibon et al. (2004).
Extraction and Assay of Metabolites
Metabolites were extracted twice with 80% (v/v) ethanol and once with 50% (v/v) ethanol as described in Geigenberger et al. (1996). Chlorophylls were then determined as in Arnon (1949), Suc, Glc, and Fru as in Geigenberger et al. (1996), and starch as in Hendriks et al. (2003). Extractions and assays were performed using a robotized platform and absorbances were read at 340 nm (carbohydrates) and at 645 and 665 nm (chlorophylls) in a Xenius reader (SAFAS). Extractions were performed using 1.1-mL Micronic tubes (VALDEA Biosciences) with screw caps and assays using 96-well polystyrene microplates (Sarstedt).
Sequence Structure and Promoter Analysis
The structure of SlARF4 was determined using in silico approaches (fancyGENE software version 1.4). Promoter sequences of SlARF4, SlAGPase, SlSTS, SlSBE, and SlGLK2 genes were analyzed using PLACE signal scan search software (http://www.dna.affrc.go.jp/PLACE/signalscan.html).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. In silico analysis of SlAGPase and SlGLK1 gene promoters.
Supplemental Table S2. PCR primers of all genes analyzed in the article.
Acknowledgments
The work benefited from the networking activities within the European Cooperation in Science and Technology Action FA1106. We thank L. Lemonier (Université de Toulouse, Institut National Polytechnique-Ecole Nationale Supérieure Agronomique de Toulouse, Laboratoire de Génomique et Biotechnologie des Fruits) and D. Saint-Martin (Université de Toulouse, Institut National Polytechnique-Ecole Nationale Supérieure Agronomique de Toulouse, Laboratoire de Génomique et Biotechnologie des Fruits) for tomato cultures and genetic transformation, the “plateforme de microscopie TRI” (Fédération de Recherches) for microscopy analyses, and D. Combes (Laboratoire d'Ingénierie des Systémes Biologiques et des Procédés-Institut National Polytechnique de Toulouse) for his help in carbohydrate analysis.
Glossary
- qPCR
quantitative PCR
- ASL
antisense line
- AuxRE
auxin response element
References
- Abel S, Theologis A. (1996) Early genes and auxin action. Plant Physiol 111: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts. polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M. (2012) Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol 53: 659–672 [DOI] [PubMed] [Google Scholar]
- Bassi D, Selli R. (1990) Evaluation of fruit quality in peach and apricot. Adv Hortic Sci 4: 107–112 [Google Scholar]
- Baumann K, De Paolis A, Costantino P, Gualberti G. (1999) The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter CJ, Carrari F, Bauke A, Overy S, Hill SA, Quick PW, Fernie AR, Sweetlove LJ. (2005) Fruit carbohydrate metabolism in an introgression line of tomato with increased fruit soluble solids. Plant Cell Physiol 46: 425–437 [DOI] [PubMed] [Google Scholar]
- Beckles DM, Craig J, Smith AM. (2001a) ADP-glucose pyrophosphorylase is located in the plastid in developing tomato fruit. Plant Physiol 126: 261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckles DM, Smith AM, ap Rees T. (2001b) A cytosolic ADP-glucose pyrophosphorylase is a feature of graminaceous endosperms, but not of other starch-storing organs. Plant Physiol 125: 818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Papadopoulos M, Schreiber U, Kaiser W, Roitsch T. (2004) Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol Plant 122: 419–428 [Google Scholar]
- Blanke MM, Lenz F. (1989) Fruit photosynthesis. Plant Cell Environ 12: 31–46 [Google Scholar]
- Bouzayen M, Latché A, Nath P, Pech JC. (2010) Mechanism of fruit ripening. In Pua EC, Davey MR, eds, Plant Developmental Biology: Biotechnological Perspectives. Springer, Berlin, pp 319–339 [Google Scholar]
- Bucheli P, Voirol E, de la Torre R, López J, Rytz A, Tanksley SD, Pétiard V. (1999) Definition of nonvolatile markers for flavor of tomato (Lycopersicon esculentum mill.) as tools in selection and breeding. J Agric Food Chem 47: 659–664 [DOI] [PubMed] [Google Scholar]
- Carrara S, Pardossi A, Soldatini GF, Tognoni F, Guidi L. (2001) Photosynthetic activity of ripening tomato fruit. Photosynthetica 39: 75–78 [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor M-I, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno DC, Osorio S, Nunes-Nesi A, Bertolo AL, Carneiro RT, Araújo WL, Steinhauser MC, Michalska J, Rohrmann J, Geigenberger P, et al. (2011) Malate plays a crucial role in starch metabolism, ripening, and soluble solid content of tomato fruit and affects postharvest softening. Plant Cell 23: 162–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD. (1996) In vitro tomato fruit cultures demonstrate a role for indole-3-acetic acid in regulating fruit ripening. J Am Soc Hortic Sci 121: 520–524 [Google Scholar]
- Davies JN, Cocking EC. (1965) Changes in carbohydrate, proteins and nucleic acids during cellular development in tomato fruit locule tissue. Planta 67: 242–253 [Google Scholar]
- Davies JN, Hobson GE. (1981) The constituents of tomato fruit—the influence of environment, nutrition, and genotype. Crit Rev Food Sci Nutr 15: 205–280 [DOI] [PubMed] [Google Scholar]
- de Jong M, Mariani C, Vriezen WH. (2009a) The role of auxin and gibberellin in tomato fruit set. J Exp Bot 60: 1523–1532 [DOI] [PubMed] [Google Scholar]
- de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH. (2009b) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57: 160–170 [DOI] [PubMed] [Google Scholar]
- Devoghalaere F, Doucen T, Guitton B, Keeling J, Payne W, Ling T, Ross JJ, Hallett IC, Gunaseelan K, Dayatilake GA, et al. (2012) A genomics approach to understanding the role of auxin in apple (Malus x domestica) fruit size control. BMC Plant Biol 12: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Lerchi J, Stitt M, Sonnewald U. (1996) Phloem-specific expression of pyrophosphatase inhibits long distance transport of carbohydrates and amino acids in tobacco plants. Plant Cell Environ 19: 43–55 [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M. (2004) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Pyl E-T, Sulpice R, Lunn JE, Höhne M, Günther M, Stitt M. (2009) Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ 32: 859–874 [DOI] [PubMed] [Google Scholar]
- Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM. (2006) AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18: 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. (2012) Getting a grasp on domain III/IV responsible for auxin response factor-IAA protein interactions. Plant Sci 190: 82–88 [DOI] [PubMed] [Google Scholar]
- Guillon F, Philippe S, Bouchet B, Devaux M-F, Frasse P, Jones B, Bouzayen M, Lahaye M. (2008) Down-regulation of an auxin response factor in the tomato induces modification of fine pectin structure and tissue architecture. J Exp Bot 59: 273–288 [DOI] [PubMed] [Google Scholar]
- Hädrich N, Hendriks JHM, Kötting O, Arrivault S, Feil R, Zeeman SC, Gibon Y, Schulze WX, Stitt M, Lunn JE. (2012) Mutagenesis of cysteine 81 prevents dimerization of the APS1 subunit of ADP-glucose pyrophosphorylase and alters diurnal starch turnover in Arabidopsis thaliana leaves. Plant J 70: 231–242 [DOI] [PubMed] [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P. (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington SE, Smillie RM, Davies WJ. (1998) Photosynthetic activities of vegetative and fruiting tissues of tomato. J Exp Bot 49: 1173–1181 [Google Scholar]
- Ho LC. (1996) Tomato. In Zamski E, Scheffer AA, eds, Photoassimilate distribution nosbisosuin plants and crops: source-sink relationship. Marcel Dekker, New York, pp 709–727 [Google Scholar]
- Ho LC, Hewitt JD. (1986) Fruit development. In Atherton JG, Rudich J, eds, The tomato crop. Chapman and Hall, London, pp 201–240 [Google Scholar]
- Inzé D, De Veylder L. (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- James MG, Denyer K, Myers AM. (2003) Starch synthesis in the cereal endosperm. Curr Opin Plant Biol 6: 215–222 [DOI] [PubMed] [Google Scholar]
- Jarret RL, Sayama H, Tigchelaar EC. (1984) Pleiotropic effects associated with the chlorophyll intensifier mutations high pigment and dark green in tomato. J Am Soc Hortic Sci 109: 873–878 [Google Scholar]
- Jones B, Frasse P, Olmos E, Zegzouti H, Li ZG, Latché A, Pech JC, Bouzayen M. (2002) Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J 32: 603–613 [DOI] [PubMed] [Google Scholar]
- Leclercq J, Ranty B, Sanchez-Ballesta M-T, Li Z, Jones B, Jauneau A, Pech J-C, Latché A, Ranjeva R, Bouzayen M. (2005) Molecular and biochemical characterization of LeCRK1, a ripening-associated tomato CDPK-related kinase. J Exp Bot 56: 25–35 [DOI] [PubMed] [Google Scholar]
- Liang J, Zhang J, Cao X. (2001) Grain sink strength may be related to the poor grain filling of indica-japonica rice (Oryza sativa) hybrids. Physiol Plant 112: 470–477 [DOI] [PubMed] [Google Scholar]
- Matas AJ, Yeats TH, Buda GJ, Zheng Y, Chatterjee S, Tohge T, Ponnala L, Adato A, Aharoni A, Stark R, et al. (2011) Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23: 3893–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury P, Mojayad F, Berger M, Planchon C. (1996) Photochemical response to drought acclimation in two sunflower genotypes. Physiol Plant 98: 57–66 [Google Scholar]
- Miyazawa Y, Sakai A, Miyagishima S, Takano H, Kawano S, Kuroiwa T. (1999) Auxin and cytokinin have opposite effects on amyloplast development and the expression of starch synthesis genes in cultured bright yellow-2 tobacco cells. Plant Physiol 121: 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Düchting P, Weiler EW. (2002) A multiplex GC-MS/MS technique for the sensitive and quantitative single-run analysis of acidic phytohormones and related compounds, and its application to Arabidopsis thaliana. Planta 216: 44–56 [DOI] [PubMed] [Google Scholar]
- Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. (2003) Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA 100: 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfini T, Rotino GL, Camerini S, Defez R, Spena A. (2002) Optimisation of transgene action at the post-transcriptional level: high quality parthenocarpic fruits in industrial tomatoes. BMC Biotechnol 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech J-C, Purgatto E, Bouzayen M, Latché A. (2012) Ethylene and fruit ripening. In McManus MT, ed, Annual Plant Reviews, Vol 44 Wiley-Blackwell, Oxford, pp 275–304 [Google Scholar]
- Piechulla B, Glick RE, Bahl H, Melis A, Gruissem W. (1987) Changes in photosynthetic capacity and photosynthetic protein pattern during tomato fruit ripening. Plant Physiol 84: 911–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrello J, Jaimes-Miranda F, Sanchez-Ballesta MT, Tournier B, Khalil-Ahmad Q, Regad F, Latché A, Pech JC, Bouzayen M. (2006) Sl-ERF2, a tomato ethylene response factor involved in ethylene response and seed germination. Plant Cell Physiol 47: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Powell ALT, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, et al. (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Ruan Y-L, Patrick JW, Bouzayen M, Osorio S, Fernie AR. (2012) Molecular regulation of seed and fruit set. Trends Plant Sci 17: 656–665 [DOI] [PubMed] [Google Scholar]
- Salles C, Nicklaus S, Septier C. (2003) Determination and gustatory properties of taste-active compounds in tomato juice. Food Chem 81: 395–402 [Google Scholar]
- Sanders DC, Pharr DM, Konsler TR. (1975) Chlorophyll content of a dark green mutant of 'Manapal' tomato. HortScience 10: 262–264 [Google Scholar]
- Schaffer AA, Levin I, Oguz I, Petreikov M, Cincarevsky F, Yeselson Y, Shen S, Gilboa N, Bar M. (2000) ADPglucose pyrophosphorylase activity and starch accumulation in immature tomato fruit: the effect of a Lycopersicon hirsutum-derived introgression encoding for the large subunit. Plant Sci 152: 135–144 [Google Scholar]
- Schaffer AA, Petreikov M. (1997) Sucrose-to-starch metabolism in tomato fruit undergoing transient starch accumulation. Plant Physiol 113: 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Smidansky ED, Clancy M, Meyer FD, Lanning SP, Blake NK, Talbert LE, Giroux MJ. (2002) Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc Natl Acad Sci USA 99: 1724–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM. (1999) Making starch. Curr Opin Plant Biol 2: 223–229 [DOI] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JH, Stitt M, Branscheidg A, Gibon Y, Farre EM, Geigenberger P. (2002) Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14: 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-Y, Nelson OE. (1966) Starch-deficient maize mutant lacking adenosine dephosphate glucose pyrophosphorylase activity. Science 151: 341–343 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. (1999) Dimerization and DNA binding of auxin response factors. Plant J 19: 309–319 [DOI] [PubMed] [Google Scholar]
- Vendrell M. (1985) Dual effect of 2, 4-D on ethylene production and ripening of tomato fruit tissue. Physiol Plant 64: 559–563 [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latché A, Pech J-C, Bouzayen M. (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latché A, Pech J-C, Fernie AR, Bouzayen M. (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21: 1428–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelle S, Hewitt JD, Robinson NL, Damon S, Bennett AB. (1988) Sink metabolism in tomato fruit. III. Analysis of carbohydrate assimilation in a wild species. Plant Physiol 87: 737–740 [DOI] [PMC free article] [PubMed] [Google Scholar]