Abstract
TAL (transcription activator–like) effectors, secreted by phytopathogenic bacteria, recognize host DNA sequences through a central domain of tandem repeats. Each repeat comprises 33 to 35 conserved amino acids and targets a specific base pair by using two hypervariable residues [known as repeat variable diresidues (RVDs)] at positions 12 and 13. Here, we report the crystal structures of an 11.5-repeat TAL effector in both DNA-free and DNA-bound states. Each TAL repeat comprises two helices connected by a short RVD-containing loop. The 11.5 repeats form a right-handed, superhelical structure that tracks along the sense strand of DNA duplex, with RVDs contacting the major groove. The 12th residue stabilizes the RVD loop, whereas the 13th residue makes a base-specific contact. Understanding DNA recognition by TAL effectors may facilitate rational design of DNA-binding proteins with biotechnological applications.
TAL (transcription activator–like) effectors (TALEs) are major virulence factors secreted by bacteria of the genus Xanthomonas that cause diseases in plants such as rice and cotton (1–4). TALEs, also known as AvrBs3/PthA family effectors (5, 6), are injected into plant cells through a type III secretion system and interfere with cellular activities through transcriptional activation of specific target genes (1, 7–9). TALEs share a common domain organization that enables them to be imported into nuclei and act as transcriptional activators (10–13).
The central DNA binding domain of TALEs consists of 1.5 to 33.5 tandem repeats (TAL repeats), with each repeat recognizing one specific DNA base pair (14, 15). Each TAL repeat contains 33 to 35, mostly 34, highly conserved amino acids (16, 17). Within each repeat, two hypervariable residues at positions 12 and 13 (also known as RVDs for repeat variable diresidues) confer DNA specificity (14, 15). The code of DNA recognition by RVDs has been deciphered by both experimental (14) and computational (15) approaches. The frequently occurring RVDs His/Asp (HD), Asn/Gly (NG), and Asn/Ile (NI) recognize cytosine (C), thymine (T), and adenine (A), respectively (1, 18). DNA binding by TAL repeats is modular, allowing engineering of DNA-binding proteins by assembly of TAL repeats with designed RVDs, for example, for use in targeted gene activation (14, 18, 19). Despite these advances how TAL repeats specifically recognize DNA remains unknown.
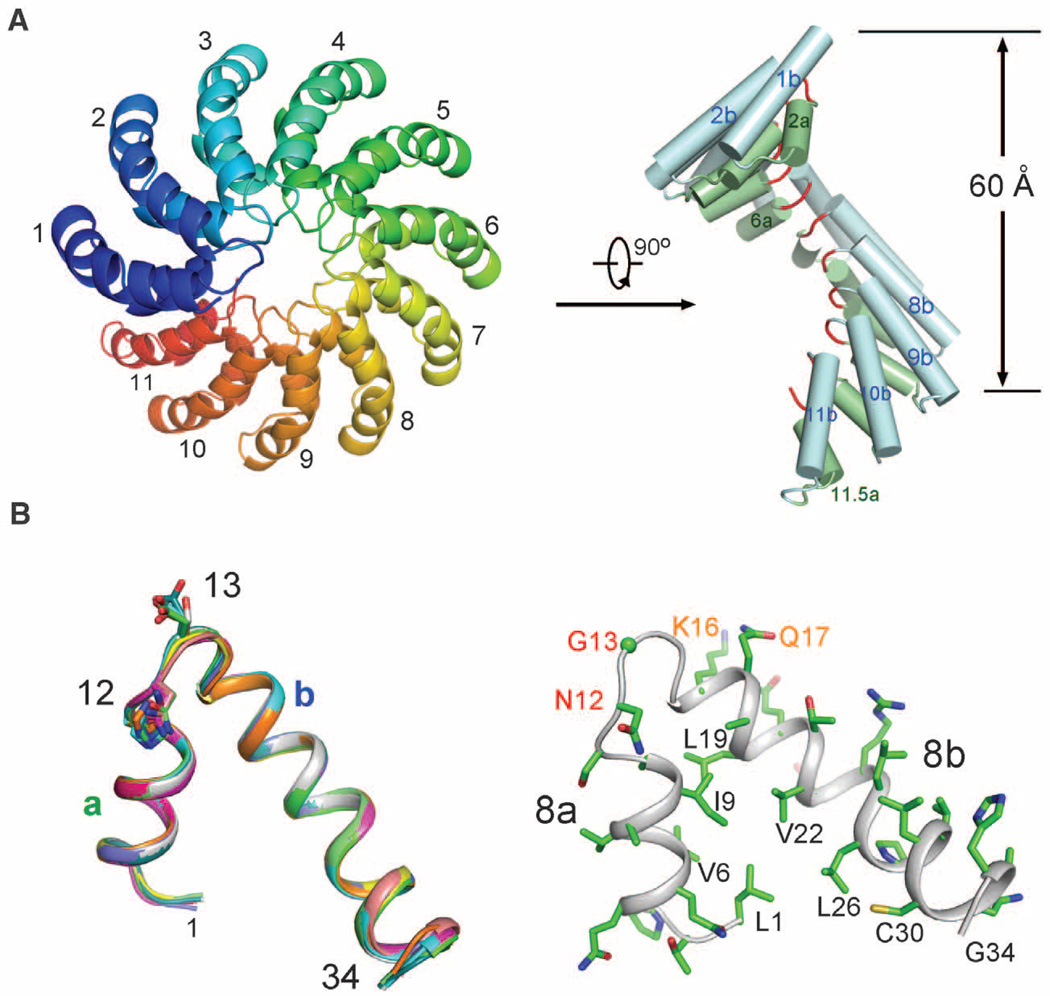
We investigated an artificially engineered TAL effector, dHax3 (20) (fig. S1). The central domain of dHax3 (residues 270 to 703), containing 11.5 TAL repeats, was crystallized in the space group C2221 (21). The structure was determined by Ta6Br12-based multiwavelength anomalous diffraction and refined to 2.4 Å resolution (tables S1 and S2 and fig. S2A). There is one molecule in each asymmetric unit. In the crystals, crystallographically independent molecules are arranged to form a continuous right-handed, superhelical assembly (fig. S2B). The structurally well-defined region of DNA-free dHax3 (residues 303 to 675) forms exactly 11 repeats, starting from the second half of repeat 1 and ending at repeat 11.5 (Fig. 1A). The superhelical assembly has an external diameter of about 60 Å.
Fig. 1.
Structure of the TAL repeats in DNA-free dHax3. (A) The 11 TAL repeats of dHax3 form a right-handed superhelical assembly. Two perpendicular views are presented with the RVDs highlighted in red in the right image. (B) All TAL repeats exhibit a nearly identical conformation. Each repeat is organized into short (a) and long (b) α helices connected by a short loop where the two (RVDs at positions 12 and 13 are located. All structure figures were prepared with PyMOL (30). Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
Each TAL repeat in dHax3 contains 34 amino acids, with residues 3 to 11 forming a short α helix (designated as “a”) and residues 15 to 33 constituting an extended, bent α helix (designated as “b”). The two helices are connected by a short loop consisting of RVD and an invariant amino acid Gly at position 14 (Fig. 1B and fig. S1). This loop is hereafter referred to as the RVD loop. Reflecting the high degree of sequence conservation (fig. S1), all 11 repeats exhibit a nearly identical conformation (Fig. 1B and fig. S2C). Helices a and b within each repeat closely stack against each other through extensive van der Waals contacts (Fig. 1B). The angle between the helices distinguishes the TAL repeat from other known α-helical repeat modules such as HEAT (22) and TPR (23), in which the two helices are nearly parallel to each other. A nuclear magnetic resonance (NMR) structure of 1.5 TAL repeats in the protein PthA was previously reported (24); however, our TAL repeat structure exhibits major differences from that in PthA (fig. S2, D and E).
The 11 TAL repeats of dHax3 complete a full helical turn; the RVD loops form the innermost spiral with a pitch of 60 Å per turn (Fig. 1A). The 11 a helices form an internal layer along the superhelical axis, whereas the 11 b helices constitute an external layer (Fig. 1A). These structural features suggest a DNA-binding model in which the DNA molecule is placed within the TAL superhelical assembly along the axis.
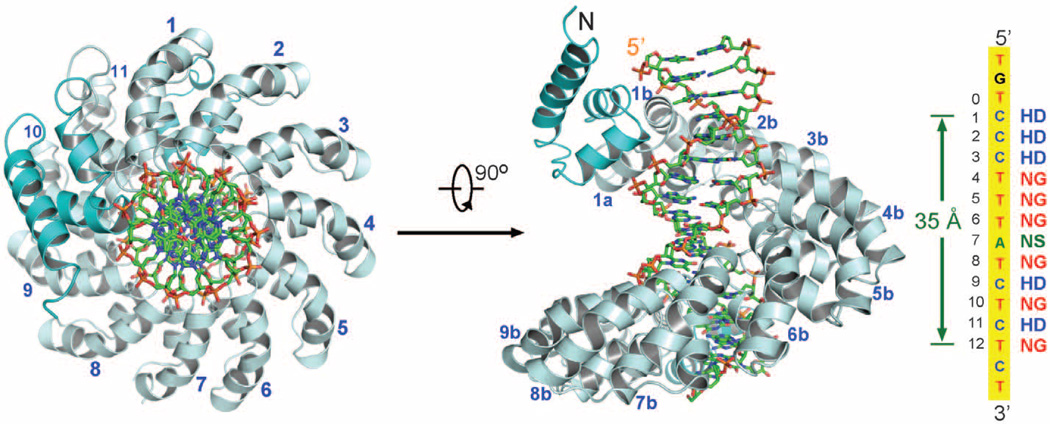
We crystallized a binary complex between dHax3 (residues 231 to 720), which encompasses the entire 11.5 TAL repeats, and a 17–base pair (bp) DNA binding element (20), with 5′-TGTCCCTTTATCTCTCT-3′ as the sense strand. The structure was determined by molecular replacement at 1.85 Å resolution (table S2 and fig. S3A). There are two complexes in each asymmetric unit (fig. S3B). The two protein molecules (designated A and B) can be superimposed with a root mean square deviation (RMSD) of 1.2 Å over 447 Cα atoms (fig. S3C). Because these two complexes exhibit identical features for most repeats, we mainly describe structural analysis on molecule A.
In the complex structure, dHax3 comprises 12 repeats (residues 289 to 691), with the C-terminal 0.5 repeat contributed by nonconserved amino acids (Fig. 2). These repeats are capped by three and two short α helices at the N and C termini, respectively (Fig. 2). Similar to DNA-free dHax3, all repeats exhibit a nearly identical conformation except RVD loops in repeat 6 of molecule A and repeat 5 of molecule B (figs. S3D and S4). The superhelical dHax3 structure tracks along the major groove of the DNA duplex. The conformation of the 17-bp DNA is largely B-form (table S3), with 11 base pairs per turn and a pitch of about 35 Å.
Fig. 2.
Overall structure of dHax3 bound to DNA. The superhelical structure of dHax3 (residues 231 to 720) binds to the major groove of DNA. Shown on the right are the DNA sequence of the sense strand and the corresponding RVDs in TAL repeats of dHax3. dHax3 contains 11.5 repeats with flanking N- and C-terminal helices shown in cyan. Two perpendicular views are presented, with the DNA duplex shown in sticks.
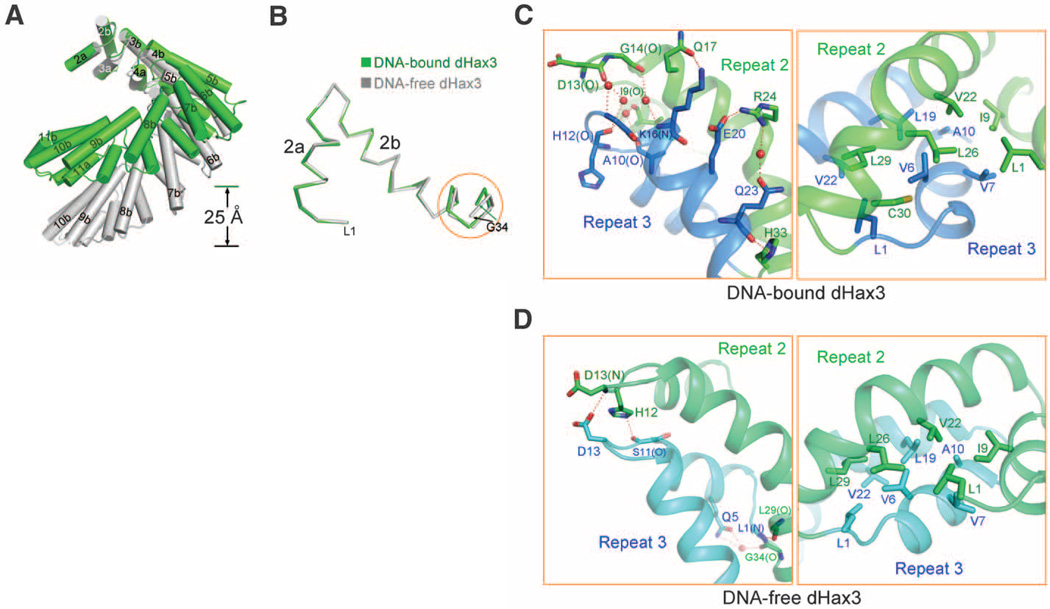
In the structures of both DNA-free and DNA-bound dHax3, there are 11 TAL repeats per helical turn (Fig. 3A). Comparison of any corresponding repeat between these two structures reveals little difference, with an RMSD of 0.25 to 0.34 Å over about 30 Cα atoms (Fig. 3B). However, the superhelical pitch is reduced from 60 Å in DNA-free form to about 35 Å in the DNA-bound form (Fig. 3A).Whereas the main chains of the first 22 amino acids are precisely superimposed, subtle conformational variations accumulate for residues 23 to 34, resulting in notable differences between the positions of the Cα atoms in Gly34 (Fig. 3B). Such differences are gradually amplified over an increasing number of repeats (fig. S5), ultimately resulting in the compression of the superhelical assembly in the DNA-bound form. Such conformational plasticity is consistent with the predominantly van der Waals interactions between adjacent TAL repeats, which can tolerate minor distance shifts (Fig. 3, C and D, and fig. S6). The ability to undergo substantial conformational changes appears to be a conserved feature for superhelical assemblies exemplified by Armadillo repeats in β-catenin (25) and HEAT repeats in keryopherin α (26) and the scaffold subunit of protein phosphatase 2A (PP2A) (27). The conformational plasticity of the TAL repeats, which was previously noted (24), is likely essential for the function of TALEs.
Fig. 3.
Structural comparison of DNA-free and DNA-bound TAL repeats in dHax3. (A) DNA-free and DNA-bound dHax3 are shown for residues 323 to 675, which comprise TAL repeats 2 to 11. The two structures are superimposed by using the N-terminal 23 amino acids, which encompass helix a and the first half of helix b of TAL repeat 2. (B) Superimposition of TAL repeat 2 from DNA-free and DNA-bound dHax3. The structures are superimposed by using the first 23 amino acids. Only the main chains are shown. The orange circle highlights where the structures exhibit variations. (C) Interrepeat interactions in the DNA-bound dHax3. TAL repeats 2 and 3 are shown here. The H bonds and van der Waals interactions are shown in the left and right images, respectively. Water molecules are shown as red spheres, and H bonds are represented by red dashed lines. (D) Interrepeat interactions in the DNA-free dHax3.
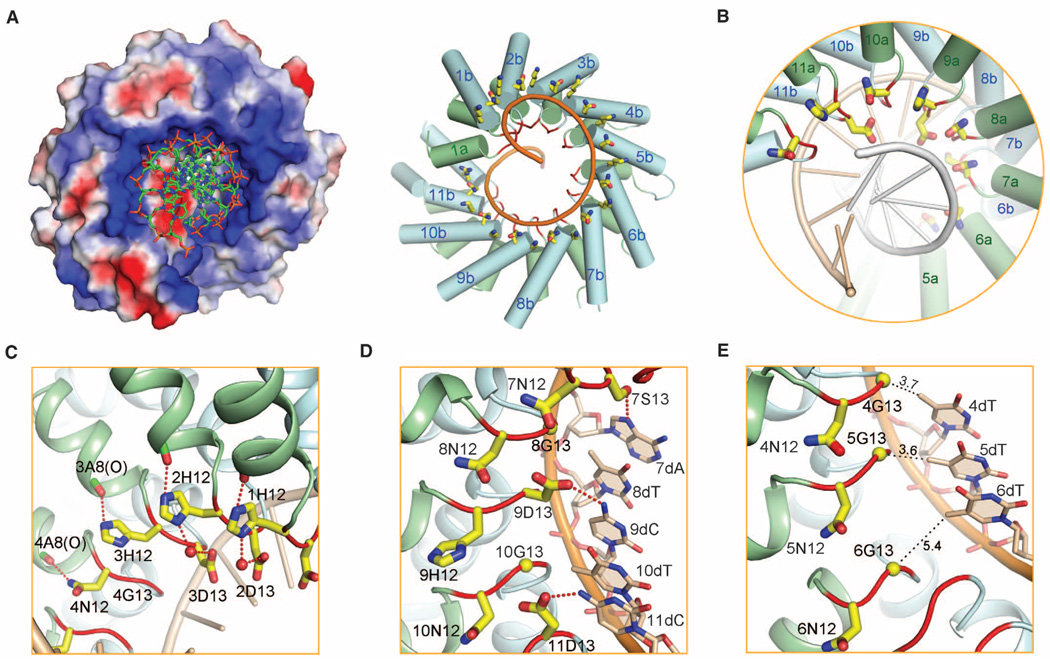
Analysis of the electrostatic surface potential reveals a stripe of positively charged amino acids along the inner ridge of the dHax3 superhelical assembly (Fig. 4A and fig. S7A). Each phosphate group in the sense strand of the DNA duplex is accommodated in a shallow surface pocket along the basic stripe (Fig. 4A, left). Lys16 and Gln17, which are located at the beginning of helix b in each repeat, contribute to the positive electrostatic potential for interaction with the negatively charged phosphate (Fig. 4A, right). Interaction with the phosphate group of DNA duplex, invariant among repeats 1 through 11, is mediated by hydrogen bonds (fig. S7, B and C).
Fig. 4.
DNA recognition by TAL repeats. (A) The phosphate groups of the DNA sense strand is embraced by the positively charged ridge of the dHax3 TAL repeats. The surface electrostatic potential was calculated with PyMOL (30) (left). The invariant residues Lys16 and Gln17 (yellow sticks), located at the beginning of helix b in each TAL repeat, contribute to the positive electrostatic potential (right). The RVD loops are highlighted in red. (B) The two hypervariable residues in each TAL repeat are placed in the major groove of DNA. The sense and antisense strands of DNA are colored gold and gray, respectively. (C) The hypervariable residues at position 12 do not contact DNA bases. These residues, either His or Asn in dHax3 repeats, form hydrogen bonds with the carbonyl oxygen of Ala8 in the same repeat, which may help stabilize the conformation of the RVD loop. When consecutive repeats containing HD are present, His12 forms a water-mediated H bond with Asp13 from the previous repeat. (D) The hypervariable residues at position 13 are direct determinants of DNA base specificity. Shown here are repeats 7 to 11 and the corresponding nucleotides from the DNA sense strand. (E) Recognition of base T by NG. A close-up view on the RVD loops in TAL repeats 4 to 6 in molecule A is shown. Note that the RVD loop of repeat 6 adopts a conformation different from all other RVD loops. All distances are shown in the unit of Å.
The two hypervariable residues in the RVD loops, positioned in close proximity to the sense strand in the DNA major groove (Fig. 4B), play different biochemical roles. Residue 12, either His or Asn in the 11.5 TAL repeats of dHax3 (fig. S1), does not directly contact DNA. Instead, the side chains of His12 and Asn12 point away from DNA bases, each making a direct H bond to the carbonyl oxygen atom of Ala8, which is invariant and located at the C-terminal end of helix a in each TAL repeat (Fig. 4C). Thus, the primary role of residue 12 in TAL repeats is not to directly recognize DNA but to stabilize the local conformation of the RVD loops. Supporting this analysis, there is a water-mediated H bond between the imidazole group of His12 in TAL repeat 1 and the carboxylate oxygen atom of Asp13 in repeat 2 (Fig. 4C). Identical interaction is observed between His12 of repeat 2 and Asp13 of repeat 3. These structural findings demonstrate that His12 or Asn12 contributes indirectly to DNA binding by stabilizing the proper conformation of the RVD loops, which allows residue 13 to specifically recognize DNA bases.
Among the more than 20 codes identified for DNA recognition by TALE RVDs, some are more frequently observed than others (1, 18). The TAL repeats in dHax3 use three codes, in which the two hypervariable residues HD, NG, and NS specifically recognize the DNA bases C, T, and A, respectively (20). These three codes account for about half of all cases reported (1). The structure of DNA-bound dHax3 provides a satisfying explanation to these codes. In the case of HD→C, the carboxylate oxygen atom of Asp13 accepts a H bond from the amine group of cytosine in TAL repeats 1 to 3, 9, and 11 (Fig. 4D). In the case of NS→A, the hydroxyl group of Ser13 in TAL repeat 7 donates a H bond to the N7 atom of adenine (Fig. 4D). Compared with HD, NS is nonselective in that it can recognize all four bases (14). Similar to adenine, guanine also contains a N7 atom, which is likely recognized by Ser13 in the same manner. Recognition of cytosine or thymine may require a slightly different conformation of the RVD loop, a scenario awaiting further structural evidence.
The correlation between NG and the base T is intriguing. Instead of providing any specific interaction, the placement of Gly at position 13 allows sufficient space to accommodate the 5-methyl group of thymine (Fig. 4E). In TAL repeats 4, 8, 10, and 12, the distance between the Cα of Gly13 and the 5-methyl group of thymine is between 3.4 and 3.7 Å, allowing van der Waals interaction. Substitution of Gly with any other residue would likely introduce steric clash with the 5-methyl group of thymine, providing a structural explanation for the observation that recognition of the base T usually requires Gly at position 13 (1). However, in repeats 5 of molecule A and 6 of molecule B, the distance between Gly-Cα and the 5-methyl group of thymine is more than 5 Å. We speculate that mutation of Gly13 to an amino acid with a short side chain may be tolerated here.
Both the structure and the mode of DNA binding by the TAL repeats differ from those of other known DNA-binding domains such as zinc-finger domain, basic leucine zipper motif, and helix-turn-helix motif (fig. S8). The modular nature of the DNA-TAL repeats is also different from that of known RNA-binding proteins such as trp RNA-binding attenuation protein (TRAP) (28). The closest entry from an exhaustive search of the Protein Data Bank (PDB) using DALI (29) is the structure of DNA-bound MTERF1 (mitochondria transcription terminator 1) (fig. S8), which also exhibits a superhelical conformation and has a Z score of 7.0 and RMSD of 3.2 Å over 184 aligned Cα atoms with dHax3. However, the MTERF motif comprises two α helices and one 310-helix, with considerable conformational variation among repeats. In addition, MTERF1 binding results in substantial unwinding of DNA duplex (fig. S8).
Our structural investigation provides explanation for about half of the frequently used codes for DNA recognition by TAL repeats. Among the remaining codes, how NI and NN recognize the bases A and G/A, respectively, remains to be elucidated. We suspect that the second Asn residue of NN may favor G/A through a specific H bond. Some of the less frequently used codes can also be explained by our available structural information. For example, explanation for the code ND→C should be similar to that for HD→C, which was observed here (Fig. 4D). On the other hand, rationalization for the code XG→T is likely the same as that for NG→T (Fig. 4E). Because 5′-methylcytosine is similar to T, we suspect that XG might also be able to recognize 5′-methylcytosine.
Our study represents a step toward comprehensive rationalization of sequence-specific DNA recognition by TAL repeats. Many questions remain. It is yet to be seen whether the arrangement of 11 repeats per turn is unique to dHax3 or a common feature of all TAL repeats. Although the base T is required for repeat “0” (14, 20), our structure of DNA-bound dHax3 does not provide an intuitive clue, because T at position zero is not particularly coordinated by either the N-terminal domain or the adjacent repeats (fig. S9). Nonetheless, visualization of the modular, base-specific recognition by the TAL repeats may greatly facilitate rational design of novel DNA-binding proteins with a range of pragmatic applications.
Supplementary Material
Acknowledgments
We thank J. He and Q. Wang at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U and K. Hasegawa and T. Kumasaka at the Spring-8 beamline BL41XU for on-site assistance. This work was supported by funds from the Ministry of Science and Technology (grant numbers 2009CB918801, 2009CB918802, and 2011CB910501); projects 30888001, 91017011, and 31070644 of the National Natural Science Foundation of China; and Tsinghua University 985 Phase II funds. Coordinates and structure factors for the DNA-free and DNA-bound structures have been deposited with the Protein Data Bank under accession codes 3V6P and 3V6T.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/science.1215670/DC1
Materials and Methods
Figs. S1 to S9
Tables S1 to S3
References (31–44)
References and Notes
- 1.Boch J, Bonas U. Annu. Rev. Phytopathol. 2010;48:419. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 2.Bai J, Choi SH, Ponciano G, Leung H, Leach JE. Mol. Plant Microbe Interact. 2000;13:1322. doi: 10.1094/MPMI.2000.13.12.1322. [DOI] [PubMed] [Google Scholar]
- 3.Gu K, et al. Nature. 2005;435:1122. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- 4.White FF, Yang B. Plant Physiol. 2009;150:1677. doi: 10.1104/pp.109.139360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonas U, Conrads-Strauch J, Balbo I. Mol. Gen. Genet. 1993;238:261. doi: 10.1007/BF00279555. [DOI] [PubMed] [Google Scholar]
- 6.Swarup S, Yang Y, Kingsley MT, Gabriel DW. Mol. Plant Microbe Interact. 1992;5:204. doi: 10.1094/mpmi-5-204. [DOI] [PubMed] [Google Scholar]
- 7.Schornack S, Meyer A, Römer P, Jordan T, Lahaye T. J. Plant Physiol. 2006;163:256. doi: 10.1016/j.jplph.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Kay S, Bonas U. Curr. Opin. Microbiol. 2009;12:37. doi: 10.1016/j.mib.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Büttner D, Bonas U. FEMS Microbiol. Rev. 2010;34:107. doi: 10.1111/j.1574-6976.2009.00192.x. [DOI] [PubMed] [Google Scholar]
- 10.Bonas U, Stall RE, Staskawicz B. Mol. Gen. Genet. 1989;218:127. doi: 10.1007/BF00330575. [DOI] [PubMed] [Google Scholar]
- 11.Hopkins CM, White FF, Choi SH, Guo A, Leach JE. Mol. Plant Microbe Interact. 1992;5:451. doi: 10.1094/mpmi-5-451. [DOI] [PubMed] [Google Scholar]
- 12.Kay S, Hahn S, Marois E, Hause G, Bonas U. Science. 2007;318:648. doi: 10.1126/science.1144956. [DOI] [PubMed] [Google Scholar]
- 13.Römer P, et al. Science. 2007;318:645. doi: 10.1126/science.1144958. [DOI] [PubMed] [Google Scholar]
- 14.Boch J, et al. Science. 2009;326:1509. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 15.Moscou MJ, Bogdanove AJ. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 16.Kay S, Boch J, Bonas U. Mol. Plant Microbe Interact. 2005;18:838. doi: 10.1094/MPMI-18-0838. [DOI] [PubMed] [Google Scholar]
- 17.Schornack S, Minsavage GV, Stall RE, Jones JB, Lahaye T. New Phytol. 2008;179:546. doi: 10.1111/j.1469-8137.2008.02487.x. [DOI] [PubMed] [Google Scholar]
- 18.Cermak T, et al. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogdanove AJ, Voytas DF. Science. 2011;333:1843. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 20.Mahfouz MM, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2623. [Google Scholar]
- 21.See supporting material on Science Online
- 22.Groves MR, Hanlon N, Turowski P, Hemmings BA, Barford D. Cell. 1999;96:99. doi: 10.1016/s0092-8674(00)80963-0. [DOI] [PubMed] [Google Scholar]
- 23.Kajander T, Cortajarena AL, Mochrie S, Regan L. Acta Crystallogr. 2007;D63:800. doi: 10.1107/S0907444907024353. [DOI] [PubMed] [Google Scholar]
- 24.Murakami MT, et al. Proteins. 2010;78:3386. doi: 10.1002/prot.22846. [DOI] [PubMed] [Google Scholar]
- 25.Huber AH, Nelson WJ, Weis WI. Cell. 1997;90:871. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 26.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Cell. 1998;94:193. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y, et al. Cell. 2006;127:1239. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 28.Antson AA, et al. Nature. 1999;401:235. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 29.Holm L, Rosenström P. Nucleic Acids Res. 2010;38:W545. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeLano WL. 2002 on www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.