Abstract
The prevalence of obesity has led to a surge of interest in understanding the detailed mechanisms underlying adipocyte development. Many protein-coding genes, mRNAs, and microRNAs have been implicated in adipocyte development, but the global expression patterns and functional contributions of long noncoding RNA (lncRNA) during adipogenesis have not been explored. Here we profiled the transcriptome of primary brown and white adipocytes, preadipocytes, and cultured adipocytes and identified 175 lncRNAs that are specifically regulated during adipogenesis. Many lncRNAs are adipose-enriched, strongly induced during adipogenesis, and bound at their promoters by key transcription factors such as peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer-binding protein α (CEBPα). RNAi-mediated loss of function screens identified functional lncRNAs with varying impact on adipogenesis. Collectively, we have identified numerous lncRNAs that are functionally required for proper adipogenesis.
Obesity is a major source of morbidity and mortality and is increasingly prevalent in many areas of the world. By 2008 the incidence of this disease was 32.2% among adult men and 35.5% among adult women in United States (1). Excess fat accumulation is a contributing factor in such severe human diseases as type 2 diabetes, certain cancers, and cardiovascular disease (2). Understanding the detailed mechanisms controlling adipogenesis and energy homeostasis is a critical part of the effort to combat obesity.
Adipogenesis is governed by a transcriptional cascade driven, in large part, by peroxisome proliferator-activated receptor γ (Pparγ), an adipocyte-enriched nuclear receptor (3). Pparγ is both necessary and sufficient for adipogenesis (4, 5); no other factor has been found to be able to induce adipogenesis in the absence of Pparγ. Pparγ cooperates with CCAAT/enhancer-binding proteins (C/EBP), including Cebpα, Cebpβ, and Cebpγ, to induce the expression of many genes important for terminal differentiation, such as Fabp4/aP2, Cd36, Lipe/Hsl, Olr1, and Me1 (6). Unlike transcriptional factors that bind to DNA directly, transcriptional cofactors often serve as molecular scaffolds to link the basal transcription machinery to either active or inactive complexes (7–9). Many cofactors, such as CBP and p300, have enzymatic activity in histone modification and can adjust chromatin environments to be more or less accessible to the transcription machinery. Indeed, dramatic changes in epigenetic signatures have been observed during adipogenesis, indicating an important role of epigenetic modifications in regulating adipogenesis (10).
Noncoding RNAs are known to play a regulatory role in many developmental contexts, including adipogenesis, and several microRNAs have been identified that positively or negatively regulate adipogenesis (11, 12). Recently we and others have identified a class of long noncoding RNAs (lncRNAs) (13–15). Previous studies have demonstrated that lncRNAs are essential regulators in a variety of biological processes, including, but not limited to, X chromatin inactivation (16), p53-mediated apoptosis (17, 18), cancer metastasis (19), and reprogramming of induced pluripotent stem cells (20), among many others (21–23). We therefore hypothesized that lncRNAs participate in the regulatory network governing adipogenesis.
To test this we used deep RNA sequencing (RNA-Seq) to identify mRNAs and polyadenylated lncRNAs that are regulated during adipogenesis. To survey the functional contributions of lncRNAs during adipogenesis we performed RNAi mediated loss of function (LOF) experiments for 20 candidate lncRNAs. In scoring each LOF assay for functional relevance, we present here a unique method that uses the Jensen-Shannon distance metric to quantify the lncRNA-dependent “transcriptome shift” from mature adipocyte to reversion back to the precursor state. Our combined results establish that lncRNAs constitute an as yet unexplored and important layer in adipogenesis regulation.
Results
Global Identification of lncRNAs Regulated During Adipogenesis.
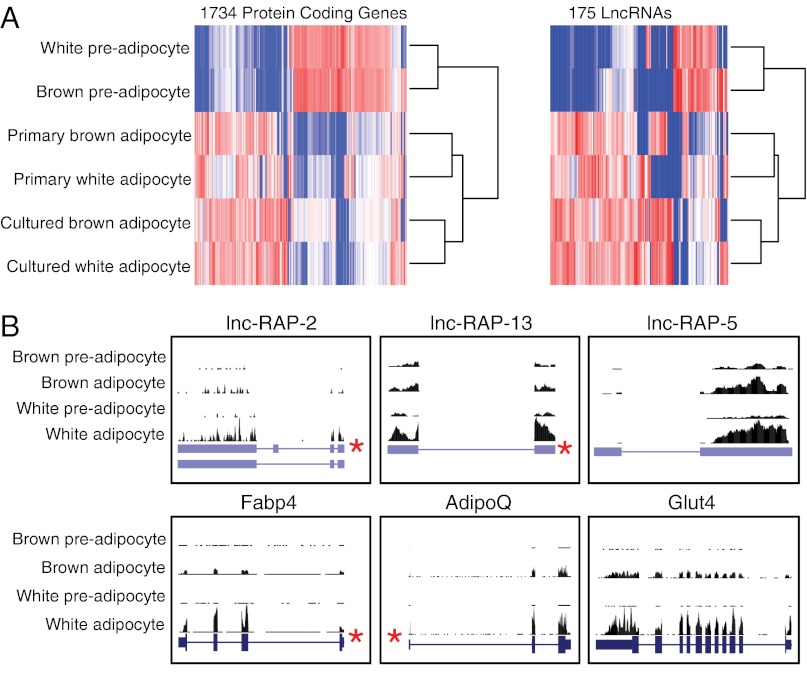
We first set out to identify global transcriptional regulation during adipogenesis for both coding and noncoding genes alike. To this end we used massively parallel RNA sequencing as previously described (24) to sequence polyA-selected RNAs from in vitro cultured brown and white preadipocytes, in vitro differentiated mature brown and white adipocytes, and primary brown and white mature adipocytes directly isolated from mice. RNA-Seq reads were mapped to the mouse genome (mm9) using TopHat (25), to which we provided gene annotations for known and/or cloned and previously undescribed transcripts to maximize spliced alignment accuracy. All annotated transcripts, corresponding to known University of California at Santa Cruz (UCSC) genes and available RIKEN cloned sequences (26), were quantified across each condition and differential expression analysis between preadipocytes and mature adipocytes using Cuffdiff (27). We identified 1,734 coding genes and 175 lncRNAs that were significantly up- or down-regulated greater than twofold during differentiation of both brown and white adipocytes [false discovery rate (FDR) <5%] (Fig. 1A).
Fig. 1.
Global transcriptome profiling of lncRNAs and protein coding genes during adipogenesis via RNA-Seq. (A) This heatmap shows 1,734 coding genes and 175 lncRNAs significantly up-regulated (red) or down-regulated (blue) between preadipocytes and differentiated adipocytes. Independent hierarchical clustering of expression values for these lncRNAs and coding genes clusters differentiated and primary adipocytes together. (B) RNA-Seq read alignment of three lncRNAs as well as three adipocyte markers—AdipoQ, Fabp4, and Glut4—is displayed. RNA-Seq read alignment coverage in brown and white preadipocytes and cultured mature adipocytes is displayed above each gene. Previously identified PPARγ ChIP-Seq binding sites (10) are identified with an asterisk (*), and exons are depicted as blue bars.
To further explore the biological validity of our derived adipocytes we examined the expression of several well-known adipogenesis markers, such as fatty acid binding protein 4 (Fabp4), adiponectin (AdipoQ), and glucose transporter type 4 (Glut4) (Fig. 1B) as well as several additional markers, such as preadipocyte factor 1 (Pref1), cell death-inducing DFFA-like effector a (Cidea), and uncoupling protein 1 (Ucp1) (Fig. S1), all of which are regulated as expected for appropriately differentiating white and/or brown preadipocytes. We next examined the protein coding gene sets up-regulated during adipogenesis by global gene ontology analysis. Consistent with bona fide adipocytes and many previous studies, we observed significant enrichments in lipid metabolism and adipocyte terms, whereas down-regulated genes are enriched in cell cycle and fibroblast terms (Fig. S2), demonstrating that our RNA-Seq data accurately reflect the known hallmarks of adipogenesis.
Expression of lncRNAs Is Tightly Regulated During Adipogenesis.
Hierarchical clustering of the RNA-Seq expression profiles determined that both lncRNAs and protein coding genes alike accurately distinguished precursors from adipocytes (Fig. 1A). Specifically, precursor cells are clustered together yet distinctly different from the gene expression patterns of both cultured adipocytes and primary adipocytes (Fig. 1A). Importantly, cultured and primary brown and white adipocytes exhibited similar expression patterns. Together these observations demonstrate that our in vitro cell cultured adipocytes share similar gene expression signatures to their primary counterparts across both protein-coding and noncoding lncRNA genes.
Several recent studies have identified ncRNAs directly regulated by key transcription factors that define and drive cellular differentiation (14, 18, 20, 28–30). Therefore, we explored whether lncRNAs regulated during adipogenesis are controlled by known adipogenic transcription factors, such as Pparγ and Cebpα. To this end we analyzed data from previous genome-wide binding site localizations of Pparγ (10) and Cebpα (31). Pparγ is physically bound within the promoter region (2 kb upstream of the transcriptional start site) of 23 (13%) of 175 lncRNAs up-regulated during adipogenesis; Cebpα was bound upstream of 34 up-regulated lncRNAs (19%). Up-regulated protein coding genes were similarly enriched for Pparγ and Cebpα binding for (215 (14%) and 352 (20%) of 1,734 genes, respectively). A permutation analysis confirmed these enrichments to be statistically significant (P < 10−3) over random chance. Collectively, these computational and experimental analyses suggest that a similar portion of regulated lncRNAs and protein coding genes are bound and activated by transcription factors responsible for coordinating adipogenesis.
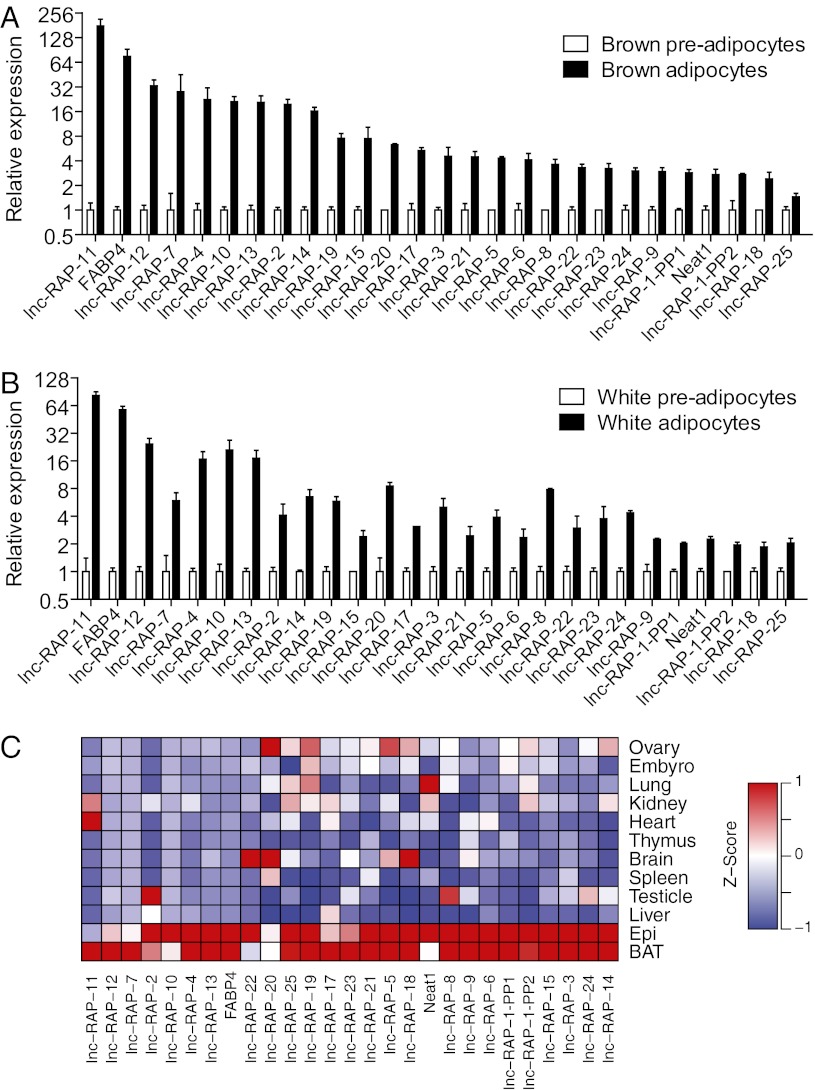
To focus our functional validation efforts, we first ranked candidate lncRNAs according to their level of up-regulation in brown and white fat, and the presence of PPARγ or CEBPα binding at the lncRNA promoter. These criteria resulted in 32 top candidates for further analysis. We performed quantitative RT-PCR (qRT-PCR) to independently measure the regulation of these 32 lncRNAs during differentiation of independently derived preadipocytes (Fig. 2 A and B). As a control, we monitored the adipogenesis marker Fabp4 as well. As assessed by qRT-PCR, 26 of 32 selected lncRNA genes were confirmed as up-regulated during adipogenesis in both primary brown and white adipocyte cultures (Fig. 2 A and B), similar to Fabp4.
Fig. 2.
Validation of selected lncRNAs. Primary brown preadipocytes (A) and primary white preadipocytes (B) were isolated, cultured, and differentiated as described in Experimental Procedures. Total RNA was extracted from D0 and D6 cells, and qRT-PCR was performed to examine the expression of selected lncRNAs. Twenty-six lncRNAs (n = 3) were validated as significantly differentially up-regulated during both brown and white fat differentiation (P < 0.05; means ± SEM). (C) RT-PCR was performed to examine the expression of lncRNAs across 12 adult mouse tissues. Their expression profiles, presented here as differences from the row mean, are plotted as a heatmap. Red represents a higher expression and blue a lower expression. BAT, brown adipose tissue; Epi, epididymal white fat.
To determine whether these 26 lncRNAs were specifically regulated in adipose tissue we monitored their expression levels using qRT-PCR across 10 additional tissues (Fig. 2C). Intriguingly, many of these lncRNAs were greatly enriched in but not totally specific to oth brown adipose tissue and white adipose tissue compared with 10 other organs. Collectively these experiments identified 26 lncRNAs that are induced during adipogenesis and exhibit adipose-enriched expression patterns; several exhibit promoter binding of Pparγ and Cebpα, suggestive of a functional role in adipogenesis.
LOF Screening Reveals Functional lncRNAs During Adipogenesis.
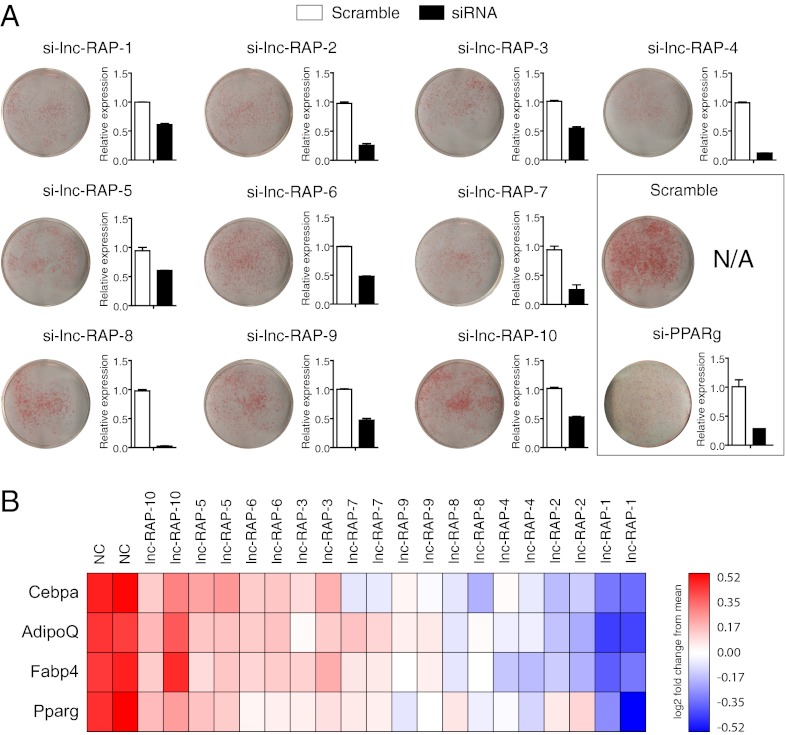
To identify lncRNAs that are functionally important for adipogenesis, we used RNAi-mediated LOF to screen the top 20 lncRNA genes identified by the above criteria of significant up-regulation in both brown and white fat cultures, PPARγ and CEPBα promoter binding, and independent validation of adipose-specific expression. Briefly, we separately transfected three siRNAs targeting each lncRNA into s.c. preadipocyte cultures 1 d before differentiation. Two siRNAs targeting Pparγ mRNA were used as positive controls and a nontargeting scrambled siRNA as a negative control. After 4 d of differentiation, lipid accumulation was evaluated by Oil Red O (ORO) staining. In addition, total RNA was subsequently isolated to monitor the siRNA knockdown efficiency of each construct. As expected, siRNA constructs targeting Pparg showed a marked reduction in lipid accumulation (Fig. 3A). Ten of the targeted lncRNAs were not effectively knocked down or did not result in detectable changes in ORO staining relative to nontargeting controls. In contrast, we identified 10 lncRNAs exhibiting moderate to strong reductions in lipid accumulation upon LOF (Fig. 3A). To further investigate the effect of lncRNA LOF on the adipogenesis process we performed qRT-PCR on four adipocyte markers: Pparγ, Cebpα, Fabp4, and AdipoQ across the subset of the lncRNAs with moderate to strong depletion in lipid accumulation (Fig. 3B). Each of the tested siRNAs caused a reduction in the expression of at least three key adipogenic marker genes relative to the scrambled control. Collectively our LOF screen of 20 lncRNAs identified 10 lncRNAs that seem to play key roles in proper differentiation of adipocyte precursors. For simplicity, we refer to these lncRNAs as Regulated in AdiPogenesis (lnc-RAP-n).
Fig. 3.
LncRNAs are essential regulators for adipogenesis. (A) Primary white preadipocytes were isolated, cultured, and differentiated as described in Experimental Procedures. One day before differentiation, cells were transfected with siRNAs targeting each lncRNA. At 4 d after differentiation induction, ORO staining was performed to detect the lipid accumulation. RT-PCR was performed to examine the expression of the targeted lncRNAs (A, bar plots) (n = 3; *P < 0.05; means ± SEM) and several adipogenesis marker genes (B).
Information-Theoretic Metric Scores Cellular Phenotype.
We next set out to quantify the LOF phenotypes by scoring the effect of each lncRNA perturbation on the global white adipocyte transcriptome. Such a scoring method should accurately measure how the expression profile from a knockdown differs from the precursor and adipocyte states, and the score should correlate with the qualitative ORO staining. Commonly used scoring functions, such as the Pearson correlation or the Euclidean distance, have been used for similar purposes. However, neither of these methods accurately corresponded with the ORO staining results (Fig. S3). This is likely due to some inherent limitations of these approaches. For example, (i) Pearson correlation is capable of identifying similar expression patterns across a range of different intensities; however, correlation is not a true distance metric and is sensitive to outliers, which can interfere with meaningful hierarchical clustering of samples; and (ii) Euclidean distance is greatly influenced by the absolute level of expression; differences in a few abundant genes may cause two profiles that are otherwise qualitatively very similar to appear distant.
To overcome these limitations, we turned to a metric of similarity between two frequency profiles based on Shannon Entropy, termed Jensen-Shannon distance (JSD) (Experimental Procedures). This metric has been used previously for simple purposes in quantifying differential splicing in high-throughput data (27, 32) and is increasingly used in machine learning applications. To test the hypothesis that JSD was a more quantitative measure to examine phenotype relative to ORO staining, we first identified the gene signature that best distinguishes the precursor state (D0) from mature cultured white adipocytes (D4). We collected total RNA from three biological replicates of D0 and D4 cells and performed gene expression analysis on the Affymetrix mouse 430A2 platform. Significance analysis between these two conditions revealed 2,200 genes that distinguish precursors from the mature adipocyte [significance analysis of microarrays (SAM), FDR <4.5%]. We further eliminated genes that exhibited differential expression due to siRNA treatment alone (Experimental Procedures). The resulting gene expression pattern comprised 1,727 genes that discretely distinguished precursors from mature adipocytes.
To monitor lncRNA-dependent perturbations to the adipogenic signature, s.c. prewhite adipocytes were transfected with siRNAs targeting each lncRNA 1 d before differentiation. Four days after differentiation, total RNA was isolated for microarray analysis. We hypothesized that if an lncRNA were required for proper adipose differentiation, then its inhibition would result in minimal differences between the expression profiles of the 1,727 genes in the lncRNA depletion samples and the profile of the same genes in the undifferentiated (D0) preadipocyte control sample.
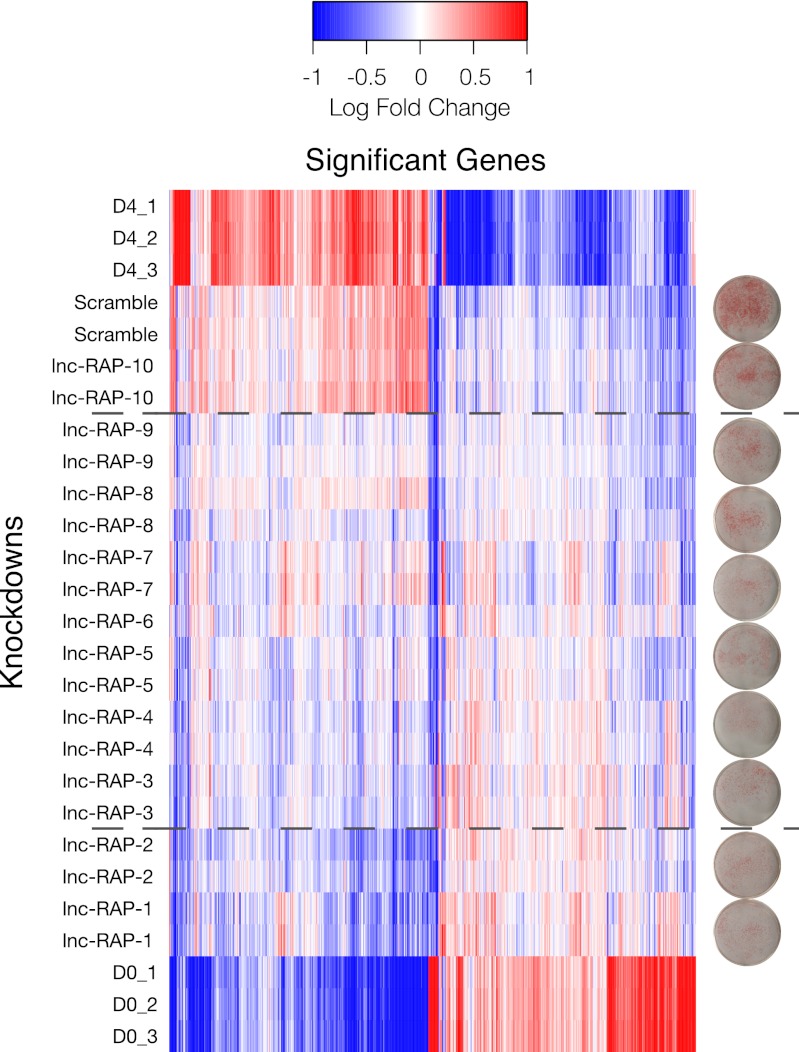
We thus quantified the “transcriptome shift” toward the mean D0 (precursor) expression profile across all 1,727 significant genes for each of the 10 lncRNA depletion expression profiles and the scrambled controls that exhibited significant to moderate ORO staining phenotypes. The mean-centered expression profiles are presented as a heatmap, where conditions are ordered by JSD to the D0 profile (Fig. 4). As expected, the gene profile of the scramble control shRNA most closely resembled that of the D4 differentiated adipocytes. Similarly, one lncRNA knockdown, RAP-10, had a differentiation profile similar to the scramble control and the D4 differentiated adipocytes, suggesting little to no effect on adipogenesis by depleting this lncRNA.
Fig. 4.
Microarray analysis of adipogenesis-regulated LncRNA knockdowns. Gene expression profiles for 1,727 genes selected as significant between undifferentiated (D0) and differentiated (D4) adipose precursors but not significantly regulated between D4 and a scrambled siRNA control. Knockdowns are rank-ordered by JSD to D0, and this ordering highlights the range of effects each LncRNA has on regulating the adipogenesis transcriptome. The dashed lines divide the knockdowns into three regions: those with little to no effect on adipogenesis (similar to D4 and scramble), those with moderate effects on the adipogenesis-regulated genes (middle block), and those ncRNAs whose absence severely affects the differentiation potential of adipocyte precursors (similar to D0). Distance from D0 correlates with lipid accumulation, as determined by Oil Red staining (plates).
Importantly, upon knockdown, nine lncRNAs exhibited a partial or near-complete reversion of the mature adipocyte (D4) to precursor (D0) expression signature. This indicates that these lncRNAs are required for proper regulation of the transcriptional network implicated in adipogenesis. Remarkably, JSD scores closely correspond to the observed levels of lipid accumulation (ORO staining) for each lncRNA knockdown as measured by the Oil Red staining (Fig. 4). Thus, the JSD metric of gene expression phenotypes proves to be a valuable means of quantifying adipogenic phenotypes that is universally applicable to other cell systems and comparisons between biological conditions.
lncRNA LOF Specifically Perturbs Adipogenic Pathways.
To further examine the specific gene networks perturbed in each of the 10 lncRNA depletions that exhibited a JSD significant phenotype, we conducted a gene set enrichment analysis (GSEA). Briefly, rank-ordered lists were generated for all genes comparing each lncRNA knockdown studied with the scrambled control. These lists were used as input to a preranked GSEA analysis (33). For each lncRNA knockdown vs. scramble control, normalized enrichment scores and significance values were determined across well-established MSigDB gene sets (33). We specifically separated gene sets involved in adipogenesis or adipocyte-related functions from all other gene sets to determine whether adipose-associated pathways were explicitly perturbed (Fig. S4). Indeed, for each knockdown, the mean of the distribution of P values for the adipose-associated gene sets was lower than the mean P values for the nonadipose gene sets (Fig. S5), indicating enrichment for reduction in adipose-associated genes at either tail of the rank-ordered lists. The enrichment for reduction in adipose-associated gene sets again corresponded to both the JSD to D0 (precursor) for each knockdown condition, as well as the ORO staining. Collectively, these results indicate a strong correlation between those lncRNAs that exhibit ORO staining defects and those that preferentially regulate adipose gene expression programs.
Discussion
Thousands of lncRNAs have been identified in mammalian genomes (13–15, 29), but the functions of the vast majority remain elusive. Our study has identified hundreds of lncRNAs that are regulated during adipogenesis, a substantial fraction of which are under the control of the same key transcription factors that activate and repress coding genes during adipocyte differentiation. Of those that we tested, most are specific to adipose tissue, and nearly half of these adipogenic up-regulated lncRNAs that we tested by knockdown are important for the development of mature adipocytes. These data suggest that lncRNAs, like protein coding genes, are likely regulated by the same mechanisms and protein factors that control protein-coding genes in adipogenesis and other developmental processes. Interestingly, loss of several lncRNAs each globally perturbed large portions of the collective adipogenic gene expression signature, suggesting that each of these plays a key role in induction of multiple adipocyte-specific genes.
Although large-scale LOF screens of lncRNAs will offer a wealth of useful information about the importance of lncRNAs in developmental processes, some inherent pitfalls of the RNAi method should be acknowledged. Some antisense lncRNAs may form RNA:RNA intermediates with their sense transcripts, and RNAi at times may trigger the degradation of both strands. Thus, for antisense lncRNAs, it should be further investigated whether the phenotype results from disruption of the sense or antisense transcript.
We have developed a unique application of the JSD metric on gene expression profiles and used it to quantify the phenotypic contributions of lncRNAs to adipogenesis. This approach circumvents shortcomings inherent to other metrics, and we expect it to be widely applicable to future LOF and gain-of-function screens. Such a metric is particularly important for screens where in vitro functional assays (e.g., the Oil Red staining used here) are unavailable or excessively laborious, expensive, or unreliable. This metric can help triage screened lncRNAs for more detailed mechanistic follow-up.
Further studies will be required to decipher the molecular mechanism by which the lncRNAs discussed in this study act to regulate adipogenesis. It is likely that some lncRNAs serve as a modular scaffold and tether protein factors to form a chromatin-modifying complex that regulates the epigenetic architectures during adipogenesis, as has been proposed for other lncRNAs (15, 22, 30, 34–36).
Although the 10 genes we discuss above were previously annotated as noncoding RNAs in the UCSC genome annotation (also based on RIKEN annotations) (26), we further scrutinized them for evidence of coding capacity. We first searched the amino acid databases SWISSPROT, Protein Data Bank, and the RefSeq protein collections (currently comprising more than 12 million amino acid sequences) for homology to all possible translations of the annotated RNAs. This analysis revealed no significant coding potential for any of the lncRNAs we knocked down except for two examples—lnc-RAP5 and lnc-RAP2—that have putative small ORFs of 56 and 36 amino acids, respectively. Notably, the small ORF in lnc-RAP5 is strongly conserved across metazoans from human to African clawed frog, yet does not resemble any of the more than 200,000 known amino acid sequences across the four kingdoms of life. This suggests the possibility of novel small peptides that might affect adipogenesis.
Experimental Procedures
Cell Isolation and Tissue Culture.
Primary adipocytes were isolated according to published methods, with few modifications (37, 38). Interscapular brown adipose tissues, epididymal fat pads, and s.c. fat pads were harvested from five 8-wk-old male mice. Fat tissues were minced, digested in collagenase, and fractioned by centrifugation. Adipocytes were collected from the top layer. Brown preadipocytes were isolated from interscapular brown fat, and white preadipocytes were isolated from s.c. fat from young mice (2 wk old). After collagenase digestion and fractionation, preadipocytes, enriched in bottom stromal vascular fractions, were resuspended and cultured to confluence in DMEM supplemented with 10% (vol/vol) newborn bovine serum. The cells were then exposed to differentiation medium: 10% (vol/vol) FBS DMEM, insulin 850 nM (Sigma), dexamethasone 0.5 μM (Sigma), 3-isobutyl-1-methylxanthine (IBMX) 250 μM (Sigma), and rosiglitazone 1 μM (Cayman Chemical) (brown adipocyte cultures, T3 1 nM and indomethacin 125 nM, were also added into the medium). After 2 d, cells were incubated in culture medium containing insulin 160 nM (supplemented with T3 for brown adipocyte cultures) for another 2 d, and then were switched to 10% FBS DMEM.
siRNA Knockdown of lncRNAs.
When cultured primary preadipocytes reached 70–80% confluence, siRNAs (200 nM) or modified DNA antisense oligo (100 nM) were transfected by DharmaFect 2 (final 5 μL/mL) according to the manufacturer’s instruction (Dharmacon). Twelve hours after transfection, cells were recovered in full culture media and grown to confluence, and then cells were induced to differentiate. Four days after differentiation, RNAs were harvested for downstream analysis. siRNAs were purchased from GenePharma. siRNA sequences are provided in Fig. S6.
Library Preparation and Sequencing.
Total RNAs were extracted using a Qiagen kit, and 10 μg of total RNAs for each sample were used to prepare the mRNA-Seq library according to the manufacturer's instruction (Illumina). cDNA libraries were prepared and sequenced by Illumina GAII according to the manufacturer’s instructions.
Differential Expression Analysis of RNA-Seq.
Reads from each sample were mapped against the mouse genome (mm9 build) using TopHat (version 1.1.0), using options “–no-novel-juncs -a 5 -F 0.0”. A splice junction index derived from the combined UCSC and RefSeq mm9 annotations together with previously discovered lncRNA transcript models built using RNA-Seq from several cell lines (39). This set of annotated transcripts was quantified in each sample using Cuffdiff (27), which estimates transcript and gene expression in each condition using a generative statistical model of RNA-Seq. Cuffdiff calculates the abundances in each condition for all transcripts that maximize the likelihood of observing the reads in the experiment under this model. Cuffdiff attaches statistical significance to observed changes to gene expression, and we restricted analysis to genes significantly differentially expressed by at least twofold between preadipocytes and adipocytes. We also required that differentially expressed genes used in downstream analysis were supported by at least 10 reads in either condition.
ORO Staining.
To prepare ORO solution, 0.5 g ORO (Sigma, catalog no. 0-0625) was dissolved in 100 mL isopropanol, mixed with H2O at a ratio 6:4, and filtered with Whatman #1 filter paper. Primary preadipocytes were differentiated in six-well plates. Cells were washed with PBS twice and fixed with formalin for 15 min at room temperature. After formalin fix, cells were washed with PBS and stained with freshly prepared ORO solution for 1 h in room temperature. Cells were washed with H2O to remove the residual ORO.
Expression Microarray Profiling.
Three replicate RNA samples each from D0 undifferentiated preadipocytes, D4 PPARg-induced adipocytes, and D4 scramble siRNA controls were labeled and hybridized to Affymetrix Mouse 430a2 arrays. Additionally, two replicate RNA samples from each of the 10 lncRNA siRNA studies were labeled and hybridized as well using standard Affymetrix protocols. Data were collected and analyzed using the “affy” package in R/Bioconductor. Briefly, probes were background corrected using the “mas” algorithm. Probe values were quantile normalized, and probe sets were summarized using the average difference of perfect matches only. All significance tests were performed using significance analysis of microarrays (SAM) (40) with Benjamini-Hochberg multiple hypothesis testing correction (MTC). Significant gene lists were selected with a Δ that constrained the FDR <4.5%.
Significant gene expression data were biclustered using the JSD, which is derived from the Shannon entropy
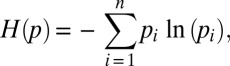 |
where p is a discrete probability distribution. The JSD is
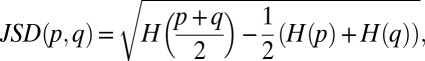 |
where p and q are two discrete probability distributions. In our array analysis, these distributions each represent either the relative abundance of all genes in a condition (for condition-level clustering) or the density of a gene’s expression across all conditions (for gene-level clustering). Pairwise condition distances thus reflect an information-theoretic summary of how similar is the program of gene expression in two cell states (e.g., preadipocyte and adipocyte). Pairwise gene distances reflect the information-theoretic similarity of expression across all conditions (potentially implying coordinated regulation).
Because only two replicates were available for each knockdown vs. scramble control analysis, rank-ordered lists of all genes were generated and used as input for the nonparametric preranked GSEA analysis (33). Rank-ordered gene lists were compared with all “curated gene sets” (C2) in MSigDB with the following parameters: collapse = True, norm = meandiv, scoring_scheme = weighted, mode = Max_probe, set_min = 12, set_max = 500, nperm = 1000, chip = Mouse430A_2.chip.
Supplementary Material
Acknowledgments
We thank Prathapan Thiru [Bioinformatics and Research Computing (BaRC) group of the Whitehead Institute], Sumeet Gupta (Genome Technology Core at the Whitehead Institute), Alla Sigova (Richard A. Young laboratory at the Whitehead Institute), and all members of the H.F.L. and J.L.R. laboratories for intellectual support and advice. This work is supported by National Institutes of Health Grants DK047618, DK068348, and 5P01HL066105 (to H.F.L.), and by Director's New Innovator Awards 1DP2OD00667, P01GM099117, and P50HG006193-01 (to J.L.R.). L.A.G. is a National Science Foundation Postdoctoral Research Fellow in Biological Informatics. C.T. is a Damon Runyon Cancer Foundation Postdoctoral Fellow. K.A.L. is supported by a National Science Scholarship from the Agency of Science, Technology and Research, Singapore. J.L.R. is a Damon Runyon-Rachleff, Searle, Smith Family, and Merkin Fellow.
Footnotes
The authors declare no conflict of interest.
Data deposition: All sequencing and related data are deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE29899).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222643110/-/DCSupplemental.
References
- 1.Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999-2008. JAMA. 2010;303(3):235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 2.Haslam DW, James WP. Obesity. Lancet. 2005;366(9492):1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 3.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7(12):885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 4.Tontonoz P, et al. Adipocyte-specific transcription factor ARF6 is a heterodimeric complex of two nuclear hormone receptors, PPAR gamma and RXR alpha. Nucleic Acids Res. 1994;22(25):5628–5634. doi: 10.1093/nar/22.25.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14(11):1293–1307. [PubMed] [Google Scholar]
- 6.Lefterova MI, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22(21):2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119(2):157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 8.Musri MM, Gomis R, Párrizas M. A chromatin perspective of adipogenesis. Organogenesis. 2010;6(1):15–23. doi: 10.4161/org.6.1.10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musri MM, Gomis R, Párrizas M. Chromatin and chromatin-modifying proteins in adipogenesis. Biochem Cell Biol. 2007;85(4):397–410. doi: 10.1139/O07-068. [DOI] [PubMed] [Google Scholar]
- 10.Mikkelsen TS, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143(1):156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander R, Lodish H, Sun L. MicroRNAs in adipogenesis and as therapeutic targets for obesity. Expert Opin Ther Targets. 2011;15(5):623–636. doi: 10.1517/14728222.2011.561317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, et al. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011;13(8):958–965. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JT. Lessons from X-chromosome inactivation: Long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23(16):1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huarte M, Rinn JL. Large non-coding RNAs: Missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loewer S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13(11):971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 25.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: Modest conservation and incompleteness. Genome Biol. 2009;10(11):R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134(3):521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 30.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacIsaac KD, et al. A quantitative model of transcriptional regulation reveals the influence of binding location on expression. PLOS Comput Biol. 2010;6(4):e1000773. doi: 10.1371/journal.pcbi.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie W, Granjeaud S, Puthier D, Gautheret D. Entropy measures quantify global splicing disorders in cancer. PLOS Comput Biol. 2008;4(3):e1000011. doi: 10.1371/journal.pcbi.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20(2):142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zappulla DC, Cech TR. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc Natl Acad Sci USA. 2004;101(27):10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cannon B, Nedergaard J. Cultures of adipose precursor cells from brown adipose tissue and of clonal brown-adipocyte-like cell lines. Methods Mol Biol. 2001;155:213–224. doi: 10.1385/1-59259-231-7:213. [DOI] [PubMed] [Google Scholar]
- 38.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6(1):38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.