Abstract
Seeds are in a natural oxidative context leading to protein oxidation. Although inevitable for proper progression from maturation to germination, protein oxidation at high levels is detrimental and associated with seed aging. Oxidation of methionine to methionine sulfoxide is a common form of damage observed during aging in all organisms. This damage is reversible through the action of methionine sulfoxide reductases (MSRs), which play key roles in lifespan control in yeast and animal cells. To investigate the relationship between MSR capacity and longevity in plant seeds, we first used two Medicago truncatula genotypes with contrasting seed quality. After characterizing the MSR family in this species, we analyzed gene expression and enzymatic activity in immature and mature seeds exhibiting distinct quality levels. We found a very strong correlation between the initial MSR capacities in different lots of mature seeds of the two genotypes and the time to a drop in viability to 50% after controlled deterioration. We then analyzed seed longevity in Arabidopsis thaliana lines, in which MSR gene expression has been genetically altered, and observed a positive correlation between MSR capacity and longevity in these seeds as well. Based on our data, we propose that the MSR repair system plays a decisive role in the establishment and preservation of longevity in plant seeds.
Keywords: seed deterioration, seed viability, redox systems, antioxidant enzymes
Successful crop implantation is crucial for maximal plant yield and relies on seed physiological quality, which is acquired at the end of development, during maturation. Quality is expressed by the capacity of seeds to germinate in a fast and homogenous manner before and after storage (seed longevity) and to provide proper seedling establishment in various environments (seed vigor). Quality is controlled by genetic traits and varies greatly among species and cultivars. The establishment of quality also depends on the prevailing environmental conditions during seed development and storage (1, 2).
During the maturation phase, mechanisms are put in place to prevent and repair desiccation damage, as well as to allow survival of seeds in the dry state (3–5). Nonetheless, drying of seeds on mother plants can occur prematurely in drought conditions, leading to low-quality seeds (6–8). Thus, in the context of a warming climate, producing good-quality seeds represents a global challenge to sustain current yield levels (9, 10).
The mechanisms underlying seed quality are controlled in part by abscisic acid (ABA), the phytohormone involved in plant responses to dehydration that is produced during seed maturation. Of note, ABA triggers the accumulation of late-embryogenesis abundant proteins, heat-shock proteins (HSPs), and storage proteins (11). Late-embryogenesis abundant proteins and HSPs are thought to primarily protect cell structures when water is removed. Reserve proteins, through their sensitivity to reactive oxygen species (ROS), play a determinant role in limiting oxidative damage on other seed components (12). Classical antioxidant systems also have an important part in seed quality (13). Thus, mutant plants deficient in tocopherol synthesis produce seeds with reduced viability when stored at high humidity and high temperature (14).
The quality of seeds ultimately depends on their capacity to repair damaged molecules when metabolism resumes on imbibition. Accordingly, protein repair at the level of aspartate/asparagine residues, spontaneously damaged over time, is required for proper seed longevity and germination (15). Repair of oxidized proteins may function in seed quality establishment, preservation, and expression as well. Indeed, seeds are in a natural oxidative context, leading to oxidation of proteins (12, 16, 17), the primary targets of ROS species owing to their high abundance (18). Sulfur-containing residues are particularly prone to oxidation (18). Oxidation of these residues is nevertheless reversible in most cases. Formation of disulfide bridges between cysteine residues occurs during seed maturation and has been proposed to provoke compaction of proteins, rendering them less sensitive to protease action or inactive (16). The reversibility of this process during germination, likely through the action of thioredoxins (Trxs), participates in the awakening of metabolism and is thought to play an important role in seed physiology (16, 19).
The role of oxidation of methionine (Met) in methionine sulfoxide (MetO) and its repair by methionine sulfoxide reductases (MSRs) has not been documented in seeds, although several lines of evidence support the possible involvement of MSRs in maturation and germination processes: (i) MSRs belong to the minimal set of proteins required for basic functions in living cells (20); (ii) Met oxidation is a hallmark of oxidative conditions and aging in all organisms (21); (iii) MSRs play a determinant role in the tolerance of oxidative stress (22–28); and (iv) MSRs control lifespan in microorganisms, insects, and mammals, including human (29–32). Moreover, one MSR gene exhibits increased expression during reinduction of desiccation tolerance in germinated seeds of Medicago truncatula (33). The participation of MSR in the establishment and preservation of seed quality remains unclear, however.
To fill this gap, we first explored whether MSRs have a function in seed longevity using the model legume M. truncatula. After characterizing the MSR family in this species, we investigated MSR abundance and activity in seeds of varying quality, using artificial aging treatments and taking advantage of the natural variability reported for this trait (34). We then analyzed the longevity of seeds in Arabidopsis thaliana lines, which exhibit altered MSR capacity owing to genetic transformation. Based on the data obtained with these two models, we propose that MSR plays a decisive role in longevity establishment and preservation in plant seeds.
Results
Identification of MSR Sequences in M. truncatula Databases.
In a search of EST libraries for M. truncatula Jemalong (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=medicago), we identified eight MSR sequences, including five MSRA sequences and three MSRB sequences (Table S1). Note that tentative consensus (TC) TC189868 sequence differs from TC188605 by only a 5′ extension, suggesting that these sequences correspond to two distinct messengers expressed from the same MSRA4.1 gene owing to alternative splicing. Genes corresponding to most TCs were identified in genomic databases (www.medicago.org) in which two additional MSRB genes for which no EST exists were found (Table S1).
Altogether, five MSRA and five MSRB isoforms were found in M. truncatula. This is similar to the numbers of isoforms identified in other plant species except for A. thaliana, which has nine MSRB isoforms. M. truncatula MSRs were aligned (Fig. S1), and their percent identity was calculated (Table S2). They were compared with the MSRs reported in A. thaliana and poplar in two phylogenic trees (Fig. S2). As described previously (24, 35), each plant MSR type is subdivided into two distinct clades based on the number of redox-active cysteines present. Thus, MSRA5 isoforms do not harbor the third cysteine found in other MSRAs. MSRB1 isoforms have one catalytic cysteine, compared with two in other MSRBs. The 10 M. truncatula isoforms identified here were named according to the classification established for poplar MSRs (24). Similar to poplar, MSRAs in M. truncatula seem to be addressed to cytosol (A2, A4.1S, and A4.2), plastids (A4.1L), and endoplasmic reticulum (A5), with A4.1S and A4.1L being respectively the short and long proteins encoded by the two alternative MSRA4.1 messengers. Regarding MSRBs, MtMSRB1 and B2 are most likely plastidial, like their orthologs in A. thaliana (36), whereas MtMSRB5.1, B5.2, and B5.3 are likely cytosolic.
Abundance of MSRs in Maturing Seeds.
The abundance of MSRs was analyzed in seeds of M. truncatula. In silico analysis (http://bioinfo.noble.org/gene-atlas) revealed expression of five MSR messengers in seeds during filling [10–24 d after pollination (DAP)] and maturation (28 DAP onward): A4.1S, A4.1L, A4.2, B1, and B2 (Fig. S3A). Three of these messengers (A4.1L, B1, and B2) encode plastidial isoforms.
The A4.1 gene is represented by two distinct probes on the M. truncatula array, the messengers encoding the short and long isoforms resulting from alternative splicing, as mentioned above. Intriguingly, the expression patterns obtained with the two probes vary in a similar way, although differing in arbitrary units (Fig. S3A). These patterns likely result from the presence of large overlapping regions in the sequences of messengers and probes (Fig. S4). The differences in levels of arbitrary units may be linked to differential stability of duplexes formed between cDNAs and probes.
To gain further insight into MSRA4.1 expression in mature seeds, we performed RT-PCR analysis (Fig. S3B) using specific primers to discriminate the two alternative forms (Table S3 and Fig. S5), and found that only the messenger encoding the long protein is present in mature seeds. The synthesis of MSRA4.1L messenger increases during seed filling from 10 to 24 DAP and decreases slightly thereafter (Fig. S3A). Regarding other messengers, MSRA4.2 and MSRB1 progressively accumulate from 10 DAP to the mature stage, whereas MSRB2 maintains an almost constant level throughout seed development, with two small peaks of abundance during filling (16 DAP) and at the mature stage (Fig. S3A).
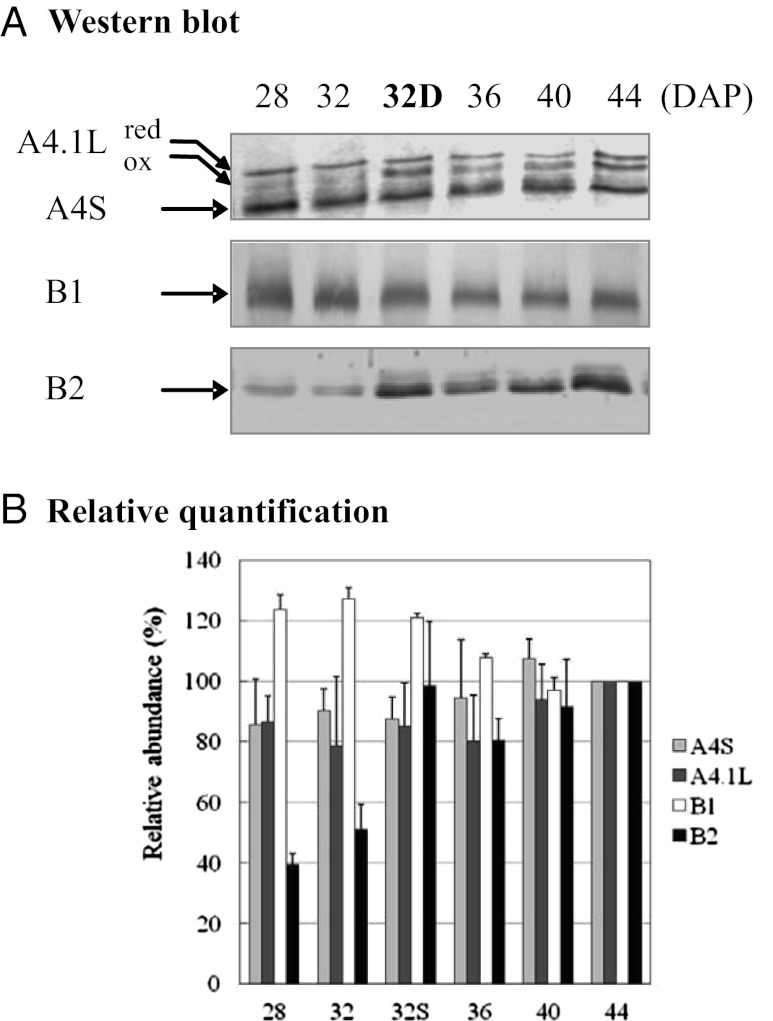
We further evaluated the abundance of MSRs by Western blot analysis using antibodies raised against MSRs from A. thaliana or poplar that could cross-react with purified recombinant M. truncatula proteins (Table S3 and Fig. S6). Two or three bands of expected size (20–25 kDa) were revealed using the anti-PtMSRA4 antibody in maturing seed extracts (Fig. 1A). The two upper bands likely correspond to reduced and oxidized forms of MtMSRA4.1L, as described previously in A. thaliana (36). The two forms were detected for the purified recombinant protein as well (Fig. S6). The lowest band detected at ∼22 kDa (A4S) in seeds might correspond to A4.2, based on the absence of A4.1S messenger and the presence of A4.2 messenger (Fig. S3). This band accumulated slightly with late maturation (Fig. 1B). The MSRA4.1L oxidized form appeared to be more abundant during natural desiccation (36 DAP onward), as well as in less mature seeds that were artificially dried (lane 32D), a treatment mimicking drying of premature seeds on mother plants in case of drought. The content in MSRB2 also increased with maturation but in a much stronger manner, given that band intensity was 2.5-fold higher in mature seeds than at 28 DAP. Interestingly, a similar increase was also observed for this isoform in seeds dried prematurely. In contrast, MSRB1 abundance decreased slightly during maturation (Fig. 1 A and B).
Fig. 1.
Abundance of MSRA4, MSRB1, and MSRB2 in seeds of M. truncatula. MSR abundance was measured by Western blot analysis in Jemalong seeds during maturation. For that purpose, soluble proteins (15 or 25 μg/lane), resolved by SDS/PAGE and transferred onto nitrocellulose membranes, were probed against anti-PtMSRA4, anti-AtMSRB1, or anti-AtMSRB2 sera (1/1,000). Quantification of bands detected on blots relative to bands stained with Coomassie blue in corresponding gels was performed using Quantity One (Bio-Rad) in two or three independent experiments. The abundance of bands in maturing seeds is expressed relative to that in mature seeds, stated as 100%.
Relationship Between MSR Activity and Longevity.
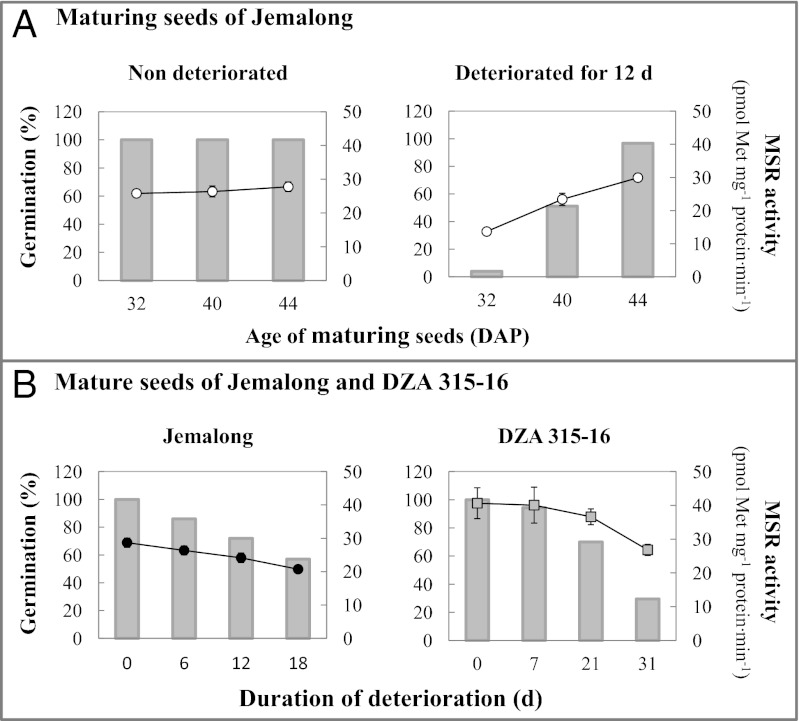
To explore whether the MSR system plays a role in seed longevity, we first analyzed MSR enzymatic capacity in different seed lots of M. truncatula. The first lot, produced in a growth chamber under controlled light and temperature conditions and dried rapidly, comprised maturing seeds of the Jemalong genotype harvested at different stages during longevity acquisition (5, 37), between 32 DAP and the mature stage (44 DAP). These seeds were able to germinate at 100% and displayed similar levels of MSR capacity, with only a slight increase, from 25.8 to 28.3 pmol Met⋅mg−1 protein⋅min−1, during maturation (Fig. 2A, Left).
Fig. 2.
Germination capacity and MSR activity in maturing and fully mature seeds before or after controlled deterioration. (A) Jemalong seeds were harvested during maturation at 32 or 40 DAP and at maturity (44 DAP), dried, and deteriorated for 0 or 12 d. (B) Fully mature Jemalong and DZA seeds were deteriorated for the indicated duration at 75% relative humidity and 35 °C. These seeds were used to determine germination capacity in percentage (gray bars) and to measure MSR capacity (white circles for maturing seeds and black circles for fully mature seeds of Jemalong; gray squares for mature seeds of DZA). Two independent experiments were performed with both immature and mature seed lots in each.
After controlled deterioration for 12 d at 75% relative humidity and 35 °C, a treatment that mimics seed aging (38), immature seeds were no longer able to germinate at 100%; seed survival rate decreased with the degree of initial seed maturity (Fig. 2A, Right). Most interestingly, the treatment induced a parallel decrease in MSR capacity, with the lowest level measured in the less mature 32 DAP seeds (13.7 pmol Met⋅mg−1 protein⋅min−1).
We also evaluated MSR capacity for fully mature seeds of the same genotype produced in a greenhouse (second seed lot). The value, 28.7 pmol Met⋅mg−1 protein⋅min−1, was similar to that of mature seeds produced in strictly controlled conditions. We then artificially aged these seeds for 0 to 18 d. The germination capacity in this lot was progressively lost with artificial aging, accompanied by decreased MSR capacity, down to 20.7 pmol Met⋅mg−1 protein⋅min−1 in seeds deteriorated for 18 d (Fig. 2B, Left). The deterioration process was not accompanied by a change in MSR abundance (Fig. S7); thus, the decreased capacity could originate from enzyme inactivation.
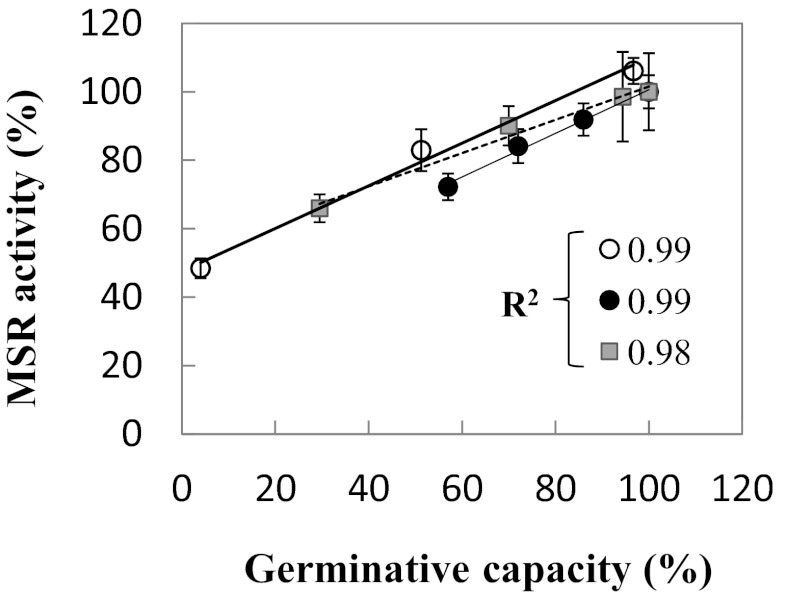
To compare the results gained from the two distinct lots under deterioration, we plotted the remaining MSR capacity, expressed as a percentage of that measured in control mature seeds, against the percentage of seed survival. Interestingly, a positive association between these two percentages was seen, with a high R2 coefficient of 0.99 in both cases (Fig. 3), highlighting the very strong correlation between MSR and germination capacities in Jemalong seeds.
Fig. 3.
Relationship between MSR activity and germination capacity of seeds. The level of MSR activity measured in seeds before and after deterioration (Fig. 2) was expressed as the percentage of activity determined in control mature seeds and plotted against the percentage of seed survival. White circles represent maturing Jemalong seeds; black circles, mature Jemalong seeds; gray squares, mature DZA315-16 seeds. The R2 correlation coefficients of linear regression for each set of values are indicated.
Because longevity is a genetic trait that varies widely within species, we analyzed mature seeds of another genotype, DZA315-16, previously shown to produce seeds of higher quality than Jemalong (34), in a similar manner. DZA seeds were produced in a greenhouse concomitantly with Jemalong seeds. As expected, the mature DZA seeds after artificial aging exhibited a higher survival rate than the Jemalong seeds (Fig. 2B, Right). Interestingly, MSR enzymatic capacity was substantially higher in mature DZA seeds (∼40 pmol Met⋅mg−1 protein⋅min−1) compared with Jemalong seeds (26–29 pmol Met⋅mg−1 protein⋅min−1). As in Jemalong, a decrease in activity accompanied the deterioration of DZA seeds from 0 to 31 d, with a remaining activity of 26.8 pmol Met⋅mg−1 protein⋅min−1 at 31 d; however, the rate of decrease was much slower in DZA. Again, plotting the percentage of remaining activity against the percentage of seed survival produced a high R2 coefficient of 0.98 (Fig. 3).
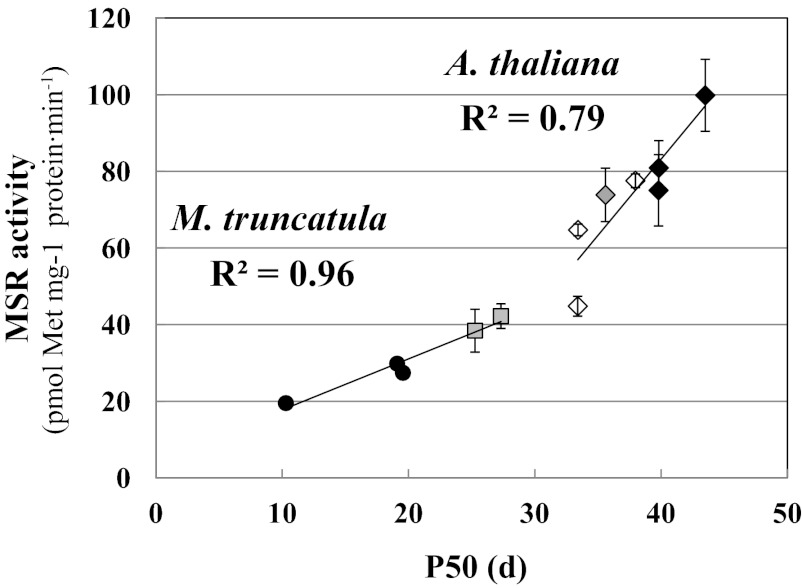
From the germination percentages gained on mature seeds of the two genotypes during deterioration, we estimated the time for the viability to fall to 50% (P50) at 75% relative humidity and 35 °C (5). We found a strong link (R2 = 0.96) between P50 and the initial MSR capacity measured before deterioration (Fig. 4). An additional lot of Jemalong seeds characterized by a very low germination capacity (P50 = 10.3 d) was included in this experiment. Collectively, these results reveal a tight relationship between MSR enzymatic capacity and seed longevity in M. truncatula.
Fig. 4.
Relationship between initial MSR capacity and longevity in M. truncatula and A. thaliana seeds. MSR activity was measured in different seed lots of mature seeds of M. truncatula (Jemalong and DZA genotypes) and A. thaliana (WT and genetically modified lines). The seeds were then subjected to deterioration at 75% relative humidity and 35 °C for 0–50 d. Seed viability, assessed by germination testing, is expressed as P50, the time for viability to fall to 50%. MSR capacity was plotted against P50 values obtained separately using M. truncatula and A. thaliana. Black circles represent M. truncatula Jemalong seeds; gray squares, M. trunculata DZA seeds. Diamonds correspond to seeds of A. thaliana: a gray diamond for the WT line, white diamonds for lines lacking MSRB (lines B1−, B2−, and B1−/B2−), and black diamonds for lines overexpressing MSRB (lines B1-8, B1-10, and B2-3).
To gain further insight into the role of MSR in seed longevity, we analyzed A. thaliana lines in which MSR capacity was altered owing to genetic modification of the expression of genes encoding the two plastidial MSRB isoforms, MSRB1 and MSRB2 (39, 40). In the lines lacking MSRB1, MSRB2, or both, designated B1−, B2−, and B1−/B2−, leaf MSR capacity is reduced by 25%, 50%, and 75%, respectively (39; Table S4). In the present work, measurement of MSR capacity in mature seeds of these lines (Table S4) revealed that this capacity indeed was substantially reduced in the line lacking MSRB2 (45 pmol Met⋅mg−1 protein⋅min−1) compared with WT (74 pmol⋅min−1) but, surprisingly, not in the line lacking MSRB1 (77 pmol⋅min−1) and to only a low extent in the double-mutant B1−/B2− line lacking both isoforms (65 pmol⋅min−1). These data suggest that MSRB1 is not functional in A. thaliana seeds or that the absence of this isoform or both plastidial MSRB isoforms leads to compensation mechanisms involving other MSR isoforms, such as plastidial MSRA4. Regarding MSRB1-overexpressing lines, designated B1-8 and B1-10, seed MSR capacity was only slightly higher than that in WT (81 and 100 pmol Met⋅mg−1 protein⋅min−1, respectively). Note that the leaf MSR capacity in these lines is fourfold higher compared with WT (40; Table S4). For MSRB2-overexpressing lines, designated B2-3 and B2-5, contrasting results were obtained, with either no change in the B2-3 line (75 pmol⋅min−1) or a dramatic increase in the B2-5 line (1,700 pmol⋅min−1). In contrast, in leaf extracts, MSR capacity was significantly increased in the former and highly increased in the latter compared with WT.
We took advantage of the varying MSR capacity in seeds of these A. thaliana lines to investigate the relationship with longevity. For this purpose, we determined P50 with the same deterioration treatment described above for M. truncatula (Table S4). Interestingly, the A. thaliana seeds had higher P50 values than the M. truncatula seeds, possibly related to the higher MSR capacity measured in A. thaliana seeds. Nonetheless, seeds of the MSRB2-overexpressing line B2-5 with the huge MSR capacity had the lowest P50. This result is likely related to the MSR capacity, which is 23-fold higher than that measured in WT seeds and indeed far outside the range of physiological values. This modification likely breaks redox homeostasis, impairing the proper establishment and expression of seed quality (41). Note that in fission yeast, the total Trx pool is limited, and there is competition among enzymes using Trxs as reducing substrates, such as MSRs and thiol-peroxidases (42). In the B2-5 line, which exhibits very high MSR capacity in seeds and leaves (Table S4), MSRB2 likely diverts reducing power from Trxs and strongly impairs other antioxidant systems involved in seed longevity. Thus, we discarded this line in our investigation of the relationship between MSR capacity and longevity. In other A. thaliana lines, unmodified (WT), down-regulated (B1−, B2−, and B1−/B2−), or up-regulated (B1-8, B1-10, and B2-3) for MSR expression, we measured MSR capacities in a range of 45–100 pmol⋅min−1. Interestingly, our analysis of the relationship between MSR capacity and P50 in all of these lines revealed a reliable positive correlation (R2 = 0.79) between MSR activity level before aging and P50 value (Fig. 4). Taken together, our data demonstrate a likely role of the MSR system in the control of longevity in seeds of M. truncatula and A. thaliana.
Discussion
Orthodox seeds in a dry and quiescent state are unable to repair or replace damaged molecules that accumulate progressively during storage. Thus, these seeds represent a stage in the plant biological cycle in which cells undergo an aging process similar to that in animal cells. In this work, using seeds of varying MSR capacity owing to either natural variation (contrasting M. truncatula ecotypes) or genetic modification (A. thaliana WT and transformant lines), we have demonstrated that seed longevity is strongly linked to MSR enzymatic capacity (Figs. 3 and 4). In M. truncatula species, this relationship, characterized by a very high R2 coefficient (≥0.96), has been established in both maturing seeds of Jemalong, harvested during longevity acquisition, and full mature seeds of Jemalong and DZA315-16, two genotypes of naturally contrasting seed quality (34). Thus, MSR capacity appears to be a good marker to predict seed performance in this species. We also found a similar relationship in A. thaliana seeds, in which MSR activity has been genetically altered, albeit with a lower R2 coefficient (0.79). Taken together, our results for natural M. truncatula ecotypes and transformed A. thaliana lines provide strong support for a role of MSR in the control of seed longevity in plants. Our findings are in agreement with the decisive role of MSRs in determining the lifespan of microorganisms and animals (29–32).
Our Western blot analysis data (Fig. 1) suggest that seed longevity may be linked to the abundance of both MSR types. Thereby, MSRB2, which accumulates significantly during maturation, might be responsible for the improved storability of mature seeds compared with less mature seeds in this species. This isoform also accumulates in immature seeds in response to anticipated drying. We also observed a slight increase in MSRA4.1L and MSRA4S abundance during maturation. MSRA4.1L may play a critical role during desiccation, as suggested by the accumulation of its transcript with reinduction of desiccation tolerance in seeds of M. truncatula (33). Of note, the abundance of oxidized MSRA.1L was substantially higher in seeds dried either naturally or artificially compared with nondried seeds. The expression of numerous MSR genes is enhanced by environmental conditions known to generate oxidative stress (24); thus, expression of MSRA4 and MSRB2 may be induced by the oxidative context of seed maturation and/or desiccation. Alternatively, MSR expression might be controlled by ABA, which plays a major role in gene expression during seed maturation. Accordingly, expression of one MSRA gene and one MSRB gene is known to be regulated by ABA in A. thaliana and tomato seedlings (26, 43).
Germination occurs in the absence of transcription and relies on stored proteins or on proteins translated from mRNA accumulated in dry seeds (4, 44). Thus, mechanisms involved in the repair of reversible biochemical alterations in proteins, critical for metabolism and seed performance, are likely required in seeds. Consistently, the repair of aspartate/asparagine residues, spontaneously damaged over time, by protein-isoaspartyl-methyltransferase is critical for proper seed longevity and germination (15, 45). Reduction of disulfide bridges in oxidized proteins by redoxins, such as thioredoxins, is a well-known key process in seeds, allowing reactivation of metabolism and mobilization of reserves after imbibition to sustain germination and seedling establishment (16, 19, 46). More than 100 proteins potentially targeted by thioredoxins have been notably identified in M. truncatula seeds, most of which are reduced during germination (19). Thus, MSRs also likely contribute in concert with redoxins to the essential function of reactivation of oxidized proteins in seeds. To date, however, few targets and putative targets of MSRs have been identified in plants. Those targets that have been identified in leaves include elongation factors, proteasome subunits, HSPs, and catalase (39, 47, 48). Interestingly, these proteins are targeted by redoxins as well (46). Proteins implicated in machineries of protein translation, such as elongation factors, and protein degradation, such as proteasome subunits, are among the most sensitive proteins inactivated under oxidative conditions (1, 49), and thus may require repair by MSRs and redoxins in seeds after imbibition. When these machineries are poorly functional and the replacement of damaged proteins is compromised, the action of MSRs and redoxins after imbibition might be the only way to restore the crucial functions required for germination and plant establishment. Catalase and HSPs are critical for resistance to oxidative stress and play important roles in seed performance (13, 50). Catalase has been shown to be inactivated by sulfoxidation in Helicobacter pylori, with reactivation of the enzyme ensured by a synergistic action of MSR and a chaperon protein (51). Similarly, a small plastidial HSP isoform from A. thaliana is inactivated by sulfoxidation, and MSRA prevents this inactivation (47). Thus, along with the functions proposed above, MSRs may play a role in the maintenance of protective mechanisms involving HSPs and catalase in seeds. MSRs also may participate in redox signaling pathways in seeds during maturation and/or germination, as has been reported for the calcium/calmodulin-dependent protein kinase II in mammalian cells (52).
MSR is known to play an important role in the response to stressful conditions in microorganisms and animals. In plants, MSRs also have important functions in response to environmental constraints such as salinity, cold temperature, and high light (24, 26–28, 39, 53). This capacity could be related to MSRs’ ability to function as ROS scavengers in concert with some of their protein targets, which would not be affected by sulfoxidation (54, 55). Such a general role is supported by the negative relationship between the level of MSR activity and the levels of proteins oxidized by carbonylation in numerous organisms (56). Thus, the involvement of MSRs in ROS scavenging could occur in seeds to limit the extent of damage resulting from the oxidative conditions reigning during maturation and imbibition. This idea is consistent with the finding that antioxidant enzymes, such as catalase and superoxide dismutase (13), and antioxidant components, such as tocopherol (14, 57), confer protection against seed deterioration and aging.
Finally, it is worth mentioning that our data suggest the likely participation of plastidial MSR isoforms in the control of plant seed longevity. This finding is consistent with the results of Bassel et al. (58), who reported that the functionality of several plastidial proteins is required for germination. Taken together, these data support the critical role of plastidial metabolism in seed physiology. It would be interesting to identify potential MSR targets in seeds, as well as proteins sulfoxidized in seeds of poor quality but not in seeds of good quality. These proteins likely play key roles in germination. In aged seeds that are unable to germinate, one can imagine that proteins are severely damaged, and MSRs and redoxins provide less efficient repair. Accordingly, MSRs are less functional in deteriorated seeds of M. truncatula (Figs. 2 and 3). Whether MSR denaturation in seeds is related to oxidation, as reported in other organisms (59), remains to be determined.
In conclusion, MSRs appear to be involved in the control of longevity in M. truncatula and A. thaliana seeds. It would be of great interest to determine whether this also holds true for seeds of species exhibiting exceptional longevity, such as those of sacred lotus and date palm, which are able to germinate after 1,000–2,000 y of storage (60, 61). It would be even more interesting to examine whether it also holds true for seeds of crops. Because seed longevity and seed vigor are tightly linked, MSR repair capacity also might play a role in seed vigor, and thus could serve as an innovative marker to predict both seed longevity and seed vigor (i.e., the likelihood that seeds will germinate and plants will emerge in various environments, the two processes underlying successful culture implantation).
Materials and Methods
Biological Materials.
Plants of M. truncatula (genotypes Jemalong and DZA315-16) were grown in a sterile mix of vermiculite and soil with regular watering, under a 16-h photoperiod (37). Maturing seeds of Jemalong were produced in a growth chamber at a day temperature of 24 °C and a night temperature of 20 °C. As flowers appeared, they were tagged, and developing seeds were removed from pods at different ages at the end of filling (28 DAP) or during maturation (32–44 DAP, with 44 DAP corresponding to fully mature seeds). Fully mature seeds of Jemalong and DZA315-16 were produced concomitantly in a greenhouse with minimum day and night temperatures of 19 °C and 16 °C, respectively. Seeds were stored at −80 °C either immediately after harvesting or after an anticipated rapid drying over 48 h at 20 °C and 44% relative humidity. This treatment applied to immature seeds mimics premature drying of the seeds on mother plants exposed to drought.
For production of seeds, A. thaliana WT (Col0) and lines modified for the expression of MSRB1 and MSRB2 genes (36, 39, 40) were grown simultaneously under a 14-h photoperiod (22/18 °C, day/night) and a photon flux density of 250 µmol photons⋅m–2⋅s–1. Seeds were stored in constant controlled conditions (15 °C, 35% hygrometry) in the dark.
Some of the dried seeds (after either natural desiccation or anticipated drying) were then subjected to controlled deterioration by exposure to 75% relative humidity and 35 °C for 0–31 d (62). For the determination of germination capacity, dry seeds were imbibed in 9-cm-diameter Petri dishes on filter paper (Whatman no. 1) soaked with 3.5 mL of distilled water for several days at 20 °C in the dark. Freshly harvested immature and mature seeds of M. truncatula were scarified by sand paper and imbibed in the presence of 83 µM of fluoridone to release dormancy (63).
Soluble Extract Preparation and Western Blot Analysis.
Seeds were ground to powder using liquid nitrogen, and soluble proteins were extracted in 25 mM Tris⋅HCl buffer supplemented with 1 mM PMSF and 1 mM EDTA (5–20 mL/g fresh weight). The resulting homogenate was centrifuged (40,000 × g for 30 min at 4 °C), and the resulting pellet was discarded. Proteins (20 µg per lane) were then resolved by SDS/PAGE using 12% (wt/vol) acrylamide gels as described previously (64). At the end of electrophoresis, proteins were either stained in gel using colloidal Coomassie blue or transferred onto nitrocellulose membranes (Schleicher & Schull) for immunodetection, as described previously (65). Membranes were probed with rabbit antibodies raised against A. thaliana MSRB1 and MSRB2 and poplar MSRA4 (36). Anti-PtMSRA4 antibodies were a kind gift from N. Rouhier (University of Nancy, Nancy, France). Immunodetection was performed with a phosphatase alkaline assay in the presence of 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium.
MSR Activity Assays.
After grounding of seeds in liquid nitrogen and suspension in extraction buffer containing 15 mM Hepes (pH 8), 10 mM MgCl2, 30 mM KCl, and 1 mM PMSF in an Eppendorf tube, the mixture was vigorously stirred for 20 min and centrifuged at 18,000 × g for 20 min at 4 °C. Soluble protein content was measured using BC Assay Reagent (Interchim). Total MSR activity in seed extracts was assayed by monitoring reduction of the synthetic substrate dabsyl-MetO in the presence of a reducing agent, DTE (36). The 100-µL reaction mixture contained 15 mM Hepes (pH 8), 10 mM MgCl2, 30 mM KCl, 20 mM DTE, 0.25 mM dabsyl-MetO, and seed-soluble proteins: 200 µg for M. truncatula and 50 µg, for A. thaliana (the lower amount owing to less-concentrated extracts).
The enzymatic assay was performed in the presence of a large excess of substrate (dabsyl-MetO), and the activity level was found to be proportional to the amount of protein during a 3-h incubation at 37 °C. After stopping using 900 µL of ethanol:acetate buffer and centrifugation, 160 µL of a supernatant aliquot was loaded onto a C18 reversed-phase 3.5-µm, 3 × 50 mm SunFire column (Waters), and HPLC separation of dabsyl-MetO and dabsyl-Met was performed as described previously (36, 39).
Protein Content Determination.
The protein content of soluble extracts was determined using Bradford reagent and BSA as a standard (66). Concentrations of pure recombinant protein preparations were estimated using εM values at 280 nm.
Supplementary Material
Acknowledgments
We thank Dr J. Buitink (Institut National de Recherche Agronomique/Institut de Recherche en Horticulture et Semences) for comments on seed longevity and B. Ksas (Commissariat à l'Energie Atomique et aux Energies Alternatives/Direction des Sciences du Vivant/Institut de Biologie Environnementale et Biotechnologie) for valuable help with HPLC maintenance. This work was funded in part by the region Les Pays de la Loire (QUALISEM 2009-2013) and a CADRES fellowship (to E.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220589110/-/DCSupplemental.
References
- 1.Walters C. Understanding the mechanisms and kinetics of seed aging. Seed Sci Res. 1998;8:223–244. [Google Scholar]
- 2.Priestley D. Seed Aging: Implication for Seed Storage and Persistence in Soil. Ithaca, NY: Cornell Univ Press; 1986. [Google Scholar]
- 3.Hoekstra FA, Golovina EA, Buitink J. Mechanisms of plant desiccation tolerance. Trends Plant Sci. 2001;6(9):431–438. doi: 10.1016/s1360-1385(01)02052-0. [DOI] [PubMed] [Google Scholar]
- 4.Rajjou L, Debeaujon I. Seed longevity: Survival and maintenance of high germination ability of dry seeds. C R Biol. 2008;331(10):796–805. doi: 10.1016/j.crvi.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Chatelain E, et al. Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ. 2012;35(8):1440–1455. doi: 10.1111/j.1365-3040.2012.02501.x. [DOI] [PubMed] [Google Scholar]
- 6.Bailly C, et al. Changes in oligosaccharide content and antioxidant enzyme activities in developing bean seeds as related to acquisition of drying tolerance and seed quality. J Exp Bot. 2001;52(357):701–708. doi: 10.1093/jexbot/52.357.701. [DOI] [PubMed] [Google Scholar]
- 7.Sanhewe AJ, Ellis RH. Seed development and maturation in Phaseolus vulgaris, II: Post-harvest longevity in air-dry storage. J Exp Bot. 1996;47(7):959–965. [Google Scholar]
- 8.Bewley JD, Black M. Seeds: Physiology of Development and Germination. 2nd Ed. New York: Plenum; 1994. [Google Scholar]
- 9.Salon C, et al. Plant N fluxes and modulation by nitrogen, heat and water stresses: A review based on comparison of legumes and non legume plants. In: Shanker A, Venkateswarlu B, editors. Abiotic Stress in Plants: Mechanisms and Adaptations. Rijeka, Croatia: InTech; 2010. pp 79–118. [Google Scholar]
- 10.Neilson KA, Gammulla CG, Mirzaei M, Imin N, Haynes PA. Proteomic analysis of temperature stress in plants. Proteomics. 2010;10(4):828–845. doi: 10.1002/pmic.200900538. [DOI] [PubMed] [Google Scholar]
- 11.Almoguera C, Jordano J. Developmental and environmental concurrent expression of sunflower dry-seed-stored low-molecular-weight heat-shock protein and Lea mRNAs. Plant Mol Biol. 1992;19(5):781–792. doi: 10.1007/BF00027074. [DOI] [PubMed] [Google Scholar]
- 12.Job C, Rajjou L, Lovigny Y, Belghazi M, Job D. Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol. 2005;138(2):790–802. doi: 10.1104/pp.105.062778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailly C. Active oxygen species and antioxidants in seed biology. Seed Sci Res. 2004;14:93–107. [Google Scholar]
- 14.Horvath G, et al. Accumulation of tocopherols and tocotrienols during seed development of grape (Vitis vinifera L. cv. Albert Lavallée) Plant Physiol Biochem. 2006;44(11-12):724–731. doi: 10.1016/j.plaphy.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Ogé L, et al. Protein repair L-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell. 2008;20(11):3022–3037. doi: 10.1105/tpc.108.058479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchanan BB, Balmer Y. Redox regulation: A broadening horizon. Annu Rev Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 17.Oracz K, et al. ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 2007;50(3):452–465. doi: 10.1111/j.1365-313X.2007.03063.x. [DOI] [PubMed] [Google Scholar]
- 18.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta. 2005;1703(2):93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Alkhalfioui F, et al. Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds. Plant Physiol. 2007;144(3):1559–1579. doi: 10.1104/pp.107.098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci USA. 1996;93(19):10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40(12):1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 22.Moskovitz J, et al. Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc Natl Acad Sci USA. 1998;95(24):14071–14075. doi: 10.1073/pnas.95.24.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moskovitz J, et al. Escherichia coli peptide methionine sulfoxide reductase gene: Regulation of expression and role in protecting against oxidative damage. J Bacteriol. 1995;177(3):502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouhier N, Vieira Dos Santos C, Tarrago L, Rey P. Plant methionine sulfoxide reductase A and B multigenic families. Photosynth Res. 2006;89(2-3):247–262. doi: 10.1007/s11120-006-9097-1. [DOI] [PubMed] [Google Scholar]
- 25.Weissbach H, Resnick L, Brot N. Methionine sulfoxide reductases: History and cellular role in protecting against oxidative damage. Biochim Biophys Acta. 2005;1703(2):203–212. doi: 10.1016/j.bbapap.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Dai C, Wang M-H. Characterization and functional analysis of methionine sulfoxide reductase A gene family in tomato. Mol Biol Rep. 2012;39(5):6297–6308. doi: 10.1007/s11033-012-1451-0. [DOI] [PubMed] [Google Scholar]
- 27.Kwon SJ, Kwon SI, Bae MS, Cho EJ, Park OK. Role of the methionine sulfoxide reductase MsrB3 in cold acclimation in Arabidopsis. Plant Cell Physiol. 2007;48(12):1713–1723. doi: 10.1093/pcp/pcm143. [DOI] [PubMed] [Google Scholar]
- 28.Oh S-K, et al. CaMsrB2, pepper methionine sulfoxide reductase B2, is a novel defense regulator against oxidative stress and pathogen attack. Plant Physiol. 2010;154(1):245–261. doi: 10.1104/pp.110.162339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moskovitz J, et al. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci USA. 2001;98(23):12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan H, et al. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci USA. 2002;99(5):2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koc A, Gasch AP, Rutherford JC, Kim H-Y, Gladyshev VN. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and -independent components of aging. Proc Natl Acad Sci USA. 2004;101(21):7999–8004. doi: 10.1073/pnas.0307929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cabreiro F, et al. Overexpression of mitochondrial methionine sulfoxide reductase B2 protects leukemia cells from oxidative stress-induced cell death and protein damage. J Biol Chem. 2008;283(24):16673–16681. doi: 10.1074/jbc.M708580200. [DOI] [PubMed] [Google Scholar]
- 33.Buitink J, et al. Transcriptome profiling uncovers metabolic and regulatory processes occurring during the transition from desiccation-sensitive to desiccation-tolerant stages in Medicago truncatula seeds. Plant J. 2006;47(5):735–750. doi: 10.1111/j.1365-313X.2006.02822.x. [DOI] [PubMed] [Google Scholar]
- 34.Vandecasteele C, et al. Quantitative trait loci analysis reveals a correlation between the ratio of sucrose/raffinose family oligosaccharides and seed vigour in Medicago truncatula. Plant Cell Environ. 2011;34(9):1473–1487. doi: 10.1111/j.1365-3040.2011.02346.x. [DOI] [PubMed] [Google Scholar]
- 35.Tarrago L, Laugier E, Rey P. Protein-repairing methionine sulfoxide reductases in photosynthetic organisms: Gene organization, reduction mechanisms, and physiological roles. Mol Plant. 2009;2(2):202–217. doi: 10.1093/mp/ssn067. [DOI] [PubMed] [Google Scholar]
- 36.Vieira Dos Santos C, Cuiné S, Rouhier N, Rey P. The Arabidopsis plastidic methionine sulfoxide reductase B proteins: Sequence and activity characteristics, comparison of the expression with plastidic methionine sulfoxide reductase A, and induction by photooxidative stress. Plant Physiol. 2005;138(2):909–922. doi: 10.1104/pp.105.062430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosnoblet C, et al. The regulatory gamma subunit SNF4b of the sucrose non-fermenting-related kinase complex is involved in longevity and stachyose accumulation during maturation of Medicago truncatula seeds. Plant J. 2007;51(1):47–59. doi: 10.1111/j.1365-313X.2007.03116.x. [DOI] [PubMed] [Google Scholar]
- 38.Rajjou L, et al. Proteome-wide characterization of seed aging in Arabidopsis: A comparison between artificial and natural aging protocols. Plant Physiol. 2008;148(1):620–641. doi: 10.1104/pp.108.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laugier E, et al. Arabidopsis thaliana plastidial methionine sulfoxide reductases B, MSRBs, account for most leaf peptide MSR activity and are essential for growth under environmental constraints through a role in the preservation of photosystem antennae. Plant J. 2010;61(2):271–282. doi: 10.1111/j.1365-313X.2009.04053.x. [DOI] [PubMed] [Google Scholar]
- 40.Laugier E, et al. Involvement of thioredoxin y2 in the preservation of leaf methionine sulfoxide reductase capacity and growth under high light. Plant Cell Environ. 2013;36(3):670–682. doi: 10.1111/pce.12005. [DOI] [PubMed] [Google Scholar]
- 41.Kranner I, Minibayeva FV, Beckett RP, Seal CE. What is stress? Concepts, definitions and applications in seed science. New Phytol. 2010;188(3):655–673. doi: 10.1111/j.1469-8137.2010.03461.x. [DOI] [PubMed] [Google Scholar]
- 42.Day AM, et al. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol Cell. 2012;45(3):398–408. doi: 10.1016/j.molcel.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 43.Oh J-E, et al. Modulation of gene expressions and enzyme activities of methionine sulfoxide reductases by cold, ABA or high salt treatments in Arabidopsis. Plant Science. 2005;169(6):1030–1036. [Google Scholar]
- 44.Rajjou L, et al. The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 2004;134(4):1598–1613. doi: 10.1104/pp.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mudgett MB, Lowenson JD, Clarke S. Protein repair L-isoaspartyl methyltransferase in plants (phylogenetic distribution and the accumulation of substrate proteins in aged barley seeds) Plant Physiol. 1997;115(4):1481–1489. doi: 10.1104/pp.115.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montrichard F, et al. Thioredoxin targets in plants: The first 30 years. J Proteomics. 2009;72:452–474. doi: 10.1016/j.jprot.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Gustavsson N, et al. A peptide methionine sulfoxide reductase highly expressed in photosynthetic tissue in Arabidopsis thaliana can protect the chaperone-like activity of a chloroplast-localized small heat shock protein. Plant J. 2002;29(5):545–553. doi: 10.1046/j.1365-313x.2002.029005545.x. [DOI] [PubMed] [Google Scholar]
- 48.Tarrago L, et al. Affinity chromatography: A valuable strategy to isolate substrates of methionine sulfoxide reductases? Antioxid Redox Signal. 2012;16:79–84. doi: 10.1089/ars.2011.4153. [DOI] [PubMed] [Google Scholar]
- 49.Davies K. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 50.Prieto-Dapena P, Castaño R, Almoguera C, Jordano J. Improved resistance to controlled deterioration in transgenic seeds. Plant Physiol. 2006;142(3):1102–1112. doi: 10.1104/pp.106.087817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahawar M, Tran V, Sharp JS, Maier RJ. Synergistic roles of Helicobacter pylori methionine sulfoxide reductase and GroEL in repairing oxidant-damaged catalase. J Biol Chem. 2011;286(21):19159–19169. doi: 10.1074/jbc.M111.223677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erickson JR, et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133(3):462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo X, Wu Y, Wang Y, Chen Y, Chu C. OsMSRA4.1 and OsMSRB1.1, two rice plastidial methionine sulfoxide reductases, are involved in abiotic stress responses. Planta. 2009;230(1):227–238. doi: 10.1007/s00425-009-0934-2. [DOI] [PubMed] [Google Scholar]
- 54.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA. 1996;93(26):15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moskovitz J, Berlett BS, Poston JM, Stadtman ER. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci USA. 1997;94(18):9585–9589. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moskovitz J, Oien D. Protein carbonyl and the methionine sulfoxide reductase system. Antioxid Redox Signal. 2010;12:405–415. doi: 10.1089/ars.2009.2809. [DOI] [PubMed] [Google Scholar]
- 57.Méne-Saffrané L, Jones AD, DellaPenna D. Plastochromanol-8 and tocopherols are essential lipid-soluble antioxidants during seed desiccation and quiescence in Arabidopsis. Proc Natl Acad Sci USA. 2010;107(41):17815–17820. doi: 10.1073/pnas.1006971107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bassel GW, et al Elucidating the germination transcriptional program using small molecules. Plant Physiol. 2008;147(1):143–155. doi: 10.1104/pp.107.110841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le H-T, et al. Impact of hydrogen peroxide on the activity, structure, and conformational stability of the oxidized protein repair enzyme methionine sulfoxide reductase A. J Mol Biol. 2009;393(1):58–66. doi: 10.1016/j.jmb.2009.07.072. [DOI] [PubMed] [Google Scholar]
- 60.Sallon S, et al. Germination, genetics, and growth of an ancient date seed. Science. 2008;320:1464. doi: 10.1126/science.1153600. [DOI] [PubMed] [Google Scholar]
- 61.Shen-Miller J, Mudgett M, Schopf J, Clarke S, Berger R. Exceptional seed longevity and robust growth: ancient sacred lotus from China. American Journal of Botany. 1995;82:1367–1380. [Google Scholar]
- 62.International Seed Testing Association . International Rules for Seed Testing. Bassersdorf, Zwitzerland: International Seed Testing Association; 2012. [Google Scholar]
- 63.Bolingue W, Ly Vu B, Leprince O, Buitink J. Characterization of dormancy behaviour in seeds of the model legume Medicago truncatula. Seed Sci Res. 2010;20:97–107. [Google Scholar]
- 64.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 65.Alkhalfioui F, Renard M, Montrichard F. Unique properties of NADP-thioredoxin reductase C in legumes. J Exp Bot. 2007;58:969–978. doi: 10.1093/jxb/erl248. [DOI] [PubMed] [Google Scholar]
- 66.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantity of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.