Abstract
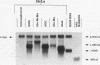
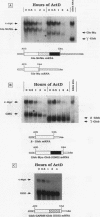
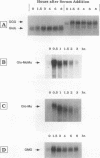
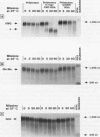
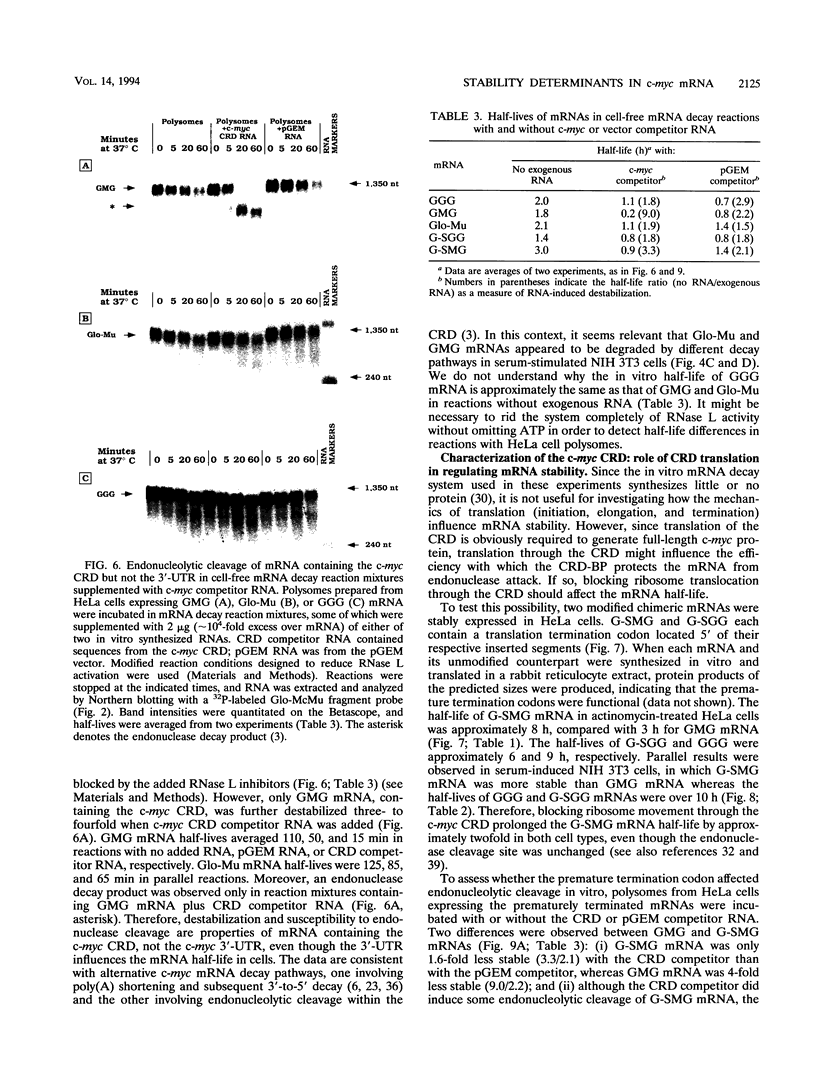
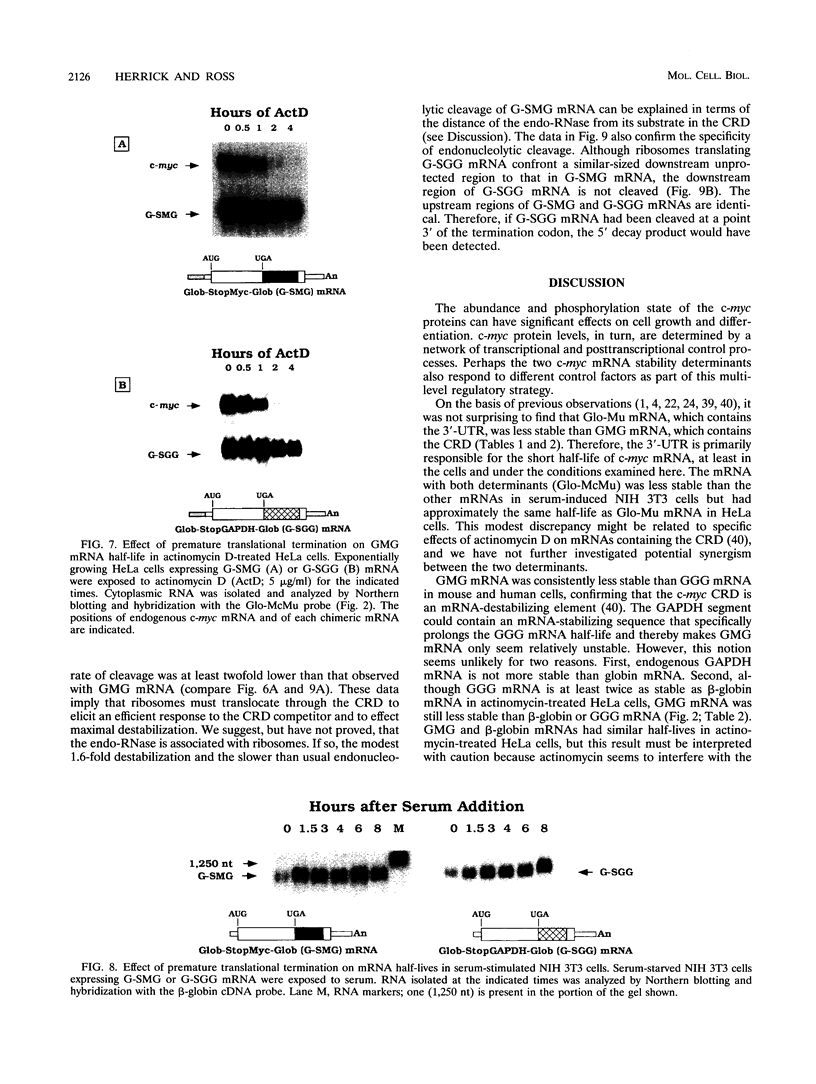
c-myc mRNA contains at least two discrete sequence elements that account for its short half-life, one in the 3' untranslated region and the other in the carboxy-terminal coding region (coding-region determinant). To investigate the function of each determinant, one or both were fused in frame to portions of a gene encoding long-lived beta-globin mRNA. Each chimeric gene was stably transfected into HeLa and NIH 3T3 cells and was transcribed from a constitutive cytomegalovirus promoter or from a serum-regulated c-fos promoter, respectively. The steady-state levels of the chimeric mRNAs in exponentially growing HeLa cells were compared, and their half-lives were measured by two independent methods: (i) in actinomycin D-treated HeLa cells and (ii) after serum addition to starved 3T3 cells. By each method, mRNAs containing either instability determinant were less stable than beta-globin mRNA. mRNA containing only the c-myc 3' untranslated region was not significantly more stable than mRNA with both determinants. In a cell-free mRNA decay system containing polysomes from transfected HeLa cells, mRNA containing the coding-region determinant was destabilized by addition of a specific RNA competitor, whereas mRNA containing only the 3' untranslated region was unaffected. When a stop codon was inserted upstream of the coding-region determinant, the chimeric mRNA was stabilized approximately twofold. These and other data suggest that degradation involving the coding-region determinant occurs most efficiently when ribosomes are translating the determinant.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aghib D. F., Bishop J. M., Ottolenghi S., Guerrasio A., Serra A., Saglio G. A 3' truncation of MYC caused by chromosomal translocation in a human T-cell leukemia increases mRNA stability. Oncogene. 1990 May;5(5):707–711. [PubMed] [Google Scholar]
- Baglioni C., De Benedetti A., Williams G. J. Cleavage of nascent reovirus mRNA by localized activation of the 2'-5'-oligoadenylate-dependent endoribonuclease. J Virol. 1984 Dec;52(3):865–871. doi: 10.1128/jvi.52.3.865-871.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P. L., Herrick D. J., Prokipcak R. D., Ross J. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes Dev. 1992 Apr;6(4):642–654. doi: 10.1101/gad.6.4.642. [DOI] [PubMed] [Google Scholar]
- Bonnieu A., Roux P., Marty L., Jeanteur P., Piechaczyk M. AUUUA motifs are dispensable for rapid degradation of the mouse c-myc RNA. Oncogene. 1990 Oct;5(10):1585–1588. [PubMed] [Google Scholar]
- Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991 May;11(5):2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G., Ross J. Poly(A) shortening and degradation of the 3' A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988 Apr;8(4):1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G., Ross J. Regulation of c-myc mRNA stability in vitro by a labile destabilizer with an essential nucleic acid component. Mol Cell Biol. 1989 May;9(5):1996–2006. doi: 10.1128/mcb.9.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., You Y., Shyu A. B. Two cellular proteins bind specifically to a purine-rich sequence necessary for the destabilization function of a c-fos protein-coding region determinant of mRNA instability. Mol Cell Biol. 1992 Dec;12(12):5748–5757. doi: 10.1128/mcb.12.12.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M. D. The myb and myc nuclear oncogenes as transcriptional activators. Curr Opin Cell Biol. 1990 Jun;2(3):502–508. doi: 10.1016/0955-0674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fort P., Marty L., Piechaczyk M., el Sabrouty S., Dani C., Jeanteur P., Blanchard J. M. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985 Mar 11;13(5):1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazal P., Nelson J. A. Enhancement of RNA polymerase II initiation complexes by a novel DNA control domain downstream from the cap site of the cytomegalovirus major immediate-early promoter. J Virol. 1991 May;65(5):2299–2307. doi: 10.1128/jvi.65.5.2299-2307.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Gritz L., Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983 Nov;25(2-3):179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- Harrold S., Genovese C., Kobrin B., Morrison S. L., Milcarek C. A comparison of apparent mRNA half-life using kinetic labeling techniques vs decay following administration of transcriptional inhibitors. Anal Biochem. 1991 Oct;198(1):19–29. doi: 10.1016/0003-2697(91)90500-s. [DOI] [PubMed] [Google Scholar]
- Jones T. R., Cole M. D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3' untranslated sequences. Mol Cell Biol. 1987 Dec;7(12):4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabnick K. S., Housman D. E. Determinants that contribute to cytoplasmic stability of human c-fos and beta-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988 Aug;8(8):3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K., Li S. W., Sobol R. W., Jr, Suhadolnik R. J., Charubala R., Pfleiderer W. Phosphorothioate analogues of 2',5'-oligoadenylate. Activation of 2',5'-oligoadenylate-dependent endoribonuclease by 2',5'-phosphorothioate cores and 5'-monophosphates. Biochemistry. 1987 Nov 3;26(22):7136–7142. doi: 10.1021/bi00396a040. [DOI] [PubMed] [Google Scholar]
- Karikó K., Sobol R. W., Jr, Suhadolnik L., Li S. W., Reichenbach N. L., Suhadolnik R. J., Charubala R., Pfleiderer W. Phosphorothioate analogues of 2',5'-oligoadenylate. Enzymatically synthesized 2',5'-phosphorothioate dimer and trimer: unequivocal structural assignment and activation of 2',5'-oligoadenylate-dependent endoribonuclease. Biochemistry. 1987 Nov 3;26(22):7127–7135. doi: 10.1021/bi00396a039. [DOI] [PubMed] [Google Scholar]
- Kruijer W., Cooper J. A., Hunter T., Verma I. M. Platelet-derived growth factor induces rapid but transient expression of the c-fos gene and protein. Nature. 1984 Dec 20;312(5996):711–716. doi: 10.1038/312711a0. [DOI] [PubMed] [Google Scholar]
- Lachman H. M., Cheng G. H., Skoultchi A. I. Transfection of mouse erythroleukemia cells with myc sequences changes the rate of induced commitment to differentiate. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6480–6484. doi: 10.1073/pnas.83.17.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird-Offringa I. A., Elfferich P., van der Eb A. J. Rapid c-myc mRNA degradation does not require (A + U)-rich sequences or complete translation of the mRNA. Nucleic Acids Res. 1991 May 11;19(9):2387–2394. doi: 10.1093/nar/19.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird-Offringa I. A., de Wit C. L., Elfferich P., van der Eb A. J. Poly(A) tail shortening is the translation-dependent step in c-myc mRNA degradation. Mol Cell Biol. 1990 Dec;10(12):6132–6140. doi: 10.1128/mcb.10.12.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn R. M., Efstratiadis A., O'Connell C., Maniatis T. The nucleotide sequence of the human beta-globin gene. Cell. 1980 Oct;21(3):647–651. doi: 10.1016/0092-8674(80)90428-6. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. New light on Myc and Myb. Part I. Myc. Genes Dev. 1990 Dec;4(12A):2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- Müllner E. W., Kühn L. C. A stem-loop in the 3' untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988 Jun 3;53(5):815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- Ross J. A precursor of globin messenger RNA. J Mol Biol. 1976 Sep 15;106(2):403–420. doi: 10.1016/0022-2836(76)90093-0. [DOI] [PubMed] [Google Scholar]
- Ross J., Kobs G. H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3' terminus and proceeds 3' to 5'. J Mol Biol. 1986 Apr 20;188(4):579–593. doi: 10.1016/s0022-2836(86)80008-0. [DOI] [PubMed] [Google Scholar]
- Ross J., Sullivan T. D. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985 Nov;66(5):1149–1154. [PubMed] [Google Scholar]
- Shyu A. B., Belasco J. G., Greenberg M. E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991 Feb;5(2):221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Greenberg M. E., Belasco J. G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989 Jan;3(1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- Suhadolnik R. J., Lee C., Karikó K., Li S. W. Phosphorothioate analogues of 2',5'-oligoadenylate. Enzymatic synthesis, properties, and biological activities of 2',5'-phosphorothioates from adenosine 5'-O-(2-thiotriphosphate) and adenosine 5'-O-(3-thiotriphosphate). Biochemistry. 1987 Nov 3;26(22):7143–7149. doi: 10.1021/bi00396a041. [DOI] [PubMed] [Google Scholar]
- Swartwout S. G., Kinniburgh A. J. c-myc RNA degradation in growing and differentiating cells: possible alternate pathways. Mol Cell Biol. 1989 Jan;9(1):288–295. doi: 10.1128/mcb.9.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5' element and c-fos 3' sequences. Cell. 1985 Oct;42(3):889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- Wellington C. L., Greenberg M. E., Belasco J. G. The destabilizing elements in the coding region of c-fos mRNA are recognized as RNA. Mol Cell Biol. 1993 Aug;13(8):5034–5042. doi: 10.1128/mcb.13.8.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisdom R., Lee W. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991 Feb;5(2):232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- Wisdom R., Lee W. Translation of c-myc mRNA is required for its post-transcriptional regulation during myogenesis. J Biol Chem. 1990 Nov 5;265(31):19015–19021. [PubMed] [Google Scholar]
- You Y., Chen C. Y., Shyu A. B. U-rich sequence-binding proteins (URBPs) interacting with a 20-nucleotide U-rich sequence in the 3' untranslated region of c-fos mRNA may be involved in the first step of c-fos mRNA degradation. Mol Cell Biol. 1992 Jul;12(7):2931–2940. doi: 10.1128/mcb.12.7.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]