Abstract
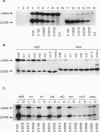
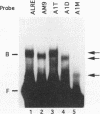
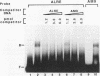
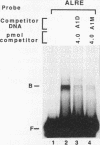
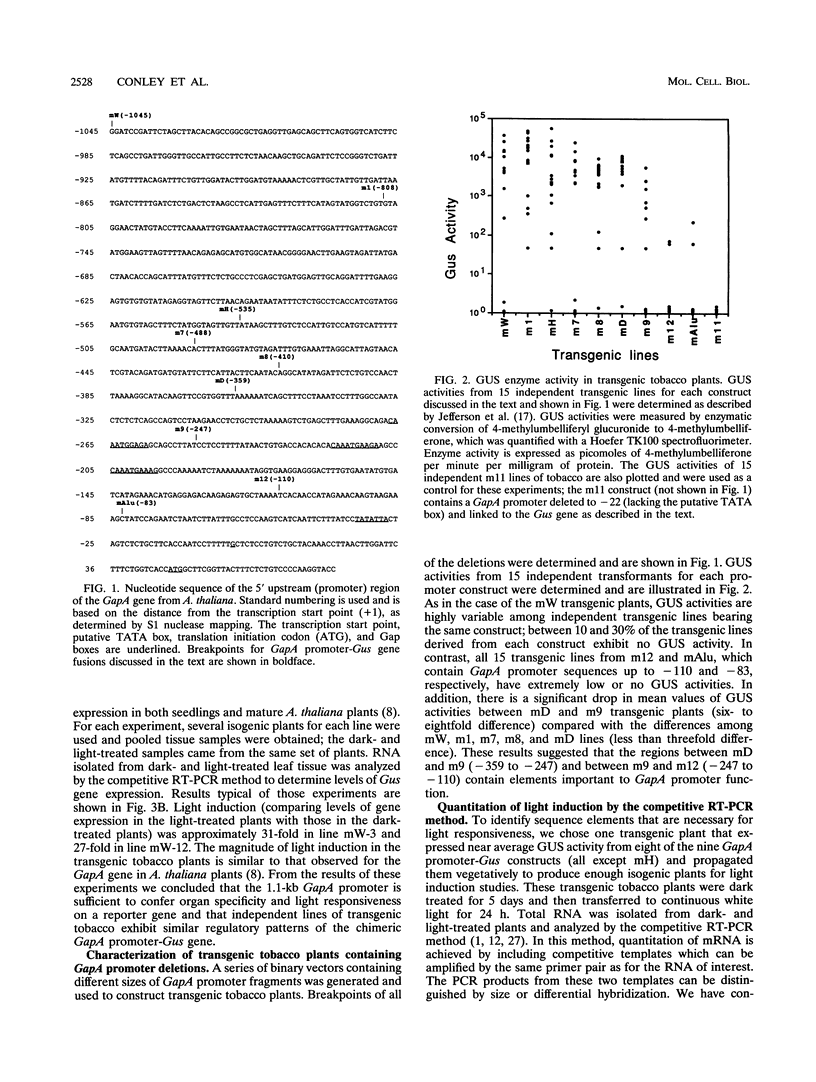
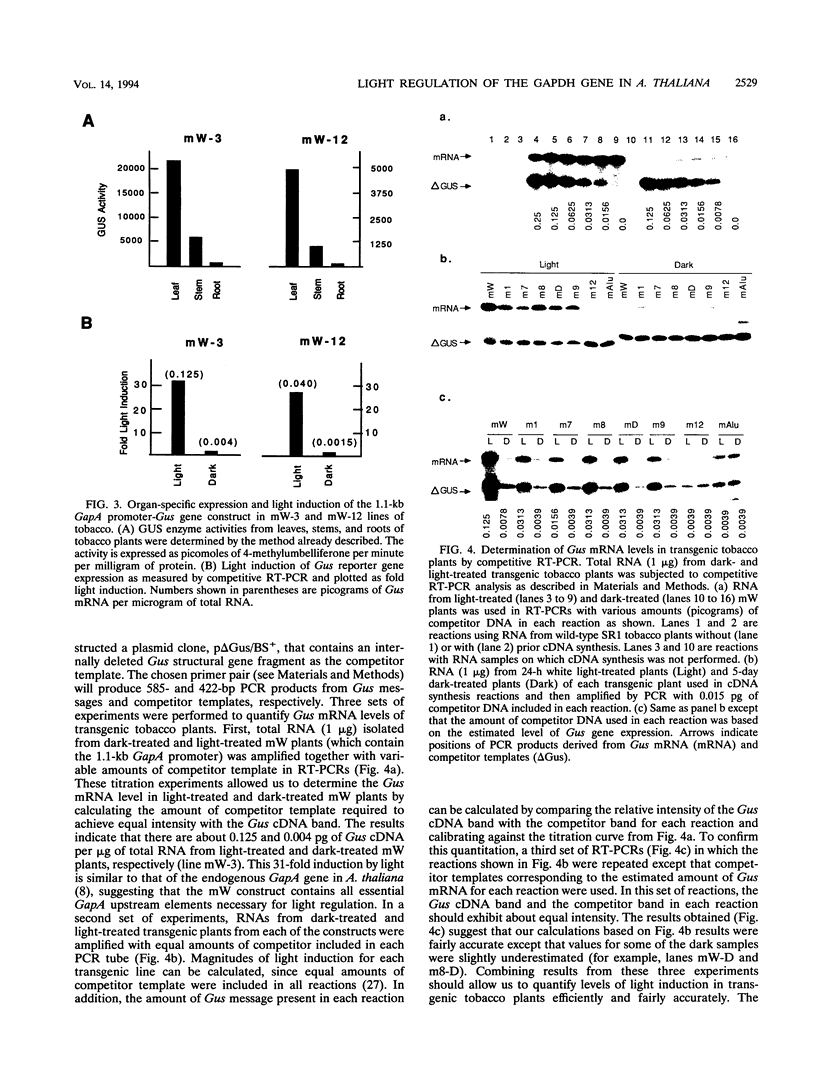
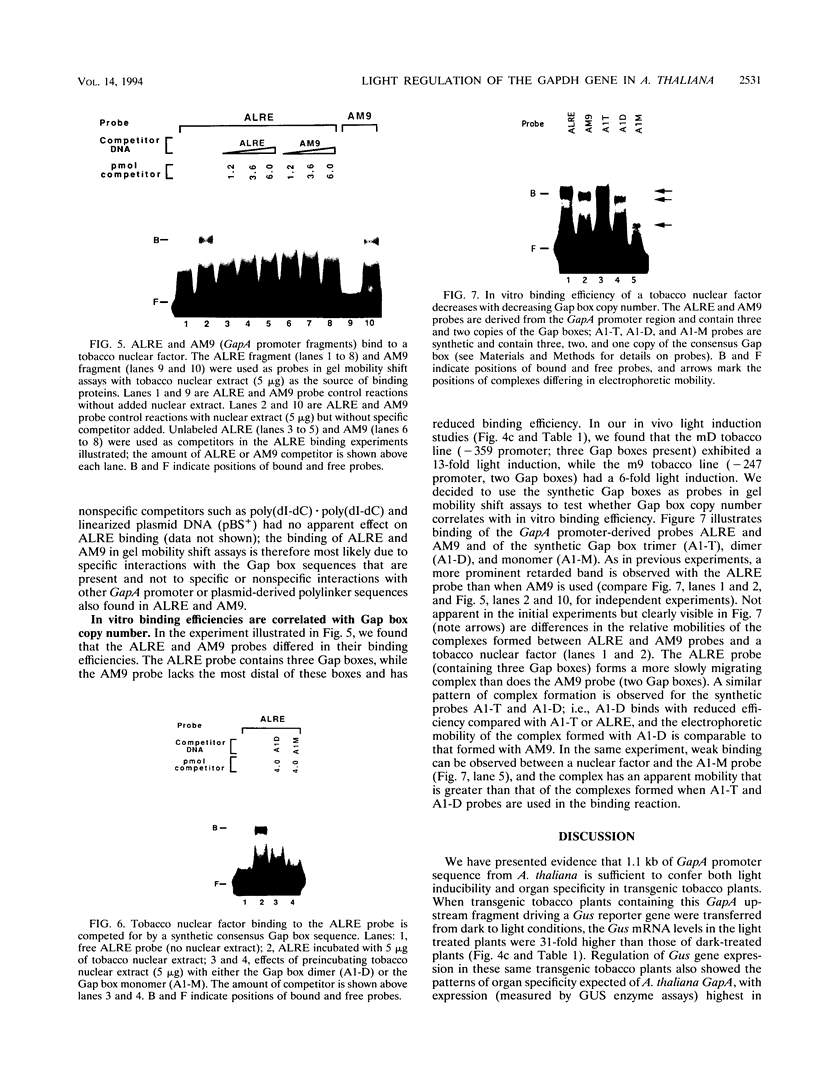
We have characterized cis-acting elements involved in light regulation of the nuclear gene (GapA) encoding the A subunit of chloroplast glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in Arabidopsis thaliana. Our results show that a 1.1-kb promoter fragment of the GapA gene is sufficient to confer light inducibility and organ specificity in transgenic Nicotiana tabacum (tobacco) plants, using the beta-glucuronidase gene of Escherichia coli as the reporter gene. Deletion analysis indicates that the -359 to -110 bp region of the GapA gene is necessary for light responsiveness. Within this region there are three copies of a decamer repeat (termed the Gap box) having the consensus sequence 5'-CAAATGAA(A/G)A-3', which has not been characterized in the promoter regions of other light-regulated genes. A deletion (to -247) producing loss of one copy of these elements from the GapA promoter reduces light induction by two- to threefold compared with a promoter deletion (to -359) with all three Gap boxes present, while deletion of all three Gap boxes (to -110) abolishes light induction completely. Gel mobility shift experiments using tobacco nuclei as the source of nuclear proteins show that GapA promoter fragments that contain these repeats bind strongly to a factor in the nuclear extract and that binding can be abolished by synthetic competitors consisting only of a monomer or dimer of the Gap box. Furthermore, a trimer, dimer, and monomer of the Gap box show binding activity and, like the authentic GapA promoter-derived probes, show binding activities that are correlated with Gap box copy number. These results strongly suggest that these repeats play important roles in light regulation of the GapA gene of A. thaliana.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker-André M., Hahlbrock K. Absolute mRNA quantification using the polymerase chain reaction (PCR). A novel approach by a PCR aided transcript titration assay (PATTY). Nucleic Acids Res. 1989 Nov 25;17(22):9437–9446. doi: 10.1093/nar/17.22.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984 Nov 26;12(22):8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Buzby J. S., Yamada T., Tobin E. M. A light-regulated DNA-binding activity interacts with a conserved region of a Lemna gibba rbcS promoter. Plant Cell. 1990 Aug;2(8):805–814. doi: 10.1105/tpc.2.8.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C., Elzen P., Tamaki S., Dunsmuir P., Bedbrook J. Differential expression of the eight genes of the petunia ribulose bisphosphate carboxylase small subunit multi-gene family. EMBO J. 1985 Dec 1;4(12):3055–3061. doi: 10.1002/j.1460-2075.1985.tb04045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewdney J., Conley T. R., Shih M. C., Goodman H. M. Effects of blue and red light on expression of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase of Arabidopsis thaliana. Plant Physiol. 1993 Dec;103(4):1115–1121. doi: 10.1104/pp.103.4.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald R. G., Cashmore A. R. Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 1990 Jun;9(6):1717–1726. doi: 10.1002/j.1460-2075.1990.tb08295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr R., Chua N. H. Developmental regulation of two genes encoding ribulose-bisphosphate carboxylase small subunit in pea and transgenic petunia plants: Phytochrome response and blue-light induction. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2358–2362. doi: 10.1073/pnas.83.8.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidoni D., Brosio P., Bond-Nutter D., Bedbrook J., Dunsmuir P. Novel cis-acting elements in Petunia Cab gene promoters. Mol Gen Genet. 1989 Jan;215(2):337–344. doi: 10.1007/BF00339739. [DOI] [PubMed] [Google Scholar]
- Gilliland G., Perrin S., Blanchard K., Bunn H. F. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin P. M., Sarokin L., Memelink J., Chua N. H. Molecular light switches for plant genes. Plant Cell. 1990 May;2(5):369–378. doi: 10.1105/tpc.2.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G., Pichersky E., Malik V. S., Timko M. P., Scolnik P. A., Cashmore A. R. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7089–7093. doi: 10.1073/pnas.85.19.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P. J., Kay S. A., Chua N. H. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 1987 Sep;6(9):2543–2549. doi: 10.1002/j.1460-2075.1987.tb02542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob U., Stüber K. Discrimination of phytochrome dependent light inducible from non-light inducible plant genes. Prediction of a common light-responsive element (LRE) in phytochrome dependent light inducible plant genes. Nucleic Acids Res. 1987 Dec 10;15(23):9957–9973. doi: 10.1093/nar/15.23.9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987 Dec 20;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin-Neumann G. A., Sun L., Tobin E. M. Expression of Light-Harvesting Chlorophyll a/b-Protein Genes Is Phytochrome-Regulated in Etiolated Arabidopsis thaliana Seedlings. Plant Physiol. 1988 Dec;88(4):1323–1331. doi: 10.1104/pp.88.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L. S., Thompson W. F., Briggs W. R. Different Red Light Requirements for Phytochrome-Induced Accumulation of cab RNA and rbcS RNA. Science. 1984 Dec 21;226(4681):1447–1449. doi: 10.1126/science.226.4681.1447. [DOI] [PubMed] [Google Scholar]
- Lam E., Chua N. H. GT-1 binding site confers light responsive expression in transgenic tobacco. Science. 1990 Apr 27;248(4954):471–474. doi: 10.1126/science.2330508. [DOI] [PubMed] [Google Scholar]
- Manzara T., Carrasco P., Gruissem W. Developmental and organ-specific changes in promoter DNA-protein interactions in the tomato rbcS gene family. Plant Cell. 1991 Dec;3(12):1305–1316. doi: 10.1105/tpc.3.12.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P. H. Phytochrome: a light-activated molecular switch that regulates plant gene expression. Annu Rev Genet. 1991;25:389–409. doi: 10.1146/annurev.ge.25.120191.002133. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih M. C., Goodman H. M. Differential light regulated expression of nuclear genes encoding chloroplast and cytosolic glyceraldehyde-3-phosphate dehydrogenase in Nicotiana tabacum. EMBO J. 1988 Apr;7(4):893–898. doi: 10.1002/j.1460-2075.1988.tb02893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert P. D., Larrick J. W. Competitive PCR. Nature. 1992 Oct 8;359(6395):557–558. doi: 10.1038/359557a0. [DOI] [PubMed] [Google Scholar]
- Ueda T., Pichersky E., Malik V. S., Cashmore A. R. Level of expression of the tomato rbcS-3A gene is modulated by a far upstream promoter element in a developmentally regulated manner. Plant Cell. 1989 Feb;1(2):217–227. doi: 10.1105/tpc.1.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Kwon H. B., Peng H. P., Shih M. C. Stress responses and metabolic regulation of glyceraldehyde-3-phosphate dehydrogenase genes in Arabidopsis. Plant Physiol. 1993 Jan;101(1):209–216. doi: 10.1104/pp.101.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]