Abstract
Three-dimensional molecular structures can provide detailed information on biological mechanisms and, in cases where molecular function impacts on human health, significantly aid in the development of therapeutic interventions. Over the past 23 years, key components of the lentivirus HIV-1, including its envelope glycoproteins and capsid, and the replication enzymes reverse transcriptase, integrase and protease, have accordingly been scrutinized to near atomic scale resolution. Structural analyses of the interactions between viral and host cell components have moreover yielded key insights into the mechanisms of virus entry, chromosomal integration, transcription and egress from cells. Here, we review recent advances in HIV-1 structural biology, focusing on the impact these results have had on our understanding of virus replication and the development of new therapeutics.
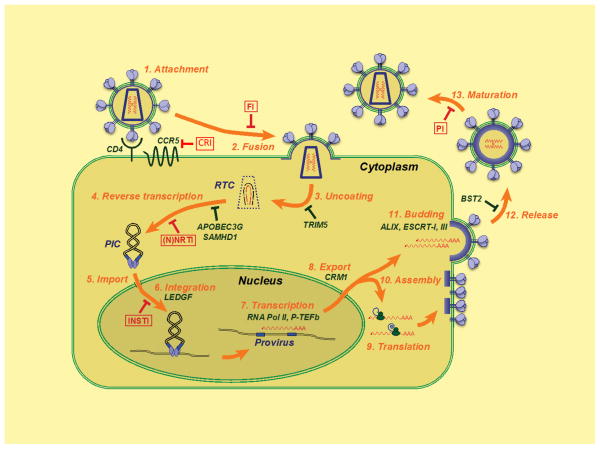
HIV-1 arose through several independent zoonotic transmissions of simian immunodeficiency viruses during the last century 1–3. Today, HIV-1, along with its less widespread cousin HIV-2, infects over 30 million people worldwide. Both viruses belong to the Retroviridae, a viral family that has left numerous scars of ancient infections in mammalian genomes, with derelict retroviral sequences comprising as much as 8% of our “own” DNA 4. The evolutionary success of this family is contrasted by its deceptive simplicity: encoding only 16 proteins, HIV-1 can persistently infect humans, subverting the innate and adaptive immune systems. Viral replication at the cellular level proceeds through a series of steps that start when a virus productively engages cell surface receptors and ends when nascent particles mature into infectious virions (Fig. 1). During this process, HIV-1 exploits a myriad of cellular factors to accomplish specific tasks at the same time as host restriction factors fight to suppress replication 5,6. The mainstream highly active anti-retroviral therapy (HAART) drug cocktails that are primarily used to target the reverse transcriptase (RT) and protease (PR) enzymes potently suppress viral loads and transmission rates, yet complications can arise from compound toxicity and the emergence of resistant strains (Box 1). Advances in structural biology can aid the development of next-generation compounds that are active against previously exploited targets, help to define new drug targets, and boost the effectiveness of vaccination strategies. This review proceeds stepwise through the HIV-1 replication cycle, highlighting the impact that major structural biology advances have had on our understanding of virus growth and the development of new anti-retroviral therapies.
Figure 1.
Schematic overview of the HIV-1 replication cycle. The infection begins when envelope glycoprotein spikes engage the CD4 receptor and the membrane spanning co-receptor (step 1; cell proteins discussed in text are indicated in green type), which leads to viral–cell membrane fusion and entry of the virus particle into the cell (step 2). Partial core shell uncoating (step 3) facilitates reverse transcription (step 4), which in turn yields the preintegration complex (PIC). Following import into the cell nucleus (step 5), PIC-associated IN orchestrates formation of the integrated provirus (step 6). Proviral transcription (step 7) yields different sizes of viral mRNAs (not shown), the larger of which require energy-dependent export to leave the nucleus (step 8). Genome-length mRNAs serve as a template for protein production (step 9) or viral particle assembly with protein components (step 10). ESCRT-mediated viral particle budding (step 11) and release (step 12) from the cell is accompanied or followed shortly thereafter by PR-mediated maturation (step 13) to create an infectious viral particle. Each step in the HIV-1 lifecycle is a potential target for antiviral intervention 165; the sites of action of clinical inhibitors (boxed) and cellular restriction factors are indicated with red and green block signs, respectively. CRI, CCR5 inhibitor; FI, fusion inhibitor; RTC, reverse transcription complex.
Box 1. Highly active anti-retroviral therapy.
Approximately thirty different drugs targeting four different steps in the HIV-1 replication cycle are currently approved for administration to HIV-positive individuals in the US (see http://www.aidsmeds.com/list.shtml). Nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs) and non-nucleoside reverse transcriptase inhibitors (NNRTIs) target the reverse transcription step that converts the viral genomic RNA into linear double stranded DNA whereas protease inhibitors (PIs) inhibit protease enzyme activity that is critical for the maturation of viral particles that bud out from infected cells. Two different inhibitors can block the entry of the virus into new target cells by thwarting either the interaction between the viral envelope glycoprotein gp120 and the CCR5 co-receptor (maraviroc) or the formation of the gp41 six-helix bundle that drives the fusion between the viral and cellular membranes (Fuzeon). The sole integrase strand transfer inhibitor (INSTI), raltegravir, blocks integrase’s strand transfer activity that is required to insert viral DNA into a host cell chromosome. Highly active anti-retroviral therapy or HAART routinely prescribes an NRTI, NNRTI and PI as a single pill or in various pill combinations. The combinatorial approach to drug treatment significantly suppresses the probability for the selection and resulting outgrowth of resistant HIV-1 strains that quickly arise during monotherapy.
Virus entry
The HIV-1 envelope spikes, which comprise trimers of non-covalently linked heterodimers of the surface gp120 and transmembrane gp41 glycoproteins 7–9, initiate a cascade of conformational changes that culminates in fusion between the viral and host cell membranes and the release of the viral core into the cytoplasm. HIV-1 primarily infects CD4-positive T lymphocytes and macrophage cells. An initial interaction between gp120 and the surface receptor CD4 induces the formation of a bridging sheet between the inner and outer domains of the gp120 monomer, exposing the binding site for a second cell surface molecule, typically the chemokine receptor CCR5 10–12 (Fig. 1, step 1). Co-receptor engagement leads to insertion of the fusion peptide located at the N-terminus of gp41 into the cell membrane, which in turn triggers significant rearrangements between trimerized N- and C-terminal heptad repeat sequences within gp41, the formation of a six helical hairpin structure, and the apposition and fusion of the viral and host cell membranes 13–15(Fig. 1, step 2).
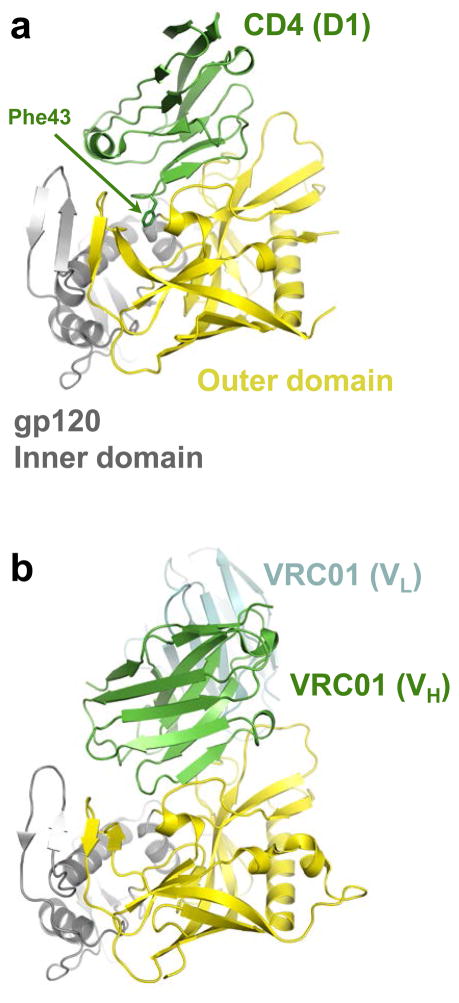
Initial cryo-electron tomography studies provided crucial glimpses of the HIV-1 envelope and its associated conformational flexibility 7,8, although the low-resolution models that were generated left many key aspects of the native structure unresolved 9,16,17. Higher-resolution crystallographic studies using engineered HIV-1 glycoprotein constructs have been instrumental in developing entry inhibitors and elucidating the mechanistic basis of virus neutralization by antibodies. Recent studies have highlighted the striking flexibility of the core gp120 structure, which allows extreme conformational changes upon CD4 engagement without destabilizing the interaction with gp41 12,18. CD4 binds gp120 at a depression formed between the inner and outer domains, where the CD4 residue Phe43 partially fills a hydrophobic cavity (Fig. 2a) 10. Small molecules designed to bind to and extend further into this pocket display antiviral activity, and increasing the gp120 binding affinity might lead to the development of clinically useful inhibitors 19.
Figure 2.
CD4 and CD4-mimicking antibody binding to the gp120 core. (a) Structure of HIV-1 gp120 (outer domain is shown in yellow and inner domain in gray) in complex with CD4 (green; pdb code 3JWD). Only immunoglobulin-like domain 1 (D1) of CD4 is shown; the Phe43 side-chain is depicted as sticks. (b) VRC01 antibody–gp120 co-crystal structure (pdb code 3NGB, heavy chain shown in green and light chain in cyan) oriented as in panel a. Only the variable domains of the heavy (VH) and light (VL) chains of the antibody are shown.
Most antibodies directed against gp120 tend to be strain-specific and moreover fail to neutralize the virus. Several groups recently described patient-derived gp120-reactive antibodies with broad HIV-1 neutralization activity 20–24. Wu and colleagues 21,22 took a structure-based approach to stabilize the CD4-bound conformation of gp120 using disulfide bonds and redesign its surface to mask positions exterior to the CD4 binding site. Using one such construct as bait, patient B cell clones producing antibodies with remarkably broad neutralizing activity were recovered. Structural characterization of these antibodies revealed that in binding to gp120, the heavy chains of the immunoglobulins mimic CD4 (Fig. 2a, b), with their epitopes almost precisely overlapping the primary CD4-binding site on gp120 22,25. These results define the structural basis for HIV-1 neutralization by antibodies that engage the CD4 binding site. Interestingly, immunoglobulins isolated from the sera of different donors using the resurfaced gp120 construct were derived from the same precursor heavy chain gene (IGHV1-2*02) that had subsequently undergone extensive affinity maturation 21,22,25. The requirement for extensive somatic mutation to achieve virus neutralization 21,22 might pose a challenge for the experimental elicitation of such antibodies. However, the recent discovery of highly potent gp120-binding antibodies with alternative modes of action suggests there are multiple genetic pathways to achieve cross-clade HIV-1 neutralization 20,23,24. These results should encourage attempts to design immunogens to elicit humoralimmunity for vaccination purposes.
Peptides derived from gp41 N-terminal 26 or C-terminal 27 sequences, which disrupt the six-helix bundle formation and hence membrane fusion, possess potent antiviral activity. A peptide based on the C-terminal sequence was licensed as Fuzeon in 2003, although the requirement for twice-daily injections and the relative ease through which drug mutations arise have limited its utility. D-peptides that target a pocket at the base of the N-terminal gp41 helical structure are also potent antivirals, and may overcome some of the limitations associated with Fuzeon use 28.
Post-entry events: uncoating to integration
The HIV core, which houses the replication enzymes RT and integrase (IN) as well as the viral genomic RNA, is encased by a cone-shaped shell 29 composed of the viral capsid (CA) protein. Recent work has highlighted interactions among composite CA molecules that underlie the structural integrity and functionalityof the protective shell 30–32.
Uncoating
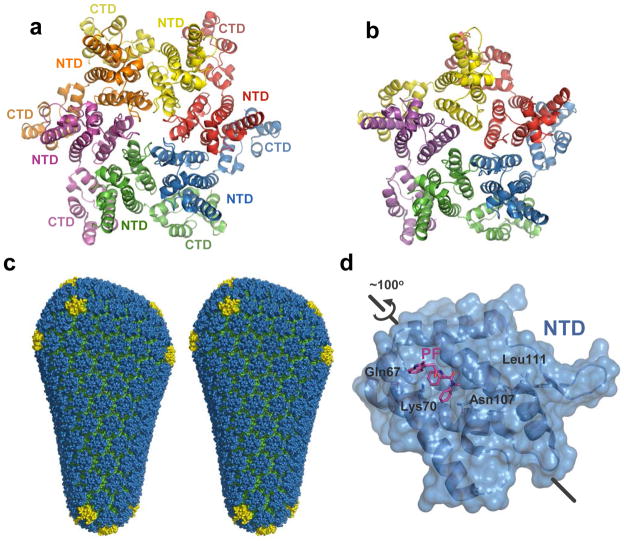
Partial CA shell dissolution, which is required for reverse transcription 33,34, is a recently verified therapeutic target 35 (Fig. 1, step 3). Moreover, the underlying features of the assembled shell seem to determine its propensity to uncoat 32. CA protein, which comprises independently folded N-terminal and C-terminal domains (NTD and CTD) connected by a flexible linker 36,37, can assemble into ring structures containing five or six protomers 31,32 (Fig. 3a, b). The rings further congregate to form a fullerene cone composed predominantly of hexamers; seven pentamers at the wide end and five at the narrow end allow for shape declinations 32,38 (Fig. 3c), and the flexibility of intramolecular NTD–CTD and intermolecular CTD–CTD interactions further contribute to the curvature of the shell lattice 30,32 (Fig. 3a, b). The relatively high concentration of penton declinations expected at the narrow end of the cone may furthermore serve to initiate uncoating 32.
Figure 3.
HIV-1 capsid structures. Crystal structures of the hexameric (a, pdb code 3H47) and pentameric (b, pdb code 3P05) full-length HIV-1 CA assemblies. Individual subunits are coloured by chain. (c) The model for the complete HIV-1 capsid, based on the crystal structures 32. NTDs of the hexameric and pentameric CA units are shown in blue and yellow, respectively; CTDs are green. (d) HIV-1 CA NTD in complex with PF-3450074 (PF, pdb code 2XDE). The orientation is related to that of the blue NTD in panel a by an ~100° rotation, as shown. Residues critical forPF -3450074 binding as revealed by resistance mutations 44 are indicated.
TRIM5α, a potent HIV-1 restriction factor isolated from rhesus macaques 39, recognizes the assembled CA structure to accelerate uncoating 40 and activate innate immune signalling pathways 41. A replacement of the N-terminal RING domain of rhesus TRIM5α with that from the related human TRIM21 protein yielded a chimera that is amenable to recombinant techniques 42. The hybrid construct forms 2D hexameric crystalline arrays in the presence of a higher-order six-fold lattice of HIV-1 CA 43. Such CA-templated multimerisation may underlie functional HIV-1 restrictionby rhesus TRIM5 α through a pattern recognition mechanism that is common to other components of the innate immune system 41. Stimulation of premature uncoating could moreover be a useful therapeutic approach; for example, PF-3450074, a small molecule inhibitor of HIV-1 replication that binds to a pocket within the NTD of CA (Fig. 3d), may work by triggering premature uncoating through destabilization of CA–CA interactions 35,44.
Viral DNA synthesis
Reverse transcription and integration of the resultant linear viral DNA molecule into a host cell chromosome occurs within the context of the nucleoprotein complex structures that are derived from the viral core (Fig. 1, steps 4–6). High-resolution HIV-1 RT structures have been available for a number of years, with initial drug-and nucleic acid template -bound crystal structures reported nearly 2 decades ago 45,46.
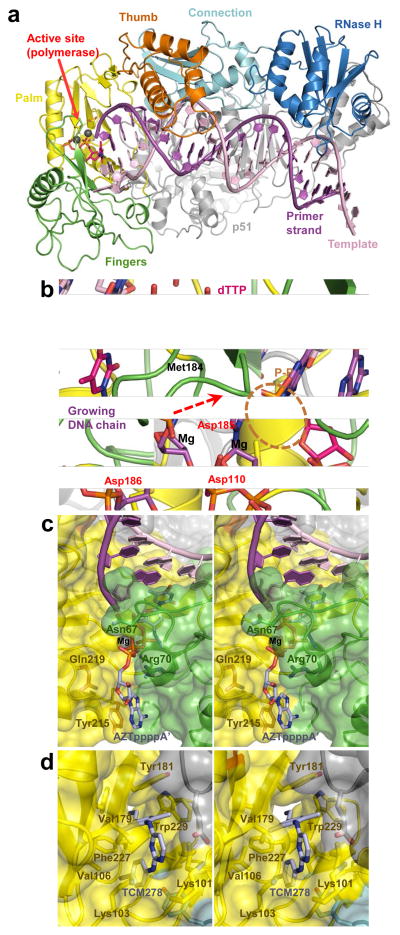
HIV-1 RT is a heterodimer composed of p66 and p51 subunits, with p66 harbouring two functional active sites: an N-terminal RNA- and DNA-dependent DNA polymerase and a C-terminal RNase H that digests the RNA component of RNA/DNA hybrids. The polymerase domain resembles a right hand with four subdomains: fingers, thumb, palm and connection (Fig. 4a) 45–48. During DNA polymerization, the catalytic residues Asp110, Asp185 and Asp186 within the palm subdomain activate the DNA primer 3′ hydroxyl and stabilize the hypothetical pentavalent α-phosphorous intermediate state within the substrate dNTP, incorporating the nucleotide into the growing DNA chain and liberating free pyrophosphate (Fig. 4b) 48.
Figure 4.
Structural analyses of HIV-1 RT function and its inhibition by small molecules. (a) Overview of the HIV-1 RT-template-primer complex (pdb code 1RTD). The protein and DNA chains are shown as cartoons. The subdomains of the active RT subunit are indicated and colour-coded; the inactive (p51) subunit is shown in gray. The structure contains a bound molecule of dTTP in the active site (pink). Grey spheres are Mg atoms. (b) Close-up of the polymerase active site (pdb code 1RTD) and DNA polymerization. The 3′-hydroxyl, absent in the original structure 48, is added for illustration purposes. The direction of nucleophilic attack is indicated by a dashed arrow. The catalytic residues, Met184 and the leaving pyrophosphate group (P–P) are shown as sticks and indicated. RT chain colours are conserved from panel a. (c) Stereo view of ATP-binding pocket in AZT-resistant HIV-1 RT (pdb code 3KLE). The excision product (AZTpppp A′) is shown as sticks with carbon atoms in light blue. Protein chains are shown as cartoons with semitransparent surfaces (colouring as in panel a); residues implicated in AZT resistance are indicated. (d) TCM278 bound to HIV-1 RT (pdb code 2ZD1). RTresidues forming the NNRTI-binding pocket are indicated.
Two classes of antiviral drugs, nucleoside and non-nucleoside RT inhibitors (NRTI and NNRTI, respectively), inhibit DNA polymerization and are core components of HAART (Box 1). Upon phosphorylation in infected cells, NRTIs mimic natural nucleoside triphosphates and are incorporated into the viral DNA by RT. Lacking the 3′-OH group needed for incorporation of the subsequent nucleotide, NRTIs act as chain terminators, and viral resistance to some of these small molecules accordingly occurs through drug exclusion mechanisms. For instance, mutations of Met184 (to Val or Ile) selectively preclude the binding of oxathiolane-containing inhibitors like 3TC over dNTPs with normal deoxyribose rings (Fig. 4b) 48,49. However, resistance to azidothymidine (AZT) and other thymidine analogues puzzled researchers for some time: inexplicably, the mutant RT from AZT-resistant virus strains efficiently incorporated AZT monophosphate into the viral DNA 50. Instead of preventing incorporation, the mutant enzyme developed the ability to excise the incorporated drug from the primer strand. Remarkably, RT accomplishes this by utilizing ATP as a pyrophosphate donor to excise the incorporated drug in the form of an AZT-adenosine tetraphosphate adduct, regenerating an active 3′-OH primer terminus in a reaction that is mechanistically equivalent to the reversal of the polymerization step 51,52. Recent structural analyses revealed that the AZT resistance mutations K70R, T215Y and K219Q create an optimal ATP-binding site between the fingers and palm subdomains of RT to promote the excision reaction 53(Fig. 4c).
NNRTIs are allosteric inhibitors that induce the formation of a flexible binding pocket through relatively large conformational changes involving Tyr181, Tyr188 and the primer grip (residues 227–235 within the palm subdomain) 45,54,55 (Fig. 4d). The mechanistic basis of inhibition may be due to displacement of the primer grip 56 or the 3-stranded β-sheet that contains the catalytic triad 55,57. Stacking interactions between the aromatic side-chains of Tyr181 and Tyr188 and first-generation NNRTIs like nevirapine contribute significantly to drug binding 45, and the associated mutations accordingly conferred resistance due to loss of aromatic character 58. K103N is also fairly widely associated with NNRTI resistance, and the Asn103-Tyr188 interaction in the mutant RT appears to restrict the movement of Tyr188 that is required for drug binding 59,60. The more recently developed diarylpyrimidine NNRTIs TMC-125 and TMC-278 retain potency in the face of first-generation NNRTI resistance mutations, with inherent drug flexibility contributing significantly to high affinity compound binding to the mutant RT 61(Fig. 4d).
Reverse transcription is inhibited by the cellular restriction factor APOBEC3G, a virion-incorporated cytidine deaminase that both impedes elongation 62,63 and converts nascent cytidines in viral cDNA to uracils 64–66. HIV-1 accordingly deploys a countermeasure, the Vif protein, which antagonizes the incorporation of APOBEC3G by binding and inducing its degradation in virus producer cells 67,68. Such observations highlight the importance of the Vif–APOBEC3G nexus for antiviral drug development, and small molecules that limit the ability of Vif to degrade APOBEC3G and, accordingly, inhibit HIV-1 infection have been described 69,70.
APOBEC3G harbours two cytidine deaminase domains: the NTD mediates virion incorporation whereas the CTD is a functional deaminase 71–73. Several NMR 74–76 and X-ray crystal 77,78 structures of the CTD revealed a 5-stranded β sheet intermixed with 5 helices, with conserved elements of the catalytic zinc-coordination motif (H/C-X-E-X23–28-PC-X2-C) contributed by a pair of α helices. These results afford important glimpses into the mechanism of HIV deamination, although additional structures that incorporate the NTD and especially the single-stranded DNA substrate will reveal a more complete picture of catalysis. Structures that include Vif should further aid the development of novel antiviral compounds.
Integration
IN possesses two catalytic activities, 3′ processing and DNA strand transfer. Each end of the HIV-1 DNA long terminal repeat (LTR) is cleaved adjacent to the invariant dinucleotide sequence CA, unveiling recessed 3′ termini. IN then uses the 3′ hydroxyls to cut chromosomal DNA strands across a major groove, at the same time joining the viral DNA ends to the target DNA 5′-phosphates. Host enzymes complete the integration process by repairing the single strand gaps abutting the unjoined viral DNA 5′ ends, resulting in establishment of a stable provirus (Fig. 1, step 6). IN-mediated reversal of integration is impossible, although rare instances of cell-mediated homologous recombination across the LTRs can excise proviral DNA 79. Site-specific recombinases can be engineered to similarly excise the HIV-1 provirus ex vivo 80, although such approaches would appear to be far from clinical application.
Although crystal and NMR structures of various fragments of HIV-1 IN were reported over several years 81, detailed views of the functional IN-viral DNA nucleoprotein complex, or intasome, were lacking until recently. Given that clinically useful HIV-1 IN inhibitors selectively interact with the intasome rather than free IN 82, this dearth of structural information limited drug development. Recent successes are owed to the tractability of the intasome derived from the related prototype foamy virus (PFV), a member of the Spumavirus retroviral genus, by X-ray crystallography 83,84. An overview of these advances is given here; for in-depth reviews see refs 85,86.
The intasome contains a dimer-of-dimers of IN, with only one subunit of each dimer binding a viral DNA end 83 (Fig. 5a, b). Thus, akin to RT, functional IN active sites are delegated to a subset of protein molecules within the multimeric complex. The intasome accommodates the target DNA within a cleft between the functional active sites in a severely bent conformation (Fig. 5b, c). The contortion in target DNA allows the intasome active sites (which are separated from one another by as much as 26.5 Å) to access their target scissile phosphodiester bonds 84. The residues of the catalytic D, D-35-E motif coordinate two divalent metal ions, revealing roles in viral DNA 3′-OH nucleophile activation and scissile phosphodiester bond destabilization during DNA strand transfer 83,84 (Fig. 5c). The reversal of the reaction appears to be restricted by a conformational change, which causes a 2.3-Å displacement of the newly formed viral-target DNA phosphodiester bond from the IN active site following transesterification 84.
Figure 5.
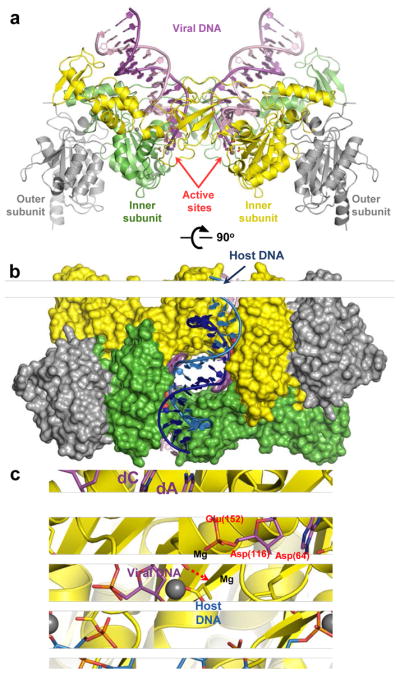
Retroviral intasome structures and mechanism of IN catalysis. (a) Overview of the PFV intasome structure (pdb code 3OY9). The active (inner) IN chains are shown as green and yellow cartoons; catalytically inactive (outer) chains are gray. The transferred and non-transferred viral DNA strands are shown in dark and light magenta, respectively. Active site carboxylates are shown as sticks and divalent metal ions as gray spheres. (b) The PFV intasome in complex with a host DNA mimic (light and dark blue; pdb code 3OS2). IN chains are shown in space-fill mode conserving colours from panel a. (c) DNA strand transfer. The model is based on structures of the Mn2+-bound intasome and target capture complex (see 84 for details). IN is shown as cartoons with D, D-35-E active site residues as sticks. DNA is shown as sticks; the invariant viral dA and dC nucleotides are indicated. Colours are conserved from panel a. Residue numbering corresponds to the HIV-1 IN sequence. Direction of nucleophilic attack is indicated by a red dashed arrow.
The clinically approved HIV-1 IN inhibitor raltegravir and similar small molecules that are in development preferentially inhibit DNA strand transfer activity, and IN strand transfer inhibitors (INSTIs) fortuitously harbour broad anti-retroviral activity 87–89. Results based on PFV intasome-INSTI co-crystal structures have been accordingly illuminating. INSTIs harbour two common moieties: co-planar heteroatoms (typically three oxygen atoms) that chelate the active site metal ions 90 and halogenated benzyl groups, whose function until recently was largely speculative. INSTIs engage the bound metal ions, only slightly influencing their positions within the IN active site. Primarily through interactions with the penultimate viral DNA G·C base pair and a 310 helix (Pro145-Gln146 in HIV-1 IN), INSTI halogenated benzyl groups assume the position of the terminal adenine ring, ejecting the viral 3′-deoxyadenosine with its associated 3′-OH nucleophile from the active site 83,88. This displacement of the DNA strand transfer nucleophile forms the mechanistic basis of INSTI action. In addition, INSTIs sterically preclude target DNA binding, explaining the competition between target DNA and the small molecules 82,84. The PFV model has provided important clues about the mechanism of drug resistance associated with HIV-1 IN mutations selected in the presence of raltegravir 88.
Analogous to RT, there is precedence that a second region of HIV-1 IN, in this case distal from the active site, affords an opportune location for allosteric inhibitor binding. Lentiviruses such as HIV-1 favour integration within active genes due to an interaction between IN and the chromatin binding protein LEDGF/p75 (reviewed in 91). The IN binding domain (IBD) of LEDGF/p75 is a pseudo HEAT repeat analogous topology domain that consists of two units of a helix-hairpin-helix repeat 92, and the LEDGF/p75 hotspot residues Ile365 and Asp366 at the tip of the N-terminal hairpin nestle into a cleft at the HIV-1 IN CCD dimer interface 93. In a remarkable example of structure-based drug design, Debyser and colleagues discovered a novel class of HIV-1 IN inhibitors capable of suppressing viral replication. These small molecules, termed LEDGINs, mimic the LEDGF/p75-IN interaction in silico and inhibit protein-protein binding in vitro 94. Given the highly conserved nature of INSTI binding at the active site 88,95 and the likelihood of considerable cross-resistance among INSTIs 96, the development of such allosteric HIV-1 IN inhibitors is highly desirable.
Viral mRNA biogenesis and transport
Integration marks the transition from the early to late phase of HIV-1 replication, in which the focus shifts to viral gene expression followed by the assembly and egress of nascent viral particles. Transcription, which initiates from the U3 promoter within the upstream LTR (Fig. 1, step 7), requires the viral Tat transactivator protein for efficient elongation. Viral mRNAs are produced as a variety of alternatively spliced species. The smaller messages are exported readily from the nucleus, whereas the unspliced and singly spliced mRNAs require the action of Rev. This small viral protein acts as an adaptor, binding to the Rev-response element (RRE) located within the mRNA env coding region and the nuclear export factor CRM1 (step 8 in Fig. 1). Recent structural biology advances yield insight into the mechanisms of Tat transactivation 97 and Rev-dependent mRNA export 98,99.
Transcriptional elongation
Tat recruits the cellular positive transcription elongation factor P-TEFb, comprising the Cdk9 kinase and cyclin T1 (CycT1) subunits, to the viral trans-activation response (TAR) element present in stalled transcripts 100,101. Subsequent phosphorylation of the heptad repeat residues Ser2 and Ser5 in the CTD of the large subunit of RNA polymerase II by activated Cdk9 stimulates transcriptional elongation.
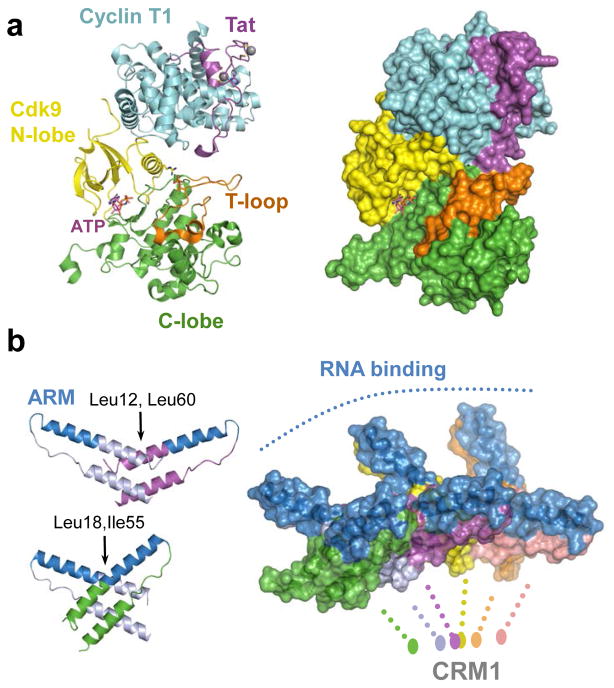
Tat is largely unstructured in the absence of binding ligands 102. TAR binding occurs primarily via an α-helical Arg-rich motif (ARM), which inserts into the RNA major groove within the stem-loop structure 103. The N-terminal activation domain of Tat, which contains acidic/Pro-rich, zinc binding motifs and core subdomains, assumes an ordered structure upon P-TEFb binding 97. Within the complex, Tat primarily interacts with the CycT1 subunit, also contacting the T loop region of Cdk9 (Fig. 6a). Tat binding stimulates phosphorylation of RNA polymerase II CTD Ser5 heptad repeat residues by Cdk9 104 and reciprocal conformation changes in the kinase accordingly alter the substrate-binding surface of P-TEFb. Crucially, the fact that Tat induces conformational changes in P-TEFb suggests that it may be possible to develop anti-HIV agents directed against P-TEFb with limited sideeffects on its normal cellular functions 97.
Figure 6.
Higher-order Tat and Rev structures. (a) Crystal structure of HIV-1 Tat in complex with ATP-bound P-TEFb (pdb code 3MIA). The protein chains are shown as cartoons (left) or in space-fill mode (right). The N-lobe of Cdk9 is shown in yellow, C-lobe in green, and the T-loop in orange; Cyclin T1 is in blue, and HIV-1 Tat is shown in magenta. ATP bound to the active site of Cdk9 is shown as sticks and indicated. Gray spheres are Zn atoms. (b) Left: dimeric assemblies of HIV-1 Rev core observed in crystals (pdb codes 2X7L and 3LPH). Rev monomers are shown as cartoons and colored by chain, expect for the ARM motifs, which are blue. The crystal structures elucidate two types of Rev-Rev hydrophobic interfaces, one involving Leu12 and Leu60 and the other Leu18 and Ile55. Right: model of the Rev hexamer based on the dimeric structures, shown in space-fill mode. The oligomer projects RNA-binding ARM domains (blue) on one side, with CRM1-binding nuclear export signals (not resolved in the current structures) emanating from the other side.
mRNA export
Rev binds to the RRE in a highly cooperative manner, forming an RNA-dependent dimer en route to a higher order Rev-RNA multimer 105,106. The structural basis for Rev multimerisation was recently elucidated by two complementary crystallographic studies 98,99. Rev adopts an amphipathic helical hairpin, which multimerizes via face-to-face and back-to-back symmetric interfaces stabilized by conserved hydrophobic interactions (Fig. 6b). Collectively, the crystal structures 98,99 describe both types of interface and allow modelling of a Rev hexamer, which projects pairs of ARMs on one side and C-terminal nuclear export signals for latching onto the cellular CRM1 nuclear export factor on the other (Fig. 6b). The relative orientations of the ARMs in the context of the oligomer are thought to dictate the selectivity of the viral protein for the RRE structure and sequence. The model also accounts for the cooperativity of RNA binding by Rev, although a more complete structure including the RRE will be required to explain the details of protein-RNA recognition.
Viral egress and maturation
The retroviral structural proteins CA, matrix (MA) and NC are synthesized as parts of the Gag precursor polypeptide, and HIV-1 Gag is sufficient to assemble virus-like particles at the plasma membrane and bud from cells 107 (Fig. 1, steps 10 and 11). MA, through an N-terminal myristic acid 108,109 and conserved basic amino acid residues 110–112, contributes to Gag membrane association. The differential exposure of the myristate through a process known as the myristyl switch 113 allows Gag to associate preferentially with the plasma membrane rather than intracellular membranes. The switch can be activated by phosphatidylinositol 4,5-bisphosphate 114, a phospholipid that is concentrated in the inner leaflet of the plasma membrane and interacts directly with MA 115. Several steps along the pathway of HIV-1 assembly and particle release from cells have been targeted for antiviral drug development.
Viral late domains and the cellular ESCRT machinery
Retroviral budding is orchestrated by interactions between Pro-rich motifs in Gag that are known as late (L) domains and cellular class E vacuolar protein sorting (Vps) proteins, the actions of which are required to form the nascent particle and sever it from the plasma membrane. The intended functions of Vps proteins are in the formation of multi-vesicular bodies (MVBs), a reaction that is topologically identical to virus budding as in each case a membrane-coated vesicle leaves the cytoplasm, and in abscission during cell division 116,117. Most class E Vps proteins function as subunits of endosomal sorting complexes required for transport (ESCRT), which come in four varieties (ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III). ESCRT-I and ESCRT-II function during membrane budding, whereas ESCRT-III is important for membrane scission. Recent advances have yielded structures of several class E proteins as well as the class E protein–L domain interactions that are crucial for virus budding from infected cells (see 118,119 for in -depth reviews).
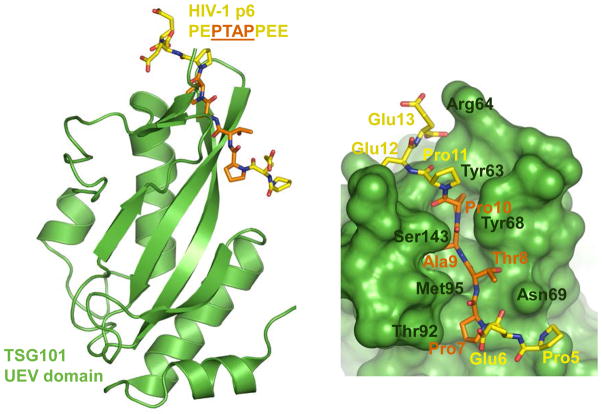
The C-terminal HIV-1 Gag cleavage product p6 harbours two L domains: P(T/S)AP and LYPx1–3L 120,121. The TSG101 component of ESCRT-I engages P(T/S)AP whereas ALIX, itself not formally an ESCRT protein, binds LYPx1–3L 121,122. ALIX contains three domains, an N-terminal Bro1 domain, an interior V domain and a C-terminal proline-rich domain (PRD). The boomerang-shaped Bro1 domain interacts with different isoforms of the ESCRT-III protein CHMP4, whereas LYPx1–3L interacts with arm 2 of the α helical V domain 123–126. The PRD within ALIX in turn interacts with TSG101 127, accounting for the direct link that ALIX provides between ESCRT-I and ESCRT-III 121,128. Highlighting one potential target for the development of inhibitors of HIV-1 budding, the P(T/S)AP domain inserts into a cleft on the N-terminal UEV domain of TSG101 (Fig. 7) 129,130.
Figure 7.
Virus-cell interactions and HIV-1 budding. The structure of the UEV domain of TSG101 bound to the PT(S)AP domain of HIV-1 p6 protein (pdb code 3OBU). TSG101 is shown as green ribbon (left) or space-fill (right) cartoons. P6 (residues 5–12; PEPTAPPEE) is shown as sticks; the carbon atoms of the core L domain PTAP and the flanking regions are orange and yellow, respectively. Some of the key TSG101 residues involved in the interaction are indicated on the right panel.
Restriction of viral egress
The type II trans-membrane (TM) protein CD317/BST2/tetherin inhibits the release of budding particles by retaining them on the plasma membrane of the virus producer cell 131,132 (step 12 in Fig. 1). Tetherin consists of a short N-terminal cytoplasmic tail followed by a TM region, an approximate 110-residue ectodomain ending on an amphipathic sequence that reconnects the protein to the plasma membrane 133. The hydrophobic C-terminal peptide of tetherin, initially thought to be a signal for glycosyl phosphatidylinositol modification, may in fact function as a second TM domain 134. Thisunusual dual membrane -bound topology of tetherin led to several models, involving extended or laterally arranged parallel or anti-parallel protein dimers at the cell surface, to explain virus tethering 131, and a number of recent X-ray crystal structures revealed that the ectodomain indeed forms a parallel dimeric α helical coiled coil 135–137. In addition, the tetherin dimers can further assemble head-to-head into tetramers via formation of a four-helix bundle 136,137. However, mutations designed to ablate tetramer formation did not eliminate tetherin function, indicating that tetramerization is not essential for HIV-1 restriction 137. These data highlight the extended ectodomain coiled coil dimer as the likely virus tethering unit. Ectodomain residues Ala88 and Gly109, which disfavoured coiled coil packing, probably impart some flexibility to the structure, perhaps facilitating terminal anchor insertion into the viral membrane 136.
HIV-1 Vpu, also a TM protein, counteracts the restriction by tetherin 131,132 through a mechanism that depends on a direct interaction between the viral and host proteins 138,139. Previously elucidated structures of Vpu fragments yielded limited insight into the mechanism of the Vpu–tetherin interaction, though a recent NMR analysis of lipid membrane-embedded TM peptides indicates a likely anti-parallel helix-helix binding interface 140.
Protease and virus maturation
The final step of the viral lifecycle, which is mediated by PR and occurs concomitant with or soon after budding, converts immature particles to infectious virions via the proteolysis of Gag and Gag-Pol precursor polypeptides to yield the structural components MA, CA and NC, and the PR, RT and IN enzymes 141 (Fig. 1, step 13). Cryo-electron tomography revealed Gag structural rearrangements that occur within immature particles during proteolysis and maturation 142,143 and characterized cellular sites of HIV-1 budding 144. Following cleavage of the MA/CA bond, a novel β hairpin formed by a salt-bridge between the liberated Pro1 N-terminus and Asp51 in CA triggers core shell assembly 145. Recent evidence indicates that the morphological transitions occuring during HIV-1 particle assembly and maturation represent druggable targets. A 12-mer peptide, selected in a phage display screen for binding to the HIV-1 CA CTD, potently restricted CA assembly in vitro 146. Bevirimat, a betulinic acid derivative of herbal origin, inhibited HIV-1 replication by specifically blocking PR cleavage of the CA-SP1 junction 147. Exposure to bevirimat leads to stabilization of the immature CA lattice in HIV-1 virions 148. CAP1 is another small molecule reported to elicit abnormal HIV-1 core morphologies 149. Binding of CAP1 to the CA NTD involves formation of a deep hydrophobic pocket, which serves as a ligand binding site 150. The binding mode of CAP1 is therefore very different from that of PF-3450074, which engages a pre-formed pocket on the CA NTD surface (Fig. 3d) 35. It seems likely that the distortion in CA structure associated with CAP1 binding interferes with CA hexamer assembly.
Unlike the previously discussed viral enzymes, the structure of full-length PR 151–153 preceded the approval of the initial clinical inhibitor that targeted the enzyme by several years 154. Accordingly, the development of PR inhibitors (PIs) has benefited more from structure-based design efforts than other anti-retroviral drugs, and readers are directed to refs 155 and 156 for historical accounts of the interplay between PR structure and the development of PIs and resistance mechanisms.
The nine different peptide bonds within Gag and Gag-Pol that are cleaved by PR display limited primary sequence homology. Co-crystallization of six peptide substrates with PR defined a common substrate volume or envelope, indicating that substrate shape rather than primary sequence is a key predictor of functionality 157. The approved PIs are competitive inhibitors that bind to the enzyme active site, and overlays of PR–PI co-crystal structures identified regions of the so-called PI envelope that protruded from the substrate envelope and contacted amino acid residues that, when changed, confer drug resistance 158. These findings led to the hypothesis that PIs designed to fit more snugly within the substrate envelope would display favourable genetic resistance barriers, and some novel amprenavir-based compounds displayed marginally improved binding profiles to drug resistant PR as compared to the wild-type enzyme in vitro 159. Because compounds with enhanced binding affinities for wild-type PR bound drug-resistant enzymes relatively less well than amprenavir, additional work is required to determine whether substrate envelope-based PIs will display beneficial profiles against drug resistant strains in the clinic.
Conclusions and perspectives
HIV-1 has been analyzed by structural biology techniques more so than any other virus, with partial or complete structures known for all 16 of its protein components and additional structures determined for substrate- and host factor-bound complexes. Structural biology will continue to have a significant impact on HIV/AIDS research moving forward by providing high-resolution glimpses of target protein–drug complexes and viral–host interactions, such as CA–TRIM5α, Vif–APOBEC3G or Vpu–tetherin, which will reveal novel druggable sites. Despite decades of research, the interactions between HIV-1 and host proteins that underlie some steps in the viral life cycle, for example the import of the preintegration complex into the nucleus (Fig. 1, step 5), are only now being illuminated. The simian immunodeficiency virus Vpx protein was moreover recently shown to counteract the SAMHD1 restriction factor that inhibits HIV-1 reverse transcription and infection of monocytic cells 160,161, indicating that these protein complexes could define new pathways for antiviral drug developmentas well.
Notwithstanding the ongoing work with PIs, it will be interesting to see if structure-based substrate/inhibitor envelope hypotheses will apply to the development of other HIV-1 inhibitors. Because NNRTIs form induced fit binding pockets, they would appear to be poor candidates for this technique. The relatively tight overlay of multiple bound drugs at the IN active site and similarities in drug positions with the ejected terminal adenosine base 88 hints that INSTIs could be another drug class to benefit from such approaches. 3D structures of new drug targets as well as inhibitor or antibody-bound targets will predictably increase the pace of antiviral development and help guide vaccine development efforts 162,163. The development of new technologies and improvements in existing methods will also significantly influence structural virology moving forward. Single-particle electron cryo-microscopy has recently yielded near-atomic resolution structures of a number of so-called naked viruses that, unlike HIV-1, lack an exterior envelope lipid bilayer 164. Although the icosahedral symmetry underlying these structures greatly facilitated their determination, ongoing developments in instrumentation and computational science may very well yield similar resolution structures for particles that possess less inherent symmetry.
The development of HAART has dramatically changed the face of the HIV/AIDS epidemic since the disease was first recognized 30 years ago. Considered virtually a death sentence prior to the advent of anti-retroviral drugs, HIV-1 infection is now a manageable chronic disease. Yet, despite these remarkable advances, there remains significant room for improvement. Some of the drugs, in particular the PIs, exert toxic side-effects. More tolerable antiviral regimens could strengthen patient compliance and consequently reduce the emergence of resistant strains. Although the recently approved INSTI raltegravir is apparently non-toxic, the relative ease by which it selects for drug resistant strains highlights the need for second-generation INSTIs with more favorable genetic barriers to the resistance. The development of compounds that inhibit functions of less explored drug targets, in particular of the accessory HIV-1 proteins and host factors, would be of obvious benefit as well. The availability and efficacy of the current arsenal of anti-retroviral drugs should not be taken for granted. It is important to bear in mind, that the majority of the HIV-infected population do not have access to the advanced treatment options. Short of an effective vaccination strategy, the ongoing race against drug resistance can best be won by sustained effort to develop novel ever more potent and tolerable antiviral approaches.
At a glance.
HIV-1 replication relies on the proper functioning of specific viral proteins and three of these, protease, integrase, and reverse transcriptase with associated RNase H activity, are enzymes. Antiviral drugs that inhibit protease, integrase and reverse transcriptase DNA polymerase activities are approved for treating AIDS patients. The highly active anti-retroviral therapy or HAART regimens utilize cocktails of three inhibitors to suppress HIV-1 replication and the outgrowth of drug resistant viral strains.
HIV-1 replication depends on a plethora of functional interactions between its proteins and those of the host. Other cellular proteins, which are referred to as restriction factors, work to counteract virus growth. TRIM5α, APOBEC3G, tetherin and SAMHD1 are examples of such restriction factors.
In addition to enzyme active sites, critical viral-host protein interactions define targets for therapeutic intervention. Drugs might block interactions between the virus and host proteins needed for replication, as is the case for the approved entry inhibitor maraviroc, or enhance the effects of cell restriction factors.
Neutralization of the viral envelope glycoprotein gp120 by the adaptive immune system underscores AIDS vaccine development strategies.
Structural biology studies yield three-dimensional glimpses of protein function at near atomic resolution. Such results form the cornerstones of modern antiviral drug and vaccine development efforts.
Acknowledgments
We thank Mark Yeager for sharing coordinates of the HIV-1 capsid model 32. This work was supported by grants AI070042 from the US National Institutes of Health (A.E.) and G1000917 from the UK Medical Research Council (P.C.). The opinions voiced herein in no way reflect those of these funding agencies. We apologize to colleagues whose work we were unable to cite or discuss due to space limitations.
Biographies
Alan Engelman
Alan Engelman received his Ph.D. in Molecular Biology and Microbiology from Tufts University School of Medicine in Boston, Massachusetts in 1990 and focused on the mechanism of HIV DNA integration as a postdoctoral fellow at the National Institute of Diabetes and Digestive and Kidney Diseases in Bethesda, Maryland. He joined the faculty of the Dana-Farber Cancer Institute and Harvard Medical School in Boston in 1995, where he studies the molecular virology of HIV-host interactions.
Peter Cherepanov
Peter Cherepanov obtained his Ph.D. in Medical Sciences from the University of Leuven, Belgium in 2000 and did postdoctoral work at the University of Leuven and Dana-Farber Cancer Institute on HIV integration. He was a lecturer and then professor at Imperial College London from 2005 to 2011. Presently, he is a group leader at Clare Hall Laboratories, Cancer Research UK. Research in his laboratory is focusing on structural biology of chromatin and retroviral integration.
References
- 1.Gao F, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 2.Korber B, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 3.Lemey P, et al. Tracing the origin and history of the HIV-2 epidemic. Proc Natl Acad Sci U S A. 2003;100:6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Evans DT, Serra-Moreno R, Singh RK, Guatelli JC. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 2010;18:388–396. doi: 10.1016/j.tim.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huthoff H, Towers GJ. Restriction of retroviral replication by APOBEC3G/F and TRIM5 alpha. Trends Microbiol. 2008;16:612–619. doi: 10.1016/j.tim.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 8.Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2006;2:e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. The X-ray crystal structure of a core HIV-1 gp120 construct revealed the 3D fold of the critical viral glycoprotein as well as details of its interaction with the cell CD4 receptor and a neutralizing antibody. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzuto CD, et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 12.Chen B, et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 13.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 14.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. Together with reference 13, the structure of the gp41 ectodomian revealed an extended triple-stranded alpha helical coiled coil that underscored similarity in mechanism of entry with other enveloped viruses and set the stage for the eventual development of the entry inhibitor Fuzeon. [DOI] [PubMed] [Google Scholar]
- 15.Buzon V, et al. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6:e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramaniam S. The SIV surface spike imaged by electron tomography: one leg or three? PLoS Pathog. 2006;2:e91. doi: 10.1371/journal.ppat.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu P, Winkler H, Chertova E, Taylor KA, Roux KH. Cryoelectron tomography of HIV-1 envelope spikes: further evidence for tripod-like legs. PLoS Pathog. 2008;4:e1000203. doi: 10.1371/journal.ppat.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pancera M, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madani N, et al. Small -molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure. 2008;16:1689–1701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. Targeted selection of B cell clones from patients led to the identification of antibodies that neutralized approximately 90% of circulating HIV-1 isolates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. High-throughput functional screening of AIDS patient B cells yielded scores of new antibodies possessing superior breadth of cross-strain neutralization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corti D, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckert DM, Kim PS. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc Natl Acad Sci USA. 2001;98:11187–11192. doi: 10.1073/pnas.201392898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welch BD, et al. Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance. J Virol. 2010;84:11235–11244. doi: 10.1128/JVI.01339-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briggs JA, et al. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Byeon IJ, et al. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pornillos O, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. X-ray crystal structures reveal how individual monomers of HIV-1 capsid protein interact to form a hexamer ring, the basic building block of the conical shell incasingthe components of the viral core. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. The crystal structure reveals the basic building block of the HIV-1 capsid pentamer that affords critical shape declinations to the conical capsid shell and accordingly leads to an atomicscale model of overallshell structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitzon T, et al. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology. 2000;268:294–307. doi: 10.1006/viro.1999.0178. [DOI] [PubMed] [Google Scholar]
- 34.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blair WS, et al. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog. 2010;6:e1001220. doi: 10.1371/journal.ppat.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamble TR, et al. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 37.Gamble TR, et al. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 38.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. Electron microscopy of recombinant HIV-1 capsid-nucleocapsid protein multimers leads to the prediction of a fullerene cone structure for the viral capsid shell. [DOI] [PubMed] [Google Scholar]
- 39.Stremlau M, et al. The cytoplasmic body component TRIM5 alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 40.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5 alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kar AK, Diaz-Griffero F, Li Y, Li X, Sodroski J. Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5 alpha protein. J Virol. 2008;82:11669–11681. doi: 10.1128/JVI.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ganser-Pornillos BK, et al. Hexagonal assembly of a restricting TRIM5 alpha protein. Proc Natl Acad Sci USA. 2011;108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi J, Zhou J, Shah VB, Aiken C, Whitby K. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J Virol. 2011;85:542–549. doi: 10.1128/JVI.01406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. The X-ray crystal structure of the HIV-1 reverse transcriptase heterodimer reveals the asymmetric nature of the protein complex and the binding site for the non-nucleoside reverse transcriptase inhibitor nevirapine. [DOI] [PubMed] [Google Scholar]
- 46.Jacobo-Molina A, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. The crystal structure of HIV-1 reverse transcriptase reveals the positioning of template nucleic acid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodgers DW, et al. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. The X-ray crystal structure of the reverse transcriptase heterodimer with covalently trapped nucleic acid template, primer and deoxynucleoside triphosphate reveals the mechanism of DNA polymerization. The protein–nucleic acid covalent linkage is adopted as a field standard technique moving forward. [DOI] [PubMed] [Google Scholar]
- 49.Sarafianos SG, et al. Lamivudine (3 TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc Natl Acad Sci USA. 1999;96:10027–10032. doi: 10.1073/pnas.96.18.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lacey SF, et al. Biochemical studies on the reverse transcriptase and RNase H activities from human immunodeficiency virus strains resistant to 3′-azido-3′-deoxythymidine. J Biol Chem. 1992;267:15789–15794. [PubMed] [Google Scholar]
- 51.Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 52.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 53.Tu X, et al. Structural basis of HIV-1 resistance to AZT by excision. Nat Struct Mol Biol. 2010;17:1202–1209. doi: 10.1038/nsmb.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tantillo C, et al. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanismsof drug inhibition and resistance. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 55.Ren J, et al. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat Struct Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 56.Das K, et al. Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant. J Mol Biol. 1996;264:1085–1100. doi: 10.1006/jmbi.1996.0698. [DOI] [PubMed] [Google Scholar]
- 57.Esnouf R, et al. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nat Struct Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 58.Ren J, et al. Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors. J Mol Biol. 2001;312:795–805. doi: 10.1006/jmbi.2001.4988. [DOI] [PubMed] [Google Scholar]
- 59.Hsiou Y, et al. The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance. J Mol Biol. 2001;309:437–445. doi: 10.1006/jmbi.2001.4648. [DOI] [PubMed] [Google Scholar]
- 60.Das K, et al. Crystal structures of clinically relevant Lys103Asn/Tyr181Cys double mutant HIV-1 reverse transcriptase in complexes with ATP and non-nucleoside inhibitor HBY 097. J Mol Biol. 2007;365:77–89. doi: 10.1016/j.jmb.2006.08.097. [DOI] [PubMed] [Google Scholar]
- 61.Das K, et al. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci USA. 2008;105:1466–1471. doi: 10.1073/pnas.0711209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo F, Cen S, Niu M, Saadatmand J, Kleiman L. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J Virol. 2006;80:11710–11722. doi: 10.1128/JVI.01038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 65.Mangeat B, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 66.Zhang H, et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 68.Yu X, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 69.Nathans R, et al. Small-molecule inhibition of HIV-1 Vif. Nat Biotechnol. 2008;26:1187–1192. doi: 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cen S, et al. Small molecular compounds inhibit HIV-1 replication through specifically stabilizing APOBEC3G. J Biol Chem. 2010;285:16546–16552. doi: 10.1074/jbc.M109.085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Navarro F, et al. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 72.Newman EN, et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 73.Hache G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 74.Chen KM, et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 75.Furukawa A, et al. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J. 2009;28:440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harjes E, et al. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J Mol Biol. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holden LG, et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shandilya SM, et al. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hughes JF, Coffin JM. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: Implications for human and viral evolution. Proc Natl Acad Sci U S A. 2004;101:1668–1672. doi: 10.1073/pnas.0307885100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 81.Jaskolski M, Alexandratos JN, Bujacz G, Wlodawer A. Piecing together the structure of retroviral integrase, an important target in AIDS therapy. FEBS J. 2009;276:2926–2946. doi: 10.1111/j.1742-4658.2009.07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Espeseth AS, et al. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc Natl Acad Sci USA. 2000;97:11244–11249. doi: 10.1073/pnas.200139397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. The X-ray crystal structure reveals the basis for retroviral DNA integration and the mechanism bywhich INSTIs block DNA strand transfer activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. X-ray crystal structures of the intasome in complex with target DNA prior to and following strand transfer that define the mechanism of retroviral DNA integration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li X, Krishnan L, Cherepanov P, Engelman A. Structural biology of retroviral DNA integration. Virology. 2011;411:194–205. doi: 10.1016/j.virol.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cherepanov P, Maertens GN, Hare S. Structural insights into the retroviral DNA integration apparatus. Curr Opin Struct Biol. 2011;21:249–256. doi: 10.1016/j.sbi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 87.Valkov E, et al. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hare S, et al. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci USA. 2010;107:20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koh Y, Matreyek KA, Engelman A. Differential sensitivities of retroviruses to integrase strand transfer inhibitors. J Virol. 2011;85:3677–3682. doi: 10.1128/JVI.02541-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grobler JA, et al. Diketo acid inhibitor mechanism and HIV-1 integrase: Implications for metal binding in the active site of phosphotransferase enzymes. Proc Natl Acad Sci USA. 2002;99:6661–6666. doi: 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 2008;4:e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cherepanov P, et al. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat Struct Mol Biol. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- 93.Cherepanov P, Ambrosio ALB, Rahman S, Ellenberger T, Engelman A. From the Cover: Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Christ F, et al. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol. 2010;6:442–448. doi: 10.1038/nchembio.370. Structure-based design of LEDGIN compounds with antiviral activity. [DOI] [PubMed] [Google Scholar]
- 95.Hare S, et al. Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572) Mol Pharmacol. 2011;80:565–572. doi: 10.1124/mol.111.073189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Métifiot M, Marchand C, Maddali K, Pommier Y. Resistance to integrase inhibitors. Viruses. 2010;2:1347–1366. doi: 10.3390/v2071347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tahirov TH, et al. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465:747–751. doi: 10.1038/nature09131. The crystal structureof the Tat –P-TEFb complexsuggests it should be explored as a target for antiviral development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Daugherty MD, Liu B, Frankel AD. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol. 2010;17:1337–1442. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.DiMattia MA, et al. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci USA. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fujinaga K, et al. The ability of positive transcription elongation factor b to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J Virol. 1998;72:7154–7159. doi: 10.1128/jvi.72.9.7154-7159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bayer P, et al. Structural studies of HIV -1 Tat protein. J Mol Biol. 1995;247:529–535. doi: 10.1006/jmbi.1995.0158. [DOI] [PubMed] [Google Scholar]
- 103.Anand K, Schulte A, Vogel-Bachmayr K, Scheffzek K, Geyer M. Structural insights into the cyclin T1-Tat-TAR RNA transcription activation complex from EIAV. Nat Struct Mol Biol. 2008;15:1287–1292. doi: 10.1038/nsmb.1513. [DOI] [PubMed] [Google Scholar]
- 104.Zhou M, et al. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jain C, Belasco JG. Structural model for the cooperative assembly of HIV-1 Rev multimers on the RRE as deduced from analysis of assembly-defective mutants. Mol Cell. 2001;7:603–614. doi: 10.1016/s1097-2765(01)00207-6. [DOI] [PubMed] [Google Scholar]
- 106.Daugherty MD, Booth DS, Jayaraman B, Cheng Y, Frankel AD. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc Natl Acad Sci USA. 2010;107:12481–12486. doi: 10.1073/pnas.1007022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gheysen D, et al. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 108.Göttlinger HG, Sodroski JG, Haseltine WA. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rao Z, et al. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 113.Zhou W, Resh MD. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Saad JS, et al. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4, 5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 117.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 120.Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Strack B, Calistri A, Craig S, Popova E, Göttlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 122.Garrus JE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 123.Fisher RD, et al. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 124.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc Natl Acad Sci USA. 2008;105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhai Q, et al. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 127.Katoh K, et al. The penta-EF-hand protein ALG-2 interacts directly with the ESCRT-I component TSG101, and Ca2+-dependently co-localizes to aberrant endosomes with dominant-negative AAA ATPase SKD1/Vps4B. Biochem J. 2005;391(Pt. 3):677–685. doi: 10.1042/BJ20050398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 129.Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol. 2002;9:812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- 130.Im YJ, et al. Crystallographic and functional analysis of the ESCRT-I/HIV-1 Gag PTAP interaction. Structure. 2010;18:1536–1547. doi: 10.1016/j.str.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 132.Van Damme N, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kupzig S, et al. Bst-2/HM1, 24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 134.Andrew AJ, Kao S, Strebel K. C-terminal hydrophobic region in human bone marrow stromal cell antigen 2 (BST-2)/tetherin protein functions as second transmembrane motif. J Biol Chem. 2011;286:39967–39981. doi: 10.1074/jbc.M111.287011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hinz A, et al. Structural basis of HIV-1 tethering to membranes by the BST-2/tetherin ectodomain. Cell Host Microbe. 2010;7:314–323. doi: 10.1016/j.chom.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang H, et al. Structural insight into the mechanisms of enveloped virus tethering by tetherin. Proc Natl Acad Sci USA. 2010;107:18428–18432. doi: 10.1073/pnas.1011485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schubert HL, et al. Structural and functional studies on the extracellular domain of BST2/tetherin in reduced and oxidized conformations. Proc Natl Acad Sci USA. 2010;107:17951–17956. doi: 10.1073/pnas.1008206107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mangeat B, et al. HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 2009;5:e1000574. doi: 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dube M, et al. Antagonism of tetherin restriction of HIV-1 release by Vpu involves binding and sequestration of the restriction factor in a perinuclear compartment. PLoS Pathog. 2010;6:e1000856. doi: 10.1371/journal.ppat.1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Skasko M, et al. HIV-1 Vpu antagonizes the innate restriction factor BST-2 via lipid-embedded helix-helix interactions. J Biol Chem. 2012;287:58–67. doi: 10.1074/jbc.M111.296772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pettit SC, et al. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Briggs JA, et al. Structure and assembly of immature HIV. Proc Natl Acad Sci USA. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.de Marco A, et al. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 2010;6:e1001215. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Carlson LA, et al. Cryo electron tomography of native HIV-1 budding sites. PLoS Pathog. 2010;6:e1001173. doi: 10.1371/journal.ppat.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.von Schwedler UK, et al. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitatesviral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sticht J, et al. A peptide inhibitor of HIV-1 assembly in vitro. Nat Struct Mol Biol. 2005;12:671–677. doi: 10.1038/nsmb964. [DOI] [PubMed] [Google Scholar]
- 147.Li F, et al. PA -457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci U S A. 2003;100:13555–13560. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Keller PW, Adamson CS, Heymann JB, Freed EO, Steven AC. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J Virol. 2011;85:1420–1428. doi: 10.1128/JVI.01926-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tang C, et al. Antiviral inhibition of the HIV-1 capsid protein. J Mol Biol. 2003;327:1013–1020. doi: 10.1016/s0022-2836(03)00289-4. [DOI] [PubMed] [Google Scholar]
- 150.Kelly BN, et al. Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein. J Mol Biol. 2007;373:355–366. doi: 10.1016/j.jmb.2007.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lapatto R, et al. X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes. Nature. 1989;342:299–302. doi: 10.1038/342299a0. [DOI] [PubMed] [Google Scholar]
- 152.Navia MA, et al. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature. 1989;337:615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- 153.Wlodawer A, et al. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989;245:616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- 154.Kitchen VS, et al. Safety and activity of saquinavir in HIV infection. Lancet. 1995;345:952–955. doi: 10.1016/s0140-6736(95)90699-1. [DOI] [PubMed] [Google Scholar]
- 155.Wlodawer A, Erickson JW. Structure-based inhibitors of HIV-1 protease. Ann Rev Biochem. 1993;62:543–585. doi: 10.1146/annurev.bi.62.070193.002551. [DOI] [PubMed] [Google Scholar]
- 156.Wensing AM, van Maarseveen NM, Nijhuis M. Fifteen years of HIV Protease Inhibitors: raising the barrier to resistance. Antiviral Res. 2010;85:59–74. doi: 10.1016/j.antiviral.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 157.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure. 2002;10:369–381. doi: 10.1016/s0969-2126(02)00720-7. [DOI] [PubMed] [Google Scholar]
- 158.King NM, Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem Biol. 2004;11:1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 159.Nalam MNL, et al. Evaluating the substrate-envelope hypothesis: Structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance. J Virol. 2010;84:5368–5378. doi: 10.1128/JVI.02531-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wlodawer A. Structure-based design of AIDS drugs and the development of resistance. Vox Sang. 2002;83 (Suppl 1):23–26. doi: 10.1111/j.1423-0410.2002.tb05261.x. [DOI] [PubMed] [Google Scholar]
- 163.Burton DR, Weiss RA. AIDS/HIV. A boost for HIV vaccine design. Science. 2010;329:770–773. doi: 10.1126/science.1194693. [DOI] [PubMed] [Google Scholar]
- 164.Grigorieff N, Harrison SC. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Curr Opin Struct Biol. 2011;21:265–273. doi: 10.1016/j.sbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Flexner C. HIV drug development: the next 25 years. Nat Rev Drug Discov. 2007;6:959–966. doi: 10.1038/nrd2336. [DOI] [PubMed] [Google Scholar]