Abstract
Reverse-genetics systems are powerful tools enabling researchers to study the replication cycle of RNA viruses, including filoviruses and other hemorrhagic fever viruses, as well as to discover new antivirals. They include full-length clone systems as well as a number of life cycle modeling systems. Full-length clone systems allow for the generation of infectious, recombinant viruses, and thus are an important tool for studying the virus replication cycle in its entirety. In contrast, life cycle modeling systems such as minigenome and transcription and replication competent virus-like particle systems can be used to simulate and dissect parts of the virus life cycle outside of containment facilities. Minigenome systems are used to model viral genome replication and transcription, whereas transcription and replication competent virus-like particle systems also model morphogenesis and budding as well as infection of target cells. As such, these modeling systems have tremendous potential to further the discovery and screening of new antivirals targeting hemorrhagic fever viruses. This review provides an overview of currently established reverse genetics systems for hemorrhagic fever-causing negative-sense RNA viruses, with a particular emphasis on filoviruses, and the potential application of these systems for antiviral research.
Keywords: reverse genetics, virus-like particles, minigenome, minireplicon, viral hemorrhagic fever, filoviruses, arenaviruses, bunyaviruses
INTRODUCTION
Viral hemorrhagic fever (VHF) is a clinical syndrome that can be caused by multiple unrelated viruses (Marty et al., 2006). Infection with a VHF agent can cause hemorrhage, multi-organ failure and high case-fatality rates. To date, all known human VHFs are caused by a range of single stranded, enveloped RNA viruses including filoviruses, arenaviruses, bunyaviruses, and flaviviruses. The requirement for biosafety level (BSL) 3 or 4 when working with these pathogens is slowing down research; however, a better understanding of the molecular pathogenesis of these agents is a prerequisite for the advancement of specific countermeasures. Reverse genetics systems are experimental tools allowing for the production and subsequent replication and transcription of virus RNA genomes, or genome analogues, from complementary DNA (cDNA). In contrast to classical genetics, in which a specific phenotype is analyzed for its underlying genotype, reverse genetics is based on manipulation of a virus genotype and analysis of the resulting changes to its phenotype. The advent of reverse genetics systems for several VHF agents has pushed the field forward by allowing for the generation of recombinant viruses, and thus providing the means to address questions about virtually every aspect of the virus replication cycle. However, reverse genetics systems also provide opportunities to study these viruses outside of BSL-3/4 laboratories by use of reverse genetics based life cycle modeling systems. Such systems are particularly useful in understanding basics of the viral life cycle, and to study and dissect viral genome replication and transcription, morphogenesis and budding, infection of target cells and primary transcription in target cells. While there has been considerable work on elucidating these steps in the viral replication cycle, many details are still not fully understood. Examples are the regulation of replication and transcription, the morphogenesis of ribonucleoprotein (RNP) complexes, and the early steps immediately after virus entry, i.e. uncoating of the RNP complexes and primary transcription. This review will discuss recent advances in the field of reverse genetics and focus on negative stranded VHF agents in general and filoviruses in particular, because the filovirus field has been on the forefront of many advances in the field of negative stranded reverse genetics for VHF agents.
CLASSIFICATION, GENOME ORGANIZATION AND REPLICATION CYCLE
Negative stranded RNA viruses with the capacity to cause VHF in humans belong to three families: Arenaviridae, Bunyaviridae and Filoviridae (Marty et al., 2006). An overview of VHF-causing viruses and closely related but non-pathogenic viruses, for which reverse genetics systems have been developed, is shown in Table 1.
Table 1.
Available reverse genetics systems for negative-sense RNA viral hemorrhagic fever agents and closely related viruses.
| Family | Genus | Virus | Human disease | Genome | Minigenome systems | trVLP systems | Full-length clone system |
|---|---|---|---|---|---|---|---|
| Filoviridae | Ebolavirus | Ebola virus (EBOV) | Ebola virus disease | non-segmented negative sense | (Mühlberger et al., 1999) | (Watanabe et al., 2004)*, (Hoenen et al., 2006)** | (Volchkov et al., 2001) |
| Reston virus (RESTV) | none | (Groseth et al., 2005) | – | – | |||
| Marburgvirus | Marburg virus (MARV) | Marburg virus disease | (Mühlberger et al., 1998) | (Wenigenrath et al., 2010)*, (Krähling et al., 2010)** | (Enterlein et al., 2006) | ||
| Arenaviridae | Arenavirus (Old World) | Lassa virus (LASV) | Lassa fever | bi-segmented ambisense | (Hass et al., 2004) | – | – |
| Arenavirus (New World) | Junín virus (JUNV) | Argentinian HF | (Albarino et al., 2009) | – *** | (Albarino et al., 2009) | ||
| Tacaribe virus (TCRV) | none | (Lopez et al., 2001) | –*** | – | |||
| Machupo virus (MACV) | Bolivian HF | (Kranzusch et al., 2010) | – | – | |||
| Bunyaviridae | Orthobunyavirus | Bunyamwera (BUNV) | none | tri-segmented ambisense or negative sense | (Dunn et al., 1995) | (Shi et al., 2006) | (Bridgen and Elliott, 1996) |
| Phlebovirus | Rift Valley fever virus (RVFV) | Rift Valley fever | (Ikegami et al., 2005) | (Habjan et al., 2009a) | (Ikegami et al., 2006) | ||
| Uukuniemi virus (UUKV) | none | (Flick and Pettersson, 2001) | (Overby et al., 2006) | – | |||
| Hantavirus | Hantaan virus (HTNV) | HF with renal syndrome | (Flick et al., 2003a) | – | – | ||
| Nairovirus | Crimean-Congo hemorrhagic fever virus (CCHFV) | Crimean Congo HF | (Bergeron et al., 2010; Flick et al., 2003b) | – | – |
System with pre-transfected target cells
System with naïve target cells
A chimeric approach using a combination of components from Tacaribe virus and Junin virus has been reported (Casabona et al., 2009)
While the specific organization of their genomes differs, the filoviruses, arenaviruses and bunyaviruses share basic features common to all negative-sense RNA viruses. Viruses of all three families have genomes that consist of linear, single-stranded, negative-sense RNA (ssRNA) whose replication takes place in the host cell cytoplasm. By their nature, purified genomes are non-infectious because the genomic RNA alone cannot serve as a template for protein synthesis. Specific viral proteins must associate with the genomic RNA to initiate transcription of positive-sense messenger RNA (mRNA). The specific life cycles for all three families are described in detail elsewhere (Buchmeier et al., 2007; Sanchez et al., 2007; Schmaljohn and Nichol, 2007), and a schematic representation of the filovirus replication cycle is depicted in Figure 1A. Filovirus genomes are linear nonsegmented single-stranded negative-sense RNA molecules approximately 19 kb in length. They contain seven genes in the order 3′-NP-VP35-VP40-GP-VP30-VP24-L-5′. The genes, respectively, encode seven structural proteins, the nucleoprotein (NP), RNA-dependent RNA polymerase cofactor (VP35), matrix protein (VP40), spike glycoprotein (GP1,2), transcriptional activator (VP30), secondary matrix protein (VP24), and the RNA-dependent RNA polymerase (L) (Sanchez et al., 2007). NP in complex with the genome associates with VP30, VP35, and L to form the tubule-like RNP complex (Mühlberger et al., 1998; Mühlberger et al., 1999). VP40 and VP24 surround the RNP complex and are wrapped in a lipid envelope derived from the host cell during virion egress (Han et al., 2003; Noda et al., 2002). GP1,2 is inserted into the virion membrane, protruding from the surface (Volchkov et al., 1998). The genomic 3′ leader and 5′ trailer regions regulate virus RNA replication and transcription. Open reading frames are flanked by intergenic regions that possess transcription initiation and termination signals (Mühlberger, 2007).
Figure 1. Virus life cycle and full-length clone systems for filoviruses.
(A) Virus life cycle. After entry and uncoating (A), free RNP-complexes are transcribed into mRNA using virus proteins that were introduced into the cell within virions (primary transcription) (B). These mRNAs are then translated into virus proteins (C), which encapsidate antigenomic cRNAs and genomic vRNAs (D) produced during genome replication (E) facilitated by these proteins. They also participate in further secondary transcription of mRNAs (F). Late in infection, RNPs and other virus structural proteins are transported to budding sites, where morphogenesis and budding of progeny virus occurs (G). (B) Full-length clone system. Virus cRNA is provided in the form of a cDNA plasmid encoding the complete virus genome (rgZ), which is by itself not infectious and can be safely manipulated in a BSL-1/2 laboratory. This cDNA is initially transcribed in cells either by endogenous RNA polymerases (RNA polymerase I or RNA polymerase II) or T7 RNA polymerase (A). The resulting “naked” cRNA then has to be complexed with the other RNP components, particularly the nucleoprotein (NP), in a process called artificial encapsidation (B). These RNP components are coexpressed from separate expression plasmids. After assembly of the RNP, virus genome replication (C) and secondary transcription (D), driven by the virus RNP proteins, occurs and the resulting mRNAs are translated to produce all of the virus proteins (E). Genomic vRNAs and antigenomic cRNAs are encapsidated by the other RNP components simultaneously to replication (F). RNPs and other virus proteins can then assemble at budding sites, which leads to morphogenesis and budding of recombinant progeny virions (G), which in the case of VHF-causing viruses have to be handled in a BSL-3/4 laboratory.
The filovirus life cycle begins with cell entry. The ectodomain of GP1,2 (GP1) mediates virion binding to an unidentified cell-surface receptor (Kuhn et al., 2006), upon which the transmembrane domain (GP2) mediates fusion of the virion envelope with a cellular membrane and the release of the virion RNP into the cytosol (Gallaher, 1996; Ruiz-Arguello et al., 1998). Once released into the cytoplasm, the negative-sense genome must be transcribed. Encapsidated vRNA (i.e. negative-sense viral RNA) acts as template for the generation of polyadenylated, monocistronic mRNAs that are transcribed sequentially from a single promoter in the 3′ end of the genome. This process called primary transcription is facilitated by the polymerase complex brought into the cell within virus particles. The host cellular machinery then translates the mRNAs leading to the accumulation of viral proteins within cells. After the translation of virus proteins there is a switch from transcription to replication that leads to the synthesis and encapsidation of full-length cRNA (i.e. positive-sense complementary RNA) also known as antigenome. Antigenome copies serve as templates for the synthesis of full-length genomic vRNA, which is rapidly encapsidated by RNP complex proteins. Further, newly translated RNP proteins can also participate in the transcription of additional mRNAs from available vRNA templates (called secondary transcription). Finally, virus egress occurs when RNP complexes in the cytoplasm traffic to the cell membrane where, together with the membrane-bound proteins (VP24, VP40, and GP1,2), they accumulate and organize themselves into new virus particles, which then bud from the plasma membrane (Hartlieb and Weissenhorn, 2006).
The life cycle of arena- and bunyaviruses is similar to that of filoviruses, although there are important differences. One major difference is that both arena- and bunyaviruses have more than one genome segment, which all have to be replicated and packaged into progeny virions. Also, arenaviruses employ an ambisense coding strategy, which results in their glycoprotein GPC and their matrix protein Z mRNAs being transcribed not from genomic vRNAs, but rather from antigenomic cRNAs, which first have to be produced during replication. This allows the virus to temporally regulate gene expression, which might be particularly important since Z is a strong negative regulator of genome replication and transcription (Cornu and de la Torre, 2001; Hass et al., 2004; Lee et al., 2000; Lopez et al., 2001). Also some bunyaviruses employ an ambisense coding strategy. Further differences for bunyaviruses include the absence of a classical matrix protein, where budding is believed to be driven by the glycoproteins (Overby et al., 2007b), and the intracellular location of the budding site. For a detailed description of the life cycle of arena- and bunyaviruses the interested reader is referred to (Buchmeier et al., 2007; Schmaljohn and Nichol, 2007).
REVERSE GENETICS SYSTEMS
While the term reverse genetics is sometimes used exclusively to describe the generation of recombinant viruses from cDNA, often a broader definition is used. According to this broader definition, which is applied in this review, reverse genetics is the generation and subsequent replication and transcription of full-length virus RNA genomes or truncated genome analogues from cDNA (Table 2). If a full-length copy of the viral genome is contained within the cDNA, reverse genetics systems allow for the generation of recombinant viruses entirely from cDNA (full-length clone systems). These systems are sometimes also called infectious clone systems, although it is important to note that the cDNA plasmid itself is not infectious, and can be safely handled in a BSL-1/2 laboratory. Alternatively, if the cDNA encodes a truncated genome analogue, reverse genetics systems facilitate the modeling of various parts of the virus replication cycle (life cycle modeling systems). These modeling systems, which include both minigenome and transcription and replication competent virus-like particle (trVLP) systems, which are also known as infectious virus-like particle (iVLP) systems, are powerful tools for understanding the biology of viruses and have significantly contributed to our understanding of the molecular biology and pathogenesis of VHF-causing viruses (Ebihara et al., 2005; Hoenen et al., 2007).
Table 2.
Types of reverse genetics systems exemplified using filoviruses.
| system | Full-length clone system | Life cycle modeling systems | |
|---|---|---|---|
| Minigenome system | trVLP system | ||
| cDNA | full length antigenome (or genome) | truncated genome (or antigenome) | truncated genome (or antigenome) |
| required viral helper proteins | NP, VP35, VP30, L | NP, VP35, VP30, L | NP, VP35, VP30, L, VP40, VP24, GP |
| modeling capacities |
|
|
|
| research applications | tissue culture and animal models | tissue culture only | tissue culture only |
| differences to live virus |
|
|
|
| BSL requirements | BSL-4 for work with recombinant viruses BSL-1/2 for work with cDNA plasmid** | BSL-1/2** | BSL-1/2** |
only in trVLP system with naïve target cells
depending on local regulations
Of particular interest are reverse genetics systems for VHF-causing viruses and closely related nonpathogenic relatives (e.g. Ebola virus (EBOV) and Reston virus (RESTV); Junin virus (JUNV) and Tacaribe virus (TCRV); Rift Valley Fever Virus (RVFV) and Uukuniemi virus (UUKV)). When available, such complementary pairs of full-length clone systems allow for the mapping of pathogenic determinants through the generation of chimeric viruses, whereas pairs of life cycle modeling systems (i.e. minigenomes and trVLP systems) facilitate the comparison of individual aspects of the virus life cycle and correlation of the observed differences with the pathogenic potential of the respective viruses.
Generation of recombinant viruses from cDNA
Full-length clone systems allow for the generation of recombinant viruses from cDNA plasmids. These cDNA plasmids are by themselves non-infectious and can be safely handled and manipulated in a BSL-1/2 laboratory, depending on local regulations, although they contain a cDNA copy of the complete viral genome under the control of a promoter for a DNA dependent RNA polymerase such as T7 RNA polymerase (T7) or RNA polymerase II. However, it is only upon cotransfection of such a cDNA plasmid, together with expression plasmids for all the viral RNP proteins and the DNA dependent RNA polymerase, that infectious viruses are produced. Therefore, only these transfections and all subsequent work have to be performed under BSL-3/4 conditions.
Upon transfection of a cDNA plasmid into cells (p0 or producer cells) an unencapsidated (naked) genomic RNA is transcribed by the coexpressed DNA dependent RNA polymerase (Figure 1B). However, since only an encapsidated genome in the form of a RNP complex is a suitable template for the viral polymerase complex, this naked RNA has to be artificially encapsidated by the nucleoprotein provided in trans, a process called artificial or illegitimate encapsidation (Conzelmann, 2004). This process is generally believed to be highly inefficient and mechanistically different from the encapsidation of nascent vRNA or cRNA molecules simultaneously with replication (Hoenen et al., 2010b; Pattnaik et al., 1992; Peeples and Collins, 2000), and does not have a natural equivalent in the viral life cycle. Once encapsidated, this genomic RNA is then recognized by the other RNP components, which have to be provided in trans either by helper virus infection or from expression plasmids, before being replicated and transcribed into mRNAs. This leads to the production of all virus proteins and starts the virus replication cycle, and ultimately results in the production of a clonal population of infectious viruses.
The first rescue of any recombinant negative-sense RNA virus (rabies virus) completely from cDNA was achieved in 1994 (Schnell et al., 1994). Virus RNA was transcribed from a plasmid using T7 RNA polymerase provided by infection with a recombinant vaccinia virus. Since the use of T7 RNA polymerase leads to additional nucleotides at the 3′ ends of transcripts when compared to authentic viral RNA, a hepatitis delta virus ribozyme (HDVrib) was used to provide an authentic 3′ end to the RNA. Critical for the success of this system was the initial synthesis of a cRNA antigenome, rather than a vRNA genome. This most likely avoided the problem of hybridization between naked negative-sense genomic vRNAs and the positive-sense mRNAs encoding for the virus proteins (Ebihara et al., 2005; Neumann et al., 2002b; Schnell et al., 1994). Although rescue of negative-sense RNA viruses from cDNA encoding a negative-sense genomic vRNA has since been reported, the efficiency of this approach is generally lower than when a positive-sense antigenomic cRNA is encoded (Durbin et al., 1997; Kato et al., 1996; Neumann et al., 2002b).
In full-length clone systems that use T7 RNA polymerase for the initial transcription of cRNA antigenomes, this polymerase has to be supplied in trans. This can be achieved either by expressing it from an expression plasmid, by using a cell line that stably expresses T7 RNA polymerase (Buchholz et al., 1999), or through superinfection with a recombinant vaccinia virus encoding T7 RNA polymerase. However, full-length clone systems have also been developed in which the virus cRNA is initially transcribed by the endogenous host-cell RNA polymerases I or II. RNA polymerase I-driven systems were first used for viruses replicating in the cell nucleus, particularly orthomyxoviruses (Fodor et al., 1999; Neumann et al., 1994), but have since been successfully used for viruses replicating in the cytoplasm (Billecocq et al., 2008; Flatz et al., 2006; Habjan et al., 2008; Ogawa et al., 2007). Similarly, viruses replicating in the cell nucleus can be successfully rescued with full-length clone systems based on T7 RNA polymerase (de Wit et al., 2007; Martin et al., 2006), which is usually located in the cytoplasm. The basis for this phenomenon remains unclear. A recent technical advance in the field of full-length clone systems has been the additional development of RNA polymerase II-driven systems, which were shown to enhance rescue efficacy both for viruses replicating in the cytoplasm and in the nucleus (Martin et al., 2006). Since transcripts are capped and poly-adenylated in these systems, both the 3′ and the 5′ end have to be processed to generate authentic genome ends. This is achieved by the use of two ribozymes, i.e. hepatitis delta virus ribozyme and a hammerhead ribozyme.
Full-length clone systems have been developed for a number of VHF viruses (Table 1). While most of these systems are based on the classical T7-driven approach, an RNA polymerase I-driven full-length clone system was also reported for RVFV (Billecocq et al., 2008; Habjan et al., 2008). These systems have been quite widely used to study virus life cycles. For example, for EBOV, recombinant viruses were used to show that while the editing of the glycoprotein mRNA plays a crucial role in down-regulating cytotoxicity during infection (Volchkov et al., 2001), the cleavage of the glycoprotein into the mature GP1 and GP2 subunits is almost completely dispensable, both for in vitro growth (Neumann et al., 2002a) and infection in a non-human primate model (Neumann et al., 2007). In contrast, work with the full-length clone system for JUNV has shown that while it is possible to substitute the normal SKI-1/S1P cleavage site in its glycoprotein with a furin cleavage site, deletion of the cleavage site at this position does not allow rescue of a viable virus (Albarino et al., 2009) indicating that, unlike EBOV, maturation cleavage of the arenavirus glycoprotein is essential. Similar studies using arenaviruses expressing chimeric glycoproteins between JUNV and Lassa virus (LASV) have further shown that is it necessary that the stable signal peptide and the transmembrane domain of GP2 are derived from the same virus, while the GP1 ectodomain can be freely substituted (Albarino et al., 2010). Full-length clone system studies with RVFV, on the other hand, have focused almost exclusively on the role of the non-structural proteins NSs and NSm in regulating the host-cell response to infection. In particular, RVFV lacking NSs or NSm have been shown to be viable (Billecocq et al., 2008; Gerrard et al., 2007; Ikegami et al., 2006; Won et al., 2006). However, the mutant lacking NSs is impaired in its ability to inhibit accumulation of interferon (IFN) mRNAs during infection and shut-off host cell protein synthesis (Billecocq et al., 2008; Ikegami et al., 2006), and has recently been used to establish a role for NSs in protein kinase R (PKR) degradation (Habjan et al., 2008; Ikegami et al., 2009a, b), while the NSm deficient virus showed an increase in apoptosis in infected cells, due to the loss of the anti-apoptotic function of NSm (Won et al., 2006, 2007).
Interestingly, findings with recombinant viruses produced using these full-length clone systems sometimes necessitate re-evaluation of findings using other more directed approaches. This has clearly been the case in studies examining the role of late domains in filovirus budding where, despite compelling evidence that late-domain motif knock-outs in the matrix protein VP40 drastically impair its budding activity (Jasenosky et al., 2001; Licata et al., 2003), viruses containing these mutations are viable and have only slight defects in growth (Neumann et al., 2005) – clearly advocating the existence of additional budding mechanisms independent of these motifs.
In addition to their utility in addressing basic research questions, full-length clone systems have led to a number of developments of applied significance. For both EBOV and JUNV attempts have been made to exploit reverse genetics for the production of vaccine candidates. In the case of EBOV this was accomplished by deleting the VP30 open reading frame, thus rendering the virus replication-deficient (Halfmann et al., 2008), while for JUNV the GPC open reading frame was deleted and replaced with either a green fluorescent protein (GFP) or luciferase (Albarino et al., 2010) resulting in a virus that produces progeny that is incapable of infecting new target cells. In a different approach, a recombinant virus based on a JUNV vaccine strain, Candid-1, was generated that expressed LASV GPC (Albarino et al., 2010), indicating the feasibility of using such existing vaccine strains to produce a vaccine against multiple pathogens via reverse genetics. Given the ability to substitute RVFV non-structural genes with foreign genes, and the existence of a full-length clone system for the MP-12 vaccine strain (Billecocq et al., 2008; Ikegami et al., 2006), a similar approach based on RVFV might also be successful, but has not yet been investigated.
Due to the fact that full-length clone systems generate infectious viruses, there are very few limitations to the scientific questions that can be addressed with them, as long as mutations introduced into the virus genome do not render the virus non-viable. Further, in some cases viruses which contain lethal mutation can also be rescued by providing the wild-type protein in trans, e.g. by means of transfection. As an example it was possible to rescue an EBOV lacking the gene for VP30 in cells that stably express VP30, which restricted growth of this virus to this cell line (Halfmann et al., 2008). However, this approach is not feasible for all viral proteins, particularly in cases were a regulated expression of the protein is required for the viral life cycle. In the case of VP40 stable expression of this protein renders cells resistant to infection with EBOV (T. Hoenen, unpublished data), most likely due to the negative regulatory effect of VP40 on viral genome replication and transcription (Hoenen et al., 2010b), making it impossible to rescue viruses containing lethal mutations in VP40 by providing this protein in trans. Similar problems will most likely also arise with other proteins that negatively regulate viral genome replication and transcription, a function that seems to be common to matrix proteins of negative stranded RNA viruses (Carroll and Wagner, 1979; Clinton et al., 1978; Finke and Conzelmann, 2003; Hoenen et al., 2010b; Iwasaki et al., 2009; Lopez et al., 2001). Another limitation of full-length clone systems is that they initially generate clonal populations of viruses, so that any effects of quasispecies which might play a role in pathogenesis or interaction with the host organism are not reflected. Nevertheless, full-length clone systems are the most authentic reverse genetics system available because they model all facets of the virus life cycle. At the same time, newly generated recombinant viruses must perform all the basic steps of the viral replication cycle, and if an introduced mutation abolishes one of these steps, no recombinant virus can be generated, and no information can be deduced at which step the viral replication cycle was interrupted. In such cases life cycle modeling systems, which allow dissection of individual steps in the viral life cycle, might be a better alternative.
Modeling viral genome replication and transcription
Minigenome or minireplicon systems are used as model systems for virus genome replication and transcription, and are frequently, although not always, generated as precursors to the development of full-length clone systems. While these systems resemble full-length clone systems, instead of a full-length genomic vRNA or antigenomic cRNA they use a genomic vRNA analogue, called a minigenome or minireplicon, in which some or all of the virus open reading frames have been removed and replaced with a reporter gene such as chloramphenicol acetyl transferase, GFP or luciferase. However, the non-coding regions, which contain the minimal signals important for replication and transcription, are retained in these vRNA analogues, thus allowing them to serve as templates for the virus polymerase. In the case of filoviruses, these signals are located within the genome termini, called leader and trailer (Mühlberger et al., 1998; Mühlberger et al., 1999), whereas in the case of arenaviruses and some bunyaviruses the intergenic region, located between virus genes, contains additional important signals and is of central importance for proper termination of transcription (Flick and Bouloy, 2005; Lopez et al., 1995; Pinschewer et al., 2005).
To establish minigenome systems, cDNA copies of these vRNA minigenomes are cloned into expression vectors under control of either T7 RNA polymerase or mammalian RNA polymerase I or II promoters, and naked vRNA is initially transcribed by the respective RNA polymerase (Figure 2). The other RNP components (nucleoprotein and RNA-dependent RNA polymerase, and in the case of filoviruses also VP35 and VP30) are provided either from expression plasmids or by superinfection with helper virus. The naked genomic RNA is artificially encapsidated by the nucleoprotein, and the resulting encapsidated genomic RNA is recognized by the RNA-dependent RNA polymerase as a template for replication via a cRNA intermediate. Further, the encapsidated vRNA serves as a template for transcription, leading to the production of mRNAs and, eventually, reporter activity.
Figure 2. Minigenome systems.
EBOV is shown as an example for VHF-causing negative-sense RNA viruses. A vRNA minigenome, encoding a reporter gene in the place of some or all of the viral open reading frames, is provided in the form of a cDNA plasmid (mg) and initially transcribed in cells either by endogenous RNA polymerases (RNA polymerase I or RNA polymerase II) or T7 RNA polymerase (A). The resulting “naked” vRNA then has to be complexed with the other RNP components, particularly the nucleoprotein (NP), in a process called artificial encapsidation (B). In a replication-competent minigenome system, virus genome replication then occurs via cRNA intermediates (C), which increases the number of vRNA templates available for use as templates for secondary transcription (D). The resulting reporter mRNAs are then translated into reporter protein (E), leading to reporter activity, which reflects both virus genome replication and transcription. In contrast, in a replication-deficient minigenome system, mutations in the antigenomic replication promoter abolish the production of additional vRNA molecules from cRNA replication intermediates so that the amount of vRNA is independent of genome replication. However, secondary transcription still occurs, resulting in reporter activity, but it is only dependent on virus transcription.
The amount of vRNA template available for transcription is dependent on genome replication and, therefore, reporter activity (as well as the amount of mRNA) reflects both genome replication and transcription. Thus, it is not possible to distinguish between these two processes with classical minigenome systems. To alleviate this problem, replication-deficient minigenomes have been developed, in which the antigenomic replication promoter has been destroyed (Hoenen et al., 2010b; Peeples and Collins, 2000). Whereas these minigenomes still allow for the production of cRNA from vRNA templates, these cRNA antigenomes cannot be copied back into vRNA genomes. Therefore, the number of vRNA genome copies available for transcription is independent of genome replication (Figure 2). Reporter activity in such replication-deficient minigenome systems is several orders of magnitude lower than in classical, replication-competent minigenome systems, highlighting the influence of genome replication on overall reporter activity (Hoenen et al., 2010b). It has to be noted that for a replication-deficient minigenome system to work it is necessary that the genomic and antigenomic replication promoters are fully separated from each other. In the case of filoviruses this has been experimentally verified (Weik et al., 2005). However, arenavirus and bunyavirus genomes form panhandle structures via interaction of the 3′ and 5′-termini of the genome segments. These panhandles are important for promoter function; therefore, replication-deficient minigenomes can most likely not be generated using the approach taken for filoviruses. Classical minigenome systems have been established for a large number of VHF-causing viruses (Table 1), and since infectious virus is not present in these systems (unless virus is used as source for the required RNP proteins), they can be used safely outside of a BSL-3/4 laboratory. Consequently, minigenome systems have been extensively used to study the life cycle of VHF viruses, particularly as it relates to the processes of transcription and replication.
Filovirus minigenome systems have been used extensively to characterize the transacting factors involved in genome replication and transcription. It was determined that for EBOV the nucleoprotein NP, the RNA-dependent RNA polymerase L and the polymerase cofactor VP35 are necessary and sufficient for genome replication (Mühlberger et al., 1999). For transcription VP30 is also required and serves to overcome a secondary structure in the viral RNA at the beginning of the first gene, that otherwise leads to premature termination of transcription (Weik et al., 2002). Interestingly, for MARV only NP, VP35, and L are necessary and sufficient for genome replication and transcription in MARV minigenome systems (Mühlberger et al., 1998). Other aspects of genome replication and transcription studied using minigenome systems include the role of VP35 oligomerization for genome replication and transcription (Möller et al., 2005), as well as the regulatory function of the matrix proteins VP40 and VP24 in filovirus genome replication and transcription (Hoenen et al., 2010a; Hoenen et al., 2010b; Watanabe et al., 2007). Further, minigenome systems were used to map the filovirus promoters (Enterlein et al., 2009; Weik et al., 2005), and to investigate the influence of the phylogenetic origin of RNP components on transcription and replication and a possible relationship to differences in virulence observed among members of different filovirus species (Boehmann et al., 2005; Groseth et al., 2005).
In the case of arenaviruses, minigenome systems were used to show that the nucleoprotein and the RNA-dependent RNA polymerase L are necessary and sufficient for genome replication and transcription, and that the matrix protein Z regulates these processes (Cornu and de la Torre, 2001; Hass et al., 2004; Lee et al., 2000; Lopez et al., 2001). This regulation of genome replication and transcription by the matrix protein, which seems to be a common feature of negative-stranded RNA viruses, was then further shown to be dependent on the interaction of Z with L (Jacamo et al., 2003; Wilda et al., 2008). Further, arenavirus minigenome systems were used for promoter mapping (Hass et al., 2006; Perez and de la Torre, 2003), and to analyze the role of the intergenic stem-loop region between the two genes on the ambisense arenavirus genome segments in transcription termination (Lopez and Franze-Fernandez, 2007; Pinschewer et al., 2005). Also, the question of whether free nucleoprotein might act as a switch between transcription and genome replication has been addressed using arenavirus minigenome systems, but no evidence supporting such a role was found (Pinschewer et al., 2003). Finally, these systems have been applied to identify a putative cap-snatching domain in L (Lelke et al., 2010).
A number of minigenome systems have also been established for VHF-causing bunyaviruses and their apathogenic relatives (Table 1). Similar to arenaviruses, the nucleoprotein and the polymerase L are necessary and sufficient for genome replication and transcription in these systems. Bunyavirus minigenomes have been extensively used to characterize cis-acting elements involved in genome replication and transcription. Exhaustive studies were undertaken using Bunyamwera virus, whose transcription promoter (Barr et al., 2005; Barr and Wertz, 2005), replication promoter (Kohl et al., 2004a), transcription terminator (Barr et al., 2006) and packaging signals (Kohl et al., 2006) were analyzed in detail. Also, the role of the small non-structural protein NSs was studied using minigenome systems, and it was shown that Bunyamwera NSs inhibits virus RNA synthesis in a minigenome system in mammalian (Weber et al., 2001), but not in mosquito cells (Kohl et al., 2004b). For the phleboviruses RVFV and UUKV, promoter (Flick et al., 2004; Flick et al., 2002; Gauliard et al., 2006) and terminator (Ikegami et al., 2007) mapping studies have been performed, and the influence of NSs on virus genome replication and transcription has been studied. In contrast to Bunyamwera NSs, the RVFV NSs seems to increase rather than inhibit virus RNA synthesis in a plasmid-driven minigenome system (Ikegami et al., 2005); however, this effect was not seen when all components of the minigenome system were expressed by means of recombinant vaccinia viruses (Lopez et al., 1995). Also, this effect was not observed in a plasmid-based UUKV minigenome system (Flick and Pettersson, 2001). Bunyavirus minigenome systems have also been used to study trans-acting factors involved in genome replication and transcription. In particular, the roles of L-oligomerization (Zamoto-Niikura et al., 2009) as well as that of nucleoprotein-nucleoprotein interactions for genome replication and transcription have been investigated (Katz et al., 2010; Leonard et al., 2005).
Despite the extensive use of minigenome systems in order to study replication and transcription, there are still many open questions about these processes. One example is the regulation of replication and transcription, where there still is a significant lack of knowledge; however, the ability to dissect replication and transcription using replication-deficient minigenome systems will help to further our knowledge in this respect. Nevertheless, when using minigenome systems it has to be kept in mind that they do not model other aspects of the viral life cycle, and also cannot be used to study primary transcription, since they necessitate the presence of viral proteins provided in trans.
Dissecting the viral life cycle
To overcome the limitation that minigenome systems cannot be used to model steps of the viral life cycle other than replication and transcription, infectious virus-like particle (iVLP) systems have been developed. The particles in these systems mediate single-cycle infection of target cells; however, they cannot establish a multi-cycle infection and are, therefore, non-infectious in a clinical sense, making it possible to work with these systems outside of a BSL-3/4 environment. In contrast to regular VLPs they contain a RNP complex that is able to undergo transcription and replication. Therefore, it has been proposed that these particles could be better termed transcription and replication competent VLPs (trVLPs), a terminology that is used for the rest of this review. trVLP systems are based on the fact that expression of the virus matrix protein of most negative-stranded RNA viruses in mammalian cells leads to the formation of virus-like particles (VLPs). If VLPs are formed in the presence of minigenome-containing RNP complexes, these RNP complexes are incorporated, and if in addition the virus glycoproteins responsible for entry are present, the RNP complex-containing VLPs are able to infect target cells and to deliver their RNP complexes into these target cells (Figure 3). Once inside target cells, the RNP complexes can then serve as templates for replication and transcription, which in most trVLP systems is driven by RNP proteins already expressed in the target cells prior to infection, either by means of helper virus infection or by transfection of expression plasmids (trVLP systems with pretransfected target cells). While reporter activity in the VLP-producing cells (producer or p0 cells) reflects replication and transcription, reporter activity in the target cells (also called p1 cells) is dependent on replication and transcription in target cells and on the production of trVLPs in producer cells and their subsequent entry into target cells. Thus, these systems mirror most of the steps in the virus life cycle, with one exception: since RNP proteins are already expressed in target cells prior to infection, primary transcription, which by definition is performed only by RNP components brought into target cells within virus particles, is not reflected in this system. However, in a few instances it has been possible to optimize trVLP systems in a way that naïve target cells can also be infected. These systems have proven useful in studying primary transcription (Habjan et al., 2009a; Hoenen et al., 2006; Krähling et al., 2010). The choice of target cells, as well as the use of a highly sensitive reporter, has been critical for establishing trVLP systems with naïve target cells, as reporter signals in trVLP assays with naïve target cells are about 20–100-fold lower than in trVLP assays with pretransfected target cells.
Figure 3. trVLP systems.
EBOV is shown as an example for VHF-causing negative-sense RNA viruses. Initial transcription (A), artificial encapsidation (B), genome replication (C), secondary transcription (D) and translation (E) take place as in a minigenome system (Figure 2) after expression of all the required components in transfected cells (p0 or producer cells). The expression of the other virus structural proteins, particularly the matrix proteins (VP40 and VP24) and glycoprotein (GP1,2) allows the formation of trVLPs (F) containing fully functional RNP complexes. These trVLPs can enter target cells (p1 or target cells) (G), which in most trVLP systems additionally express RNP proteins by means of pre-transfection with expression plasmids or superinfection with helper virus. These additional RNP proteins allow genome replication and secondary transcription to occur, resulting in reporter activity that is dependent on genome replication in both producer and target cells, trVLP production and entry, as well as secondary transcription in target cells. In contrast, in trVLP systems with naïve target cells after entry (G) only primary transcription occurs (H), resulting in reporter activity that is dependent on genome replication in producer cells only, trVLP production and entry, as well as primary transcription in target cells.
trVLP systems for VHF viruses are less commonly used than minigenome systems, but systems are available for filoviruses, arenaviruses, and bunyaviruses. The first plasmid-based trVLP system established for a VHF-causing virus was a trVLP system with pretransfected target cells for EBOV, which was used to study the role of VP24 in morphogenesis and budding (Watanabe et al., 2004). This system has since been advanced into an trVLP system with naïve target cells, which was used to show that VP24 is essential for primary transcription and/or the formation of RNP complexes capable of supporting primary transcription (Hoenen et al., 2006). Since then, similar systems have also been developed for MARV (Krähling et al., 2010; Wenigenrath et al., 2010). Currently, only a chimeric trVLP system exists for arenaviruses, which uses TCRV RNPs together with JUNV Z and GPC. This system was used to study the requirements for recruitment of the nucleoprotein and GPC into virions (Casabona et al., 2009). In contrast, a number of trVLP systems exist for bunyaviruses. The first of these systems was an trVLP system with pretransfected target cells for Bunyamwera virus and was used to investigate the role of the non-structural protein NSm in virus assembly and morphogenesis (Shi et al., 2006). However, the majority of the work with trVLP systems has been done for phleboviruses. For the human apathogenic UUKV, an trVLP system with pretransfected target cells exists (Overby et al., 2006) that has been used for detailed studies of the contributions of the glycoproteins to RNP packaging and budding (Overby et al., 2007a; Overby et al., 2007b). For RVFV a trVLP system with naïve target cells has been developed and revealed that the cellular MxA protein can impair both primary and secondary transcription of RVFV (Habjan et al., 2009a).
trVLP systems model a complete single infectious cycle, and thus have a tremendous potential to further our understanding of fundamental aspects of the virus life cycle. However, since they do not model multiple infectious cycles, and can only be used in tissue culture, it is very difficult to use these systems for studies addressing pathogenesis and interactions with host organisms.
Differences between reverse-genetics-based modeling systems and the viral life cycle
Reverse genetics systems have been extremely valuable in elucidating the life cycles of VHF viruses, but they do not reflect the replication cycles perfectly in every detail. A key difference, for example, is that minigenome and rcVLP systems require plasmid-derived expression of virus proteins and/or minigenomes. Other differences include the need for artificial encapsidation of naked vRNA and the constant expression of virus proteins rather than the temporally organized expression that occurs in the virus life cycle. Finally, particularly in the case of filoviruses, minigenomes are considerably shorter than full-length genomes. Whereas in most instances these differences appear to be of little consequence, they have to be kept in mind when interpreting data obtained in minigenome and trVLP systems. They can also help to explain the differences that are sometimes observed when comparing findings from minigenome and trVLP systems to results from experiments conducted with infectious virus. A salient example for this is the phenomenon that MARV VP30 is not required for replication and transcription in a minigenome system (Mühlberger et al., 1998), but has to be provided in trans for rescue of recombinant MARV virus (Enterlein et al., 2006). During rescue of infectious virus VP30 is expressed from the viral genome as soon as secondary transcription occurs, thus suggesting that in the context of a full length genome VP30 is required for genome replication, secondary transcription, or the processes of initial transcription and artificial encapsidation, all of which are also present in a minigenome system. Thus, the only difference between a full-length clone system at these early stages and the minigenome system are the respective lengths of the full-length genome and the minigenome, leading to the conclusion that VP30 might act as an elongation factor during replication or transcription. Alternatively, it might also act as a structural component that is required for the formation of RNP complexes containing a full-length genome, but not a minigenome (Enterlein et al., 2006).
In most current minigenome and trVLP systems virus proteins are expressed from plasmids, requiring transcription by the cellular RNA polymerase II. The minigenome is either transcribed by endogenous RNA polymerases, or by T7 RNA polymerase, which in most cases is itself either transcribed from an expression plasmid or the cell genome (in the case of stable expression cell lines) by RNA polymerase II. Thus effects of transfected virus proteins on cellular RNA polymerases, which would not directly affect virus genome replication and transcription during an infection with infectious virus, will affect the expression of the minigenome system components and, therefore, reporter activity. This could lead to the misinterpretation that these virus proteins directly affect virus genome replication and/or transcription, which is not necessarily the case. Somewhat alleviating this problem is the common practice of using a control plasmid that encodes a second reporter under the control of a eukaryotic promoter. If no changes are observed in expression of this control reporter in the presence of the virus protein in question, any differences observed in minigenome reporter activity are then most likely due to a direct effect on genome replication and/or transcription. However, if an influence of the virus protein in question on control reporter activity is observed, the interpretation of any differences observed in minigenome reporter activity becomes problematic, particularly since the correlation between levels of plasmid-derived protein expression (i.e. expression of minigenome components as well as the control reporter) and the resulting effects on minigenome reporter activity has been shown to be non-linear (Figure 4) (Hoenen et al., 2010b). In a study using a filovirus minigenome system it was observed, for example, that reduction of plasmid-derived protein expression by 50% through the use of RNA polymerase II-specific inhibitors (actinomycin D or α-amanitin) reduced minigenome reporter activity by only about 20% (Figure 4) (Hoenen et al., 2010b).
Figure 4. Influence of plasmid-derived expression levels on minigenome-derived reporter activity.
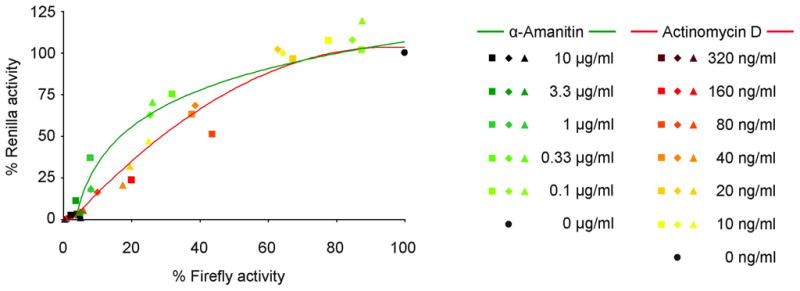
Filovirus minigenome assays were performed in presence or absence of RNA polymerase II inhibitors (α-amanitin and actinomycin D) at varying concentrations. The minigenome was based on EBOV and encoded renilla luciferase, whereas the control reporter for the level of plasmid-derived expression encoded firefly luciferase. Plotted are the renilla luciferase activity (minigenome-derived reporter activity) measurements as a function of firefly luciferase activity (plasmid derived gene expression), with both renilla and firefly luciferase activity in absence of inhibitors set to 100%. Shown are results from three independent experiments. Figure modified from (Hoenen et al., 2010b) with permission.
Indeed, an effect of virus proteins on plasmid-derived protein expression has been shown for a number of viruses, emphasizing the importance of such considerations. For example, EBOV VP40 impairs plasmid-derived protein expression by an as of yet unknown mechanism (Hoenen et al., 2010b), whereas EBOV VP35 increases it by inhibition of PKR (Schümann et al., 2009). In the case of RVFV, the small non-structural protein NSs leads to a specific degradation of PKR (Habjan et al., 2009b), which in its activated form is responsible for translational arrest of cellular and virus mRNAs (Garcia et al., 2006). NSs also inhibits cellular transcription by targeting the cellular TFIIH transcription factor (Le May et al., 2004). Interestingly, Bunyamwera virus NSs has no effect on PKR (Streitenfeld et al., 2003), but still inhibits host cell transcription (Leonard et al., 2006). It is currently unclear whether this difference in PKR antagonism is responsible for the observation that RVFV NSs increases minigenome reporter activity (Ikegami et al., 2005), whereas Bunyamwera NSs does not do so (Weber et al., 2001), or whether RVFV NSs really has a direct influence on genome replication and/or transcription.
Following the expression of virus proteins and naked minigenomes, artificial encapsidation has to occur to reconstitute a viable RNP complex that can serve as a template for further transcription and replication. This step is particular to reverse genetics systems and has no equivalent in the virus life cycle. Further, it is generally thought to be a very inefficient process (Conzelmann, 2004), which has been experimentally demonstrated for respiratory syncytial virus, vesicular stomatitis Indiana virus and EBOV minigenomes (Hoenen et al., 2010b; Pattnaik et al., 1992; Peeples and Collins, 2000). Based on comparisons of vRNA levels and reporter activity in replication-deficient and replication-competent EBOV minigenome systems it can be estimated that only about 3% of the initially transcribed EBOV-minigenomes are able to form a suitable encapsidated template for mRNA production (T. Hoenen, unpublished data). The need for artificial encapsidation can cause two problems. First, naked vRNA can form double-stranded RNA structures usually obscured during virus infection due to encapsidation by the nucleoprotein. These double-stranded RNA structures may then activate PKR (Garcia et al., 2006), thus leading to unwanted downstream effects not necessarily seen in virus infection. Second, expression levels of NP not only influence replication and transcription, but most likely also artificial encapsidation, making it very problematic to titrate the amounts of nucleoprotein in a minigenome system to study its influence on replication and transcription. This is of particular importance since the amount of nucleoprotein has long been suggested to be a deciding factor in regulating the switch from transcription to genome replication (Pinschewer et al., 2003). However, until now there is no experimental proof for this concept. The availability of trVLP systems for several viruses now provides an opportunity to avoid this problem, since it is possible to study replication and transcription in target cells, and thus to provide minigenomes in the form of fully encapsidated RNP-complexes within trVLPs and eliminating the need for artificial encapsidation.
trVLP systems also made it possible to study parts of the virus life cycle other than replication and transcription. Particularly, they have been used to study morphogenesis and budding. However, one fundamental problem with trVLPs is the ratio of infectious trVLPs to non-infectious VLPs, which are always produced as well. In the case of EBOV, infectious trVLP titers only reach 5×102 infectious particles per ml, despite the fact that very high amounts of VLPs are produced (Watanabe et al., 2004). In contrast, an infection with EBOV usually yields titers of 1×106 pfu/ml or more (Calain et al., 1999). Morphologically it can be observed for the related MARV that, in addition to short filamentous trVLPs, which are highly infectious, long filamentous as well as spherical VLPs are also produced and that these particles have minimal infectivity (Wenigenrath et al., 2010). Also, when examined by electron microscopy, the vast majority of particles in filovirus trVLP preparations do not contain RNP-complexes (L. Kolesnikova, personal communication)(Wenigenrath et al., 2010). Therefore, morphological and biochemical studies of trVLPs face the inherent problem that only a small percentage of the analyzed material is actually infectious. The reason for this low efficiency of filovirus trVLP systems is currently unknown. Interestingly, RVFV trVLP systems produce up to 1×106 infectious particles per ml, which is close to the titer reached during infection with wild-type virus (Habjan et al., 2009a). One obvious difference to filoviruses is that bunyaviruses do not encode a classical matrix protein, as budding is driven by their glycoproteins (Overby et al., 2006; Overby et al., 2007b). In contrast, filoviruses encode two matrix proteins, both of which are strong inhibitors of replication and transcription (Hoenen et al., 2010b). Thus it is plausible that their constant expression in a trVLP system, while required for morphogenesis and budding, might be detrimental to the production of infectious particles due to these effects on minigenome replication. Timed expression of filovirus matrix proteins in trVLP systems is an obvious approach to address this issue, and may help in future to improve the ratio of infectious to non-infectious particles that can be achieved in trVLP preparations.
APPLICATIONS FOR DRUG SCREENING/DEVELOPMENT OF ANTIVIRAL APPROACHES
Reverse genetics systems have a great potential for the development of antivirals targeting VHF-causing viruses. Full-length clone systems allow the generation of recombinant viruses expressing reporter proteins such as GFP, which makes their detection very easy and should allow the use of these viruses in high-throughput screenings of drug libraries. Such GFP-expressing viruses have been developed for a number of VHF-causing viruses. The first of these viruses were recombinant EBOVs expressing a GFP from additional open reading frames either located between the genes encoding NP and VP35 or VP24 and L (Ebihara et al., 2007; Towner et al., 2005). Infection with these viruses led to an easily detectable GFP-signal in infected cells. The viruses were not attenuated in VeroE6 cells, although attenuation in vivo was observed.
For arenaviruses, trisegmented Lymphocytic Choriomeningitis Viruses expressing GFP have been developed, and these viruses are not attenuated in tissue culture, although again they were attenuated in vivo (Emonet et al., 2009). To generate these trisegmented viruses, two recombinant S segments were generated, in each of which one of the virus genes was replaced by a reporter gene, thus separating the virus genes encoded by the S segment onto two segments. Since both virus genes on the arenavirus S segment are absolutely essential for the life cycle, the trisegmented virus was forced to retain both recombinant S segments. A similar trisegmented virus has not yet been reported for VHF-causing arenaviruses, but the principal strategy should be applicable to them as well.
GFP-expressing viruses are also available for bunyaviruses; however, in these cases the non-structural protein NSs was replaced with GFP, and the resulting viruses were highly attenuated both in tissue culture and in vivo (Billecocq et al., 2008; Bird et al., 2008). One future challenge will be to generate GFP-expressing bunyaviruses that show little or no attenuation in tissue culture to be reasonable surrogates for infection with wild-type viruses. Due to their coding strategy it might be difficult to generate a bunyavirus stably packaging an additional segment, as was successful in the case of trisegmented arenaviruses. Therefore, an easier approach might be the expression of GFP fused to a virus protein. While this has not yet been achieved with VHF-causing viruses, the successful generation of recombinant paramyxoviruses expressing GFP fused to the virus polymerase shows the general feasibility of this approach (Chambers and Takimoto, 2010; Duprex et al., 2002). Despite the availability of GFP-expressing VHF viruses and their potential for high-throughput screening there are currently no reports of this application being successfully exploited. However, a as a proof-of-principle it was shown for the trisegmented LCMV that its growth (and reporter activity) is inhibited by compounds known to interfere with arenavirus infection (Emonet et al., 2009).
While GFP-expressing viruses show great potential benefit for screening antivirals, they still have to be handled under BSL-3/4 conditions. In contrast, minigenome systems and trVLP systems make it possible to model virus genome replication and transcription, particle production, and entry of these particles into target cells in a BSL-1/2 environment, and allow screening for compounds interfering with these steps. Since these systems use reporter genes as a readout, they are very well suited for the development of high throughput assays provided reporters are chosen that can be easily detected. However, despite their great potential, studies using minigenome or trVLP systems for screening purposes are rare. In the case of filoviruses, siRNAs directed against different targets in the coding and noncoding regions of the genome have been screened for their biological activity using a minigenome assay (Groseth et al., 2007). The most efficacious siRNA was then chosen for further characterization, and was validated in an in vitro assay with infectious EBOV, reducing titers to the same extent as other siRNAs that had been previously shown to be protective in guinea pigs. Similarly, siRNAs directed against the conserved LASV genome termini were first evaluated in a minigenome system, before the inhibitory function of the most active ones was confirmed with infectious LASV and related arenaviruses in tissue culture (Müller and Günther, 2007). However, neither of these approaches has been further validated for therapeutic use in animal models or applied in a wider context. Nevertheless, these studies demonstrate that minigenomes can be used to screen for compounds inhibiting virus genome replication and transcription, and that findings from such studies may be used to predict the effectiveness of compounds in an infection with live virus.
Also, the principal usefulness of several other minigenome systems for VHF-causing viruses with respect to antiviral screening has been shown by confirming the activity of substances already known to show antiviral properties during infection in the context of a minigenome assay. For instance, ribavirin, a known inhibitor of Crimean-Congo hemorrhagic fever virus (CCHFV) infection in vitro (Watts et al., 1989) that is also used for clinical treatment of patients, although there is some controversy as to its efficacy (Keshtkar-Jahromi et al., 2011; Soares-Weiser et al., 2010), was shown to inhibit CCHFV minigenome replication at a physiologically relevant concentration (Bergeron et al., 2010). Similar experiments were also performed for LASV, which in addition to ribavirin was also inhibited by interferon alpha (Hass et al., 2004). Finally, the zinc finger antiviral protein, which is a potent inhibitor of filovirus infection in tissue culture, reduced reporter activity in a minigenome system, most likely through the degradation of L mRNAs (Müller et al., 2007).
The only experiments regarding antiviral efficacy performed with trVLPs until now are proof-of-principle experiments. For instance, in the case of MARV a trVLP system was used to determine the neutralizing titer of an antiserum, and this titer matched the titer determined using infectious virus (Wenigenrath et al., 2010). An effect of neutralizing antibodies can also be seen against UUKV trVLPs (Overby et al., 2006), RVFV trVLPs (Habjan et al., 2009a), and EBOV trVLPs (Watanabe et al., 2004). Nevertheless, trVLP systems are the most comprehensive model systems for the virus life cycle and since they allow modeling of almost all of its aspects outside of a BSL-3/4 laboratory, their exploitation during future drug discovery efforts is highly promising.
BIOSAFETY CONCERNS AND REVERSE GENETICS SYSTEMS
One sometimes misunderstood aspect of using reverse genetics systems relates to the biosafety concerns associated with them. This problem is augmented by the fact that these systems have sometimes misleading names, which can generate unjustified concerns.
Full-length clone systems are sometimes called infectious clone systems; however, in contrast to positive sense RNA viruses the cDNA clones used in these systems are not infectious by themselves, and only produce infectious viruses under a very specific set of circumstances. To achieve this, it is necessary to express the full-length RNA genome encoded by these plasmids in mammalian cells, which in the case of T7-driven full-length clone systems requires coexpression of the non-endogenous T7 RNA polymerase in these cells. Also, the viral polymerase and the nucleoprotein, and for some viruses other viral proteins, have to be provided in trans, usually from expression plasmids. Only a combination of all these components can lead to the production of infectious virus, which then has to be handled under BSL-3/4 conditions. Also, at least for filoviruses it has been shown that the ratio of the individual RNP components is crucial for efficient minigenome replication and transcription (Mühlberger et al., 1998; Mühlberger et al., 1999), and most likely this ratio is also critical for efficient rescue of viruses from cDNA clones. Therefore, it is safe and justified to handle full-length cDNA clones alone under BLS-1/2 conditions, depending on local biosafety regulations. However, it is a reasonable precautionary measure to physically separate work with cDNA clones from work with the other components required for rescue of virus, such as work with viral protein expression plasmids and/or mammalian tissue culture, e.g. by utilizing a separate room for all cloning work with full-length cDNA clones.
Maybe even more problematic is the name infectious VLP system. While iVLPs are able to infect target cells in the sense that they enter these cells and deliver their RNP complex into them, they do not cause an infection in organisms, since they are non-replicating, so that in the latter sense they are actually non-infectious. As an alternative and in order to solve this dilemma the term transcription and replication competent VLPs (trVLPs) has been proposed. This term clearly describes the main functional difference of iVLPs as compared to regular VLPs, i.e. their ability to undergo genome transcription and replication, while avoiding the ambiguity of the word infectious. Regardless of terminology, from a biosafety standpoint iVLP/trVLP systems can be handled at the same biosafety level as minigenome systems and regular VLP systems, which is depending on the regulations of the country these studies are performed in either BSL-1 or BSL-2.
CONCLUSION AND OUTLOOK
Reverse genetics systems have contributed considerably to our understanding of VHF virus biology and pathogenesis. Full-length clone systems allow for the generation of recombinant viruses, whereas life cycle modeling systems such as minigenomes and trVLPs enable one to study the replication cycle of VHF viruses outside of BSL-3/4 laboratories. This makes these systems perfectly suited for application as screening tools for novel antivirals; however, this potential has so far not been realized to any great extent. The development of GFP-expressing VHF viruses will allow for screening of novel antivirals under BSL-3/4 conditions and/or confirmation of antiviral efficacy. One future challenge in this direction will be the generation of viruses that show as little attenuation as possible when compared to wild-type viruses. Whereas this has already been achieved for filoviruses and arenaviruses in vitro, for bunyaviruses this still remains to be achieved, and for none of these families recombinant viruses currently exist which express fluorescent proteins but show no attenuation in vivo.
Additional future challenges will lie in modifying the reverse genetics based life cycle modeling systems available so that they resemble the viral replication cycle even more closely, without losing their advantage, which is that they can be used outside of a BSL-3/4 laboratory, and in optimizing them for use in high throughput assays. However, perhaps the most important challenge for the future is to increase awareness of these systems, and to bring experts on the development of antivirals and experts on reverse genetics systems together to fully exploit the potential of these systems.
Acknowledgments
The authors thank Friedemann Weber, Hideki Ebihara, Heinz Feldmann, Peter Jahrling and Stephan Becker for their ideas and discussion. This research was supported in part by the Intramural Research Program of the NIH, NIAID, as well as by fellowships from the Schering Foundation (TH) and the Canadian Institutes of Health Research (AG).
Footnotes
Disclaimer: The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services or of the institutions and companies affiliated with the authors. JHK and VW-J performed this work as an employee of Tunnell Consulting, Inc., a subcontractor to Battelle Memorial Institute under its prime contract with NIAID, under Contract No. HHSN272200200016I
References
- Albarino CG, Bergeron E, Erickson BR, Khristova ML, Rollin PE, Nichol ST. Efficient reverse genetics generation of infectious junin viruses differing in glycoprotein processing. J Virol. 2009;83:5606–5614. doi: 10.1128/JVI.00276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarino CG, Bird BH, Chakrabarti AK, Dodd KA, White DM, Bergeron E, Shrivastava-Ranjan P, Nichol ST. Reverse genetics generation of chimeric infectious Junin/Lassa virus is dependent on interaction of homologous glycoprotein stable signal peptide and G2 cytoplasmic domains. J Virol. 2010 doi: 10.1128/JVI.01837-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JN, Rodgers JW, Wertz GW. The Bunyamwera virus mRNA transcription signal resides within both the 3′ and the 5′ terminal regions and allows ambisense transcription from a model RNA segment. J Virol. 2005;79:12602–12607. doi: 10.1128/JVI.79.19.12602-12607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JN, Rodgers JW, Wertz GW. Identification of the Bunyamwera bunyavirus transcription termination signal. J Gen Virol. 2006;87:189–198. doi: 10.1099/vir.0.81355-0. [DOI] [PubMed] [Google Scholar]
- Barr JN, Wertz GW. Role of the conserved nucleotide mismatch within 3′- and 5′-terminal regions of Bunyamwera virus in signaling transcription. J Virol. 2005;79:3586–3594. doi: 10.1128/JVI.79.6.3586-3594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron E, Albarino CG, Khristova ML, Nichol ST. Crimean-Congo hemorrhagic fever virus-encoded ovarian tumor protease activity is dispensable for virus RNA polymerase function. J Virol. 2010;84:216–226. doi: 10.1128/JVI.01859-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billecocq A, Gauliard N, Le May N, Elliott RM, Flick R, Bouloy M. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology. 2008;378:377–384. doi: 10.1016/j.virol.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird BH, Albarino CG, Hartman AL, Erickson BR, Ksiazek TG, Nichol ST. Rift valley fever virus lacking the NSs and NSm genes is highly attenuated, confers protective immunity from virulent virus challenge, and allows for differential identification of infected and vaccinated animals. J Virol. 2008;82:2681–2691. doi: 10.1128/JVI.02501-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmann Y, Enterlein S, Randolf A, Mühlberger E. A reconstituted replication and transcription system for Ebola virus Reston and comparison with Ebola virus Zaire. Virology. 2005;332:406–417. doi: 10.1016/j.virol.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Bridgen A, Elliott RM. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc Natl Acad Sci U S A. 1996;93:15400–15404. doi: 10.1073/pnas.93.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier MJ, de la Torre JC, Peters CJ. Arenaviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott, Williams & Wilkins; Philadelphia: 2007. pp. 1791–1828. [Google Scholar]
- Calain P, Monroe MC, Nichol ST. Ebola virus defective interfering particles and persistent infection. Virology. 1999;262:114–128. doi: 10.1006/viro.1999.9915. [DOI] [PubMed] [Google Scholar]
- Carroll AR, Wagner RR. Role of the membrane (M) protein in endogenous inhibition of in vitro transcription by vesicular stomatitis virus. J Virol. 1979;29:134–142. doi: 10.1128/jvi.29.1.134-142.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casabona JC, Levingston Macleod JM, Loureiro ME, Gomez GA, Lopez N. The RING domain and the L79 residue of Z protein are involved in both the rescue of nucleocapsids and the incorporation of glycoproteins into infectious chimeric arenavirus-like particles. J Virol. 2009;83:7029–7039. doi: 10.1128/JVI.00329-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers R, Takimoto T. Trafficking of Sendai virus nucleocapsids is mediated by intracellular vesicles. PLoS One. 2010;5:e10994. doi: 10.1371/journal.pone.0010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton GM, Little SP, Hagen FS, Huang AS. The matrix (M) protein of vesicular stomatitis virus regulates transcription. Cell. 1978;15:1455–1462. doi: 10.1016/0092-8674(78)90069-7. [DOI] [PubMed] [Google Scholar]
- Conzelmann KK. Reverse genetics of mononegavirales. Curr Top Microbiol Immunol. 2004;283:1–41. doi: 10.1007/978-3-662-06099-5_1. [DOI] [PubMed] [Google Scholar]
- Cornu TI, de la Torre JC. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J Virol. 2001;75:9415–9426. doi: 10.1128/JVI.75.19.9415-9426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Spronken MI, Vervaet G, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. A reverse-genetics system for Influenza A virus using T7 RNA polymerase. J Gen Virol. 2007;88:1281–1287. doi: 10.1099/vir.0.82452-0. [DOI] [PubMed] [Google Scholar]
- Dunn EF, Pritlove DC, Jin H, Elliott RM. Transcription of a recombinant bunyavirus RNA template by transiently expressed bunyavirus proteins. Virology. 1995;211:133–143. doi: 10.1006/viro.1995.1386. [DOI] [PubMed] [Google Scholar]
- Duprex WP, Collins FM, Rima BK. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J Virol. 2002;76:7322–7328. doi: 10.1128/JVI.76.14.7322-7328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin AP, Hall SL, Siew JW, Whitehead SS, Collins PL, Murphy BR. Recovery of infectious human parainfluenza virus type 3 from cDNA. Virology. 1997;235:323–332. doi: 10.1006/viro.1997.8697. [DOI] [PubMed] [Google Scholar]
- Ebihara H, Groseth A, Neumann G, Kawaoka Y, Feldmann H. The role of reverse genetics systems in studying viral hemorrhagic fevers. Thromb Haemost. 2005;94:240–253. doi: 10.1160/TH05-05-0335. [DOI] [PubMed] [Google Scholar]
- Ebihara H, Theriault S, Neumann G, Alimonti JB, Geisbert JB, Hensley LE, Groseth A, Jones SM, Geisbert TW, Kawaoka Y, Feldmann H. In vitro and in vivo characterization of recombinant Ebola viruses expressing enhanced green fluorescent protein. J Infect Dis. 2007;196(Suppl 2):S313–322. doi: 10.1086/520590. [DOI] [PubMed] [Google Scholar]
- Emonet SF, Garidou L, McGavern DB, de la Torre JC. Generation of recombinant lymphocytic choriomeningitis viruses with trisegmented genomes stably expressing two additional genes of interest. Proc Natl Acad Sci U S A. 2009;106:3473–3478. doi: 10.1073/pnas.0900088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enterlein S, Schmidt KM, Schümann M, Conrad D, Krähling V, Olejnik J, Mühlberger E. The marburg virus 3′ noncoding region structurally and functionally differs from that of ebola virus. J Virol. 2009;83:4508–4519. doi: 10.1128/JVI.02429-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enterlein S, Volchkov V, Weik M, Kolesnikova L, Volchkova V, Klenk HD, Mühlberger E. Rescue of recombinant Marburg virus from cDNA is dependent on nucleocapsid protein VP30. J Virol. 2006;80:1038–1043. doi: 10.1128/JVI.80.2.1038-1043.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke S, Conzelmann KK. Dissociation of rabies virus matrix protein functions in regulation of viral RNA synthesis and virus assembly. J Virol. 2003;77:12074–12082. doi: 10.1128/JVI.77.22.12074-12082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatz L, Bergthaler A, de la Torre JC, Pinschewer DD. Recovery of an arenavirus entirely from RNA polymerase I/II-driven cDNA. Proc Natl Acad Sci U S A. 2006;103:4663–4668. doi: 10.1073/pnas.0600652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick K, Hooper JW, Schmaljohn CS, Pettersson RF, Feldmann H, Flick R. Rescue of Hantaan virus minigenomes. Virology. 2003a;306:219–224. doi: 10.1016/s0042-6822(02)00070-3. [DOI] [PubMed] [Google Scholar]
- Flick K, Katz A, Overby A, Feldmann H, Pettersson RF, Flick R. Functional analysis of the noncoding regions of the Uukuniemi virus (Bunyaviridae) RNA segments. J Virol. 2004;78:11726–11738. doi: 10.1128/JVI.78.21.11726-11738.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick R, Bouloy M. Rift Valley fever virus. Curr Mol Med. 2005;5:827–834. doi: 10.2174/156652405774962263. [DOI] [PubMed] [Google Scholar]
- Flick R, Elgh F, Pettersson RF. Mutational analysis of the Uukuniemi virus (Bunyaviridae family) promoter reveals two elements of functional importance. J Virol. 2002;76:10849–10860. doi: 10.1128/JVI.76.21.10849-10860.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick R, Flick K, Feldmann H, Elgh F. Reverse genetics for crimean-congo hemorrhagic fever virus. J Virol. 2003b;77:5997–6006. doi: 10.1128/JVI.77.10.5997-6006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick R, Pettersson RF. Reverse genetics system for Uukuniemi virus (Bunyaviridae): RNA polymerase I-catalyzed expression of chimeric viral RNAs. J Virol. 2001;75:1643–1655. doi: 10.1128/JVI.75.4.1643-1655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher WR. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell. 1996;85:477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauliard N, Billecocq A, Flick R, Bouloy M. Rift Valley fever virus noncoding regions of L, M and S segments regulate RNA synthesis. Virology. 2006;351:170–179. doi: 10.1016/j.virol.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Gerrard SR, Bird BH, Albarino CG, Nichol ST. The NSm proteins of Rift Valley fever virus are dispensable for maturation, replication and infection. Virology. 2007;359:459–465. doi: 10.1016/j.virol.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A, Feldmann H, Theriault S, Mehmetoglu G, Flick R. RNA polymerase I-driven minigenome system for ebola viruses. J Virol. 2005;79:4425–4433. doi: 10.1128/JVI.79.7.4425-4433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A, Hoenen T, Alimonti JB, Zielecki F, Ebihara H, Theriault S, Ströher U, Becker S, Feldmann H. In vitro evaluation of antisense RNA efficacy against filovirus infection, by use of reverse genetics. J Infect Dis. 2007;196(Suppl 2):S382–389. doi: 10.1086/520604. [DOI] [PubMed] [Google Scholar]
- Habjan M, Penski N, Spiegel M, Weber F. T7 RNA polymerase-dependent and -independent systems for cDNA-based rescue of Rift Valley fever virus. J Gen Virol. 2008;89:2157–2166. doi: 10.1099/vir.0.2008/002097-0. [DOI] [PubMed] [Google Scholar]
- Habjan M, Penski N, Wagner V, Spiegel M, Overby AK, Kochs G, Huiskonen JT, Weber F. Efficient production of Rift Valley fever virus-like particles: The antiviral protein MxA can inhibit primary transcription of bunyaviruses. Virology. 2009a;385:400–408. doi: 10.1016/j.virol.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J Virol. 2009b;83:4365–4375. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P, Kim JH, Ebihara H, Noda T, Neumann G, Feldmann H, Kawaoka Y. Generation of biologically contained Ebola viruses. Proc Natl Acad Sci U S A. 2008;105:1129–1133. doi: 10.1073/pnas.0708057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Boshra H, Sunyer JO, Zwiers SH, Paragas J, Harty RN. Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J Virol. 2003;77:1793–1800. doi: 10.1128/JVI.77.3.1793-1800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlieb B, Weissenhorn W. Filovirus assembly and budding. Virology. 2006;344:64–70. doi: 10.1016/j.virol.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Hass M, Gölnitz U, Müller S, Becker-Ziaja B, Günther S. Replicon system for Lassa virus. J Virol. 2004;78:13793–13803. doi: 10.1128/JVI.78.24.13793-13803.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass M, Westerkofsky M, Müller S, Becker-Ziaja B, Busch C, Günther S. Mutational analysis of the lassa virus promoter. J Virol. 2006;80:12414–12419. doi: 10.1128/JVI.01374-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Biedenkopf N, Zielecki F, Jung S, Groseth A, Feldmann H, Becker S. Oligomerization of Ebola virus VP40 is essential for particle morphogenesis and regulation of viral transcription. J Virol. 2010a;84:7053–7063. doi: 10.1128/JVI.00737-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Groseth A, Kolesnikova L, Theriault S, Ebihara H, Hartlieb B, Bamberg S, Ströher U, Feldmann H, Becker S. Infection of naive target cells with virus-like partices - Implications for the function of Ebola virus VP24. J Virol. 2006;80:7260–7264. doi: 10.1128/JVI.00051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Jung S, Herwig A, Groseth A, Becker S. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology. 2010b;403:56–66. doi: 10.1016/j.virol.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Hoenen T, Kolesnikova L, Becker S. Recent advances in filovirus- and arenavirus-like particles. Future Virology. 2007;2:193–203. [Google Scholar]
- Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. Dual functions of Rift Valley fever virus NSs protein: inhibition of host mRNA transcription and post-transcriptional downregulation of protein kinase PKR. Ann N Y Acad Sci. 2009a;1171(Suppl 1):E75–85. doi: 10.1111/j.1749-6632.2009.05054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 2009b;5:e1000287. doi: 10.1371/journal.ppat.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Peters CJ, Makino S. Rift valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J Virol. 2005;79:5606–5615. doi: 10.1128/JVI.79.9.5606-5615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Won S, Peters CJ, Makino S. Rescue of infectious rift valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J Virol. 2006;80:2933–2940. doi: 10.1128/JVI.80.6.2933-2940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Won S, Peters CJ, Makino S. Characterization of Rift Valley fever virus transcriptional terminations. J Virol. 2007;81:8421–8438. doi: 10.1128/JVI.02641-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki M, Takeda M, Shirogane Y, Nakatsu Y, Nakamura T, Yanagi Y. The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J Virol. 2009;83:10374–10383. doi: 10.1128/JVI.01056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacamo R, Lopez N, Wilda M, Franze-Fernandez MT. Tacaribe virus Z protein interacts with the L polymerase protein to inhibit viral RNA synthesis. J Virol. 2003;77:10383–10393. doi: 10.1128/JVI.77.19.10383-10393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasenosky LD, Neumann G, Lukashevich I, Kawaoka Y. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J Virol. 2001;75:5205–5214. doi: 10.1128/JVI.75.11.5205-5214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Sakai Y, Shioda T, Kondo T, Nakanishi M, Nagai Y. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells. 1996;1:569–579. doi: 10.1046/j.1365-2443.1996.d01-261.x. [DOI] [PubMed] [Google Scholar]
- Katz A, Freiberg AN, Backström V, Schulz AR, Mateos A, Holm L, Pettersson RF, Vaheri A, Flick R, Plyusnin A. Oligomerization of Uukuniemi virus nucleocapsid protein. Virol J. 2010;7:187. doi: 10.1186/1743-422X-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshtkar-Jahromi M, Kuhn JH, Christova I, Bradfute SB, Jahrling PB, Bavari S. Crimean-Congo hemorrhagic fever: Current and future prospects of vaccines and therapies. Antiviral Res. 2011 doi: 10.1016/j.antiviral.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Kohl A, Dunn EF, Lowen AC, Elliott RM. Complementarity, sequence and structural elements within the 3′ and 5′ non-coding regions of the Bunyamwera orthobunyavirus S segment determine promoter strength. J Gen Virol. 2004a;85:3269–3278. doi: 10.1099/vir.0.80407-0. [DOI] [PubMed] [Google Scholar]
- Kohl A, Hart TJ, Noonan C, Royall E, Roberts LO, Elliott RM. A bunyamwera virus minireplicon system in mosquito cells. J Virol. 2004b;78:5679–5685. doi: 10.1128/JVI.78.11.5679-5685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl A, Lowen AC, Leonard VH, Elliott RM. Genetic elements regulating packaging of the Bunyamwera orthobunyavirus genome. J Gen Virol. 2006;87:177–187. doi: 10.1099/vir.0.81227-0. [DOI] [PubMed] [Google Scholar]
- Krähling V, Dolnik O, Kolesnikova L, Schmidt-Chanasit J, Jordan I, Sandig V, Günther S, Becker S. Establishment of Fruit Bat Cells (Rousettus aegyptiacus) as a Model System for the Investigation of Filoviral Infection. PLoS Negl Trop Dis. 2010;4 doi: 10.1371/journal.pntd.0000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Schenk AD, Rahmeh AA, Radoshitzky SR, Bavari S, Walz T, Whelan SP. Assembly of a functional Machupo virus polymerase complex. Proc Natl Acad Sci U S A. 2010;107:20069–20074. doi: 10.1073/pnas.1007152107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JH, Radoshitzky SR, Guth AC, Warfield KL, Li W, Vincent MJ, Towner JS, Nichol ST, Bavari S, Choe H, Aman MJ, Farzan M. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J Biol Chem. 2006;281:15951–15958. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- Le May N, Dubaele S, Proietti De Santis L, Billecocq A, Bouloy M, Egly JM. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell. 2004;116:541–550. doi: 10.1016/s0092-8674(04)00132-1. [DOI] [PubMed] [Google Scholar]
- Lee KJ, Novella IS, Teng MN, Oldstone MB, de La Torre JC. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol. 2000;74:3470–3477. doi: 10.1128/jvi.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelke M, Brunotte L, Busch C, Günther S. An N-terminal region of Lassa virus L protein plays a critical role in transcription but not replication of the virus genome. J Virol. 2010;84:1934–1944. doi: 10.1128/JVI.01657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard VH, Kohl A, Hart TJ, Elliott RM. Interaction of Bunyamwera Orthobunyavirus NSs protein with mediator protein MED8: a mechanism for inhibiting the interferon response. J Virol. 2006;80:9667–9675. doi: 10.1128/JVI.00822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard VH, Kohl A, Osborne JC, McLees A, Elliott RM. Homotypic interaction of Bunyamwera virus nucleocapsid protein. J Virol. 2005;79:13166–13172. doi: 10.1128/JVI.79.20.13166-13172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol. 2003;77:1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez N, Franze-Fernandez MT. A single stem-loop structure in Tacaribe arenavirus intergenic region is essential for transcription termination but is not required for a correct initiation of transcription and replication. Virus Res. 2007;124:237–244. doi: 10.1016/j.virusres.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Lopez N, Jacamo R, Franze-Fernandez MT. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J Virol. 2001;75:12241–12251. doi: 10.1128/JVI.75.24.12241-12251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez N, Müller R, Prehaud C, Bouloy M. The L protein of Rift Valley fever virus can rescue viral ribonucleoproteins and transcribe synthetic genome-like RNA molecules. J Virol. 1995;69:3972–3979. doi: 10.1128/jvi.69.7.3972-3979.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Staeheli P, Schneider U. RNA polymerase II-controlled expression of antigenomic RNA enhances the rescue efficacies of two different members of the Mononegavirales independently of the site of viral genome replication. J Virol. 2006;80:5708–5715. doi: 10.1128/JVI.02389-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty AM, Jahrling PB, Geisbert TW. Viral hemorrhagic fevers. Clin Lab Med. 2006;26:345–386. viii. doi: 10.1016/j.cll.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Möller P, Pariente N, Klenk HD, Becker S. Homo-oligomerization of Marburgvirus VP35 is essential for its function in replication and transcription. J Virol. 2005;79:14876–14886. doi: 10.1128/JVI.79.23.14876-14886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlberger E. Filovirus replication and transcription. Future Virol. 2007;2:205–215. doi: 10.2217/17460794.2.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlberger E, Lotfering B, Klenk HD, Becker S. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J Virol. 1998;72:8756–8764. doi: 10.1128/jvi.72.11.8756-8764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlberger E, Weik M, Volchkov VE, Klenk HD, Becker S. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Günther S. Broad-spectrum antiviral activity of small interfering RNA targeting the conserved RNA termini of Lassa virus. Antimicrob Agents Chemother. 2007;51:2215–2218. doi: 10.1128/AAC.01368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Möller P, Bick MJ, Wurr S, Becker S, Günther S, Kümmerer BM. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol. 2007;81:2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Ebihara H, Takada A, Noda T, Kobasa D, Jasenosky LD, Watanabe S, Kim JH, Feldmann H, Kawaoka Y. Ebola Virus VP40 Late Domains Are Not Essential for Viral Replication in Cell Culture. J Virol. 2005;79:10300–10307. doi: 10.1128/JVI.79.16.10300-10307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Feldmann H, Watanabe S, Lukashevich I, Kawaoka Y. Reverse genetics demonstrates that proteolytic processing of the Ebola virus glycoprotein is not essential for replication in cell culture. J Virol. 2002a;76:406–410. doi: 10.1128/JVI.76.1.406-410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Geisbert TW, Ebihara H, Geisbert JB, Daddario-DiCaprio KM, Feldmann H, Kawaoka Y. Proteolytic processing of the Ebola virus glycoprotein is not critical for Ebola virus replication in nonhuman primates. J Virol. 2007;81:2995–2998. doi: 10.1128/JVI.02486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Whitt MA, Kawaoka Y. A decade after the generation of a negative-sense RNA virus from cloned cDNA - what have we learned? J Gen Virol. 2002b;83:2635–2662. doi: 10.1099/0022-1317-83-11-2635. [DOI] [PubMed] [Google Scholar]
- Neumann G, Zobel A, Hobom G. RNA polymerase I-mediated expression of influenza viral RNA molecules. Virology. 1994;202:477–479. doi: 10.1006/viro.1994.1365. [DOI] [PubMed] [Google Scholar]
- Noda T, Sagara H, Suzuki E, Takada A, Kida H, Kawaoka Y. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol. 2002;76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Sugiura K, Kato K, Tohya Y, Akashi H. Rescue of Akabane virus (family Bunyaviridae) entirely from cloned cDNAs by using RNA polymerase I. J Gen Virol. 2007;88:3385–3390. doi: 10.1099/vir.0.83173-0. [DOI] [PubMed] [Google Scholar]
- Overby AK, Pettersson RF, Neve EP. The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J Virol. 2007a;81:3198–3205. doi: 10.1128/JVI.02655-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overby AK, Popov V, Neve EP, Pettersson RF. Generation and analysis of infectious virus-like particles of uukuniemi virus (bunyaviridae): a useful system for studying bunyaviral packaging and budding. J Virol. 2006;80:10428–10435. doi: 10.1128/JVI.01362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overby AK, Popov VL, Pettersson RF, Neve EP. The cytoplasmic tails of Uukuniemi Virus (Bunyaviridae) G(N) and G(C) glycoproteins are important for intracellular targeting and the budding of virus-like particles. J Virol. 2007b;81:11381–11391. doi: 10.1128/JVI.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattnaik AK, Ball LA, LeGrone AW, Wertz GW. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- Peeples ME, Collins PL. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J Virol. 2000;74:146–155. doi: 10.1128/jvi.74.1.146-155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M, de la Torre JC. Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2003;77:1184–1194. doi: 10.1128/JVI.77.2.1184-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinschewer DD, Perez M, de la Torre JC. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J Virol. 2003;77:3882–3887. doi: 10.1128/JVI.77.6.3882-3887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinschewer DD, Perez M, de la Torre JC. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J Virol. 2005;79:4519–4526. doi: 10.1128/JVI.79.7.4519-4526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Arguello MB, Goni FM, Pereira FB, Nieva JL. Phosphatidylinositol-dependent membrane fusion induced by a putative fusogenic sequence of Ebola virus. J Virol. 1998;72:1775–1781. doi: 10.1128/jvi.72.3.1775-1781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, Geisbert TW, Feldmann H. Filoviridae: Marburg and Ebola Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1409–1448. [Google Scholar]
- Schmaljohn C, Nichol ST. Bunyaviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 1741–1790. [Google Scholar]
- Schnell MJ, Mebatsion T, Conzelmann KK. Infectious rabies viruses from cloned cDNA. Embo J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümann M, Gantke T, Mühlberger E. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. J Virol. 2009;83:8993–8997. doi: 10.1128/JVI.00523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kohl A, Leonard VH, Li P, McLees A, Elliott RM. Requirement of the N-terminal region of orthobunyavirus nonstructural protein NSm for virus assembly and morphogenesis. J Virol. 2006;80:8089–8099. doi: 10.1128/JVI.00579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares-Weiser K, Thomas S, Thomson G, Garner P. Ribavirin for Crimean-Congo hemorrhagic fever: systematic review and meta-analysis. BMC Infect Dis. 2010;10:207. doi: 10.1186/1471-2334-10-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitenfeld H, Boyd A, Fazakerley JK, Bridgen A, Elliott RM, Weber F. Activation of PKR by Bunyamwera virus is independent of the viral interferon antagonist NSs. J Virol. 2003;77:5507–5511. doi: 10.1128/JVI.77.9.5507-5511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci U S A. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volchkov VE, Volchkova VA, Mühlberger E, Kolesnikova LV, Weik M, Dolnik O, Klenk HD. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science. 2001;291:1965–1969. doi: 10.1126/science.1057269. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Noda T, Halfmann P, Jasenosky L, Kawaoka Y. Ebola virus (EBOV) VP24 inhibits transcription and replication of the EBOV genome. J Infect Dis. 2007;196(Suppl 2):S284–290. doi: 10.1086/520582. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Watanabe T, Noda T, Takada A, Feldmann H, Jasenosky LD, Kawaoka Y. Production of novel ebola virus-like particles from cDNAs: an alternative to ebola virus generation by reverse genetics. J Virol. 2004;78:999–1005. doi: 10.1128/JVI.78.2.999-1005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DM, Ussery MA, Nash D, Peters CJ. Inhibition of Crimean-Congo hemorrhagic fever viral infectivity yields in vitro by ribavirin. Am J Trop Med Hyg. 1989;41:581–585. doi: 10.4269/ajtmh.1989.41.581. [DOI] [PubMed] [Google Scholar]
- Weber F, Dunn EF, Bridgen A, Elliott RM. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology. 2001;281:67–74. doi: 10.1006/viro.2000.0774. [DOI] [PubMed] [Google Scholar]
- Weik M, Enterlein S, Schlenz K, Mühlberger E. The Ebola virus genomic replication promoter is bipartite and follows the rule of six. J Virol. 2005;79:10660–10671. doi: 10.1128/JVI.79.16.10660-10671.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weik M, Modrof J, Klenk HD, Becker S, Mühlberger E. Ebola virus VP30-mediated transcription is regulated by RNA secondary structure formation. J Virol. 2002;76:8532–8539. doi: 10.1128/JVI.76.17.8532-8539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenigenrath J, Kolesnikova L, Hoenen T, Mittler E, Becker S. Establishment and application of an infectious virus-like particle system for Marburg virus. J Gen Virol. 2010;91:1325–1334. doi: 10.1099/vir.0.018226-0. [DOI] [PubMed] [Google Scholar]
- Wilda M, Lopez N, Casabona JC, Franze-Fernandez MT. Mapping of the tacaribe arenavirus Z-protein binding sites on the L protein identified both amino acids within the putative polymerase domain and a region at the N terminus of L that are critically involved in binding. J Virol. 2008;82:11454–11460. doi: 10.1128/JVI.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S, Ikegami T, Peters CJ, Makino S. NSm and 78-kilodalton proteins of Rift Valley fever virus are nonessential for viral replication in cell culture. J Virol. 2006;80:8274–8278. doi: 10.1128/JVI.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S, Ikegami T, Peters CJ, Makino S. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J Virol. 2007;81:13335–13345. doi: 10.1128/JVI.01238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamoto-Niikura A, Terasaki K, Ikegami T, Peters CJ, Makino S. Rift valley fever virus L protein forms a biologically active oligomer. J Virol. 2009;83:12779–12789. doi: 10.1128/JVI.01310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]