Abstract
The rapamycin-inducible gene regulation system was designed to minimize immune reactions in man and may thus be suited for gene therapy. We assessed whether this system indeed induces no immune responses. The protein components of the regulation system were produced in the human cell lines HEK 293T, D407, and HER 911 following lentiviral transfer of the corresponding genes. Stable cell lines were established, and the peptides presented by major histocompatibility complex class I (MHC I) molecules on transduced and wild-type (wt) cells were compared by differential mass spectrometry. In all cell lines examined, expression of the transgenes resulted in prominent changes in the repertoire of MHC I-presented self-peptides. No MHC I ligands originating from the transgenic proteins were detected. In vitro analysis of immunogenicity revealed that transduced D407 cells displayed slightly higher capacity than wt controls to promote proliferation of cytotoxic T cells. These results indicate that therapeutic manipulations within the genome of target cells may affect pathways involved in the processing of peptide antigens and their presentation by MHC I. This makes the genomic modifications visible to the immune system which may recognize these events and respond. Ultimately, the findings call attention to a possible immune risk.
Keywords: gene therapy, genotoxicity, HLA antigens, immune responses
Introduction
The use of gene transfer as a therapeutic tool requires a regulatory system allowing control of the expression of the therapeutic gene by the administration of a small inducer molecule. The treatment could then be adapted to the needs of the patients and, should complications arise, the therapy could be stopped or interrupted. Several regulatory systems have been developed.1 Tetracycline-dependent regulatory systems2 perform very well in transgenic animals.3 However, the transactivators used include peptide sequences from Escherichia coli and Herpes simplex and may therefore provoke immune reactions in patients. Immune responses against the transactivator of a recent version of the tetracycline-dependent regulatory system were observed after expression in the muscles of nonhuman primates.4 Moreover, an immunodominant HLA-A*0201 epitope was detected in the reverse tetracycline-dependent transactivator. This epitope caused cytolytic responses and compromised transgene expression under the control of the tetracycline-on system.5 As a consequence, adverse immunity may interfere, in this case, with gene transfer protocols and prevent gene therapy achieving its aims.
Recently, two novel regulatory systems have been developed which are induced by nonimmunosuppressive derivatives of rapamycin. The first system interferes with transcription and exploits the inducer-dependent interaction between Frap kinase and the Frap kinase binding protein for the reversible in situ assembly of a functional transcription factor which activates transcription from a minimal promoter.6 The second system interferes with the secretory pathway and is adapted to controlling the production of secreted therapeutic factors. It exploits the ability of the inducer to control, in the endoplasmic reticulum, aggregation of a mutated Frap kinase binding protein fusion protein harboring the secreted polypeptide.7 Rapamycin-inducible transcription allows very tightly regulated expression of transgenes.8 Inducer-dependent secretion may be used in combination with inducible transcription for optimized control of the production of therapeutic factors such as the glia derived neurotrophic factor (GDNF).9
The key advantage of rapamycin-inducible systems is that they involve fusion proteins of human origin and consequently immune reactions in humans are minimized. They are therefore expected to be safe. However, the protein components include short peptide sequences that link the various peptide domains. These sequences may themselves constitute novel epitopes or may affect the proteasome cleavage pattern thereby generating novel peptide antigens from the fusion proteins. Any such novel antigens may be presented by the major histocompatibility complex (MHC). This possibility can readily be assessed by application of algorithms for the prediction of proteasome cleavage10 and MHC class I (MHC I) ligand motifs.11 MHC I ligands may also emerge after production of the fusion proteins due to the changes in phenotype which may result from transactivatory effects on transcription and/or competitive effects on translation and protein degradation. Moreover, the transgenic protein components of the regulatory systems may directly interfere with pathways involved in antigen processing and thereby modulate the presentation of peptide antigens.
The immunogenic potential of rapamycin-inducible transcription is of particular importance because of the diverse possible clinical applications of small molecule-inducible gene regulation. We addressed this issue in transduced human cell lines using a mass spectrometry protocol allowing differential analysis of MHC I peptide ligands after stable isotope labeling.12 We compared the presentation of MHC I peptides by cells expressing the protein components required for rapamycin-regulated transcription with the antigen presentation on the respective wild-type (wt) control cells. Production of the transgenic proteins was associated with major changes in the presentation of antigens by MHC I in all cell lines analyzed. Allogeneic in vitro immunogenicity assays provide first evidence that these changes may affect immune tolerance towards transduced cells, though in an individual manner.
Results
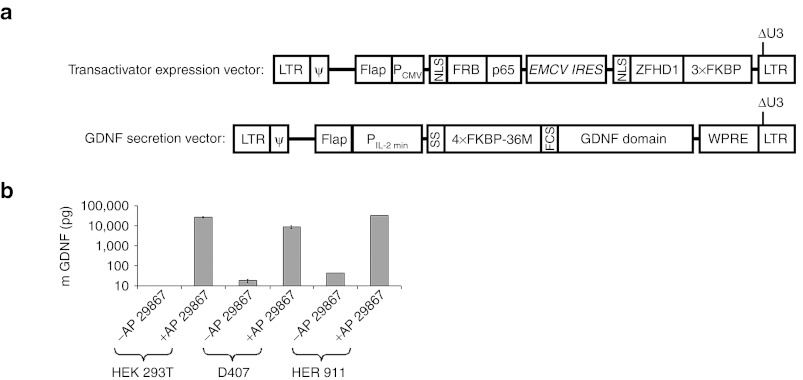
The human cells lines HEK 293T, D407,13 and HER 91114 were treated with lentiviral vectors mediating rapamycin-inducible production and secretion of GDNF as described previously9 (Figure 1a). Selected transduced clones displaying rapamycin-inducible production and secretion of GDNF (Figure 1b), and the corresponding wt cell lines were then characterized for their HLA genotype and their expression of immune relevant molecules. HEK 293T cells were HLA-A*02/02, HLA-B*07/07, HLA-C*07/07, and HLA-DRB1*15/15, HLA-DQB1*06/06; D407 cells were HLA-A*68:02/68:02, HLA-B*15:03/15:03 (B72), HLA-C*12, and HLA-DRB1*01:02/01:02, HLA-DQB1*05/05; and HER 911 cells were HLA-A*02/24:03, HLA-B*37/51, HLA-C*06/15, and HLA-DRB1*10:01/13:01, HLA-DQB1*05/06. Immune phenotyping revealed that all cell lines expressed MHC I, and low levels of MHC II molecules, but were negative for the costimulatory molecules CD80 (B7.1) and CD86 (B7.2) (Supplementary Figure S1).
Figure 1.
Rapamycin-inducible production and secretion of GDNF. (a) Lentiviral constructs: The transactivator expression vector (TEV) mediates constitutive expression of the fusion proteins NLS-FRB-p65 and NLS-ZFHD1-3xFKBP allowing inducible transcription from an engineered minimal interleukin 2 promoter (PIL-2 min). PIL-2 min is used in the glia derived neurotrophic factor (GDNF) expression vector to produce the fusion protein SS-4xFKBP-36M-FCS-GDNF providing rapamycin-regulated secretion of GDNF.9 LTR, ψ, and Flap sequences are derived from HIV-1 (the long terminal repeats, the packaging sequence, and the central Flap element, respectively). PCMV is the CMV promoter, WPRE the Woodchuck hepatitis virus responsive element, and ECMV IRES an internal ribosome entry sequence from the encephalomyocarditis virus. Under the conditions used for cell amplification (i.e., in the absence of inducer) only the fusion proteins NLS-FRB-p65 and NLS-ZFHD1-3xFKBP were produced from TEV. (b) Cell clones derived from HEK 293T, D407, and HER 911 cells displaying inducible production of GDNF. Cells (5 × 104) were cultivated in the presence and absence of 10 nmol/l of the rapamycin derivative AP 21967. Values (±SE, n = 3) indicate the amounts of GDNF in the medium after 2 days of culture.
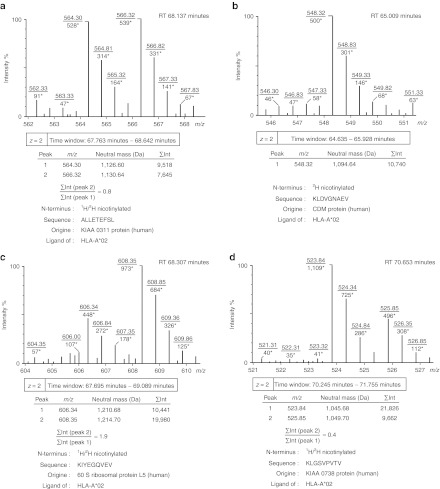
To focus on the effect of the constitutive expression of the transgenic fusion proteins mediating regulation of transcription, both transduced and wt cells of each cell line were amplified in the absence of inducers of the production and secretion of GDNF using parallel cultures maintained under identical rigorously controlled conditions. MHC I molecules were isolated from transduced and wt cells, and the peptides presented were extracted. To distinguish glutamine and lysine residues in mass spectrometry and to ensure specificity of the subsequent N-terminal labeling step, the ε-amino groups of lysine residues were modified by guanidination. A source-dependent difference in neutral mass of m = 4.03 was obtained by N-terminal nicotinylation using a 2H and 1H nicotinoyloxy-succinimide reagent, maintaining the same physico-chemical properties for identical peptide sequences. Aliquots of labeled peptides from both cell populations were combined and analyzed by liquid chromatography linked to electrospray ionization mass spectrometry (LC-ESI-MS). The spectra included double signals (with peak constituents displaying a difference in neutral mass of m = 4.03) and single signals (Figure 2a,b) representing peptides presented in both and only one of the two specimens, respectively.12 Single and double signals were counted for each comparative analysis (Table 1). Single signals revealing the appearance or disappearance of particular MHC I peptides in association with the expression of the transgenic proteins accounted for 40% of all signals for HEK 293T, 21% for D407, and 43% for HER 911 cells. In addition, integration of the peak intensities over time revealed differences in intensity between the peak constituents of double signals (Figure 2a,c,d). These variations indicated two- to threefold up- and downregulation of various MHC I peptides and provided evidence of more subtle changes in the presentation of MHC I antigens associated with the expression of the protein components mediating rapamycin-inducible transcription.
Figure 2.
Typical signals obtained by LC-ESI-MS analysis of combined aliquots of 2H and 1H nicotinylated MHC I peptides from transduced and wild-type cells, respectively. (a) Double signal revealing presentation of approximately equal quantities of the peptide antigen on transduced and wild-type cells. (b) Single signal representing a peptide antigen only presented on transduced cells as assessed by analysis of corresponding LC-ESI-MS/MS data. (c) Double signal revealing upregulation of the peptide antigen on transduced cells. (d) Double signal revealing downregulation of the peptide antigen on transduced cells. Within the spectra, m/z values are underlined, and intensity values are marked with asterisks. RT is the chromatographic retention time, and ∑Int are intensity values integrated over the given time window. Sequence information was obtained by analysis of corresponding LC-ESI-MS/MS data. All examples were taken from the comparative analysis of transduced and wild-type HEK 293T cells.
Table 1. Differential analysis of MHC I peptides on transduced and wild-type cells: Quantification of total signals, single signals, and double signals.

To obtain information about the sequences of peptides presented at the cellular surface by MHC I, aliquots of labeled peptides from transduced and wt cell populations were analyzed individually by liquid chromatography linked to electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS). The results were first screened for peak constituents of double signals on the basis of chromatographic retention time and total mass data from LC-ESI-MS analysis. As could be expected from the labeling protocol (see Materials and Methods section), the peak constituents that corresponded to the lower masses within double signals were from wt cells, whereas peak constituents corresponding to the higher masses were from transduced cells. For a significant number of double signals, MS/MS fragment spectra of both peak constituents were analyzed and gave identical peptide sequences. This confirms that the double signals represent peptides presented at the surface of both transduced and wt cells.
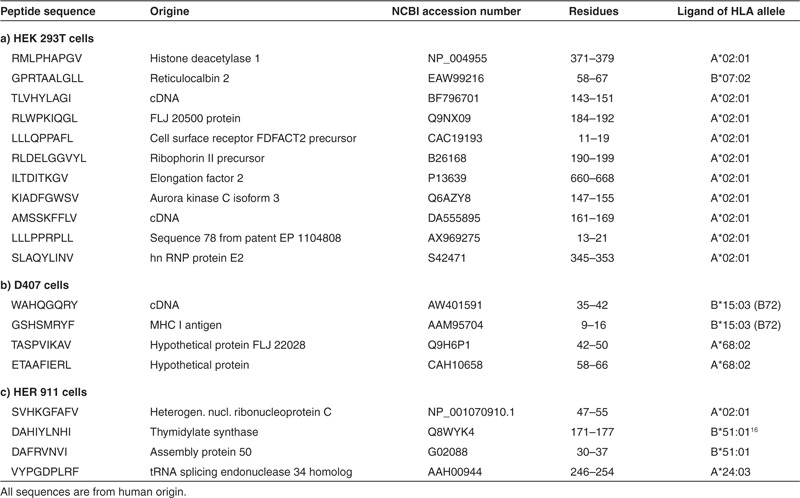
We next identified MHC I peptides that appeared or disappeared after expression of the transgenic proteins. Information on chromatographic retention time and total mass was again used to screen the LC-ESI-MS/MS data for the single signals detected by mass spectrometry in combined aliquots of labeled peptides from transduced and wt cells. The single signals were detected exclusively in either the transduced or the wt cell samples. Signal intensities were sufficiently robust to allow sequencing of a significant number of peptides, and the masses of the respective b1-fragments (representing the 2H or 1H nicotinylated N-terminal amino acid residues) provided controls confirming the presence of the respective peptides in the transduced or wt cell samples. MHC I peptides that appeared or disappeared after expression of the transgenic proteins are listed in Tables 2 and 3, respectively. The sequences correspond to structural motifs required for binding to MHC I molecules. Epitope prediction using the NetMHCpan algorithm11 indicated, in HEK 293T cells, the presence of peptides binding to MHC I molecules derived from the HLA alleles A*02:01 and B*07:02. In D407 cells, peptides were observed that bind to MHC I molecules from the HLA alleles A*68:02 and B*15:03. In HER 911 cells, peptides were found that bind to MHC I derived from the HLA alleles A*02:01, A*24:03, B*51:01, and C*06:02. Two of the peptides – DVANKIGII originating from the ribosomal protein L23a and DAHIYLNHI originating from thymidylate synthase – have previously been described as ligands of MHC I derived from the HLA allele B*51:01.15,16 The peptides identified were derived from a wide variety of cellular proteins, i.e., self-proteins. No HLA peptide antigens originating from the expressed transgenic proteins were detected by the analytical method used.
Table 2. MHC I peptide ligands presented at the surface of HEK 293T, D407, and HER 911 cells after expression of the protein components required for rapamycin-regulated transcription.
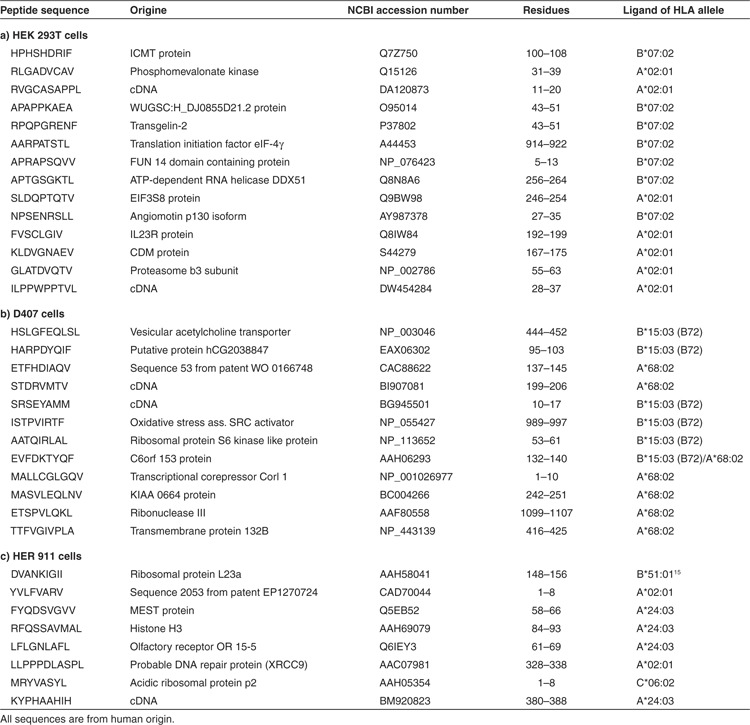
Table 3. MHC I peptide ligands disappearing from the surface of HEK 293T, D407, and HER 911 cells after expression of the protein components required for rapamycin-regulated transcription.
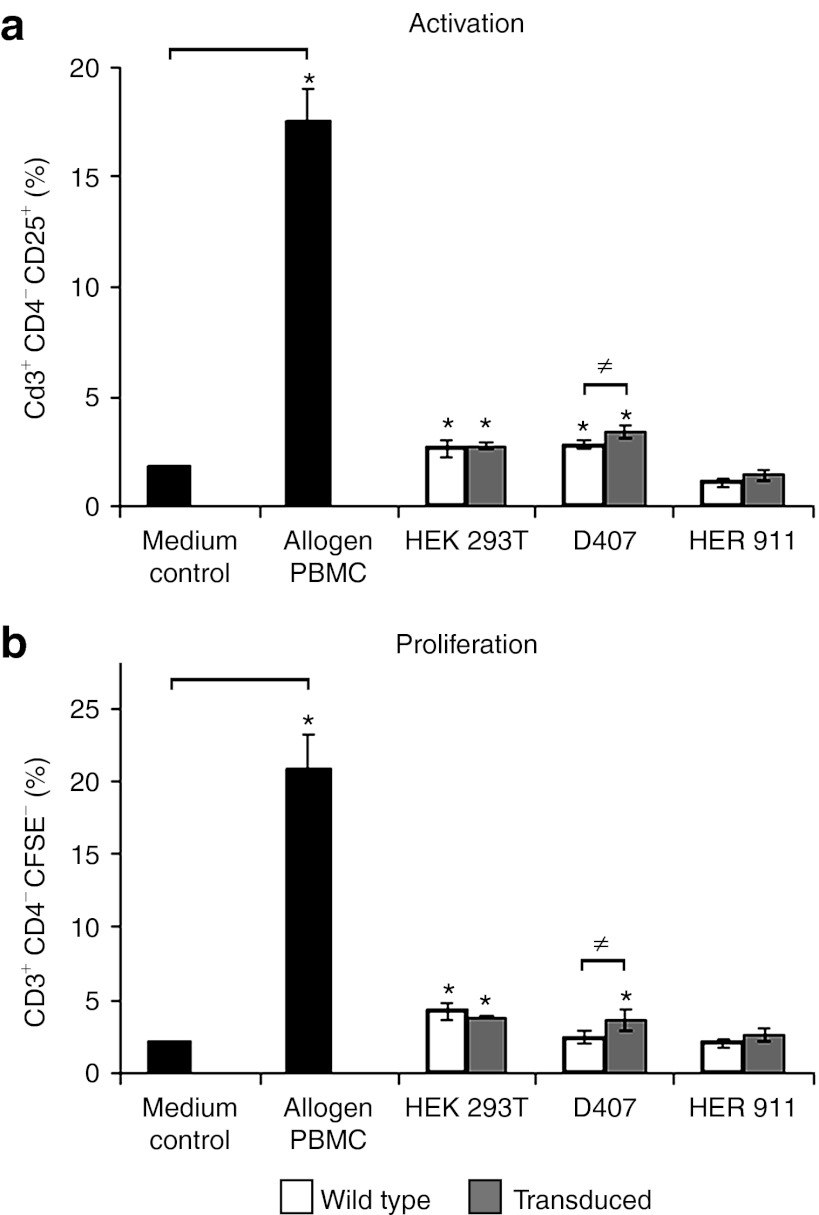
In one-way allogeneic mixed lymphocyte reactions, we then investigated the capacity of transduced and wt HEK 293T, HER 911, and D407 cells to elicit an allogeneic response from CD3+CD4− T cells which mainly comprise cytotoxic CD8+ T cells. The expression of CD25 was used as a marker of activation and CFSE labeling was used to assess proliferation. Transduced D407 cells activated more strongly (1.7-fold) CD3+CD4− T cells than wt D407 cells (Figure 3a), and this significantly increased activation was followed by a significantly increased proliferation of CD3+CD4− T cells (Figure 3b). No significant differences compared with the respective wt controls were observed for HEK 293T and HER 911 cells suggesting that immunogenicity is limited to individual cell clones.
Figure 3.
Allogeneic immune responses elicited by wild-type and transduced cells. CSFE-labeled peripheral blood mononuclear cells (PBMCs) were incubated with medium alone, or cocultured with control allogeneic PBMCs, wild-type or transduced cells. (a) Activation of CD3+CD4− T cells as determined by the expression of CD25. (b) Proliferation of CD3+CD4− T cells as evaluated by determining the percentages of dividing T cells. Results are expressed as mean values ± SD for three independent experiments each performed in triplicate at different times of culture (*P < 0.05). Note: Control allogeneic PBMCs triggered large responses both in terms of activation and proliferation, as expected (positive control).
Discussion
The regulatory system allowing rapamycin-inducible transcription is promising for clinical applications in part, because it is composed of fusion proteins comprising polypeptide sequences that all originate from human proteins thereby minimizing the risk of immune reactions in patients. However, the sequences at the junctions between the various domains could create sites that are potentially immunogenic. Therefore, we assessed the potential immunogenicity associated with rapamycin-inducible transcription.
We assessed changes in the presentation of MHC I peptides associated with the production of the fusion proteins allowing rapamycin-inducible expression of transgenes and observed variations in the presentation of peptide antigens. A significant number of peptides appeared and disappeared, and the amounts of some peptides were affected. Surprisingly, all these peptides were derived from self-proteins. We detected no peptide antigens originating from the two transgenic proteins allowing inducible transcription of a therapeutic transgene. Using an in vitro immunogenicity assay, we demonstrated that the expression of these transgenic proteins and the associated changes in the repertoire of MHC I-presented peptide antigens could affect the immunogenicity of the cells.
The mass spectrometry protocol used was developed for direct and quantitative comparison of MHC-presented peptides in pairs of samples.12 In contrast to approaches addressing changes in the transcriptome and the proteome, this technique is able to reveal events after protein degradation and the binding of peptides to distinct MHC I molecules. Interestingly, there is only a weak correlation between mRNA copy number and the density of corresponding MHC ligands17 indicating that the presentation of peptide antigens has to be investigated directly to obtain pertinent information about the immunological consequences of the expression of transgenes. No peptides originating from the transgenic proteins were detected. The detection limit of the method is about 100 fmol, so peptides present at 6–10 copies per cell could be detected; nevertheless, we have to consider the possibility that peptides from the two transgenic proteins are presented by MHC I molecules in amounts too low to be detected. Also, peptides derived from the degradation of the transgenic proteins may have low affinity for the MHC I molecules tested. Moreover, we cannot exclude the possibility that these peptides are presented by MHC II despite its low abundance in the cell lines used. In contrast to the absence of MHC ligands of transgenic origin, numerous changes in the presentation of peptides of self-origin were apparent, although, most probably, we only detected the largest effects because more subtle changes may have escaped detection for the reasons discussed above. Abundant MHC ligands such as those detected by the analytical protocol can repeatedly be found in batches from cultured cells harvested at different time points15,18,19,20,21,22 which underlined evidence for the observed impact of transgene expression on the presentation of peptide antigens by MHC I.
The presentation of self-peptides may result from the phenomenon of insertional mutagenesis, commonly associated with integrative vectors. Sequences randomly inserted into the genome can modify the expression of genes in the proximity of the integration sites. Indeed, such insertions may be deleterious and even lead to the activation of oncogenes. In a clinical gene therapy trial for X-linked severe combined immune deficiency, 3 of 13 patients developed leukemia 30 months after treatment as a consequence of oncogene activation by the integrating vector genome.23 Activation of oncogenes is accompanied by substantial changes at the proteome level24 which may ultimately provoke appearance and disappearance of peptide antigens of self-origin. In the context of infection with HIV, di Marzo Veronese et al. showed that infected cells from HIV-1 positive subjects overexpressed vinculin and that three peptides originating from this self-protein are presented by MHC I at the surface of these infected cells, leading to the activation of specific cytotoxic T-lymphocytes.25 Kane et al. recently reported that lentiviral vectors may provoke somatic cell reprogramming as a result of dysregulated host gene expression in the vicinity of integration sites.26
Mechanisms other than those associated with genotoxicity may also contribute to the changes in the presentation of peptide antigens, including particular mechanisms involving the fusion proteins allowing rapamycin-inducible expression of transgenes. In our model, these transgenic proteins were strongly expressed under the control of a CMV promoter. This could lead to interference with the proteome by competing for the protein synthesis and degradation machinery. Furthermore, specific interference of these transgenic proteins might trigger changes in the presentation of self-peptides by MHC I. The transgenic proteins include domains from the human Frap kinase and the Frap kinase binding protein, which may interfere with the endogenous Frap kinase and Frap kinase binding protein in the regulation of autophagy.27 Recently, it has emerged that autophagy is a pathway for the processing of antigens presented by MHC I.28 Consistent with this possibility, we detected several peptide antigens originating from membrane proteins that could not have been produced by proteasome-mediated proteolysis but might have got access to MHC I by autophagy-mediated digestion of membrane components and mitochondria.29 However, these peptides may also be produced by translation of mRNA using alternative translational reading frames.30
Regardless of the molecular mechanism involved in the changes in the repertoire of MHC I-presented peptides, our coculture experiments revealed that transduction of cells with lentiviral vectors might affect the immunogenicity of individual transduced cells. In particular, transduced cells were able to activate and trigger proliferation of T cells. Cytotoxic T cells may arise, if the presentation of the respective MHC peptide complexes in the thymus is insufficient during the selection process leading to self-tolerance. In clinical settings, this phenomenon could lead to an immunological rejection of the transduced cells and thereby prevent a gene therapy protocol achieving its aims.
Any adverse immune reaction probably depends on a variety of factors including cell type, the site of vector integration and the HLA haplotype. Furthermore, the changes in the presentation of peptides by MHC I may only be recognized, if cytotoxic T cells bearing appropriate T cell receptors are available in the repertoire. We studied three cell lines, and only one, the transduced D407-derived cell clone, induced proliferation of CD4− T cells (comprising cytotoxic CD8+ T and NKT cells). It is therefore likely that immune reactivity to transduced cells depends on the details of each individual case and context. Most probably, following administration of lentiviral vectors to patients, only a subset of the broad diversity of transduced cells could be eliminated by the immune system. Interestingly, in a recent trial of gene therapy for β-thalassemia, one particular clone of transduced cells came to predominate over time and provided therapeutic efficacy,31 suggesting that the majority of the transduced cells were probably successively eliminated by the immune system.
The effects on the presentation of peptide epitopes might not be restricted to MHC I. They might also be found in the repertoire of peptides presented by MHC II as transduced HER 911 cells (but not D407 or HEK 293T cells) revealed, compared with wt controls, significantly enhanced capacity to activate CD3+CD4+ allogeneic T cells, although no significant proliferation was observed (Supplementary Figure S2). It would therefore be of interest to assess the effect of the vectors on the presentation of MHC II peptides. These studies should also address whether activated CD4+ cells belong to effector or regulatory subtypes which induce adverse immunity and immune tolerance, respectively. Indeed, particular individual subsets of transduced cells might escape elimination by the immune system by activation of tolerance-mediating CD4+ regulatory T cells, a scenario frequently observed in studies monitoring the progression of tumors in patients.32
Taken together, our findings provide the first evidence (i) that pathways involved in processing of peptide antigens and their presentation by MHC I render therapeutic manipulations within the genome of particular target cells visible to the immune system and (ii) that the immune system may recognize these events and respond.
Materials and Methods
Cell culture, lentiviral gene transfer, and generation of stable cell lines. All cell lines were maintained at 37 °C under a water-saturated atmosphere of 5% CO2/95% air. HER 91114 and HEK 293T cells were cultivated in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 20 U/ml penicillin G, and 20 µg/ml streptomycin sulfate; D407 cells13 were cultivated in Dulbecco's modified Eagle medium containing 5% fetal calf serum, and antibiotics as above.
For gene transfer, 20,000 cells were incubated overnight with aliquots of the transactivator expression vector and the GDNF secretion vector9 (Figure 1a), each corresponding to 60 ng of capsid protein p24. Single viable cells were then seeded one in each well of 96-well culture dishes. After 3 weeks, wells were screened for cell clones which were tested for inducible production of GDNF using the GDNF Emax immunoassay system (Promega, Charbonnières, France). Appropriate clones released GDNF into the medium after addition of the rapamycin derivative AP 21967 (10 nmol/l, ARIAD, Cambridge, MA) but not in the absence of the inducer. These clones were amplified in the absence of inducer to 1010 cells.
Extraction and modification of MHC I-presented peptides. MHC I-presented peptides were obtained by immune precipitation of MHC I molecules from the amplified cell lines as described33 using the HLA-A, -B, and -C specific antibody W6/32 immobilized on sepharose, acid treatment and ultrafiltration. As described previously,17 peptides from wt and transduced cells were concentrated by lyophilization, dissolved in 500 μl water, modified by guanidination, and then nicotinylated with 1H4 and 2H4 nicotinoyloxy-succinimide (LGC Promochem, Molsheim, France), respectively.
LC-ESI-MS and LC-ESI-MS/MS analyses. The modified peptide extracts were analyzed as described12 using a reversed phase nanoLC-2D system (Eksigent, Darmstadt, Germany), coupled to a hybrid quadrupole orthogonal acceleration time-of-flight MS/MS (Q-TOF Ultima; Micromass/Waters, Saint-Quentin en Yvelines, France) equipped with a micro-ESI source. Results for mixed transduced and wt samples were recorded in an LC-ESI-MS experiment without fragmentation. For sequence analysis, results for transduced and wt samples were recorded separately in individual LC-ESI-MS/MS experiments. Fragment spectra were analyzed manually and database searches (Proteomics Department at the Hammersmith Campus of Imperial College London, National Center for Biotechnology Information, Expressed Sequence Tag) involved using the Multiple Alignment System for Protein Sequences Based on Three-way Dynamic Programming (MASCOT, http://www.matrixscience.com). The NetMHCpan 2.3 Server (http://www.cbs.dtu.dk/services/NetMHCpan)11 was used to search for MHC I binding partners of the peptide sequences found.
HLA-class I and class II genotyping. DNA was extracted from HEK293T, HER911, and D407 cells using a salting-out technique. HLA medium resolution typing for HLA-A, -B, -C, DRB1, and DQB1 was performed using PCR-sequence specific oligonucleotide (SSO) Luminex kits (LABType SSO A Locus, LABType SSO B Locus, LABType SSO DRB1, and LABType SSO DQB1; One Lambda, Canoga Park, CA).
Immunogenicity assay. Unfractionated allogeneic peripheral blood mononuclear cells (HLA-A*02/29:02, HLA-B*40:02/44:03, HLA-C*02:02/16:01, HLA-DRB1*07:01/11:01, HLA-DQB1*02:02/03:01, 1 × 105 cells) purified from a blood sample from a healthy donor were labeled with the carboxyfluorescein succinimidyl ester (CFSE, 10 μmol/l for 10 minutes at 37 °C) and then incubated with 1 × 104 target cells (transduced cells, wt cells, or allogeneic stimulatory peripheral blood mononuclear cells for control). Before incubation with peripheral blood mononuclear cells, target cells were treated with mitomycin C (50 μg/ml) for 30 minutes. After 6 days of coculture, cells were stained with eFluor780-conjugated anti-CD3 (UCHT1; eBiosciences, Paris, France), phycoerythrin-conjugated anti-CD25 and allophycocyanin-conjugated anti-CD4 antibodies, and with 7-aminoactinomycin D to exclude dead cells (BD Biosciences, Le Pont de Claix, France). T cell activation and proliferation was then monitored by flow cytometry using a Canto II flow cytometer (BD Biosciences). Results were analyzed with BD Diva software (BD Biosciences) and are reported as mean values ± SE of at least three independent experiments. One-way analysis of variance followed by the Student-Newman–Keuls test (SigmaStat software; Systat Software, Chicago, IL) were used for statistical analyses and a P of <0.05 was considered significant.
SUPPLEMENTARY MATERIAL Figure S1. (a) Flow cytometric analysis of the expression of MHC I, MHC II (HLA-DR, HLA-DQ, and HLA-DP), CD80, and CD86 molecules and transduced HER 911 cells. (b) Expression of immune relevant molecules by wt and transduced HEK 293T, HER 911 and D407 cells. Figure S2. Allogeneic immune responses to wt and transduced cells mediated by MHC II.
Acknowledgments
We thank ARIAD Inc. (Cambridge, MA) for providing the regulation systems allowing rapamycin-inducible transcription and secretion. We thank the “Association Française contre les Myopathies (AFM)”, the “Institut pour la Recherche sur la Moelle Épinière (IRME)”, and “Rétina France” for financial support. The authors declared no conflict of interest.
Supplementary Material
(a) Flow cytometric analysis of the expression of MHC I, MHC II (HLA-DR, HLA-DQ, and HLA-DP), CD80, and CD86 molecules and transduced HER 911 cells. (b) Expression of immune relevant molecules by wt and transduced HEK 293T, HER 911 and D407 cells.
Allogeneic immune responses to wt and transduced cells mediated by MHC II.
References
- Fussenegger M. The impact of mammalian gene regulation concepts on functional genomic research, metabolic engineering, and advanced gene therapies. Biotechnol Prog. 2001;17:1–51. doi: 10.1021/bp000129c. [DOI] [PubMed] [Google Scholar]
- Baron U., and, Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Meth Enzymol. 2000;327:401–421. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Zheng T, Lee CG, Homer RJ., and, Elias JA. Tetracycline-controlled transcriptional regulation systems: advances and application in transgenic animal modeling. Semin Cell Dev Biol. 2002;13:121–128. doi: 10.1016/s1084-9521(02)00018-6. [DOI] [PubMed] [Google Scholar]
- Latta-Mahieu M, Rolland M, Caillet C, Wang M, Kennel P, Mahfouz I.et al. (2002Gene transfer of a chimeric trans-activator is immunogenic and results in short-lived transgene expression Hum Gene Ther 131611–1620. [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Turbant S, Gross DA, Poupiot J, Marais T, Lone Y.et al. (2004HLA-A*0201-restricted cytolytic responses to the rtTA transactivator dominant and cryptic epitopes compromise transgene expression induced by the tetracycline on system Mol Ther 10279–289. [DOI] [PubMed] [Google Scholar]
- Rivera VM, Clackson T, Natesan S, Pollock R, Amara JF, Keenan T.et al. (1996A humanized system for pharmacologic control of gene expression Nat Med 21028–1032. [DOI] [PubMed] [Google Scholar]
- Rivera VM, Wang X, Wardwell S, Courage NL, Volchuk A, Keenan T.et al. (2000Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum Science 287826–830. [DOI] [PubMed] [Google Scholar]
- Pollock R, Issner R, Zoller K, Natesan S, Rivera VM., and, Clackson T. Delivery of a stringent dimerizer-regulated gene expression system in a single retroviral vector. Proc Natl Acad Sci USA. 2000;97:13221–13226. doi: 10.1073/pnas.230446297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel R, Mammeri H., and, Mallet J. Lentiviral vectors mediate nonimmunosuppressive rapamycin analog-induced production of secreted therapeutic factors in the brain: regulation at the level of transcription and exocytosis. Hum Gene Ther. 2008;19:167–178. doi: 10.1089/hum.2007.125. [DOI] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Lund O., and, Kesmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S.et al. (2007NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence PLoS ONE 2e796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmel C, Weik S, Eberle U, Dengjel J, Kratt T, Becker HD.et al. (2004Differential quantitative analysis of MHC ligands by mass spectrometry using stable isotope labeling Nat Biotechnol 22450–454. [DOI] [PubMed] [Google Scholar]
- Davis AA, Bernstein PS, Bok D, Turner J, Nachtigal M., and, Hunt RC. A human retinal pigment epithelial cell line that retains epithelial characteristics after prolonged culture. Invest Ophthalmol Vis Sci. 1995;36:955–964. [PubMed] [Google Scholar]
- Fallaux FJ, Kranenburg O, Cramer SJ, Houweling A, Van Ormondt H, Hoeben RC.et al. (1996Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors Hum Gene Ther 7215–222. [DOI] [PubMed] [Google Scholar]
- Weinzierl AO, Rudolf D, Hillen N, Tenzer S, van Endert P, Schild H.et al. (2008Features of TAP-independent MHC class I ligands revealed by quantitative mass spectrometry Eur J Immunol 381503–1510. [DOI] [PubMed] [Google Scholar]
- Falk K, Rötzschke O, Takiguchi M, Gnau V, Stevanovic S, Jung G.et al. (1995Peptide motifs of HLA-B51, -B52 and -B78 molecules, and implications for Behcet's disease Int Immunol 7223–228. [DOI] [PubMed] [Google Scholar]
- Weinzierl AO, Lemmel C, Schoor O, Müller M, Krüger T, Wernet D.et al. (2007Distorted relation between mRNA copy number and corresponding major histocompatibility complex ligand density on the cell surface Mol Cell Proteomics 6102–113. [DOI] [PubMed] [Google Scholar]
- Thommen DS, Schuster H, Keller M, Kapoor S, Weinzierl AO, Chennakesava CS.et al. (2012Two preferentially expressed proteins protect vascular endothelial cells from an attack by peptide-specific CTL J Immunol 1885283–5292. [DOI] [PubMed] [Google Scholar]
- Wölk B, Trautwein C, Büchele B, Kersting N, Blum HE, Rammensee HG.et al. (2012Identification of naturally processed hepatitis C virus-derived major histocompatibility complex class I ligands PLoS ONE 7e29286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliger B, Dressler SP, Massa C, Recktenwald CV, Altenberend F, Bukur J.et al. (2011Identification and characterization of human leukocyte antigen class I ligands in renal cell carcinoma cells Proteomics 112528–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer VS, Drews O, Günder M, Hennenlotter J, Rammensee HG., and, Stevanovic S. Identification of natural MHC class II presented phosphopeptides and tumor-derived MHC class I phospholigands. J Proteome Res. 2009;8:3666–3674. doi: 10.1021/pr800937k. [DOI] [PubMed] [Google Scholar]
- Meyer VS, Kastenmuller W, Gasteiger G, Franz-Wachtel M, Lamkemeyer T, Rammensee HG.et al. (2008Long-term immunity against actual poxviral HLA ligands as identified by differential stable isotope labeling J Immunol 1816371–6383. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E.et al. (2003A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency N Engl J Med 348255–256. [DOI] [PubMed] [Google Scholar]
- Oh WJ, Rishi V, Pelech S., and, Vinson C. Histological and proteomic analysis of reversible H-RasV12G expression in transgenic mouse skin. Carcinogenesis. 2007;28:2244–2252. doi: 10.1093/carcin/bgm127. [DOI] [PubMed] [Google Scholar]
- di Marzo Veronese F, Arnott D, Barnaba V, Loftus DJ, Sakaguchi K, Thompson CB.et al. (1996Autoreactive cytotoxic T lymphocytes in human immunodeficiency virus type 1-infected subjects J Exp Med 1832509–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane NM, Nowrouzi A, Mukherjee S, Blundell MP, Greig JA, Lee WK.et al. (2010Lentivirus-mediated reprogramming of somatic cells in the absence of transgenic transcription factors Mol Ther 182139–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118 Pt 1:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D.et al. (2009Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection Nat Immunol 10480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Müller M.et al. (2005Autophagy promotes MHC class II presentation of peptides from intracellular source proteins Proc Natl Acad Sci USA 1027922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malarkannan S, Horng T, Shih PP, Schwab S., and, Shastri N. Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism. Immunity. 1999;10:681–690. doi: 10.1016/s1074-7613(00)80067-9. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F.et al. (2010Transfusion independence and HMGA2 activation after gene therapy of human ß-thalassaemia Nature 467318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries W, Wei J, Sampson JH., and, Heimberger AB. The role of Tregs in glioma-mediated immunosuppression: potential target for intervention. Neurosurg Clin N Am. 2010;21:125–137. doi: 10.1016/j.nec.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzierl AO, Maurer D, Altenberend F, Schneiderhan-Marra N, Klingel K, Schoor O.et al. (2008A cryptic vascular endothelial growth factor T-cell epitope: identification and characterization by mass spectrometry and T-cell assays Cancer Res 682447–2454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Flow cytometric analysis of the expression of MHC I, MHC II (HLA-DR, HLA-DQ, and HLA-DP), CD80, and CD86 molecules and transduced HER 911 cells. (b) Expression of immune relevant molecules by wt and transduced HEK 293T, HER 911 and D407 cells.
Allogeneic immune responses to wt and transduced cells mediated by MHC II.