Abstract
Sialoadhesin (Sn) is a sialic acid–binding Ig-like lectin expressed selectively on macrophage subsets. In a model of experimental autoimmune encephalomyelitis, Sn interacted with sialylated ligands expressed selectively on CD4+Foxp3+ regulatory T cells (Tregs) and inhibited their proliferation. In this study, we examined the induction of Sn ligands (SnL) on all splenic CD4+ T cells following in vitro activation. Most CD4+ Tregs strongly upregulated SnL, whereas only a small subset of ∼20% CD4+Foxp3− T cells (effector T cells [Teffs]) upregulated SnL. SnL+ Teffs displayed higher levels of activation markers CD25 and CD69, exhibited increased proliferation, and produced higher amounts of IL-2 and IFN-γ than corresponding SnL− Teffs. Coculture of activated Teffs with Sn+ macrophages or Sn+ Chinese hamster ovary cells resulted in increased cell death, suggesting a regulatory role for Sn–SnL interactions. The key importance of α2,3-sialylation in SnL expression was demonstrated by increased binding of α2,3-linkage–specific Maackia amurensis lectin, increased expression of α2,3-sialyltransferase ST3GalVI, and loss of SnL following treatment with an α2,3-linkage–specific sialidase. The induction of SnL on activated CD4+ T cells was dependent on N-glycan rather than O-glycan biosynthesis and independent of the mucin-like molecules CD43 and P-selectin glycoprotein ligand-1, previously implicated in Sn interactions. Induction of ligands on CD4+Foxp3− Teffs was also observed in vivo using the New Zealand Black × New Zealand White F1 murine model of spontaneous lupus and SnL levels on Teffs correlated strongly with the degree of proteinuria. Collectively, these data indicate that SnL is a novel marker of activated CD4+ Teffs that are implicated in the pathogenesis of autoimmune diseases.
Introduction
Sialoadhesin (Sn), also known as Siglec-1 or CD169, is a founding member of the family of sialic acid (Sia)–binding Ig superfamily lectins (siglecs) and normally restricted in its expression to macrophage subsets. Siglecs are type I membrane proteins containing a V-set Ig domain that binds Sia and variable numbers of C2-set Ig domains. Sn is one of the largest members of the Ig superfamily, with 17 extracellular Ig domains that are well conserved in mammals (1). Ig domains 4-17 appear to constitute a stem region made of alternating short and long C2-set domains, which are thought to be derived from an ancestral two-domain precursor that expanded via gene duplication. Unlike most other siglecs, Sn lacks intracellular tyrosine-based signaling motifs, and the cytoplasmic tail is poorly conserved (2). These features are consistent with a predominant role of Sn in mediating cell–cell interactions rather than intrinsic cell signaling. The elongated extracellular region of Sn is thought to be important for the adhesive function of Sn, as it allows the Sia binding V-set Ig domain to project away from the macrophage glycocalyx, thereby reducing inhibitory cis-interactions with neighboring Sia (3). Sn is rather promiscuous, and although it prefers Sias in α2-3 linkage, it can also bind to α2-6 and α2-8 glycosidic linkages with low affinity (1, 4). To achieve stable, high-avidity interactions with ligands on cells, clustering of both the receptors and ligands is necessary (1). The detection of Sn ligands (SnL) on cells can be achieved by using preformed complexes containing a mixture of recombinant Sn fused to the Fc region of human IgG1 and a polyclonal anti-human IgG Ab (5).
Under homoeostatic conditions, Sn is found on discrete subsets of macrophages with highest expression in secondary lymphoid tissues (1, 4, 6). In lymph nodes, resident macrophages within the subcapsular sinus are strongly positive, and these cells are specialized in transfer of Ags to follicular B cells and invariant NKT cells, playing a key role as gatekeepers in viral infections (7, 8). Lower levels of expression are seen on other resident macrophages such as Kupffer cells in the liver, alveolar and interstitial macrophages in the lung, and macrophages in colonic lamina propria (6). Under inflammatory conditions, Sn can be strongly upregulated on macrophages, as observed in autoimmune diseases such as experimental autoimmune encephalomyelitis (EAE) (9) and human rheumatoid arthritis (6). Sn is normally undetectable on circulating monocytes, but is induced on these cells in patients with HIV infection (10), systemic lupus erythematosus (SLE) (11), and systemic sclerosis (12) in a type I IFN–dependent manner.
The biological functions of Sn have been investigated using Sn-deficient mice. Although these mice are viable and healthy (13), a proinflammatory role for Sn has been demonstrated in a number of mouse models of T cell–dependent autoimmune diseases in which Sn-deficient mice show reduced disease severity that correlates with reduced numbers of infiltrating pathogenic T cells and macrophages (13–15). In a recent study of EAE, Sn specifically interacted with sialylated SnL upregulated in disease on CD4+CD25+Foxp3+ regulatory T cells (Tregs) (9). Upregulation of SnL was not observed on CD4+CD25-Foxp3− effector T cells (Teffs). The interaction between SnL+ Tregs and Sn exerted a negative effect on the frequency of Tregs in the inflamed CNS and peripheral lymphoid tissues (9). Thus, one pathway through which Sn could function as a proinflammatory molecule would be through suppression of Treg expansion via interactions with the upregulated SnL.
In this study, we demonstrate that SnL can be readily induced in vitro on both CD4+CD25+Foxp3+ Tregs and a subset of highly activated CD4+CD25−Foxp3− Teffs. We show that exposure of activated CD4+ T cells to Sn expressed by bone marrow–derived macrophages (BMDM) leads to increased cell death and that SnL induced on Teffs is associated with α2-3-linked Sias on N-glycans rather than O-glycans. Finally, we demonstrate that SnL can be induced on Teffs in vivo in a model of murine lupus in which SnL levels correlate strongly with proteinuria.
Materials and Methods
Mice
C57BL/6 and New Zealand Black × New Zealand White F1 (NZBWF1) mice were purchased from Harlan (Oxford, U.K.). The generation of Sn-deficient mice was reported previously (13). All wild-type (WT) and Sn−/− mice used were intercross offspring of heterozygotes backcrossed for >20 generations onto a C57BL/6 background and used at 8–16 wk of age in all in vitro experiments. NZBWF1 mice were monitored for the development of proteinuria using Uristix (Bayer). Diseased mice were defined as having persistent proteinuria of >100 mg/dl on at least two occasions. Splenocytes from C2GnT 1-3 triple-knockout (16), CD43−/− (17), and P-selectin glycoprotein ligand-1 (PSGL-1)−/− (18) mice were provided by the Ziltener Laboratory (University of British Columbia, Vancouver, British Columbia, Canada). All mice were housed under specific pathogen-free conditions. The animal protocols used in this study were approved by the Ethical Review Committee of the University of Dundee. All procedures involving living animals were conducted according to the requirements of the United Kingdom Home Office Animals Scientific Procedures Act (1986), under PPL 60/3856.
Cell culture
Complete RPMI (RPMI 1640 [Life Technologies] plus 10% v/v FCS [PAA Laboratory] plus 2 mM l-glutamine [Life Technologies] plus 0.2 M 2-ME [Life Technologies] plus 10 mM HEPES (Life Technologies) plus 100 U/ml penicillin/0.1 mg/ml streptomycin [Life Technologies]) was used with splenocytes, CD4+ T cell cultures, and CD4+/BMDM cocultures. DMEM (Life Technologies) plus 10% v/v FCS plus 100 U/ml penicillin and 0.1 mg/ml streptomycin was used in BMDM cultures (without CD4+ T cells). Serum-free UltraCHO (BioWhittaker) was used in cultures of Sn–Fc-secreting Chinese hamster ovary (CHO) cells.
In vitro T lymphocyte activation
Activation of enriched CD4+ T cells and Tregs was carried out using anti-CD3/CD28 Dynabeads (Invitrogen). Beads were washed with PBS plus 2 mM EDTA plus 0.5% v/v FCS and then used at a bead/cell ratio of 1:1 to activate cells for 48–72 h. At the end of the period of activation, beads were removed using a magnetic particle concentrator (Invitrogen). Activated cells were then either used for flow cytometry studies or cocultured with BMDM or CHO cells. In proliferation experiments of enriched CD4+ T cells in coculture with BMDM, soluble anti-CD3 (clone 2C11; BD Biosciences) at 5 μg/ml was used. Whole splenocytes were stimulated with plate-bound anti-CD3, clone 2C11 (BD Biosciences). The Ab was used at 5 μg/ml unless indicated otherwise. Twenty-four– or six-well plates were coated with the Ab diluted in carbonate-bicarbonate buffer (pH 9.6; Sigma-Aldrich) for 2 h at 37°C. Plates were then washed with PBS to remove unbound Ab and blocked with RPMI 1640 plus 10% v/v FCS for 20 min. Subsequently, media was aspirated, and cell suspensions were added at 1 × 106/2 ml (24-well plate) or 3 × 106/6 ml (six-well plates). Activation was carried out for 48–72 h unless otherwise indicated.
Sialidase treatment
Two forms of sialidase were used in this project. Firstly, Vibrio cholerae sialidase (Sigma-Aldrich) was used, which cleaves Sias in α2-3, α2-6, and α2-8 glycosidic linkages. Cells were washed three times with serum-free DMEM, suspended at 106 cells per 60 μl serum-free DMEM, and incubated with 0.1 U/ml of Vibrio cholera sialidase for 1 h at 37°C. Cells were then washed three times with DMEM plus 10% v/v FCS and used in further analysis. Secondly, sialidase L (Vector Laboratories) from Macrobdella leech exhibits α2,3-specific sialidase activity (19). Cells were washed three times in HBSS and suspended at 106 cells/ml HBSS. Twenty-five units sialidase L was used per 106 cells for 2 h of incubation at 37°C. Cells were then washed with HBSS three times and used in further analysis. In addition, cells were labeled with the plant lectin SNA to assess the retention of α2,6-linked Sias following sialidase L treatment. Untreated cells were kept in medium alone for the duration of the incubation.
Flow cytometry
Anti-CD4 allophycocyanin (clone L3T4), -CD4 PerCPCy5.5 (clone RM4.5), -CD25 PE (clone PC61.5), -CD69 allophycocyanin (clone H1.2F3), -CD62L PE or allophycocyanin (clone MEL-14), -Foxp3 PE or allophycocyanin (clone FJK-16), -CTLA4 allophycocyanin or biotin (clone UC10-4B9), -GITR allophycocyanin (clone DTA-1), -CD95L biotin (clone MFL3), -CD95 biotin (clone 15A7), –IL-2 PE (clone JES6-5H4), –IFN-γ PE (clone XMG1.2), -CD45RB FITC (clone C363.16a), and rat IgG controls were purchased from eBioscience. Anti-CD43 FITC (clone 1B11) and rat IgG FITC isotype control were purchased from BioLegend. Maackia amurensis lectin (MAL)-biotin, Sambucus nigra agglutinin lectin (SNA)-biotin, peanut agglutinin-biotin, and Phaseolus vulgaris leucoagglutinin-biotin were all purchased from Vector Laboratories. Streptavidin-FITC and streptavidin–allophycocyanin were purchased from eBioscience. All staining and washes were carried out in FACS buffer (PBS plus 2% v/v FCS plus 2 mM EDTA) on ice. Cells were FcR blocked with 0.25 μg anti-CD16/CD32 mAb 2.4G2 per 106 cells in 25 μl for 20 min, washed, and then stained for 1 h with optimal dilutions of relevant Abs based on prior Ab titrations. Cells were washed twice with FACS buffer and acquired using a BD FACSCalibur flow cytometer (BD Biosciences). FlowJo software (Tree Star) was used for data analysis.
SnL detection by flow cytometry
SnL was detected using Sn–Fc fusion protein, which consists of the first three Ig domains of Sn fused to the Fc portion of human IgG1 (5). Sn–Fc at 10 μg/ml, produced as a tissue-culture supernatant from stably transfected CHO cells (1), was preincubated for 1 h on ice with fluorescent (Alexa Fluor 488 [Invitrogen] or DyLight 649 [Jackson ImmunoResearch Laboratories]) goat anti-human IgG Fc Ab. Staining was initially optimized by staining human RBCs with complexes made of different ratios of goat anti-human Fc Ab to Sn–Fc (Supplemental Fig. 1). A ratio of 5 μg/ml Sn–Fc to 1/100 goat anti-human Fc was typically used in analyses of T cell subsets.
Intracellular staining
Following surface labeling, intracellular Foxp3 was detected using a Foxp3 staining kit (eBioscience) as per the manufacturer’s instructions. Briefly, cells were fixed, washed twice in permeabilization buffer, and then incubated with the recommended concentration of PE- or allophycocyanin-conjugated anti-mouse Foxp3 (clone FJK-16s; eBioscience) for 1 h at 4°C. Cells were then washed twice with permeabilization buffer and analyzed by flow cytometry. Intracellular cytokines IFN-γ and IL-2 were detected using MACS-enriched CD4+CD25− T cells. Following enrichment, cells were stimulated with anti-CD3/CD28 Dynabeads (Invitrogen) at a 1:1 ratio for 48 h. Cells were washed following the removal of beads and restimulated with 10 ng/ml phorbol 12,13-dibutyrate (Sigma-Aldrich) and ionomycin at 10 ng/ml (Sigma-Aldrich) for 3 h in the presence of 3 μg/ml brefeldin A (eBioscience). Following surface labeling with Sn–Fc precomplexes, cells were fixed, and intracellular staining was performed as above.
Cell isolation
CD4+ T cells were enriched using MACS (Miltenyi Biotec). Single-cell suspensions from spleen were prepared by mashing the spleen through a 100-μm cell strainer (BD Biosciences). RBCs were removed using RBC Lysing Buffer (Sigma-Aldrich). This was followed by DNase treatment (2 μg/ml) (DNase I; Roche) to remove any cell clumps. CD4+, CD4+CD25+, or CD4+CD25− cells were selected using a mouse CD4+ T cell isolation kit II (Miltenyi Biotec) and a mouse CD4+CD25+ Regulatory T cell isolation kit (Miltenyi Biotec), respectively, according to the manufacturer’s protocol. A purity of >94% was obtained in all experiments.
Proliferation assays
In vitro proliferation assays were performed with Click-iT EdU (Invitrogen). 5-ethynyl-2′-deoxyuridine (EdU) is a nucleoside analog to thymidine and is incorporated in DNA during active DNA synthesis. Detection of incorporated EdU is based on a click reaction, a copper-catalyzed reaction between an alkyne (EdU) and an azide (detection dye). Activated cells were incubated with 10 mM EdU over the last 12 h of activation. Cells were then harvested and surface labeled with Sn–Fc precomplexes followed by intracellular staining for nuclear-incorporated EdU.
BMDM
To obtain a homogeneous Sn+ BMDM population, a two-phase enrichment protocol was employed. First, BMDM were prepared from bone marrow collected from the femora and tibiae of age- and sex-matched WT and Sn−/− C57BL/6 mice between 8 and 16 wk of age. The marrow was flushed out using a 25-gauge needle and syringe filled with DMEM plus 10% v/v FCS. Cells were collected by centrifugation at 350 × g for 5 min. A single-cell suspension was obtained by pipetting, followed by passage through a 70-μm cell strainer (BD Biosciences). Cells from one mouse were suspended in 20 ml DMEM plus 10% v/v FCS supplemented with macrophage colony-stimulating factor 1 (25 ng/ml) purchased from PeproTech. Cell suspensions were then plated on a 100-mm bacterial Petri dish (BD Biosciences). Cell cultures were incubated at 37°C in 8% CO2 incubator. The second phase involved stimulation of enriched BMDMs with IFN-α (R&D Systems) to induce Sn expression. On day 7, nonadherent cells were gently removed by washing with PBS twice. Adherent cells were lifted by incubation with 4 mg/ml lidocaine (Sigma-Aldrich) plus 5 mM EDTA in PBS for 2 to 3 min in an 8% CO2 incubator at 37°C. Cells were then harvested and washed with DMEM plus 10% v/v FCS. Next, cells were counted and plated at 2 × 105 cells/well of a 24-well plate in the presence of 25 U/ml of mouse IFN-α. On day 3 post–IFN-α treatment, dead cells were removed by gentle washing with DMEM plus 10% v/v FCS. Next, cells were either lifted with 4 mg/ml lidocaine plus 5 mM EDTA in PBS or used for flow cytometry studies or in coculture experiments with activated CD4+ T cells.
Cell death assays
Enriched BMDM from WT and Sn−/− mice were washed three times with RPMI 1640 plus 10% v/v FCS, following 3 d stimulation with IFN-α. Enriched CD4+ T cells previously stimulated for 48 h with anti-CD3/CD28 Dynabeads (Invitrogen) were added at a 1:1 ratio to BMDM in complete RPMI. At 12 h, total cells were lifted with 4 mg/ml lidocaine in 5 mM EDTA plus PBS. The viability of CD4+ T cells was assessed after 12 h by flow cytometry using Yo-Pro viability dye (Invitrogen) and CD4 labeling to distinguish CD4+ T cells from macrophages.
In death assays in which CHO cells were used instead of BMDM, the former were cultured initially in Ham’s F12 medium supplemented with 10% v/v FCS, until >80% confluent. CHO cells were then lifted with 4 mg/ml lidocaine plus 5 mM EDTA in PBS, washed, and plated in flat-bottom 96-well plates in medium with 2 mM sodium butyrate for 24 h. Wells were then washed three times with RPMI 1640 plus 10% v/v FCS, and activated CD4+ T cells were added at a 2:1 (CD4/CHO cells) ratio. At 12 h, total cells were lifted, and viability was measured by flow cytometry as above.
Real-time PCR
Three samples each comprising pooled spleens from five female C57BL/6 mice between the age of 8 and 10 wk were used in these experiments. CD4+CD25− T cells (purity >95%) were isolated from spleens as described above. CD4+CD25− T cells were then activated with anti-CD3/CD28 Dynabeads (cell/bead ratio 1:1; Invitrogen) for 72 h in complete RPMI. Cells were then washed with complete RPMI and beads removed using a magnetic particle concentrator (Invitrogen). Cells were blocked for nonspecific binding with 0.25 μg/106 cells/100 μl azide-free low-endotoxin anti-CD16/CD32 (BD Biosciences) for 20 min on ice and then surface-labeled with Sn–Fc precomplexes (Alexa Fluor 488) for 1 h on ice. Cells were washed three times with PBS plus 2% v/v FCS and counterstained with DAPI. Subsequently, viable activated cells (DAPI-negative and forward scatter high) were sorted by FACS into SnL+ (Alexa Fluor 488–positive) and SnL− (Alexa Fluor 488–negative). Cells were counted and pelleted by centrifugation at 350 × g for 10 min. Supernatants were aspirated completely, and samples were lysed with RLT buffer (Qiagen). RNA extraction was accomplished using RNeasy micro kit (Qiagen) according to the manufacturer’s instructions. The quality and quantity of isolated RNA was measured with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). cDNA synthesis was performed with QuantiTect reverse transcription kit (Qiagen) with integrated removal of genomic DNA contamination, according to the manufacturer’s instructions. All cDNA samples were stored at −20°C until used in RT-PCR experiments. Primers used in RT-PCR experiments were as follows: ST3Gal I: 5′-TCGCCTGGTGCCTGGCAATG-3′ and 5′-TGGTCCGGCTCCCAACGTCT-3′; ST3Gal III: 5′-CATCTTCCCCAGGTTCTCCAA-3′ and 5′-ACTTGCGAAAGGAGTCATCCA-3′; ST3Gal IV: 5′-CTGAGCCTGCCCATACAACA-3′ and 5′-GGCTAGCAGACCCGTGGTT-3′; ST3Gal VI: 5′-CTGGTGGCCATATTCCTGAG-3′ and 5′-CCGGACATGGCAGCAACC-3′; C2GnT1: 5′-CTGCTGTGATTTTGGGTAGTGAGT-3′ and 5′-GAGCCATTCGTGCCAAACAT-3′; 1,4Gal T: 5′-ACAAGAAAAATGAGCCCAATCC-3′ and 5′-CGAAGCGCATCGTTTCCTT-3′; and FUT 7 Bcl2: 5′-CGATAGCTCAGCACCCAGTTG-3′, 5′-GAGGCTGGGTAGGTGCATGT-3′, 5′-CATGGAATCGCCCAGTAATACC-3′, and 5′-GCATCTTGGCCTTGAGATCAA-3′.
Statistical analysis
Statistical significance was determined by Student t test, one-way ANOVA, two-way ANOVA, or by Mann–Whitney U test. A p value <0.05 was considered to be significant.
Results
SnL are upregulated on CD4+ T cells following TCR ligation
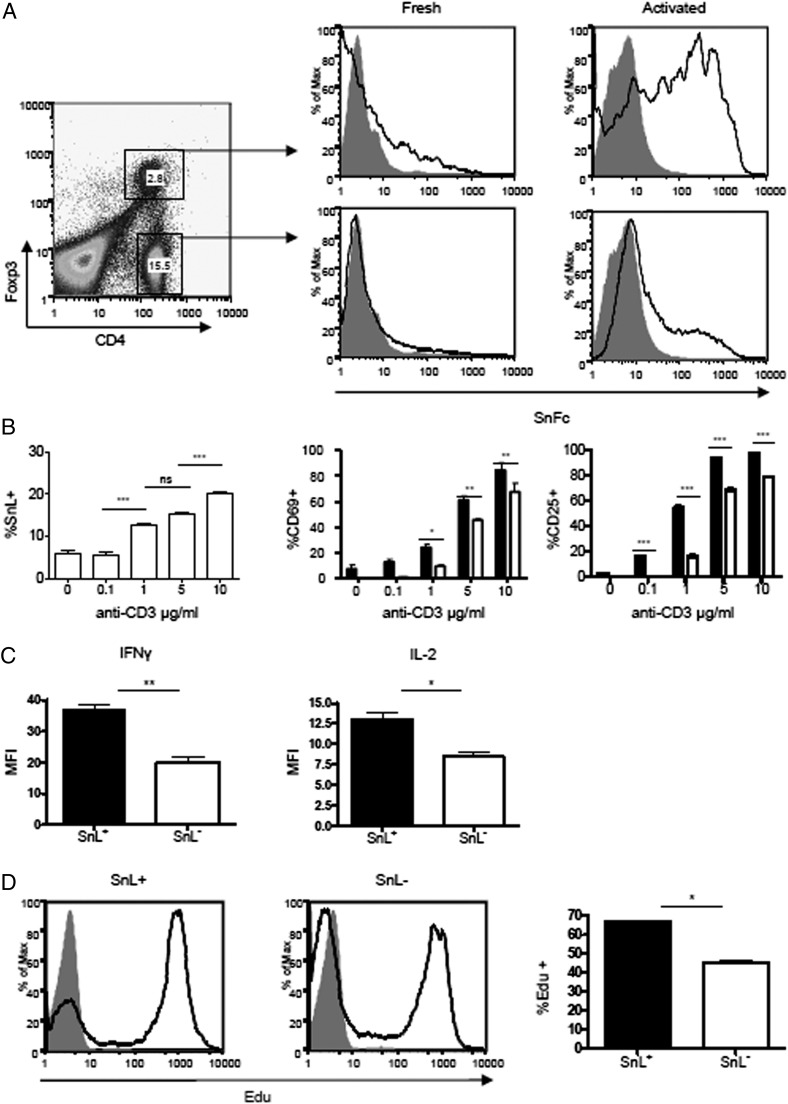
To identify SnL on T cell subsets activated in vitro, we used a soluble recombinant form of Sn containing the first three Ig domains fused to the Fc region of human IgG1, precomplexed with a fluorescently labeled anti-human IgG. Freshly harvested splenocytes from C57BL/6 mice showed low binding to Sn–Fc complexes (Fig. 1A). Following activation with plate-bound anti-CD3 mAb for 48 h, strong upregulation of SnL was seen for most CD4+Foxp3+ T cells (Tregs), consistent with the previous observation of SnL expression induced on a subset of these cells in EAE (9). Interestingly, a subset of ∼20% CD4+Foxp3− T cells (Teffs) also upregulated expression of SnL (Fig. 1A, 1B). The binding of Sn–Fc on both Tregs and Teffs was Sia dependent, as evidenced by loss of Sn–Fc binding with sialidase-treated cells (Fig. 1A). These findings suggested that SnL induction is an event that follows TCR ligation and that the expression was biased toward TCR-triggered Tregs compared with Teffs.
FIGURE 1.
Upregulation of SnL on CD4+ T cells. (A) Splenocytes from C57BL/6 mice were activated with plate-bound anti-CD3 (5 μg/ml) for 72 h and compared with freshly isolated splenocytes for the expression of SnL. Intracellular staining for Foxp3 was undertaken to examine SnL on both Tregs and Teffs in both freshly isolated and activated groups. Dot plots of activated splenocytes stained for CD4 and intracellular Foxp3. CD4+Foxp3+ (Tregs) and CD4+Foxp3− (Teffs) are gated with percentages of total splenocytes shown for each gate. Histograms of SnL expression (black lines) and sialidase-treated controls (solid gray) are presented. (B) SnL+CD4+ T cells display a higher state of activation following TCR ligation compared with SnL−CD4+ T cells. Percentages of CD69+ and CD25+ on SnL+ (black bars) and SnL− Teffs (unfilled bars) following 48 h of stimulation with different concentrations of plate-bound anti-CD3. Data represent means ± SEM of triplicate samples for each anti-CD3 titration. Data are representative of three independent experiments. Statistical analysis was done using two-way ANOVA with Bonferroni posttest analysis. (C) Intracellular cytokine expression in SnL+ and SnL− Teffs. MACS-sorted CD4+CD25− T cells from three C57BL/6 mice were activated with anti-CD3/CD28 beads for 48 h to induce SnL. Activated cells were restimulated with PdBU and ionomycin in the presence of brefeldin A (as described in Materials and Methods). Following surface labeling with Sn–Fc, intracellular staining for IFN-γ and IL-2 was carried out. (D) SnL+ Teffs display a hyperproliferative response following TCR ligation. CD4+CD25− T cells were activated with anti-CD3/CD28 beads for 72 h. EdU was added at 10 μM in the last 12 h of culture. Histograms represent EdU expression on SnL+ (left panel) and SnL− (middle panel) as compared with resting unstimulated Teffs (solid gray histogram). Bar graph (right panel) shows the percentage of EdU+ cells in SnL+ (black bar) and SnL− (unfilled bar). Data represent means ± SEM of triplicate samples. *p < 0.05, **p < 0.01, ***p < 0.001.
To determine whether the upregulation of SnL on the subset of activated Teffs was associated with an altered state of activation compared with SnL− Teffs, the expression of early activation markers CD25 and CD69 was examined by flow cytometry following 48 h of exposure to different coating concentrations of plate-bound anti-CD3 Ab. This showed that the percentages of CD69+ and CD25+ cells were consistently higher in SnL+ CD4+ cells compared with SnL− counterparts (Fig. 1B). We also compared SnL+ and SnL− subsets for other indicators of activation, including intracellular expression of IL-2 and IFN-γ (Fig. 1C) and proliferative responses (Fig. 1D). In both cases, SnL+ Teffs showed hallmarks of being more activated than the SnL− subset. Taken together, these data indicated that SnL positivity identifies a subset of activated CD4+ T cells with a heightened state of activation following TCR ligation.
Engaging SnL on CD4+ T cells induces cell death
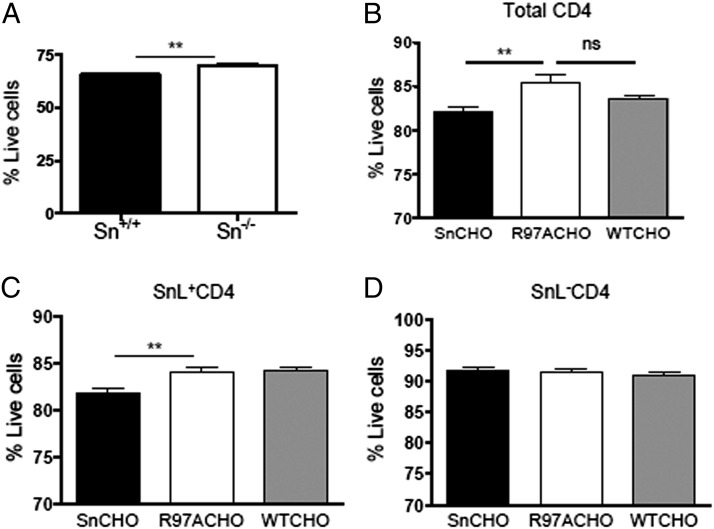
To investigate the potential functional consequences of engaging SnL on activated CD4+ Teffs with Sn on macrophages, coculture experiments were performed. BMDM were treated with IFN-α for 3 d to upregulate Sn expression in WT macrophages (Supplemental Fig. 2), and BMDM from Sn−/− mice were treated identically as a control. Following coculture with activated CD4+ T cells for 12 h, a significant reduction in viability was seen in CD4+ T cells cultured with Sn+/+ BMDM compared with CD4+ T cells cocultured with Sn−/− BMDM (Fig. 2A). These findings suggested that engaging SnL on CD4+ T cells leads to enhanced cell death. To explore whether Sn is directly involved in inducing cell death, CHO cells expressing Sn, a mutant form of Sn (SnR97A), and WT CHO were used (20). SnR97A lacks sugar-binding properties due to the substitution of arginine 97 with alanine (21). The viability of CD4+ T cells was significantly lower in cocultures with Sn CHO cells as compared with SnR97A and WT CHO groups. Interestingly, the reduced viability was due to loss of SnL+CD4+ T cells rather than their SnL− counterparts. Taken together, these data indicate a novel function for Sn in inducing CD4+ T cell death.
FIGURE 2.
Sn induces cell death of activated CD4+ T cells. Activated enriched CD4+ T cells (from three C57BL/6 mice) were cocultured for 12 h with IFN-α–stimulated BMDM from WT and Sn-deficient mice. Total cells were lifted and stained for surface expression of CD4 and with the Yo-Pro membrane permeability dye. (A) The percentage of live cells was defined as being Yo-Pro negative. Data represent means ± SEM of triplicate samples. Data are representative of three independent experiments. Statistical analysis was done using Student t test. (B) Coculture experiments using CHO cells expressing Sn, SnR97A, or WT-CHO. Total cells were lifted and stained for expression of SnL and Yo-Pro. The percentage of total (B), SnL+ (C), and SnL− (D) cells are shown. Statistical analysis was done using Mann–Whitney U test. **p < 0.01.
To investigate the potential mechanisms by which SnL engagement enhances cell death, we analyzed expression of key members of the intrinsic (Bcl2) and extrinsic (Fas and Fas ligand) apoptotic pathways. These showed a similar mRNA expression of Bcl2 (Supplemental Fig. 3A) and similar levels of surface expression of Fas (Supplemental Fig. 3B) between SnL+ and SnL− CD4+ T cells. Interestingly, the surface expression of Fas ligand was found to be consistently higher on SnL+ compared with SnL− CD4+ T cells (Supplemental Fig. 3D). These findings suggest that SnL+CD4+ T cells might have the capacity to induce cell death on neighboring SnL−CD4+ T cells in a fratricidal manner (22).
Upregulation of SnL on CD4+ T cells is associated with increased α2,3 sialylation
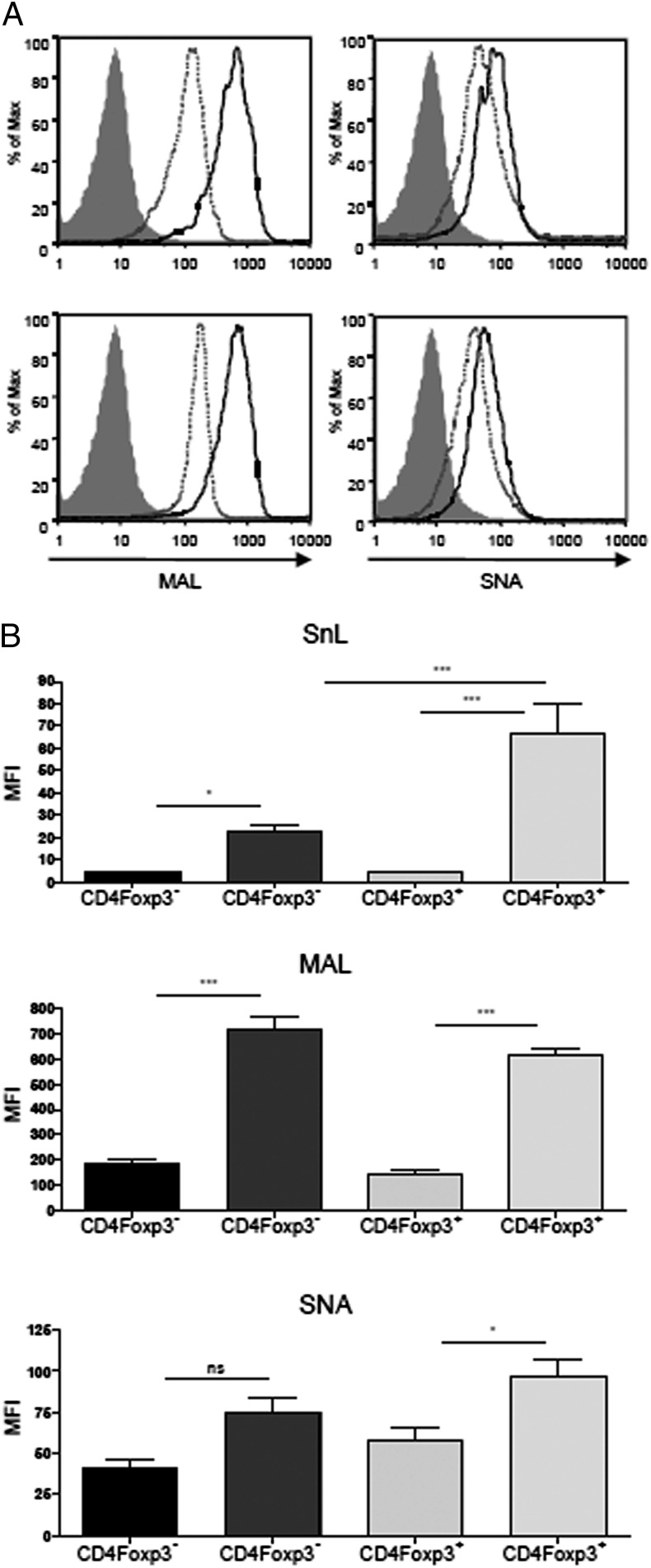
Sn has a preference for α2,3- over α2,6-linked Sias and these glycosidic linkages can be discriminated by plant lectins MAL and Sambucus nigra lectin (SNA), respectively. SnL upregulation on both Tregs and Teffs was associated with a similar strong upregulation of α2,3 sialylation, as evidenced by increased binding of MAL (Fig. 3A, 3B). In contrast, only small increases in α2,6 sialylation were observed for both Teffs and Tregs based on binding to SNA (Fig. 3A, 3B). Binding of Sn–Fc to Teffs paralleled the increased α2,3 sialylation. However, for Tregs, the increased binding of Sn–Fc was disproportionately higher than the increase observed with MAL (Fig. 3B), raising the possibility that additional ligands besides α2,3-linked Sias on Tregs were involved in Sn–Fc binding.
FIGURE 3.
SnL upregulation on CD4+ T cells is associated with increased α2,3 sialylation. (A) Splenocytes from C57BL/6 mice (n = 4) were activated for 72 h with plate-bound anti-CD3 (5 μg/ml). Activated cells (solid lines) were compared with freshly isolated splenocytes (dashed lines) for biotinylated MAL and biotinylated SNA binding. Streptavidin only was used as a control for MAL and SNA (solid gray histograms). (B) Mean fluorescence intensities (MFI) of Sn–Fc (top panel), MAL (middle panel), and SNA (bottom panel). CD4+Foxp3− T cells are depicted as dark bars and MFIs of fresh and 72-h postactivation CD4+Foxp3− T cells are depicted as light bars. Data represent means ± SEM from four mice per activation status. Data are representative of two independent experiments. Statistical analysis was done using one-way ANOVA with Bonferroni posttest. *p < 0.05, **p < 0.001.
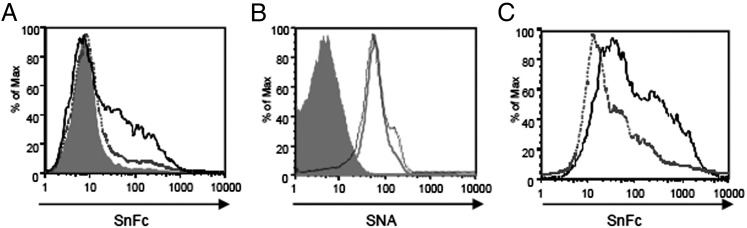
To confirm the importance of α2,3 sialylation on Tregs and Teffs for Sn–Fc binding, we treated cells with sialidase L, which specifically cleaves α2,3-linked Sias (Fig. 4A). Although SNA binding was not affected by such treatment, as expected, binding of both Sn–Fc and MAL was reduced to background levels (Fig. 4B and data not shown). Finally, to determine if the α2,3-linked Sias were associated with glycoproteins or glycolipids, we treated cells with proteinase K and observed a complete loss of Sn–Fc binding (Fig. 4C). In conclusion, the upregulation of SnL on activated CD4+ T cells is due to increased α2,3-sialylation of cell-surface glycoproteins.
FIGURE 4.
Sn–Fc binds SnL on activated CD4+ T cells in α2,3-specific manner. (A) Activated CD4+ T cells were treated with α2,3-specifc sialidase L (dashed gray histograms), nonspecific sialidase (solid gray histogram), or left untreated (black histogram). (B) Similar treatments to (A) but with surface labeling for SNA. (C) Proteinase K (gray unfilled histogram) compared with buffer-only treatment (black histogram) abolishes Sn-Fc binding to activated CD4+ T cells. Data are representative of two independent experiments.
Altered expression of glycosyltransferases on SnL+CD4+Foxp3− T cells
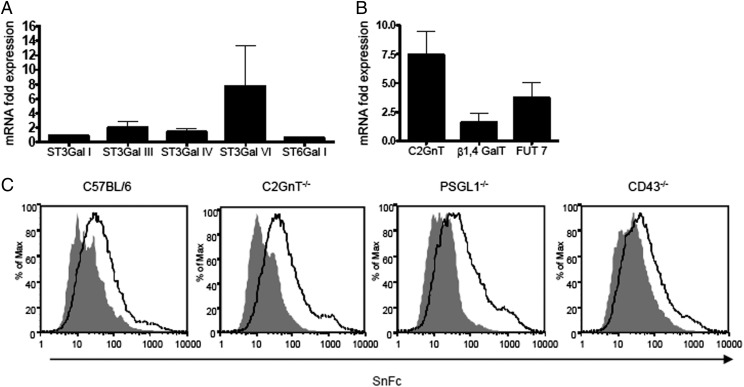
To gain insight into glycosyltransferases that might be involved in upregulating SnL expression, we analyzed the relative levels of candidate enzymes’ mRNAs by quantitative PCR using FACS-sorted SnL+ and SnL− Teffs. In view of the requirement of α2,3 sialylation on glycoproteins for Sn binding to activated CD4+ T cells, the expression of four members of the ST3Gal family of polypeptide α2,3 sialyltransferases were analyzed, namely: ST3Gal I, ST3Gal III, ST3Gal IV, and ST3Gal VI (23, 24). For comparison, the ST6Gal I that transfers Sias to N-glycans in α2,6 linkage (23) was also included in the analysis. Compared to SnL−CD4+ T cells, SnL+CD4+ T cells exhibited a significant upregulation in the mRNA expression of ST3Gal VI and to a lesser extent that of ST3Gal III (Fig. 5A). No significant change in the expression of ST3Gal I or ST3Gal IV was observed. In addition, ST6Gal I was not upregulated on the SnL+ population as expected, based on the small increases in α2,6 sialylation noted above.
FIGURE 5.
The mRNA expression of relevant sialyl (A) and glycosyl transferases (B) on SnL+ CD4+CD25− Teffs. Bars represents fold differences between FACS-enriched SnL+ and SnL− samples. Data represent means ± SEM of triplicate biological samples. Each sample consists of activated enriched CD4+CD25− Teffs pooled from spleens of six WT mice. (C) Sn–Fc binds to activated CD4+ T cells independent of Core 2 O-glycan, PSGL-1, or CD43. Histograms represent Sn–Fc binding to activated CD4+ T cells (gated) from mice lacking C2GnT (all three isoforms), PSGL-1, and CD43. Solid gray histograms represent sialidase-treated samples from each genotype.
T cell activation has been associated with an increase in Core 2 O-glycosylation, which is mediated by the Core 2 β1,6 N-acetylglucosaminyltransferase 1, or C2GnT1 (25, 26). Expression of the C2GnT1 gene was >6-fold higher in SnL+ Teffs compared with SnL− Teffs (Fig. 5B). Moreover, two other key glycosyltransferases, β1,4GalT and FucT7, which were shown previously to be involved in the biosynthesis of sLex motifs on activated T cells (27), were also found to be upregulated on SnL+ Teffs (Fig. 5B).
The strong upregulation of C2GnT1 in SnL+ Teffs suggested an involvement of Core 2 O-glycans in the synthesis of SnL. To address this possibility, splenic CD4+ T cells from triple C2GnT-deficient mice, which lack all three isoforms of the core two enzymes, were examined for Sn–Fc binding before and after activation. Interestingly, no reduction in Sn–Fc binding on activated cells was observed when compared with control WT T cells (Fig. 5C). This suggests that the upregulation of C2GnT1 on SnL+ Teffs is reflective of the higher activation status of this subset of Teffs, rather than being a binding requirement for Sn–Fc. Previously, CD43 and PSGL-1 were identified as putative counterreceptors for Sn using Sn–Fc pulldown methods with the TK1 T lymphoma cell line (28). These mucin-like glycoproteins contain multiple O-linked glycans terminating in α2,3-linked Sias (26, 27, 29). However, our analysis of SnL induction on splenocytes from mice deficient in either CD43 or PSGL-1 showed that Sn–Fc binding to activated CD4+ T cells was independent of either of these cell surface mucins (Fig. 5C).
SnL requires N-linked glycan biosynthesis
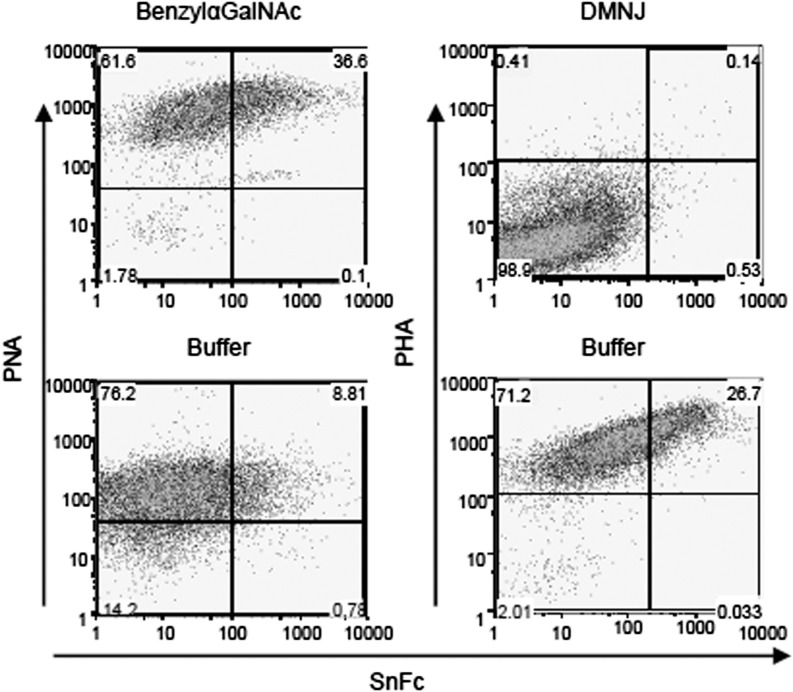
As CD43 and PSGL-1 are major carriers of O-glycans on T cells (26, 27), the failure to see reduced Sn–Fc binding to T cells lacking these glycoproteins suggested that O-glycans were not required for SnL expression. We investigated the nature of carbohydrate structures involved in Sn binding further using inhibitors of N- and O- glycan biosynthesis. BenzylαGalNAc is an O-glycan elongation inhibitor for which efficiency can be demonstrated by staining with peanut agglutinin (PNA), which recognizes the exposed Galβ1,4GalNAc sequence (19, 30, 31). 1-deoxymannojirimycin (DMNJ), in contrast, inhibits α-mannosidase I, leading to the accumulation of high mannose-type N-glycan precursor that cannot be processed further into complex N-glycans. N-glycan inhibition with DMNJ can be verified by reduced staining with Phaseolus vulgaris agglutinin (PHA) that binds complex N-glycans (30, 32). Inhibition of O-glycan elongation with benzylαGalNAc during T cell activation resulted in a concentration-dependent increase in the percentage of SnL+CD4+ T cells (Fig. 6, left panel). The inhibition of sialylated O-linked glycans by benzylαGalNAc was confirmed by the strong dose-dependent enhancement of PNA binding (Fig. 6, left panel). These findings suggested that O-glycans are not required for display of SnL on activated CD4+ T cells.
FIGURE 6.
N-glycans but not O-glycans are required for the synthesis of SnL on activated CD4+ T cells. Splenocytes from WT mice were stimulated for 72 h in the presence of O-glycan elongation inhibitor (BenzylαGalNAc, top left panel), N-glycan inhibitor (DMNJ, top right panel), or relevant buffers. Cells were harvested and stained for Sn–Fc and biotinylated PNA (to confirm O-glycan inhibition in the presence of benzylαGalNAc) or biotinylated PHA (to confirm N-glycan inhibition).
In striking contrast, inhibition of N-glycan maturation with DMNJ during T cell activation resulted in a complete block in SnL induction on activated CD4+ T cells (Fig. 6, right panel). The inhibitory effect of DMNJ was confirmed with negative staining for PHA on activated CD4+ T cells (Fig. 6, right panel). These findings indicated that N-glycans are required for the formation of SnL on activated CD4+ T cells.
The frequency of SnL+CD4+ Teffs correlates with disease state in murine lupus nephritis
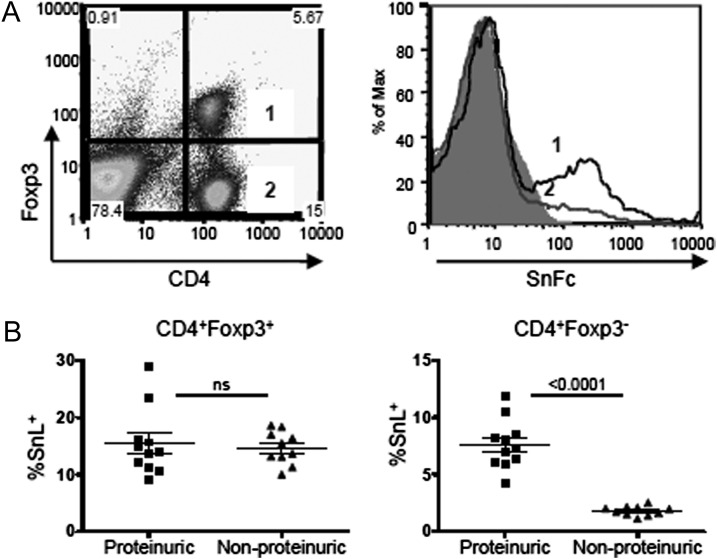
Finally, it was important to see if SnL on Teffs could be observed in a disease model in vivo to rule out the possibility that the upregulation described above was an in vitro phenomenon. We selected the NZBWF1 model of spontaneous murine SLE (33) for this analysis, as lupus disease is associated with a significant upregulation of IFN-α, which is a key player in inducing the expression of Sn. The disease in NZBWF1 often starts at 24 wk of age and is manifest by increasing proteinuria, which heralds the onset of glomerulonephritis. We analyzed the percentages of SnL+ CD4+ T cells isolated from individual NZBWF1 mice at 32 wk, and, consistent with the in vitro data, we saw higher levels of SnL on Tregs compared with Teffs (Fig. 7A). Interestingly, when the percentages SnL+ CD4+ T cells were stratified according to the presence or absence of proteinuria, the frequency of SnL+ Teffs was highly correlated with the disease status in these mice, but no correlation was seen between SnL expression on Tregs and degree of proteinuria (Fig. 7B). These data confirm that upregulation of SnL can occur on activated CD4+ Teffs in vivo and furthermore suggest that even a modest increase in the frequency of this subset of CD4+ T cells can distinguish between disease and nondisease status.
FIGURE 7.
The frequency of SnL+CD4 Teffs correlates positively with the presence of proteinuria in murine lupus nephritis. (A) SnL expression on splenic Tregs and Teffs. Dot plot (left panel) showing Treg and Teff populations with corresponding SnL expression as histograms (right panel) and compared with sialidase-treated control. (B) The percentage of SnL+ Tregs (left panel) and Teffs (right panel) in diseased NZBWF1 and nondiseased mice based on the presence of >300 mg/dl of proteinuria. Each dot represents an individual mouse. Data represent the means ± SEM. Statistical analysis was done using Student t test.
Discussion
The recent demonstration of selective expression of SnL on Tregs in EAE was a major step toward identifying a mechanism by which Sn can modulate CD4+ T cell function and promote inflammatory responses (9). The data presented in this study have shown that SnL induction can occur on both Tregs and Teffs following TCR triggering in vitro as well as in vivo in a murine model of spontaneous SLE. However, in both cases, there was a clear bias toward Tregs when the percentage of SnL+ cells among activated Tregs and Teffs was examined. Interestingly, a low level of SnL expression could be seen on Tregs, but not Teffs, prior to activation in vitro. Peripheral Tregs comprise both thymic-derived natural Tregs (nTregs) and induced Tregs (iTregs). The requirements for the generation of the latter subset include TCR stimulation, IL-2, and TGF-β (34). iTregs can be generated in secondary lymphoid organs and other tissues, particularly the gut. They have also been found in transplant tissues (35). One potential explanation for the presence of SnL+ Tregs in fresh splenocytes is that this subset could have already been activated in the periphery in vivo. Whether this SnL+ subset of Tregs belongs to nTreg or iTregs is a question that warrants further investigation. Examining the expression of markers such as Helios, which was found to be a marker for nTregs (36), might be also useful to identify the nature of the SnL+ Tregs.
Following TCR ligation with anti-CD3, SnL+CD4+ T cells exhibited a higher state of activation, as evidenced by their raised expression of early activation markers, CD25 and CD69. Thus, SnL positivity may be a useful marker for cells at the higher end of the activation spectrum. This differential state of activation might be reflective of a degree of TCR hyperresponsiveness in SnL+CD4+ T cells. This was evident when the effect of titrating TCR signal strength with various concentrations of anti-CD3 Abs was investigated. On the Teffs, even at low concentrations of the anti-CD3, there was a significant difference in the expression of activation markers between SnL+ and SnL− subsets. Moreover, the percentage of SnL+ cells and the percentage of CD69+ and CD25+ cells were proportional to the degree of TCR triggering by different concentrations of anti-CD3 Ab. The analysis of intracellular cytokine expression, especially with IFN-γ, was also in agreement with the higher state of activation of SnL+ Teffs. Although the hyperproliferative state of these cells could be related to their response to TCR stimulation, it could also be due to their higher CD25 expression, allowing these cells to use more endogenous IL-2 compared with SnL− Teffs. This possibility could be investigated further by analyzing the expression of an IL-2R–related protein such as phosphorylated STAT5 (37) following exposure of SnL+ and SnL− Teffs to exogenous IL-2.
Our observation that coculture of activated CD4+ T cells with Sn-expressing BMDMs or Sn-expressing CHO cells led to enhanced CD4+ T cell death suggests a novel death signaling pathway triggered by Sn–SnL engagement. This function is consistent with previous findings in EAE that Sn-deficient mice have higher numbers of SnL+ Tregs (9). The finding in this study, that both Tregs and Teffs can upregulate SnL, raises the possibility of dual, and potentially opposite, outcomes of interactions between Sn+ macrophages and SnL+ Tregs or Teffs. On the one hand, Sn-induced death of SnL+ Tregs can lead to reduced suppression of Teffs and enhanced inflammation as demonstrated in EAE (9). On the other hand, Sn-induced death of SnL+ Teffs could lead to elimination of a hyperactive subset of Teffs and reduced inflammation. Recently, Sn+-resident and inflammatory monocytes were found in PBMCs of patients with SLE and correlated to disease severity and other established biomarkers of the disease (11). Despite such a close association between Sn and disease activity, it remains unclear whether Sn might actively be involved in the pathogenesis of SLE. Our observation of a highly significant correlation between SnL+ Teffs and proteinuria, as a hallmark of disease status, in murine lupus nephritis, suggests a possible role of this subset of CD4+ T cells in inflammation. Future work will be directed toward answering whether Sn can play a protective role in murine lupus nephritis via eliminating potentially pathogenic SnL+ Teffs similar to our observations in vitro.
The induction of SnL on activated CD4+ T cells was associated with an increase in α2,3- versus α2,6- sialylation as evidenced by increased binding to plant lectins MAL compared with SNA. This pattern correlates well with the Sn binding preference of α2,3-linked Sia residues reported previously (1). The requirement of α2,3-linked Sia for Sn binding was demonstrated by the use of the α2,3-specific sialidase L treatment, which was as effective as a Pan-sialidase treatment in abolishing Sn ligands. Comparison of Sn–Fc and MAL binding to activated Tregs and Teffs revealed an interesting discrepancy. Although MAL binding to Tregs versus Teffs was similar, the binding of Sn–Fc to Tregs was proportionately much higher than to Teffs. This suggests the presence of additional features of SnL on Tregs that result in higher Sn–Fc binding compared with Teffs.
Consistent with the pattern of sialylation observed with plant lectins, there was increased mRNA expression of a α2,3 sialyltransferase ST3Gal VI and to a lesser extent of ST3Gal III in SnL+ compared with SnL− CD4+ T cells. The former has been reported to mediate terminal Sia capping on N- glycans and Core 2 O-glycans (38, 39), with the latter transferring Sias to N-linked glycans only (31). Consistent with the higher activation status of the SnL+CD4+ T cells, there was also an increase in C2GnT, which mediates the synthesis of Core 2 structures found on activated T cells (26). These findings were in line with previous studies of a shift in the O-glycan pattern from Core 1 to Core 2 structure following T cell activation (25). In addition, the increased mRNA expression of FUT7 and β1,4 Gal T were further evidence for the dominant Core 2 rather than Core 1 O-glycosylation on SnL+ Teffs. These findings provide evidence that T cell activation is accompanied by dynamic changes in surface glycosylation that promote or hinder binding to different lectins with multiple effects on different systems. An example of this is highlighted in the findings of enhanced Core 2 synthesis, together with a number of enzymes required for sialyl lewis X motifs, despite the fact that binding of Sn–Fc is N-glycan dependent.
Previously, CD43 and PSGL-1 were identified as counterreceptors for Sn on the TK1 lymphoma cell line using Sn–Fc pulldown approaches from whole-cell lysates. However, our data showed that CD43, PSGL-1, and Core 2 O-glycans are all dispensable in Sn–Fc binding to activated CD4+ T cells. This apparent contradiction may be related to the low affinity of Sn for its glycan ligands. Thus, biochemical isolation of Sn counterreceptors in solution may strongly favor elongated mucin-like proteins that bind to clustered Sn–Fc on protein A affinity beads with high avidity. The binding interaction between Sn and SnL may be completely different at the cell surface and depend on overall surface density of α2,3-linked Sias and the way that these Sias are displayed.
There are interesting parallels between the work reported in this study for Sn and previous studies on galectin-1, a β-galactoside-binding protein that has been widely studied for its T cell death–promoting functions (40, 41). The binding of galectin-1 to N-glycans on CD45RB was found to be dependent on the balance between α2,3- and α2,6-sialylation. Given that α2,3 sialylation was essential for Sn binding to activated T cells, it is possible that Sn and galectin-1 can bind simultaneously to the N-glycans on glycoprotein carriers such as CD45 and synergize to promote T cell death. Further in vitro and in vivo studies are required to explore this possibility.
In conclusion, this study has demonstrated that SnL can be induced differentially on subsets of CD4+ T cells, and, in the case of Teffs, they mark a subset of highly activated T cells. Exposure to Sn was associated with enhanced cell death. SnL induction was found to be associated with an increase in α2,3 sialylation, and cleavage of α2,3-linked Sia abrogated Sn–Fc binding. Similarly, SnL induction was dependent on N- and not O-linked glycans. Although the nature of the protein carriers of SnL remains unknown, data shown in this study demonstrate that previously described counterreceptors CD43 and PSGL-1 are both dispensable. Finally, SnL expression correlated with disease status in a murine mode of SLE, suggesting a potential role of this subset in inflammation. Studies using Sn−/− mice are underway to examine the role of Sn and SnL in murine SLE as well as other disease models in which SnL is upregulated on Teffs.
Supplementary Material
Acknowledgments
We thank Dr. Douglas Carlow (University of British Columbia) for helpful discussions.
This work was supported by Wellcome Trust Clinical Fellowship 087078 (to D.K.) and Wellcome Trust Senior Fellowship 081882MA (to P.R.C.).
The online version of this article contains supplemental material.
- BMDM
- bone marrow–derived macrophage
- CHO
- Chinese hamster ovary
- DMNJ
- 1-deoxymannojirimycin
- EdU
- 5-ethynyl-2′-deoxyuridine
- EAE
- experimental autoimmune encephalomyelitis
- iTreg
- induced regulatory T cell
- MAL
- Maackia amurensis lectin
- nTreg
- natural regulatory T cell
- NZBWF1
- New Zealand Black × New Zealand White F1
- PHA
- Phaseolus vulgaris agglutinin
- PNA
- peanut agglutinin
- PSGL-1
- P-selectin glycoprotein ligand-1
- Sia
- sialic acid
- SLE
- systemic lupus erythematosus
- Sn
- sialoadhesin
- SNA
- Sambucus nigra agglutinin
- SnL
- sialoadhesin ligand
- Teff
- effector T cell
- Treg
- regulatory T cell
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Crocker P. R., Paulson J. C., Varki A. 2007. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7: 255–266 [DOI] [PubMed] [Google Scholar]
- 2.Crocker P. R., Mucklow S., Bouckson V., McWilliam A., Willis A. C., Gordon S., Milon G., Kelm S., Bradfield P. 1994. Sialoadhesin, a macrophage sialic acid binding receptor for haemopoietic cells with 17 immunoglobulin-like domains. EMBO J. 13: 4490–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munday J., Floyd H., Crocker P. R. 1999. Sialic acid binding receptors (siglecs) expressed by macrophages. J. Leukoc. Biol. 66: 705–711 [DOI] [PubMed] [Google Scholar]
- 4.Crocker P. R., Kelm S., Dubois C., Martin B., McWilliam A. S., Shotton D. M., Paulson J. C., Gordon S. 1991. Purification and properties of sialoadhesin, a sialic acid-binding receptor of murine tissue macrophages. EMBO J. 10: 1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crocker P. R., Freeman S., Gordon S., Kelm S. 1995. Sialoadhesin binds preferentially to cells of the granulocytic lineage. J. Clin. Invest. 95: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartnell A., Steel J., Turley H., Jones M., Jackson D. G., Crocker P. R. 2001. Characterization of human sialoadhesin, a sialic acid binding receptor expressed by resident and inflammatory macrophage populations. Blood 97: 288–296 [DOI] [PubMed] [Google Scholar]
- 7.Barral P., Polzella P., Bruckbauer A., van Rooijen N., Besra G. S., Cerundolo V., Batista F. D. 2010. CD169(+) macrophages present lipid antigens to mediate early activation of iNKT cells in lymph nodes. Nat. Immunol. 11: 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura K., Yamaji T., Crocker P. R., Suzuki A., Hashimoto Y. 2002. Lymph node macrophages, but not spleen macrophages, express high levels of unmasked sialoadhesin: implication for the adhesive properties of macrophages in vivo. Glycobiology 12: 209–216 [DOI] [PubMed] [Google Scholar]
- 9.Wu C., Rauch U., Korpos E., Song J., Loser K., Crocker P. R., Sorokin L. M. 2009. Sialoadhesin-positive macrophages bind regulatory T cells, negatively controlling their expansion and autoimmune disease progression. J. Immunol. 182: 6508–6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pulliam L., Sun B., Rempel H. 2004. Invasive chronic inflammatory monocyte phenotype in subjects with high HIV-1 viral load. J. Neuroimmunol. 157: 93–98 [DOI] [PubMed] [Google Scholar]
- 11.Biesen R., Demir C., Barkhudarova F., Grün J. R., Steinbrich-Zöllner M., Backhaus M., Häupl T., Rudwaleit M., Riemekasten G., Radbruch A., et al. 2008. Sialic acid-binding Ig-like lectin 1 expression in inflammatory and resident monocytes is a potential biomarker for monitoring disease activity and success of therapy in systemic lupus erythematosus. Arthritis Rheum. 58: 1136–1145 [DOI] [PubMed] [Google Scholar]
- 12.York M. R., Nagai T., Mangini A. J., Lemaire R., van Seventer J. M., Lafyatis R. 2007. A macrophage marker, Siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type I interferons and toll-like receptor agonists. Arthritis Rheum. 56: 1010–1020 [DOI] [PubMed] [Google Scholar]
- 13.Jiang H. R., Hwenda L., Makinen K., Oetke C., Crocker P. R., Forrester J. V. 2006. Sialoadhesin promotes the inflammatory response in experimental autoimmune uveoretinitis. J. Immunol. 177: 2258–2264 [DOI] [PubMed] [Google Scholar]
- 14.Kobsar I., Oetke C., Kroner A., Wessig C., Crocker P., Martini R. 2006. Attenuated demyelination in the absence of the macrophage-restricted adhesion molecule sialoadhesin (Siglec-1) in mice heterozygously deficient in P0. Mol. Cell. Neurosci. 31: 685–691 [DOI] [PubMed] [Google Scholar]
- 15.Ip C. W., Kroner A., Crocker P. R., Nave K. A., Martini R. 2007. Sialoadhesin deficiency ameliorates myelin degeneration and axonopathic changes in the CNS of PLP overexpressing mice. Neurobiol. Dis. 25: 105–111 [DOI] [PubMed] [Google Scholar]
- 16.Stone E. L., Ismail M. N., Lee S. H., Luu Y., Ramirez K., Haslam S. M., Ho S. B., Dell A., Fukuda M., Marth J. D. 2009. Glycosyltransferase function in core 2-type protein O glycosylation. Mol. Cell. Biol. 29: 3770–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlow D. A., Corbel S. Y., Ziltener H. J. 2001. Absence of CD43 fails to alter T cell development and responsiveness. J. Immunol. 166: 256–261 [DOI] [PubMed] [Google Scholar]
- 18.Rossi F. M., Corbel S. Y., Merzaban J. S., Carlow D. A., Gossens K., Duenas J., So L., Yi L., Ziltener H. J. 2005. Recruitment of adult thymic progenitors is regulated by P-selectin and its ligand PSGL-1. Nat. Immunol. 6: 626–634 [DOI] [PubMed] [Google Scholar]
- 19.Hernandez J. D., Klein J., Van Dyken S. J., Marth J. D., Baum L. G. 2007. T-cell activation results in microheterogeneous changes in glycosylation of CD45. Int. Immunol. 19: 847–856 [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto Y., Suzuki M., Crocker P. R., Suzuki A. 1998. A streptavidin-based neoglycoprotein carrying more than 140 GT1b oligosaccharides: quantitative estimation of the binding specificity of murine sialoadhesin expressed on CHO cells. J. Biochem. 123: 468–478 [DOI] [PubMed] [Google Scholar]
- 21.Vinson M., van der Merwe P. A., Kelm S., May A., Jones E. Y., Crocker P. R. 1996. Characterization of the sialic acid-binding site in sialoadhesin by site-directed mutagenesis. J. Biol. Chem. 271: 9267–9272 [DOI] [PubMed] [Google Scholar]
- 22.Green D. R., Droin N., Pinkoski M. 2003. Activation-induced cell death in T cells. Immunol. Rev. 193: 70–81 [DOI] [PubMed] [Google Scholar]
- 23.Takashima S. 2008. Characterization of mouse sialyltransferase genes: their evolution and diversity. Biosci. Biotechnol. Biochem. 72: 1155–1167 [DOI] [PubMed] [Google Scholar]
- 24.Varki A., Crocker P. R. 2009. I-type lectins. In Essentials of Glycobiology, 2nd Ed., Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E., eds. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, p. 459–474 [PubMed] [Google Scholar]
- 25.Priatel J. J., Chui D., Hiraoka N., Simmons C. J., Richardson K. B., Page D. M., Fukuda M., Varki N. M., Marth J. D. 2000. The ST3Gal-I sialyltransferase controls CD8+ T lymphocyte homeostasis by modulating O-glycan biosynthesis. Immunity 12: 273–283 [DOI] [PubMed] [Google Scholar]
- 26.Jones A. T., Federsppiel B., Ellies L. G., Williams M. J., Burgener R., Duronio V., Smith C. A., Takei F., Ziltener H. J. 1994. Characterization of the activation-associated isoform of CD43 on murine T lymphocytes. J. Immunol. 153: 3426–3439 [PubMed] [Google Scholar]
- 27.Carlow D. A., Gossens K., Naus S., Veerman K. M., Seo W., Ziltener H. J. 2009. PSGL-1 function in immunity and steady state homeostasis. Immunol. Rev. 230: 75–96 [DOI] [PubMed] [Google Scholar]
- 28.van den Berg T. K., Nath D., Ziltener H. J., Vestweber D., Fukuda M., van Die I., Crocker P. R. 2001. Cutting edge: CD43 functions as a T cell counterreceptor for the macrophage adhesion receptor sialoadhesin (Siglec-1). J. Immunol. 166: 3637–3640 [DOI] [PubMed] [Google Scholar]
- 29.Fukuda M. 1991. Leukosialin, a major O-glycan-containing sialoglycoprotein defining leukocyte differentiation and malignancy. Glycobiology 1: 347–356 [DOI] [PubMed] [Google Scholar]
- 30.Varki A., Etzler M. E., Cummings R. D., Esko J. D. 2009. Discovery and classification of glycan-binding proteins. In Essentials of Glycobiology, 2nd Ed., Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E., eds. Cold Spring Harbor Laboratory Press, Cold Springs Harbor, NY, p. 375–386 [PubMed] [Google Scholar]
- 31.Earl L. A., Bi S., Baum L. G. 2010. N- and O-glycans modulate galectin-1 binding, CD45 signaling, and T cell death. J. Biol. Chem. 285: 2232–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walzel H., Fahmi A. A., Eldesouky M. A., Abou-Eladab E. F., Waitz G., Brock J., Tiedge M. 2006. Effects of N-glycan processing inhibitors on signaling events and induction of apoptosis in galectin-1-stimulated Jurkat T lymphocytes. Glycobiology 16: 1262–1271 [DOI] [PubMed] [Google Scholar]
- 33.Xu Z., Morel L. 2010. Genetics of systemic lupus erythematosus: contributions of mouse models in the era of human genome-wide association studies. Discov. Med. 10: 71–78 [PubMed] [Google Scholar]
- 34.Fantini M. C., Becker C., Tubbe I., Nikolaev A., Lehr H. A., Galle P., Neurath M. F. 2006. Transforming growth factor beta induced FoxP3+ regulatory T cells suppress Th1 mediated experimental colitis. Gut 55: 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waldmann H., Graca L., Cobbold S., Adams E., Tone M., Tone Y. 2004. Regulatory T cells and organ transplantation. Semin. Immunol. 16: 119–126 [DOI] [PubMed] [Google Scholar]
- 36.Thornton A. M., Korty P. E., Tran D. Q., Wohlfert E. A., Murray P. E., Belkaid Y., Shevach E. M. 2010. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184: 3433–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heltemes-Harris L. M., Willette M. J., Vang K. B., Farrar M. A. 2011. The role of STAT5 in the development, function, and transformation of B and T lymphocytes. Ann. N. Y. Acad. Sci. 1217: 18–31 [DOI] [PubMed] [Google Scholar]
- 38.Comelli E. M., Sutton-Smith M., Yan Q., Amado M., Panico M., Gilmartin T., Whisenant T., Lanigan C. M., Head S. R., Goldberg D., et al. 2006. Activation of murine CD4+ and CD8+ T lymphocytes leads to dramatic remodeling of N-linked glycans. J. Immunol. 177: 2431–2440 [DOI] [PubMed] [Google Scholar]
- 39.Underhill G. H., Zisoulis D. G., Kolli K. P., Ellies L. G., Marth J. D., Kansas G. S. 2005. A crucial role for T-bet in selectin ligand expression in T helper 1 (Th1) cells. Blood 106: 3867–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen J. T., Evans D. P., Galvan M., Pace K. E., Leitenberg D., Bui T. N., Baum L. G. 2001. CD45 modulates galectin-1-induced T cell death: regulation by expression of core 2 O-glycans. J. Immunol. 167: 5697–5707 [DOI] [PubMed] [Google Scholar]
- 41.Rabinovich G. A., Ilarregui J. M. 2009. Conveying glycan information into T-cell homeostatic programs: a challenging role for galectin-1 in inflammatory and tumor microenvironments. Immunol. Rev. 230: 144–159 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.