Abstract
To study homologous recombination between repeated sequences in an in vitro simian virus 40 (SV40) replication system, we constructed a series of substrate DNAs that contain two identical fragments of monkey alpha-satellite repeats. Together with the SV40-pBR322 composite vector encoding Apr and Kmr, the DNAs also contain the Escherichia coli galactokinase gene (galK) positioned between two alpha-satellite fragments. The alpha-satellite sequence used consists of multiple units of tandem 172-bp sequences which differ by microheterogeneity. The substrate DNAs were incubated in an in vitro SV40 DNA replication system and used to transform the E. coli galK strain DH10B after digestion with DpnI. The number of E. coli galK Apr Kmr colonies which contain recombinant DNAs were determined, and their structures were analyzed. Products of equal and unequal crossovers between identical 172-bp sequences and between similar but not identical (homeologous) 172-bp sequences, respectively, were detected, although those of the equal crossover were predominant among all of the galK mutant recombinants. Similar products were also observed in the in vivo experiments with COS1 cells. The in vitro experiments showed that these recombinations were dependent on the presence of both the SV40 origin of DNA replication and SV40 large T antigen. Most of the recombinant DNAs were generated from newly synthesized DpnI-resistant DNAs. These results suggest that the homologous recombination observed in this SV40 system is associated with DNA replication and is suppressed by mismatches in heteroduplexes formed between similar but not identical sequences.
Full text
PDF

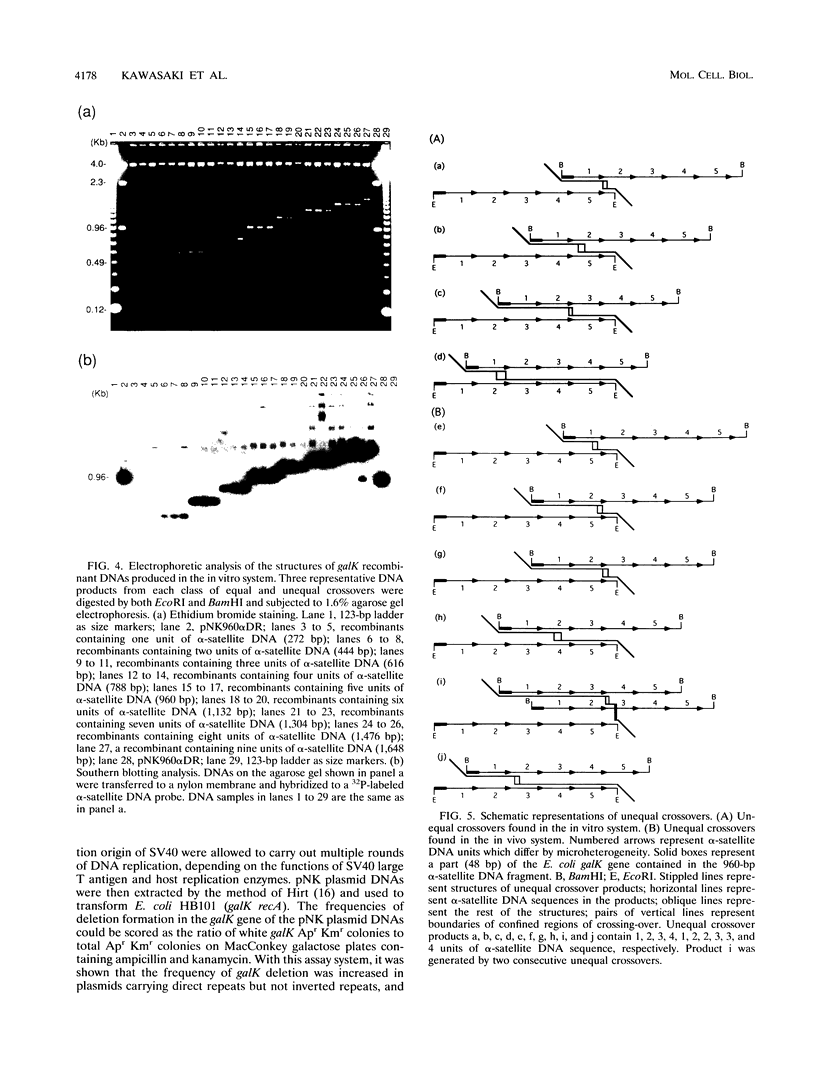
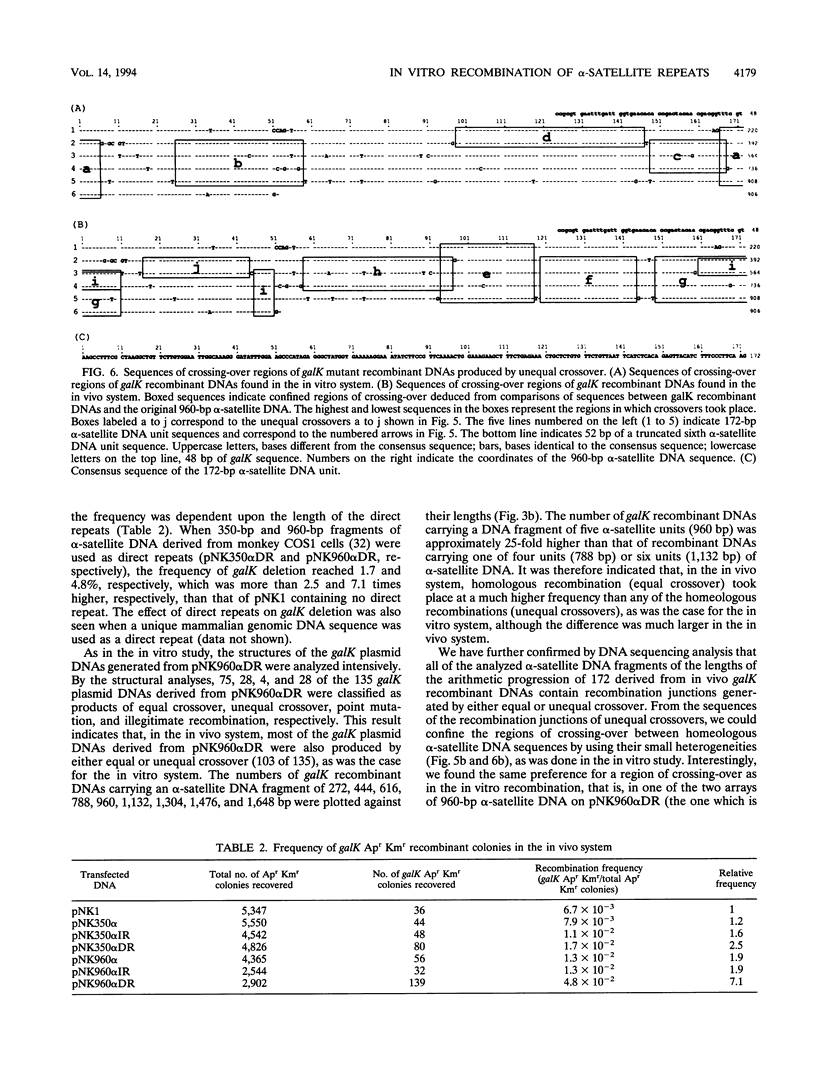



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aaltonen L. A., Peltomäki P., Leach F. S., Sistonen P., Pylkkänen L., Mecklin J. P., Järvinen H., Powell S. M., Jen J., Hamilton S. R. Clues to the pathogenesis of familial colorectal cancer. Science. 1993 May 7;260(5109):812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Bae Y. S., Chiba M., Ohira M., Ikeda H. A shuttle vector for analysis of illegitimate recombination in mammalian cells: effects of DNA topoisomerase inhibitors on deletion frequency. Gene. 1991 May 30;101(2):285–289. doi: 10.1016/0378-1119(91)90425-b. [DOI] [PubMed] [Google Scholar]
- Bailis A. M., Rothstein R. A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination between homeologous genes by an excision repair dependent process. Genetics. 1990 Nov;126(3):535–547. doi: 10.1093/genetics/126.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook J. D., McCurrach M. E., Harley H. G., Buckler A. J., Church D., Aburatani H., Hunter K., Stanton V. P., Thirion J. P., Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992 Feb 21;68(4):799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Christman M. F., Dietrich F. S., Fink G. R. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988 Nov 4;55(3):413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- Chu G., Sharp P. A. SV40 DNA transfection of cells in suspension: analysis of efficiency of transcription and translation of T-antigen. Gene. 1981 Mar;13(2):197–202. doi: 10.1016/0378-1119(81)90008-1. [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov G. L., Kuzminov A. V., Mazin A. V., Salganik R. I. Molecular mechanisms of deletion formation in Escherichia coli plasmids. I. Deletion formation mediated by long direct repeats. Mol Gen Genet. 1991 Aug;228(1-2):153–159. doi: 10.1007/BF00282460. [DOI] [PubMed] [Google Scholar]
- Fishel R., Lescoe M. K., Rao M. R., Copeland N. G., Jenkins N. A., Garber J., Kane M., Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993 Dec 3;75(5):1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J., Rudin N., Haber J. E. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992 Mar;12(3):1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Kuhl D. P., Pizzuti A., Pieretti M., Sutcliffe J. S., Richards S., Verkerk A. J., Holden J. J., Fenwick R. G., Jr, Warren S. T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991 Dec 20;67(6):1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Pizzuti A., Fenwick R. G., Jr, King J., Rajnarayan S., Dunne P. W., Dubel J., Nasser G. A., Ashizawa T., de Jong P. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992 Mar 6;255(5049):1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- Goldberg Y. P., Rommens J. M., Andrew S. E., Hutchinson G. B., Lin B., Theilmann J., Graham R., Glaves M. L., Starr E., McDonald H. Identification of an Alu retrotransposition event in close proximity to a strong candidate gene for Huntington's disease. Nature. 1993 Mar 25;362(6418):370–373. doi: 10.1038/362370a0. [DOI] [PubMed] [Google Scholar]
- Gottlieb S., Esposito R. E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989 Mar 10;56(5):771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Ionov Y., Peinado M. A., Malkhosyan S., Shibata D., Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993 Jun 10;363(6429):558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- Kim R. A., Wang J. C. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell. 1989 Jun 16;57(6):975–985. doi: 10.1016/0092-8674(89)90336-x. [DOI] [PubMed] [Google Scholar]
- Kremer E. J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., Warren S. T., Schlessinger D., Sutherland G. R., Richards R. I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991 Jun 21;252(5013):1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- Kupiec M., Petes T. D. Meiotic recombination between repeated transposable elements in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2942–2954. doi: 10.1128/mcb.8.7.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada A. R., Wilson E. M., Lubahn D. B., Harding A. E., Fischbeck K. H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991 Jul 4;352(6330):77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Intermolecular recombination between DNAs introduced into mouse L cells is mediated by a nonconservative pathway that leads to crossover products. Mol Cell Biol. 1990 Jan;10(1):103–112. doi: 10.1128/mcb.10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Letsou A., Stachelek J. L. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics. 1987 Jan;115(1):161–167. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barceló J., O'Hoy K. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992 Mar 6;255(5049):1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: evidence for a nonconservative mechanism of homologous recombination. Mol Cell Biol. 1991 Jun;11(6):3278–3287. doi: 10.1128/mcb.11.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mautner V., Mackay N. Recombination in adenovirus: analysis of crossover sites in intertypic overlap recombinants. Virology. 1984 Nov;139(1):43–52. doi: 10.1016/0042-6822(84)90328-3. [DOI] [PubMed] [Google Scholar]
- Mézard C., Pompon D., Nicolas A. Recombination between similar but not identical DNA sequences during yeast transformation occurs within short stretches of identity. Cell. 1992 Aug 21;70(4):659–670. doi: 10.1016/0092-8674(92)90434-e. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Singer J. D., Hoekstra M. F., Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J Mol Biol. 1989 Jun 5;207(3):527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- Oberlé I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boué J., Bertheas M. F., Mandel J. L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991 May 24;252(5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Ohira M., Bae Y. S., Ikeda H. A new type of insertion mutation in monkey cells: insertion accompanied by long target site duplication. Mol Gen Genet. 1991 Oct;229(3):325–333. doi: 10.1007/BF00267452. [DOI] [PubMed] [Google Scholar]
- Ozenberger B. A., Roeder G. S. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol. 1991 Mar;11(3):1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R., Li G. M., Longley M. J., Fang W. H., Papadopoulos N., Jen J., de la Chapelle A., Kinzler K. W., Vogelstein B., Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993 Dec 17;75(6):1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- Petes T. D. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 1980 Mar;19(3):765–774. doi: 10.1016/s0092-8674(80)80052-3. [DOI] [PubMed] [Google Scholar]
- Rayssiguier C., Thaler D. S., Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989 Nov 23;342(6248):396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984 Nov;4(11):2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N., Sugarman E., Haber J. E. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989 Jul;122(3):519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield M. A., Agbunag R., Michaels M. L., Miller J. H. Cloning and sequencing of Escherichia coli mutR shows its identity to topB, encoding topoisomerase III. J Bacteriol. 1992 Aug;174(15):5168–5170. doi: 10.1128/jb.174.15.5168-5170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Huang H. V. Effect of base pair mismatches on recombination via the RecBCD pathway. Mol Gen Genet. 1989 Aug;218(2):358–360. doi: 10.1007/BF00331291. [DOI] [PubMed] [Google Scholar]
- Shen P., Huang H. V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986 Mar;112(3):441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls W. P., Wallace L. J., Moore P. D. Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell. 1990 Jan 12;60(1):95–103. doi: 10.1016/0092-8674(90)90719-u. [DOI] [PubMed] [Google Scholar]
- Wahls W. P., Wallace L. J., Moore P. D. The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol Cell Biol. 1990 Feb;10(2):785–793. doi: 10.1128/mcb.10.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Liskay R. M. Differential effects of base-pair mismatch on intrachromosomal versus extrachromosomal recombination in mouse cells. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5340–5344. doi: 10.1073/pnas.84.15.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989 Jul 28;58(2):409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Ward P., Berns K. I. In vitro rescue of an integrated hybrid adeno-associated virus/simian virus 40 genome. J Mol Biol. 1991 Apr 20;218(4):791–804. doi: 10.1016/0022-2836(91)90267-a. [DOI] [PubMed] [Google Scholar]
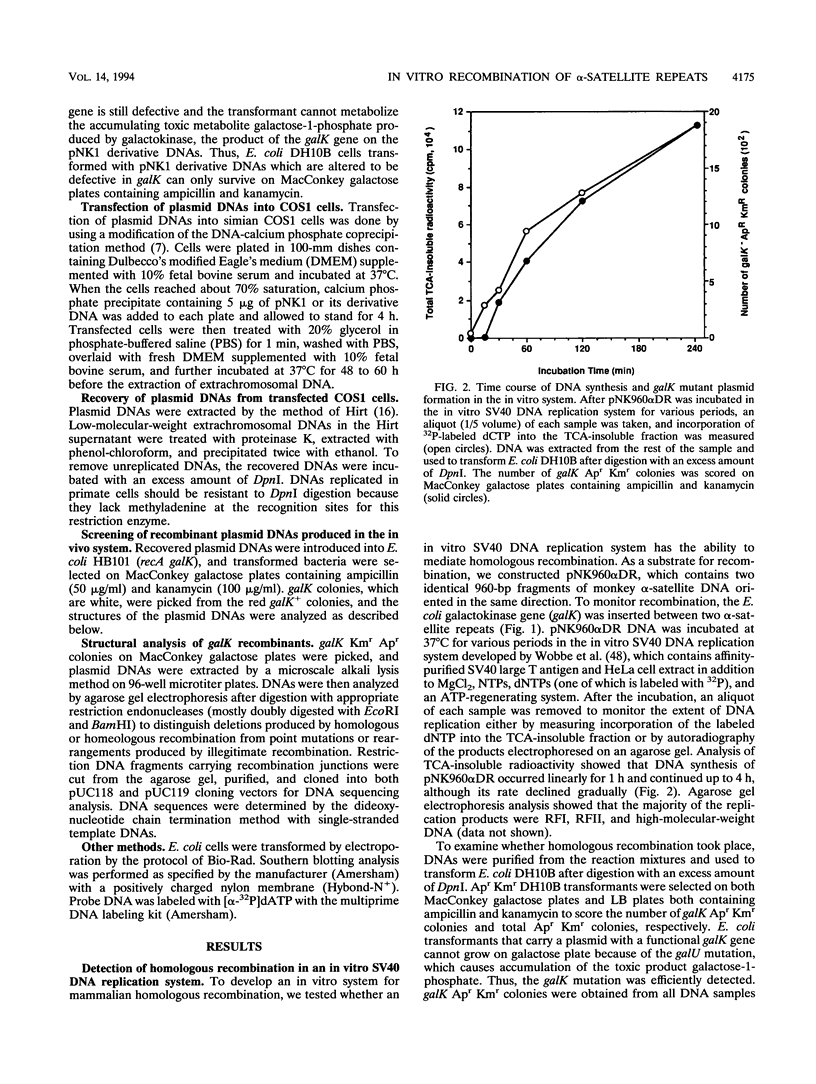
- Wobbe C. R., Dean F., Weissbach L., Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C. S., Cachianes G., Munz P., Silverstein S. Replication and recombination in adenovirus-infected cells are temporally and functionally related. J Virol. 1984 Sep;51(3):571–577. doi: 10.1128/jvi.51.3.571-577.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]