Abstract
T cells play an important role in the pathogenesis of allergic diseases. However, the proteins considered as potential immunogens of allergenic T-cell responses have traditionally been limited to those that induce IgE responses. Timothy grass (TG) pollen is a well-studied inhaled allergen for which major IgE-reactive allergens have also been shown to trigger T helper 2 (Th2) responses. Here we examined whether other TG pollen proteins are recognized by Th2 responses independently of IgE reactivity. A TG pollen extract was analyzed by 2D gel electrophoresis and IgE/IgG immunoblots using pooled sera from allergic donors. Mass spectrometry of selected protein spots in combination with de novo sequencing of the whole TG pollen transcriptome identified 93 previously undescribed proteins for further study, 64 of which were not targeted by IgE. Predicted MHC binding peptides from the previoulsy undescribed TG proteins were screened for T-cell reactivity in peripheral blood mononuclear cells from allergic donors. Strong IL-5 production was detected in response to peptides from several of the previously undescribed proteins, most of which were not targeted by IgE. Responses against the dominant undescribed epitopes were associated with the memory T-cell subset and could even be detected directly ex vivo after Th2 cell enrichment. These findings demonstrate that a combined unbiased transcriptomic, proteomic, and immunomic approach identifies a greatly broadened repertoire of protein antigens targeted by T cells involved in allergy pathogenesis. The discovery of proteins that induce Th2 cells but are not IgE reactive may allow the development of safer immunotherapeutic strategies.
Allergic diseases such as rhinitis and asthma pose a significant burden to both patients and society as a whole (1). Recent studies have estimated that up to 20% of the population in the United States and Western Europe suffers from these diseases (2, 3). Despite this high incidence, existing therapy is mostly symptomatic, and immunotherapy treatments are successful in only a fraction of patients and can be associated with significant safety concerns (4). Consequently, much effort in allergy research has been devoted to the development of safer and more effective immunological treatments. Allergic respiratory diseases are associated with high levels of IgE antibodies to certain allergenic proteins and elevated levels of eosinophils that infiltrate the target tissue (5). Production of T helper 2 (Th2) cytokines [IL-4, -5, and -13 (6)] regulates these events because they are critical for the switch to IgE production by differentiating B cells and promote the influx of eosinophils and other inflammatory cells that contribute to airway pathology.
Despite the importance of Th2 cells and their associated cytokines in the pathogenesis of allergic respiratory disease, studies of antigens considered as triggers of T-cell responses have so far been mostly limited to those known to bind IgE antibodies (7, 8) and induce IgE-mediated immediate hypersensitivity reactions (9). However, several clues suggest that T-cell and IgE reactivity might not exclusively be linked to each other. Studies conducted in mice have demonstrated the development of allergic airway hyperresponsiveness mediated by T cells in the absence of IgE (10, 11). Furthermore, data obtained from human studies have demonstrated a lack of correlation between antigen-specific IgE levels and T-cell responses (12–16).
The issue of whether T-cell recognition is always necessarily linked to antibody recognition has broader significance in terms of the classic notion of linked recognition of an antigen by both T helper cells and antigen-specific B cells. According to this notion, specific B cells internalize and process the antigen, leading to the presentation of antigen fragments bound by surface MHC class II molecules that can be recognized by specific T cells guaranteeing that the T cells deliver help to B cells specific for the same antigen (linked help). Although in some instances it has been shown that T cells can only or preferentially provide help to B cells specific for the same protein (17, 18), in other systems this restriction was not the case (19, 20). It was found that two proteins that are present on the same particle could function together and that T cells specific for one protein could provide help for B cells specific for the second protein (20). Therefore, it may be possible that, as long as the antigen recognized by T cells is in some physical association with the target of B-cell recognition (as in the case of a small virus or a pollen particle), the integrity of the “antigenic bridge” is preserved.
In our previous study of T-cell responses against Timothy grass (TG) allergens (12), no correlation was detected between IgE levels and T-cell responses in TG pollen-allergic individuals. Furthermore, we found that one-third of the patients studied had no Th2 cell response against any of the known IgE-reactive proteins, despite strong responses against whole TG extract. Based on these observations, we hypothesized that TG pollen extract may contain previously undescribed T-cell antigens in addition to the known IgE-inducing allergens. Here, we investigated the targets of allergic T-cell responses in the TG system, independently of IgE reactivity, and with a specific focus on antigens that stimulate a Th2 response because of the role of this T-cell subset in mediating allergic reactions.
Results
Significant Fraction of TG-Reactive T Cells in Allergic Donors Does Not Target the Major IgE-Reactive Allergens.
In a previous study (12), we determined the T-cell epitopes in the 10 known major IgE-reactive allergens present in TG pollen. Using 15-mer peptides overlapping by 10 residues that spanned the entire protein sequence of each of these allergens, we identified 20 dominant epitopic regions accounting for the majority of T-cell responses. However, despite this comprehensive panel of peptides and the sensitivity of the T-cell assays used, we failed to identify any IL-5–producing T cells responding to these 10 allergens in 33% of the allergic donors tested despite strong responses [≥100 spot forming cells (SFCs)] to the whole TG extract (Fig. 1). These data suggested that a significant portion of the T-cell response to TG was directed against antigens other than the known major IgE-reactive allergens.
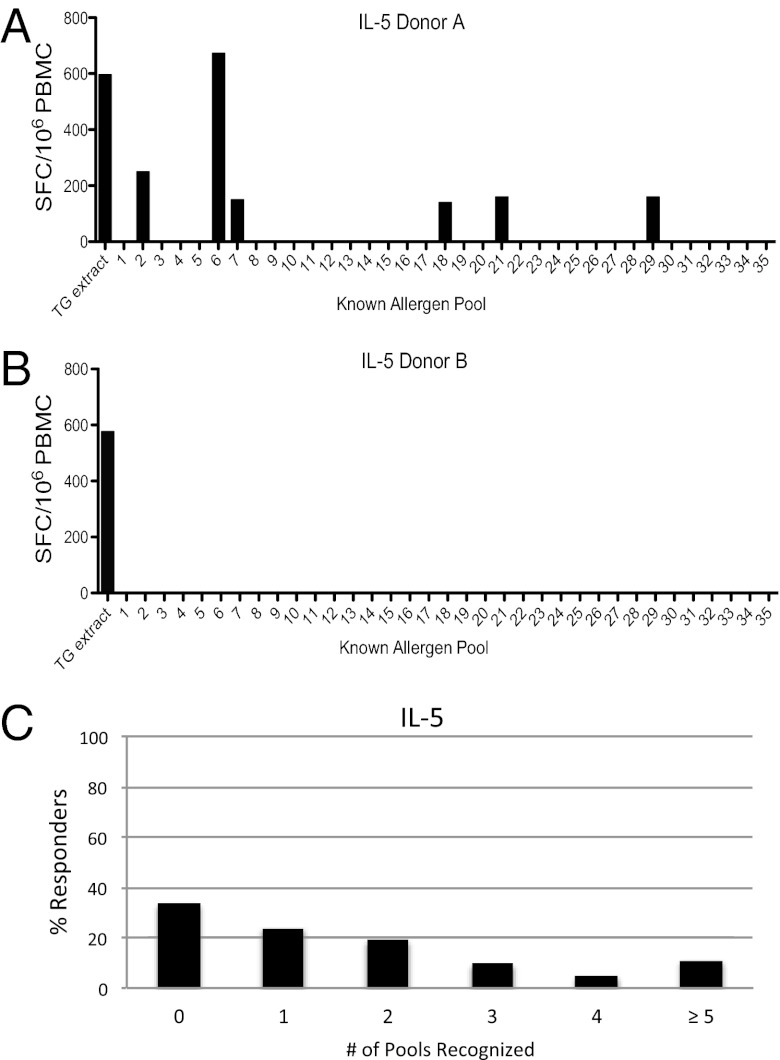
Fig. 1.
Known allergens do not account for the total T-cell response against whole pollen extract. Thirty-five pools (average 20 peptides per pool) of overlapping peptides spanning the 10 major TG allergens along with whole TG extract were screened for recognition by PBMCs from allergic donors by using IL-5 ELISPOT assays. The majority of donors who had an IL-5 T-cell response to TG extract of ≥100 SFCs per million input PBMCs showed a response pattern similar to that shown in A, with several pools eliciting strong IL-5 responses. However, some donors showed a response pattern as depicted in B, in which a vigorous response was detected against extract, but no response was detected against peptides from known allergens. In total, as shown in C, 33% of donors reacted to none of the peptide pools despite strong extract responses (n = 21).
Identification of Previously Undescribed TG Pollen Proteins Through a Combined Transcriptomics and Proteomics Analysis.
Because few proteins apart from IgE-reactive allergens have been identified in TG (or any other allergenic grasses), we pursued the identification of all proteins with significant expression in TG pollen so that a more comprehensive analysis of T-cell responses to TG could be performed. mRNA of TG pollen was isolated and analyzed by high-throughput sequencing on an Illumina Genome Analyzer. A total of 1,016,285 unique, putative transcripts (including allelic variants and isoforms) were identified. Of the 10 known TG allergen proteins considered in our previous study (Phl p 1, 2, 3, 4, 5, 6, 7, 11, 12, and 13), all but 1 (Phl p 6) were identified in our transcriptome analysis with large fragments or entire sequences matching at >90% identity. Interestingly, one transcript matched the published Phl p 6 sequence with very high significance (Blast E-value of 4E-16) but much lower sequence identity (61%), thus our results demonstrated that the transcripts we identified from TG pollen mRNA recovered the vast majority of known allergens, but our approach also identified putative variants of known allergens that had not been described previously.
Next, we used a proteomics approach to determine which of the newly identified transcripts encoded proteins detectable in TG pollen. For this purpose, TG pollen extract was separated by 2D gel electrophoresis and either stained with Coomassie blue or immunoblotted by using a pool of sera from eight TG allergic donors (Fig. 2) to detect proteins reactive with IgE and/or IgG antibodies. Spots were cut out from unstained gels, trypsin digested, and analyzed by peptide fingerprint mass mapping (using MS data) and peptide fragmentation mapping [using tandem MS (MS/MS) data] to obtain amino acid sequences. Of 131 spots picked from the 2D gels, 119 could be assigned with high confidence to one or more ORFs from our transcript sequences. After clustering ORFs with >90% sequence identity, we found that the 119 identifiable spots corresponded to 89 nonredundant protein sequences, including 6 of the known TG allergens, thus leaving 83 distinct protein sequences corresponding to previously unidentified proteins identified by spots on the gel. To these, we added another 10 proteins not identified in any spots cut out of the gel, but rather in a MS analysis of the whole TG extract. In total, a set of 93 previously undescribed proteins from TG pollen was generated and further analyzed for T-cell reactivity (Dataset S1).
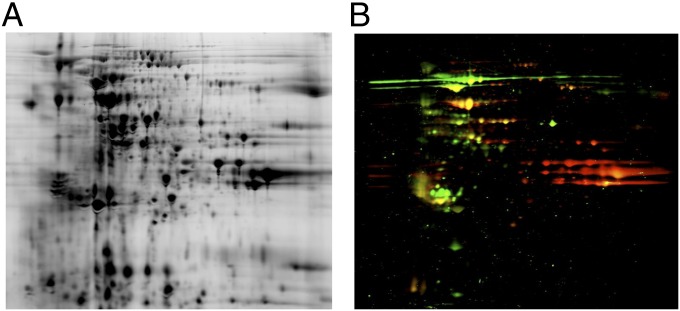
Fig. 2.
Two-dimensional gel electrophoresis and immunoblot analysis of TG extract using pooled sera from allergic donors. (A) TG pollen extract was run on a 2D gel and stained with Coomassie brilliant blue. (B) A second gel run in parallel was blotted and probed with a serum pool from eight TG-allergic donors to identify proteins reactive with human IgE and IgG. Anti IgE, red; anti IgG, green; dual reactivity, yellow.
To test whether any of the proteins identified were derived from contaminants (such as bacteria) possibly present in the TG pollen extract, we ran the amino acid sequences through National Center for Biotechnology Information BLAST. Two proteins (nos. 27 and 38) had high homology to previously identified TG proteins that are not considered allergens. Of the remaining proteins, no. 76 had highly homologous (BLAST E value < 10−10) matches in the rice genome (the closest to TG of all fully sequenced genomes available). Of the remaining proteins, none had similarly high homology to any publicly available sequence. Overall, based on similarity to known protein sequences, it was unlikely that our sets of protein sequences contained nonplant contaminants, suggesting that our sequences were from bona fide TG proteins.
T Cells Respond Vigorously to Epitopes from Previously Undescribed TG Pollen Proteins.
We scanned the 93 previously undescribed protein sequences for peptides predicted to bind to multiple human leukocyte antigen (HLA) class II molecules. For each protein, we analyzed all overlapping 15-mer peptides and predicted their binding affinity to a panel of 25 HLA DR, DP, and DQ molecules (Table S1) using a consensus approach of multiple machine learning methods (21, 22). Nonoverlapping peptides that were predicted to promiscuously bind multiple HLA class II molecules were selected from each protein (Materials and Methods). In total, this process of prediction resulted in the selection of 822 peptides from a total of 21,506 distinct 15-mers (Dataset S2). This prediction approach (12) was previously found to capture the most dominant epitopes, accounting for ∼50% of the total response.
The 822 peptides were assembled into 42 pools of ∼20 peptides each, which were tested for recognition by peripheral blood mononuclear cells (PBMCs) from 20 TG allergic and 20 normal donors (Dataset S3). We used IL-5 enzyme-linked immunospot (ELISPOT) assays to measure T-cell reactivity because we were especially interested in Th2 responses due to their importance in allergy pathogenesis. In our hands, IL-5 is a more sensitive readout in ELISPOT assays compared with other Th2 cytokines, namely IL-4 and -13 (Fig. S1) (15). In addition to peptide pools from previously undescribed proteins, we tested five peptide pools from the 10 known TG allergens assembled using the same prediction strategy. Of the previously undescribed peptide pools, 13 of 42 pools stimulated IL-5 production in PBMCs in >20% of allergic donors. For the known allergen pools, four of five pools were recognized by >20% of allergic donors. In contrast, none of the peptide pools elicited an IL-5 response in >10% of the nonallergic donors (Fig. 3A).
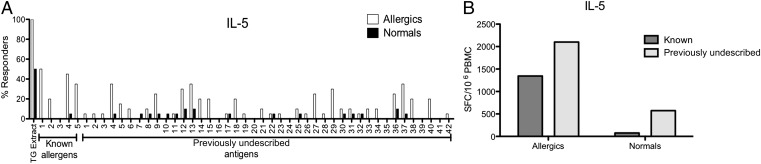
Fig. 3.
A majority of TG-specific T cells target previously undescribed antigens. (A) Bar chart indicating the IL-5 response rate of PBMCs from allergic and nonallergic donors to peptide pools from known and previously undescribed antigens. Open bars indicate response rates from allergic donors; filled bars indicate responses from normal donors (n = 20 per donor group). (B) Bar chart showing the total number of IL-5–producing T cells targeting previously undescribed antigens (light gray) vs. known allergens (dark gray) in normal and allergic patients.
When comparing the combined magnitude of responses directed against the known allergens vs. the previously undescribed TG antigens (PUTGAs), it was evident that a very significant fraction of the T-cell response to TG targeted the PUTGAs. In fact, 61% of responding T cells (2,101 SFC; Fig. 3B) recognized PUTGA peptides, whereas the response directed against the control panel of peptides from major known allergens only accounted for 39% of the T-cell responses (1,345 SFCs; Fig. 3B). In the nonallergic population, the IL-5 responses were much weaker. However, the difference in response magnitude between the known allergens and PUTGAs was even more pronounced, with 88.4% of the response targeting the PUTGAs (574 SFCs) and 11.6% targeting the known allergens (75 SFCs). Overall, these data showed that a majority of Th2 cell responses against TG extract were directed against antigens other than the known IgE-reactive allergens.
Identification of T-Cell–Reactive Antigens and Correlation with Antibody Reactivity.
Next, we deconvoluted the positive peptide pools to determine which individual peptides elicited the responses. IL-5 production was detected in at least one allergic donor for peptides from 52 of the 93 proteins tested with a total magnitude of 44,204 SFCs (Fig. S2). To exclude spurious responses, we define T-cell antigens as those proteins recognized in 20% or more of allergic donors, which is an established response threshold to define allergenic proteins when using skin test reactivity testing (23, 24). Based on this cutoff, we identified a total of 13 T-cell antigens. Three of these T-cell antigens were proteins targeted by both IgE and IgG; two proteins were targeted by IgE only; five proteins were targeted by IgG only; and three proteins were not targeted by antibodies of either Ig class (Fig. 4). Therefore eight of the previously undescribed T-cell antigens were not targeted by IgE compared with five that were. The same holds true when comparing responses on the peptide level or when considering the total number of responding T cells (Table S2). Overall, these data suggested that a sizeable fraction of the response against PUTGAs is directed at proteins not recognized by IgE.
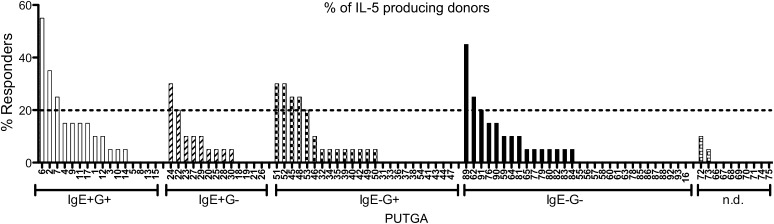
Fig. 4.
Percentage recognition of PUTGAs by TG-allergic donors. Bar chart shows percentage of donors producing IL-5 in response to PUTGA-derived peptide stimulation. PUTGAs are categorized according to antibody reactivity as indicated on the x axis. The dashed line indicates the minimum threshold of ≥20% recognition to be considered an allergen.
IL-5 Responses to PUTGAs Are Made by Memory T Cells That Can Be Detected Directly ex Vivo.
The data presented above suggest that almost two-thirds (61%) of IL-5–producing T-cell responses directed against pollen proteins were directed against PUTGAs and that a majority of those responses targeted antigens not recognized by IgE. These responses were presumably the result of in vivo priming of T cells in the allergic patients following pollen exposure. To exclude the possibility that these T-cell responses could have been due to in vitro priming of T cells cultured with TG extract, we examined whether the responding T cells were associated with either a memory or naïve phenotype. For this purpose, we assembled two peptide pools: the TG P20 pool comprising the 20 most dominant peptides from known TG allergens identified in our previous study (12)—accounting for ∼90% of IL-5 responses detected against conventional IgE-reactive TG allergens in our study population—and a PUTGA P19 pool comprising 19 of the most dominant peptides selected from IgE-unreactive PUTGAs. These 19 peptides account for ∼40% of the total IL-5 response against all PUTGAs tested in our study population.
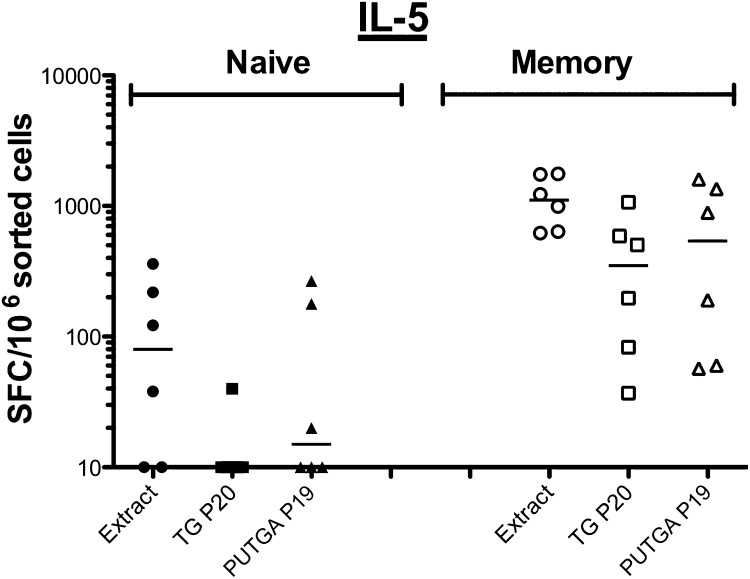
PBMCs from six allergic donors were sorted based on their chemokine receptor expression into memory (CD45RA−) and naïve (CD45RA+CCR7+) T-cell subsets. Subsequently, cells from both subpopulations were stimulated in vitro with TG pollen extract and tested after expansion for IL-5 production (Fig. 5). Responses to each stimulus were significantly higher in the memory population compared with the naïve population. In response to TG extract stimulation, the median SFCs detected was 80 and 1,108 in the naïve and memory populations, respectively. The TG P20 elicited no response in the naïve population and a median of 350 SFCs in the memory population. Stimulation with PUTGA P19 resulted in a median of 12 SFCs in the naïve and 540 in the memory subset. As expected, high amounts of IL-5 were detected in the naïve and memory population in response to phytohemagglutinin (PHA), indicating that the cells were viable and responsive to T-cell stimulation. Thus, overall far fewer antigen-specific IL-5–producing cells were found in the naïve T-cell subset compared with the memory subset (P = 0.0002), signifying that the T-cell responses to both the TG P20 and PUTGA P19 peptide pools were derived from memory T cells. Of note, the two pools of peptides cover different fractions of the total response against the sets of antigens from which they were derived. The dominant known peptide pool (TG P20) is made up of the 20 most dominant peptides, which account for >90% of the total IL-5 response detected against all known peptides. In contrast, the dominant PUTGA peptide pool (PUTGA P19) only accounts for 40% of the total IL-5 response directed against all PUTGA peptides screened. Thus, a direct quantitative comparison of the response against them is not possible.
Fig. 5.
Memory T cells are the source of IL-5 T-cell responses after in vitro expansion. IL-5 production in naïve and memory T cells in response to TG extract (Extract), the dominant known (TG P20), and PUTGA peptide pools (PUTGA P19) were measured after 14 d of expansion following TG stimulation in vitro (n = 6).
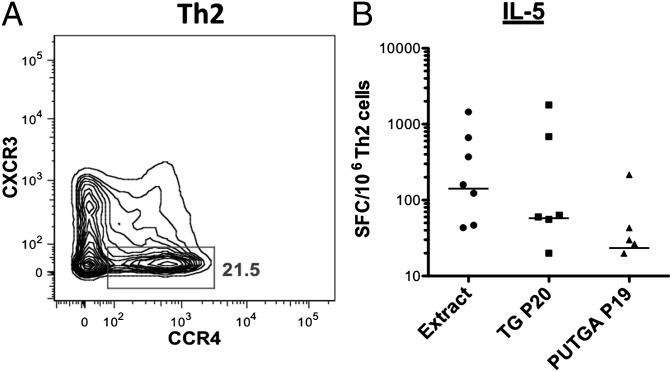
To further substantiate that the reactivity against PUTGAs was reflective of in vivo exposure, and not from primary responses induced in vitro, we examined whether responses against PUTGAs could be detected directly ex vivo without any in vitro culture expansion. Because the frequency of antigen-specific precursor cells is very low, PBMCs from eight donors were presorted to obtain enriched Th1 or Th2 subpopulations (Materials and Methods), and cells were assessed for IL-5 production in response to TG extract and the dominant known and PUTGA peptide pools in ELISPOT assays. As expected, IL-5 production for the Th1 subpopulation was not detectable for any of the stimuli except PHA. In contrast, as shown in Fig. 6, the Th2 subpopulation (CCR4+, CXCR3−) showed significant responses to each allergen stimulus compared with medium alone (extract: P = 0.01; TG20: P = 0.03; PUTGA19: P = 0.01; Wilcoxon signed rank tests).
Fig. 6.
T-cell responses against conventional allergens and PUTGAs can be detected directly ex vivo. (A) FACS plot showing the Th2 T-cell subset sorted based on expression of CXCR3 (Th1) and CCR4 (Th2). (B) IL-5 production by Th2 cells from allergic donors in response to TG extract, the dominant known (TG P20) and previously undescribed (PUTGA P19) peptide pools as measured ex vivo (n = 8).
Discussion
Our original motivation to search for previously undescribed targets of allergen-specific T-cell responses in TG was based on the finding that approximately one-third of TG-allergic donors did not recognize any peptides derived from the 10 known IgE-reactive TG allergens. This lack of T cell responses was puzzling because T cells from the same donors gave strong responses against the whole TG extract in the same sensitive ELISPOT assays. As a first explanation, we considered the possibility that posttranslational modifications might be essential for the recognition of some epitopes. Such modifications would not be present in the peptides we synthesized for our screening. However, a pilot study of peptides with hydroxylated prolines, which are thought to be prominent in some TG allergens (25), did not show any T-cell reactivity at all. Although this finding does not exclude the possibility that other posttranslational modifications are present in T-cell epitopes of TG, we instead pursued another hypothesis, namely that T cells target conventional peptides from antigens other than the 10 known IgE-reactive allergens. Our approach led to the identification of 93 previously undescribed TG proteins, of which 52 elicited Th2 responses. The recognition of these previously undescribed T-cell antigens provides an explanation for the originally observed gap in reactivity between known allergens and whole TG extract.
Immunological characterization of the previously undescribed TG proteins revealed that a majority were both antibody and T-cell reactive. Of prime interest, we demonstrated that T-cell responses against peptides derived from these PUTGAs were potent inducers of IL-5 responses in PBMCs from TG-allergic patients. The T-cell population that produced IL-5 against these PUTGAs originated from the memory Th2 cell subset and could be detected by direct ex vivo analysis demonstrating the relevance of these T-cell antigens as targets of in vivo allergic responses. Interestingly, in contrast to the findings in allergic individuals, nonallergic donors had no or very weak IL-5 responses to both the known TG allergens and the PUTGAs. Future studies are needed to determine whether the same is true for all types of T-cell responses or whether nonallergic individuals actually show an increased magnitude of “tolerogenic” responses against PUTGAs such as IL-10–producing regulatory T cells (Tregs).
Remarkably, strong IL-5 production was seen not only in response to IgE-reactive antigens but also to several antigens that were not targeted by IgE suggesting that Th2 responses to an antigen were not necessarily linked to IgE reactivity. The idea of unlinked T-cell help—meaning that T cells display a different antigen specificity than the B cells they affect—has been discussed in previous studies (18, 20) and is also of great relevance to the immune response directed against the PUTGAs. The natural structure of a pollen particle (or microsized particles released upon hydration) provides physical linkage of various proteins, which are recognized by the immune system. Therefore, the way in which pollen proteins are presented to B and T cells is likely to allow for unlinked T-cell help because the T-cell–specific epitope derived from one antigen can be present on the same physical pollen particle as a second antigen recognized by B cells. As a result, the Th2 cell immune response directed against PUTGAs may provide help for the allergic response directed against the major known IgE-reactive allergens present on the same pollen particle as the PUTGAs.
Although the direct mechanisms by which such an immune modulation may occur are unknown at this point, there are several hypotheses regarding the events by which unlinked T-cell help could potentially lead to the improvement of allergic symptoms. First, our data indicate that the majority of IL-5 responses are directed against PUTGAs. Therefore, down-regulating these responses should be of significant benefit in terms of reducing Th2-induced pathogenic effects. Secondly, induction of PUTGA-specific Tregs or Th1 cells may help in the regulation, through bystander mechanisms (26), of Th2 responses directed against known TG allergens present in the same pollen particle. Thirdly, the regulation of PUTGA-specific Th2 responses may also result in downstream regulation of IgE and induction of blocking IgG antibody responses to known TG allergens, a hallmark of specific immunotherapy (SIT) treatment (27, 28). Finally, induction of PUTGA-specific Th1 or Treg cells may lead to PUTGA-specific IgG production, which may interfere with IgE-induced mediator release and other immediate-type reactions by mechanisms such as steric hindrance, competition for antigen binding, and inhibitory signaling through FcγRIIB (29, 30). Overall, our data suggest that the PUTGAs described in this work could present promising targets for a T-cell–focused specific immunotherapy, which could be safe for administration to higher-risk patients such as asthmatics. As a result, therapies that specifically target T-cell reactivity may provide a more efficacious and safer way to treat allergic patients, especially those suffering from severe asthma, for whom current SIT regimens pose a significant risk of deleterious reactions.
There are several caveats to our findings. First, screening for T-cell reactivity by using only IL-5 production as the readout may well miss epitopes recognized by Th2 cytokine-producing cells producing IL-4 or -13 but not IL-5. We chose IL-5 for the large-scale screen because, in our hands, it is the most sensitive readout for ELISPOT assays and because we could detect IL-4 and -13 production to the peptides identified in our IL-5 screen (Fig. S1). Second, the analysis of antibody reactivity to TG pollen extract with a serum pool comprising eight different patient sera poses the risk of obfuscating correlations between T-cell and antibody reactivity for antigens on an individual level. However, we previously tested for such correlations extensively for known TG allergens (12) and could not detect them, so we find it unlikely that they are present for the PUTGAs.
Of note, in an in vitro setting, T-cell responses directed against the PUTGAs appeared to be of stronger magnitude than responses against the known allergens. However, this trend was reversed in the ex vivo T-cell response. A likely explanation for this observation is the difference of immunodominance of the individual peptides contained in the respective pools. The dominant known peptide pool accounts for ∼90% of the total response against all known allergens, whereas the dominant PUTGA peptide pool accounts for 40% of the total IL-5 response directed against all PUTGAs.
The analysis of antibody reactivity to TG pollen extract with a serum pool comprising eight different patient sera does pose the risk of diluting out low antibody levels in individual donors below detection level. Antibody reactivity determined for individual patients would be likely to give a more accurate reflection of antigen-specific antibody reactivity and its relation to IL-5 production. However, because of limited serum availability and high cost, antibody reactivity was not determined for each individual donor.
This study is a comprehensive transcriptomic and proteomic analysis of an inhaled allergen. We identified 93 previously undescribed TG proteins for which we could establish expression in the pollen itself, expanding the previously known set of such proteins by approximately an order of magnitude. The fact that we could identify 24 TG proteins targeted by IgE that do not overlap with the known IgE-reactive allergens demonstrates that the technique is a powerful tool for conventional IgE-reactive allergen discovery. The proteomic and transcriptomics approach used here is generally applicable, and the rapidly decreasing cost of DNA sequencing suggests that it could be applied to all allergens. Promisingly, such a broad application could enable a large-scale and unbiased analysis of the protein properties that give rise to allergenicity.
Materials and Methods
Donor Population.
Each donor was recruited following Institutional Review Board (La Jolla Institute for Allergy and Immunology, La Jolla, CA) approval (Federal Wide Assurance no. 00000032) and informed consent and was assigned a study identification number. Donors were tested for allergen reactivity by skin prick tests to extracts from 32 common allergens. TG-allergic donors were identified as having a skin reaction with a wheal of ≥5 mm in diameter to TG and a clinical history consistent with seasonal grass pollen allergy. Donors that had received specific immunotherapy were excluded. Nonallergic donors were identified as having negative skin prick tests to all allergens and no clinical history of allergy.
Identification of Previously Undescribed TG Pollen Proteins.
Detailed methods are given in SI Materials and Methods. Briefly, total RNA of TG pollen extract was isolated (31) and analyzed by high-throughput sequencing on an Illumina Genome Analyzer IIx. Replicate samples were run in seven lanes, producing >280 million paired-end raw sequence reads of 72 bp in length. High-quality reads were assembled into 1,016,285 nonredundant transcripts by using Velvet (32) and Oases (Version 0.18.1; D. R. Zerbino, European Bioinformatics Institute, Hinxton, United Kingdom). To identify proteins expressed in TG pollen, a TG extract was run on two 2D gels [3–10 pH range, 12% (vol/vol) acrylamide] and followed by MS analysis of individual protein spots (performed by Applied Biomics). One gel was stained with Coomassie blue, and another was blotted onto a nitrocellulose membrane and probed with serum from TG-allergic individuals to detect IgE and IgG antibody binding proteins. Spots of interest were identified, cut out, digested in gel at 37 °C, and extracted with a TFA buffer (pH 2). To ensure that the correct spots were identified on the unstained gel, all gels (gel that was blotted, gel that was stained with Coomassie, and the unstained gel from which the spots were eventually picked) were run at the same time and under the exact same conditions to allow the assumption that the proteins run in the same x–y positions of the gels. Furthermore, for the spots selected from the blot, specialized software was used. Proteins on both the Western blot and the unstained gel were labeled with CyDye, which the software was able to align to ensure that the automated spot picker selected the right spots. MS/MS analyses of the peptides in each sample were obtained by using an Applied Biosystems Proteomics Analyzer, and hits against our TG pollen transcripts were evaluated by the Mascot software package (Matrix Science). Hits with highly similar amino acid sequences (90% or higher) were grouped together and were assigned one putative protein ID (Dataset S1).
Assembly of a Peptide Set Predicted to Promiscuously Bind HLA Class II Molecules.
For each 15-mer peptide encoded in the ORFs in Dataset S2, we predicted the binding affinity to a panel of 25 HLA class II molecules (Table S1) using a consensus prediction approach (21, 22). Peptides with predicted binding scores in the top 20% for a given allele were considered potential binders. Peptides predicted to bind 13 or more HLA molecules at this threshold were considered promiscuous binders and selected for synthesis (after eliminating peptides overlapping by more than nine contiguous residues). If less than five peptides from a given protein met this threshold, we chose the top five peptides, and we selected up to four peptides in proteins where length was prohibitive. In total, this process resulted in the selection of 822 peptides from a total of 21,506 distinct 15-mers encoded in 620 ORFs (Dataset S2). As a control, a set of 105 peptides was derived from the known TG allergens by using the same prediction approach. The selected peptides were purchased from A and A as crude material on a small (1 mg) scale. Peptides that tested positive and were included in the dominant epitope pools were purchased as purified material (>95% pure) on a 5- to 10-mg scale.
PBMC Isolation and in Vitro Expansion of TG Extract-Specific T Cells.
PBMCs were isolated by density gradient centrifugation from one unit of blood (∼450 mL) and cryopreserved as described (12). For in vitro expansions, PBMCs were thawed and cultured in RPMI 1640 (Omega Scientific) supplemented with 5% human AB serum (Cellgro) at a density of 2 × 106 cells per mL in 24-well plates (BD Biosciences) and stimulated with TG pollen extract (50 µg/mL; Greer). Cells were kept at 37 °C in 5% CO2, and additional IL-2 and -7 (10 U/mL; eBioscience) was added every 3 d after initial antigenic stimulation. On day 14, cells were harvested and screened for reactivity against TG-specific peptide pools (16–25 peptides per pool, averaging 20 peptides per pool). On day 17, peptides from positive pools were tested individually to identify the reactive epitopes.
ELISPOT Assays.
The production of IL-5 from cultured PBMCs in response to antigen stimulation was measured by ELISPOT as described (12). Briefly, 1 × 105 cells per well were incubated with peptide, peptide pool, or TG extract (10, 5, and 50 µg/mL, respectively). After 24 h, cells were removed, and plates were incubated with 2 µg/mL biotinylated anti-human IL-5 Ab (Mabtech) at 37 °C. After 2 h, plates were washed, and avidin–peroxidase complex was added (Vector Laboratories) for 1 h at room temperature. Peroxidase-conjugated spots were developed with 3-amino-9-ethylcarvazole solution (Sigma-Aldrich). Criteria for peptide pool positivity were 100 SFCs per 106 PBMCs, P ≤ 0.05, and a stimulation index ≥ 2. Criteria for individual peptides were the same except a minimum of 20 SFCs was counted as positive.
FACS Sorting of T-Cell Subpopulations.
PBMCs were thawed, washed, and counted. CD4+ T cells were isolated by using a CD4+ T-Cell Isolation Kit II (Miltenyi Biotec) according to manufacturer’s instructions and subsequently stained with antibodies to sort desired T-cell subpopulations. Memory and naïve T-cell populations were sorted from CD4+ cells stained with CD45RA–FITC and CCR7–BL421. The naïve population was sorted as CD45RA+CCR7+, and the memory population was sorted as CD45RA−CCR7−/+. CD45RA+CCR7− cells were not collected. Subsequently, these subpopulations were put into culture with irradiated PBMCs and stimulated with TG pollen extract and cultured for 14 d with IL-2 and -7 added every 3 d. IL-5 production in response to stimuli was assessed by ELISPOT. For Th1 and Th2 subpopulations, cells were sorted from CD4+ cells stained with CCR4–PECy7, CCR6–PE, and CXCR3–APC. Th2 cells were sorted as CCR4+CCR6−CXCR3− T cells, and Th1 cells were sorted as CCR4−CCR6+CXCR3+ T cells. Sorted cells were plated into ELISPOT plates with stimulus and CD4-depleted, irradiated PBMC as antigen-presenting cell. IL-5 production was assessed after 24-h incubation.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JAA00001–JAA00463).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300512110/-/DCSupplemental.
References
- 1.Brozek JL, et al. Global Allergy and Asthma European Network Grading of Recommendations Assessment, Development and Evaluation Working Group Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines: 2010 revision. J Allergy Clin Immunol. 2010;126(3):466–476. doi: 10.1016/j.jaci.2010.06.047. [DOI] [PubMed] [Google Scholar]
- 2.Nathan RA. The burden of allergic rhinitis. Allergy Asthma Proc. 2007;28(1):3–9. doi: 10.2500/aap.2007.28.2934. [DOI] [PubMed] [Google Scholar]
- 3.Bauchau V, Durham SR. Prevalence and rate of diagnosis of allergic rhinitis in Europe. Eur Respir J. 2004;24(5):758–764. doi: 10.1183/09031936.04.00013904. [DOI] [PubMed] [Google Scholar]
- 4.Rolland JM, Douglass J, O’Hehir RE. Allergen immunotherapy: Current and new therapeutic strategies. Expert Opin Investig Drugs. 2000;9(3):515–527. doi: 10.1517/13543784.9.3.515. [DOI] [PubMed] [Google Scholar]
- 5.Laprise C, Boulet LP. Airway responsiveness and atopy in families of patients with asthma. Clin Invest Med. 1996;19(6):461–469. [PubMed] [Google Scholar]
- 6.Romagnani S. The role of lymphocytes in allergic disease. J Allergy Clin Immunol. 2000;105(3):399–408. doi: 10.1067/mai.2000.104575. [DOI] [PubMed] [Google Scholar]
- 7.Andersson K, Lidholm J. Characteristics and immunobiology of grass pollen allergens. Int Arch Allergy Immunol. 2003;130(2):87–107. doi: 10.1159/000069013. [DOI] [PubMed] [Google Scholar]
- 8.Basketter DA, Kimber I. Assessing the potency of respiratory allergens: Uncertainties and challenges. Regul Toxicol Pharmacol. 2011;61(3):365–372. doi: 10.1016/j.yrtph.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Gieras A, et al. Molecular determinants of allergen-induced effector cell degranulation. J Allergy Clin Immunol. 2007;119(2):384–390. doi: 10.1016/j.jaci.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 10.Vijayanand P, et al. Interleukin-4 production by follicular helper T cells requires the conserved Il4 enhancer hypersensitivity site V. Immunity. 2012;36(2):175–187. doi: 10.1016/j.immuni.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehlhop PD, et al. Allergen-induced bronchial hyperreactivity and eosinophilic inflammation occur in the absence of IgE in a mouse model of asthma. Proc Natl Acad Sci USA. 1997;94(4):1344–1349. doi: 10.1073/pnas.94.4.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oseroff C, et al. Molecular determinants of T cell epitope recognition to the common timothy grass allergen. J Immunol. 2010;185(2):943–955. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benítez D, et al. Specific immune response to Phleum pratense plant profilin in atopic patients and control subjects. Allergol Immunopathol (Madr) 2001;29(1):9–15. doi: 10.1016/s0301-0546(01)79009-x. [DOI] [PubMed] [Google Scholar]
- 14.Würtzen PA, van Neerven RJ, Arnved J, Ipsen H, Sparholt SH. Dissection of the grass allergen-specific immune response in patients with allergies and control subjects: T-cell proliferation in patients does not correlate with specific serum IgE and skin reactivity. J Allergy Clin Immunol. 1998;101(2 Pt 1):241–249. doi: 10.1016/S0091-6749(98)70389-6. [DOI] [PubMed] [Google Scholar]
- 15.Oseroff C, et al. T cell responses to known allergen proteins are differently polarized and account for a variable fraction of total response to allergen extracts. J Immunol. 2012;189(4):1800–1811. doi: 10.4049/jimmunol.1200850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oseroff C, et al. Analysis of T cell responses to the major allergens from German cockroach: Epitope specificity and relationship to IgE production. J Immunol. 2012;189(2):679–688. doi: 10.4049/jimmunol.1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sette A, et al. Selective CD4+ T cell help for antibody responses to a large viral pathogen: Deterministic linkage of specificities. Immunity. 2008;28(6):847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanzavecchia A. Antigen-specific interaction between T and B cells. Nature. 1985;314(6011):537–539. doi: 10.1038/314537a0. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, et al. Identification of CD4+ T cell epitopes in C. burnetii antigens targeted by antibody responses. PLoS ONE. 2011;6(3):e17712. doi: 10.1371/journal.pone.0017712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chain BM, Mitchison NA, Mitchison TJ, Davies DH, Marcinkiewicz J. Antigen processing: Current issues, exceptional cases (Thy 1 alloantigen, MHC class-II-restricted cytolytic T cells), and implications for vaccine development. J Autoimmun. 1989;2(Suppl):45–53. doi: 10.1016/0896-8411(89)90116-9. [DOI] [PubMed] [Google Scholar]
- 21.Wang P, et al. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLOS Comput Biol. 2008;4(4):e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang P, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Løwenstein H. Quantitative immunoelectrophoretic methods as a tool for the analysis and isolation of allergens. Prog Allergy. 1978;25:1–62. doi: 10.1159/000314432. [DOI] [PubMed] [Google Scholar]
- 24.Chapman MD. Allergen nomenclature. Clin Allergy Immunol. 2008;21:47–58. [PubMed] [Google Scholar]
- 25.Petersen A, Schramm G, Schlaak M, Becker WM. Post-translational modifications influence IgE reactivity to the major allergen Phl p 1 of timothy grass pollen. Clin Exp Allergy. 1998;28(3):315–321. doi: 10.1046/j.1365-2222.1998.00221.x. [DOI] [PubMed] [Google Scholar]
- 26.Schuhbauer DM, Mitchison NA, Mueller B. Interaction within clusters of dendritic cells and helper T cells during initial Th1/Th2 commitment. Eur J Immunol. 2000;30(5):1255–1262. doi: 10.1002/(SICI)1521-4141(200005)30:5<1255::AID-IMMU1255>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 27.Wachholz PA, Durham SR. Mechanisms of immunotherapy: IgG revisited. Curr Opin Allergy Clin Immunol. 2004;4(4):313–318. doi: 10.1097/01.all.0000136753.35948.c0. [DOI] [PubMed] [Google Scholar]
- 28.Skripak JM, et al. A randomized, double-blind, placebo-controlled study of milk oral immunotherapy for cow’s milk allergy. J Allergy Clin Immunol. 2008;122(6):1154–1160. doi: 10.1016/j.jaci.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cady CT, et al. IgG antibodies produced during subcutaneous allergen immunotherapy mediate inhibition of basophil activation via a mechanism involving both FcgammaRIIA and FcgammaRIIB. Immunol Lett. 2010;130(1-2):57–65. doi: 10.1016/j.imlet.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terada T, Zhang K, Belperio J, Londhe V, Saxon A. A chimeric human-cat Fcgamma-Fel d1 fusion protein inhibits systemic, pulmonary, and cutaneous allergic reactivity to intratracheal challenge in mice sensitized to Fel d1, the major cat allergen. Clin Immunol. 2006;120(1):45–56. doi: 10.1016/j.clim.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 31.Wallner M, et al. Immunologic characterization of isoforms of Car b 1 and Que a 1, the major hornbeam and oak pollen allergens. Allergy. 2009;64(3):452–460. doi: 10.1111/j.1398-9995.2008.01788.x. [DOI] [PubMed] [Google Scholar]
- 32.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.