Abstract
Dietary carotenoids are absorbed in the intestine and delivered to various tissues by circulating lipoproteins; however, the mechanism underlying selective delivery of different carotenoid species to individual tissues remains elusive. The products of the Yellow cocoon (C) gene and the Flesh (F) gene of the silkworm Bombyx mori determine the selectivity for transport of lutein and β-carotene, respectively, to the silk gland. We previously showed that the C gene encodes Cameo2, a CD36 family member, which is thought to function as a transmembrane lipoprotein receptor. Here, we elucidated the molecular identity of the F gene product by positional cloning, as SCRB15, a paralog of Cameo2 with 26% amino acid identity. In the F mutant, SCRB15 mRNA structure was severely disrupted, due to a 1.4 kb genomic insertion in a coding exon. Transgenic expression of SCRB15 in the middle silk gland using the binary GAL4-UAS expression system enhanced selective β-carotene uptake by the middle silk gland, while transgenic expression of Cameo2 enhanced selective lutein uptake under the same GAL4 driver. Our findings indicate that divergence of genes in the CD36 family determines the selectivity of carotenoid species uptake by silk gland tissue and that CD36-homologous proteins can discriminate among carotenoid species.
Keywords: β-carotene, carotenoid absorption, carotenoid transport, cocoon color, lutein, positional cloning, SR-BI
Carotenoids are a group of more than 600 compounds that consist of hydrophobic pigments, with a central chain of conjugated double bonds (Fig. 1A) (1). Animals appear to be incapable of synthesizing these molecules, although they can chemically modify certain types (2). Thus, carotenoids must be acquired from dietary sources and transported to target tissues where they perform their diverse physiological functions (3–7).
Fig. 1.
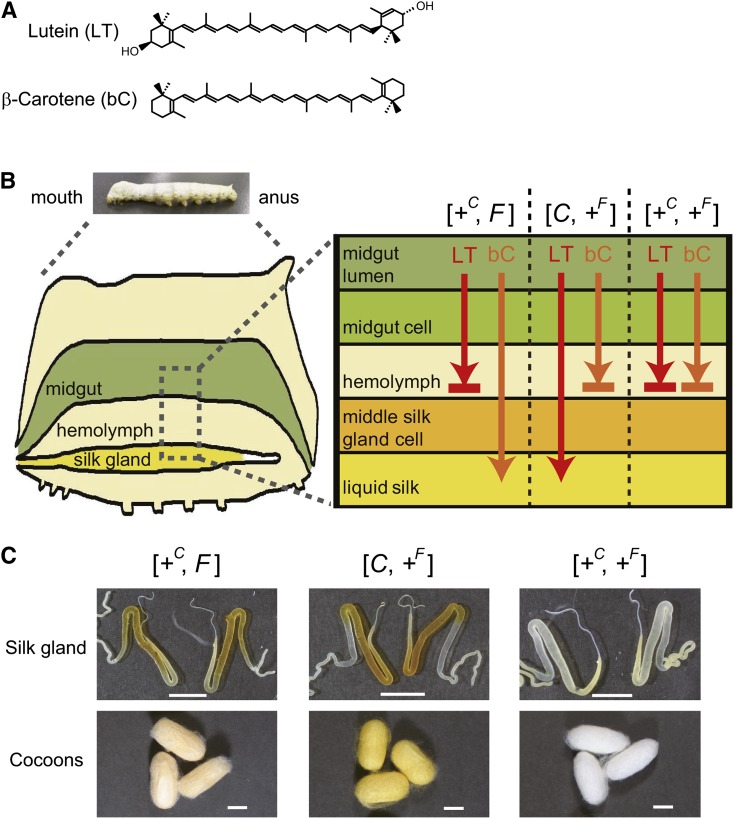
Selective transport of lutein and β-carotene by the Yellow cocoon (C) gene and the Flesh (F) gene. (A) Chemical structures of the two major carotenoids from dietary mulberry leaves, lutein (LT) and β-carotene (bC). (B) Schematic diagram of the functions of the C and F genes. +C and +F represent a recessive allele of the C and F genes, respectively. In the larvae homozygous for the +C allele ([+C, F] or [+C, +F]), lutein rarely migrates into the middle silk gland and instead accumulates in the hemolymph. Conversely, in larvae homozygous for the +F allele ([C, +F] or [+C, +F]), β-carotene rarely migrates into the middle silk gland and instead accumulates in the hemolymph. (C) Color phenotype of the silk gland and cocoons. The silk glands with [+C, F], [C, +F], and [+C, +F] genotypes were from strains c43 (+C/+C, F/F), c10 (C/C, +F/+F), and e09 (+C/+C, +F/+F), respectively. The cocoons with [+C, F], [C, +F], and [+C, +F] genotypes were from strains c43 (+C/+C, F/F), c10 (C/C, +F/+F), and w06 (+C/+C, +F/+F), respectively. Silk glands were obtained at day 0 of the wandering stage (W0). The silk glands images of [C, +F] and [+C, +F] genotypes are the same as those shown in a previous report (17). Scale bar: 1 cm.
Carotenoids are transported through the circulation in associated with lipoproteins. Interestingly, their uptake by tissues is often a selective process (i.e., individual tissues tend to acquire only specific carotenoid species). For example, the human macula in the retina acquires lutein and zeaxanthin, to the exclusion of other carotenoids, such as α-carotene, β-carotene, and lycopene (8). Selective transport of specific carotenoids is important, as molecular properties (e.g., manifesting the optical absorption spectrum, provitamin A activity, and antioxidant activity) vary among the carotenoid species. While several carotenoid-binding proteins with differential carotenoid species affinities have recently been identified (9), the molecular genetic mechanism underlying the selectivity for the transport of carotenoid species to the relevant tissue has remained elusive.
The domesticated silkworm, Bombyx mori, offers a model system to study the transport selectivity of carotenoid species to target tissues (Fig. 1B, C). Wild-type larvae feed on mulberry leaves that are rich in lutein and β-carotene. Carotenoids are absorbed in the intestine of the caterpillar, transferred to the hemolymph lipoprotein, lipophorin, and ultimately, accumulate in the middle silk gland, resulting in the formation of golden yellow cocoons containing both lutein and β-carotene (10–12). By studying spontaneous mutants obtained over the 4,000-year history of sericulture (12, 13), it was revealed that lipophorin-mediated lutein and β-carotene transport to the middle silk gland are controlled by the Yellow cocoon (C) gene (14, 15) and the Flesh (F) gene (16), respectively. The middle silk gland of the C mutant, homozygous for the recessive +C allele, has a defect in the cellular uptake of lutein, resulting in the formation of cocoons of creamier yellow (also called flesh), containing mainly β-carotene ([+C, F] in Fig. 1B, C). Conversely, the middle silk gland of the F mutant, homozygous for the recessive +F allele, has a defect in the cellular uptake of β-carotene, resulting in the formation of light yellow cocoons containing mainly lutein ([C, +F] in Fig. 1B, C). The double mutant of the C and F genes, which is homozygous for both the +C allele and the +F allele, produces white cocoons containing limited amounts of lutein and β-carotene ([+C, +F] in Fig. 1B, C). Identification and comparison of the molecular identities of the products of the C gene and the F gene is therefore expected to provide molecular genetic insights into the selectivity of transport of carotenoid species to target tissues by circulating lipoproteins.
The C gene product was previously identified as Cameo2, a transmembrane protein-encoding gene belonging to the CD36 family, which is found in species ranging from mammals to insects (17). Some CD36 family proteins, such as the mammalian SR-BI and the fruit fly NinaD, have been implicated in cellular uptake of carotenoids (3–7). SR-BI also functions as a lipoprotein receptor and facilitates cellular uptake of cholesteryl ester from high-density lipoproteins (18).
In this study, we obtained genetic evidence that the F gene encodes SCRB15, another member of the CD36 family.
EXPERIMENTAL PROCEDURES
Silkworms
Nontransgenic silkworm strains used in this study were preserved in the silkworm stock center at Kyushu University, Fukuoka, Japan, and the Genetic Resources Conservation Research Unit of the National Institute of Agrobiological Sciences, Ibaraki, Japan. The larvae were reared on mulberry leaves. The transgenic strains were produced and maintained in the Transgenic Silkworm Research Unit, National Institute of Agrobiological Sciences, Ibaraki, Japan. Larvae were reared on an artificial diet derived from mulberry leaves until the fourth instar, and on mulberry leaves during the fifth instar. The first days corresponding to the developmental stages of the fourth to fifth larval ecdysis, and wandering, a characteristic behavior with enhanced locomotory activity just prior to the commencement of the spinning of cocoons, were designated as V0 and W0, respectively. In this article, we designate the strain w06 with an F/+F genotype as “w06F” for clarity.
Mapping of the F gene using single nucleotide polymorphisms
In a previous crossing, during positional cloning of the C gene (17), we used strain c11 that is homozygous for the dominant C allele (wild-type allele) of the C gene and strain number 925 that is homozygous for the recessive +C allele (mutant allele) of the C gene. As strain c11 and number 925 were also homozygous for the dominant F allele (wild-type allele) of the F gene and the recessive +F allele (mutant allele) of the F gene, respectively, the BF1 progeny produced cocoons in which the appearance was divided into the following four colors according to the combination of the alleles of the C and the F genes: i) deep yellow, derived from the genotype [C, F] (C/+C, F/+F); ii) light yellow, derived from the genotype [C, +F] (C/+C, +F/+F); iii) flesh, derived from the genotype [+C, F] (+C/+C, F/+F); and iv) white, derived from the genotype [+C, +F] (+C/+C, +F/+F) (supplementary Fig. I).
After the visual assessment of the cocoon colors, genomic DNA was extracted from pupae in cocoons and used to map the C gene. In the BF1 individuals, there were no discrepancies between the single nucleotide polymorphism (SNP) marker pattern and cocoon color phenotype for the C gene. Subsequently, we used the BF1 progeny with the flesh-colored [+C, F] and white [+C, +F] cocoons to map the F gene. We did not use [C, F] and [C, +F] because rigorous discrimination between individuals with these genotypes was difficult.
For F gene mapping, 16 SNP markers previously reported on chromosome 6, and 11 new SNP markers were used. The PCR primers for the SNP markers are listed in supplementary Table I or were published previously (19). PCR products were treated with ExoSAP-It (USB Corp., Cleveland, OH) and subjected to direct sequencing.
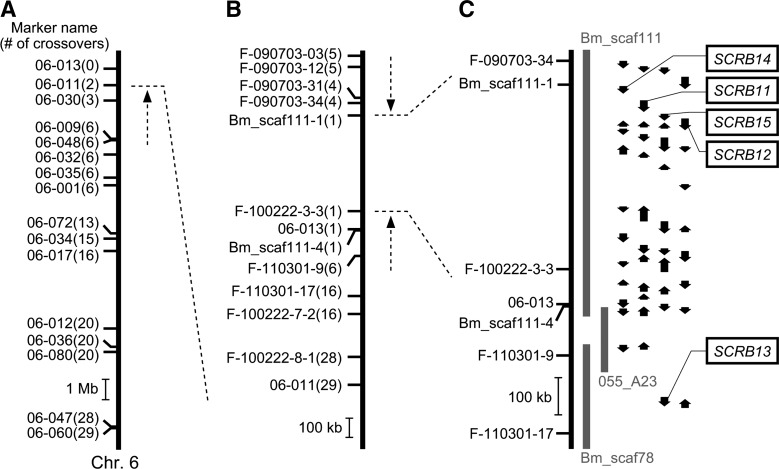
Analysis of 789 BF1 individuals by SNP genotyping indicated that 9 individuals had been misjudged by their cocoon colors; 8 BF1 individuals whose cocoons were judged to be white were in fact heterozygous for the SNP markers F-090703-03, F-100222-3-3, 06-013, F-100222-7-2, F-100222-8-1, and 06-011; and 1 BF1 individual whose cocoon had been judged to be flesh-colored was in fact homozygous for these markers (supplementary Table II). We therefore omitted these 9 individuals from Fig. 2 for the sake of simplicity. Even if these 9 individuals were taken into consideration, logarithm10 of the odds (LOD) scores for the SNP markers Bm_scaf111-1, F-100222-3-3, 06-013, and Bm_scaf111-4 were calculated to exceed 200, strongly supporting the linkage between these SNP markers and the F locus.
Fig. 2.
Mapping of the F gene on chromosome 6. (A) Rough mapping using 58 individuals. Small horizontal lines on the vertical bars of chromosome 6 denote the positions of crossover events, showing the name of the SNP marker and the number of recombinants. (B) Fine mapping using 789 individuals. Nine individuals that were considered to have been misjudged for their cocoon colors were omitted from the number of recombinants (see Experimental Procedures for details). (C) Physical map of chromosome 6 near the F locus harboring the predicted F gene. Vertical arrows indicate the orientation and relative size of the putative genes predicted by the China gene model in the silkworm genome database KAIKObase (20). Sequence of BAC clone 055_A23 was determined in this study to fill the gap between Bm_scaf111 and Bm_scaf78. No sequences homologous to Cameo2 were found in 055_A23. SCRB11–15 were named in previous reports (17, 49).
To determine the sequence of BAC clone 055_A23, derived from the genomic DNA of strain p50, shotgun sequencing data obtained with a Roche 454 DNA sequencer by Hokkaido System Science Ltd. (Sapporo, Hokkaido, Japan) were combined with sequence data from the silkworm genome database KAIKObase (URL: sgp.dna.affrc.go.jp/KAIKObase/) (20).
Quantification of transcripts by quantitative PCR
Total RNA was isolated from tissues as described previously (17). Single-stranded cDNAs from various tissue samples were synthesized from total RNAs with Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) and oligo-dT primer and were then treated with RNaseH (Takara, Kyoto, Japan). Quantification of transcripts was carried out by quantitative PCR using the cDNAs as templates with LightCycler FastStartDNA MasterPLUS SYBR Green I reagent (Roche, Mannheim, Germany) and a LightCycler DX400 thermocycler (Roche).
The primer pairs used for the detection of SCRB14, SCRB11, SCRB15, SCRB12, SCRB13, and rpL3 were Primer14-7 (5′-TGAAGAAGCTGAAGAAATGAGGTA-3′) and Primer14-6 (5′-AAGGCTAGTTGAGGAAAGTAGGAG-3′), Primer11-3 (5′-GAAGCACCAGTAATCAGGTTTACAC-3′) and Primer11-2 (5′-GGTTCCATTAAAGTCGACACAGTC-3′), Primer15-3 (5′-GAGAAGATGTTCGAGAACAATCTTTGC-3′) and Primer15-2 (5′-TCGAACAGTTTTGGATCAGCATCC-3′), Primer12-3 (5′-CCATAACGACCATCGATGTATACAG-3′) and Primer12-2 (5′-TTCCCACCCTCATTCTGTAAATGG-3′), Primer13-5 (5′-AGATCCACCTCTTCAACTACACAA-3′) and Primer13-4 (5′-TACCCTCATCGACTGTTATGTTTT-3′), and Primer-rpL3-real-cDNA1 (5′-TTCCCGAAAGACGACCCTAG-3′) and Primer-rpL3-real-cDNA2 (5′-CTCAATGTATCCAACAACACCGAC-3′), respectively.
Serial dilutions of plasmids containing the cDNA sequences were used as copy-number standards. Transcript levels of the genes other than rpL3 were normalized to the level of the rpL3 transcript in the same samples, as described previously (21).
Comparison of SCRB11, SCRB15, and SCRB12 cDNA sequences between individuals carrying the F allele and those carrying the +F allele
For SCRB11 and SCRB12, full-length cDNA sequences from strain p50 were obtained from the KAIKObase database. SCRB11 and SCRB12 were amplified from the middle silk gland of each of strains c44 (F/F) and w06 (+F/+F) via reverse transcription (RT)-PCR and were then directly sequenced. The PCR primers used for SCRB11 were Primer11-5 (5′-GCAATCATCTCACAACCCTTTACTG-3′) for the 5′-UTR and Primer11-4 (5′-GAGGCACTATCTAAGGCCAGGTTTA-3′) for the 3′-UTR. For SCRB12, we used Primer12-7 (5′-GCTAGCTAGTGCGTTTTCAATTCTA-3′) for the 5′-UTR and Primer12-4 (5′-GCGAGTACACAGACTGACAAACTGA-3′) for the 3′-UTR.
For SCRB15, the cDNA sequence containing the full-length coding sequence from the F allele was determined from strain c44 (F/F). First, a partial SCRB15 cDNA fragment was amplified from the middle silk gland via RT-PCR with the primer pair Primer15-1 (5′-GATAAGAACGTACACTATCGCATGG-3′) and Primer15-2 (5′-TCGAACAGTTTTGGATCAGCATCC-3′), both of which were designed based on the predicted SCRB15 fragment sequence (BGIBMGA013438 in the China gene model in the KAIKObase). The sequence of amplified fragments was determined by direct sequencing. Subsequently, four more primers, Primer15-11 (5′-CACCTATGAAGGCATGGCATACCCACCG-3′), Primer15-13 (5′-CACGGCAATAAGACGTCGGAATACGGCC-3′), Primer15-8 (5′-CCTTTAGGGCAAGCCCCCTTAACATCGC-3′), and Primer15-10 (5′-GCCCCTTTGATAATCCCATGGGCAAACC-3′), were designed based on the determined partial sequence.
The 5′- and 3′-ends of the SCRB15 sequence were obtained via rapid amplification of cDNA ends (RACE) using a SMART RACE cDNA amplification kit (Clontech, Mountain View, CA) with Primer15-10 for the first 5′-RACE product, Primer15-8 for the nested 5′-RACE product, Primer15-11 for the first 3′-RACE product, and Primer15-13 for the nested 3′-RACE product. The determined sequence was then combined to obtain the full-length SCRB15 cDNA sequence from strain c44 (F/F).
SCRB15 cDNA sequences from strain w06 (+F/+F) were amplified from the middle silk gland via RT-PCR with two primer pairs, Primer15-5 (5′-GATATATGAATTTTCGCAAAAGAGACTAAG-3′) for the 5′-UTR and Primer15-12 (5′-TCAATGCAGTCAATCCTTAC-3′) for the 3′-UTR, and Primer15-23 (5′-ATAGGGAAGCCGGAGTAATGAGGTA-3′) and Primer15-22 (5′-CAGACTCAGGGTGGAGTCGAGTAT-3′) for the coding exon, and directly sequenced. The SCRB15 cDNA sequence from strain w06 (+F/+F) was identical to that from strain c44 (F/F) except for a splicing out of exon 6 or a 1.4 kb insertion into exon 6 (for details, see Results). The SCRB15 cDNA sequence from strain e09 (+F/+F) was amplified via RT-PCR with the primer pair Primer15-5 and Primer15-12, and directly sequenced. The SCRB15 cDNA sequence from strain e09 (+F/+F) was identical to that from strain w06 (+F/+F).
Northern blotting analysis
A 32P-labeled riboprobe was synthesized from a plasmid containing both the 3′ part (621 bp) of the coding sequence and the 5′ part (39 bp) of the 3′-UTR of SCRB15. No repetitive sequence was found in the insert of this plasmid. Total RNA was electrophoresed on 1% agarose gels containing formaldehyde and was then transferred onto Hybond N+ membrane (GE Healthcare UK, Buckinghamshire, England). Hybridization was performed with Ultrahyb (Ambion, Austin, TX).
SCRB15 genomic sequence analysis
As a part of the SCRB15 genomic sequence, including exons 4–6, was absent from the publicly available silkworm genome sequence data (22), we determined the sequence of genomic fosmid clone RO0299-D04, derived from strain p50 by a shotgun method (23, 24), to reveal the overall genomic structure of SCRB15.
To compare the genomic sequence of exon 6 between the F and the +F alleles, the genomic sequence of each of strains c44 (F/F), w06 (+F/+F), and e09 (+F/+F) was amplified via PCR with the primer pair Primer15-25 (5′-ATCGCCCGTCTTCATTCGTAATATC-3′) and Primer15-20 (5′-TAAAATATACGAGACCAGGCACCAA-3′) and then directly sequenced. The sequences amplified from strain w06 (+F/+F) and strain e09 (+F/+F) were identical.
For genotyping by PCR, the following primers were used in addition to those described above: Primer15-18 (5′-ATTCTTCAGGAACAAACACAGATCG-3′), Primer15-21 (5′-AATTTCGTCTTGCCAATGGAAGTAT-3′), Primer15-24 (5′-CGTAACACCAAGGTTTTGGAAGGAA-3′), Primer15-25 (5′-ATCGCCCGTCTTCATTCGTAATATC-3′), Primer15-30 (5′-GAATGAAGACGGGCGATGAT-3′), and Primer15-31 (5′-TGCCTGGTCTCGTATATTTTAACAC-3′).
For Southern blotting analysis, a digoxigenin-labeled DNA probe for SCRB15 exon 6 was amplified from a cDNA-containing vector by PCR using the primer pair Primer15-25 and Primer15-20. The hybridization and detection procedures with this probe were described previously (25).
Silkworm transgenesis
Transgenic SCRB15 expression using the binary GAL4/upstream-activating sequence (UAS) system (26) was performed as described previously (17). To construct the effector vector, pBacMCS[UAS-SCRB15-3xP3-EGFP], SCRB15 was amplified by RT-PCR from the middle silk gland of strain c44 (F/F) with Primer15-17 (5′-ATGCACTAGTTTTTCGCAAAAGAGACTAAG-3′) for the 5′-UTR and Primer15-14 (5′-ATGCACTAGTCAATGCAGTCAATCCTTACA-3′) for the 3′-UTR, both of which contain an SpeI site. The amplified fragment was digested with SpeI and ligated into a pBacMCS[UAS-3xP3-EGFP] vector (27), which was previously digested with BlnI. For the effector strains, the effector construct and the helper plasmid pHA3PIG (28) were injected into preblastoderm embryos of the strain w1-pnd-925, a nondiapausing strain with the phenotype of yellow hemolymph and white cocoons (17). The existence of the transgene was identified by the fluorescence in the eye, EGFP for UAS-SCRB15 and DsRed2 for Ser1-GAL4 (29). Individuals exhibiting colorless hemolymph in the larval stage, which had colorless silk glands and produced white cocoons, were not used to examine the color phenotype or carotenoid composition of the middle silk gland and cocoons.
Analysis of carotenoid composition
Larval hemolymph was collected into an Eppendorf tube, frozen in liquid nitrogen, and stored at –70°C until use. Thawed hemolymph was centrifuged at 800 g for 5 min to remove hemocytes. Twenty microliters of the supernatant was transferred into a glass tube with 3.0 ml of dimethyl sulfoxide (DMSO), mixed, and sonicated at 50–60°C for 30 min. The middle silk gland was frozen in liquid nitrogen and broken into fine pieces; some of these pieces (20–170 mg) were then transferred into a glass tube with 3.0 ml of DMSO, mixed, and sonicated at 50–60°C for 30 min. A cocoon (6–14 mg) was cut into small pieces of less than 1 mm width using scissors; these pieces were then transferred into a glass tube with 2.0 ml of DMSO, mixed, and sonicated at 50–60°C for 30 min. After collection of the extracts, residual cocoon pieces were extracted twice more with 2.0 ml of DMSO, and the resulting three extracts were pooled. After filtration through a polyvinylidene difluoride membrane, the extracted carotenoid composition was analyzed by high-performance liquid chromatography (HPLC). A reverse-phase column (DOCOSIL SP100 [6 × 250 mm]; Senshu Kagaku, Tokyo, Japan) was used under the following conditions: mobile phase, isocratic solvent, 70% methanol, and 30% ethyl acetate; room temperature; flow rate, 2 ml/min; and detection, 445 nm. Carotenoid standards were purchased from Sigma (St. Louis, MO).
Data deposition
The cDNA sequence of SCRB11 from c44 (F/F), and of that from w06 (+F/+F), the cDNA sequence of SCRB12 from c44 (F/F), and of that from w06 (+F/+F), the cDNA sequence of SCRB15 from strain c44 (F/F), the sequence of the insertion in exon 6 of SCRB15 from strain w06 (+F/+F), the sequence of the BAC clone 055_A23, and the sequence of genomic fosmid clone RO0299-D04 have been deposited in GenBank under accession nos. AB742534, AB742535, AB742536, AB742537, AB742538, AB742539, AB742540, and AB742541, respectively.
RESULTS
Mapping of the F gene on chromosome 6 of the silkworm
To specify a candidate genomic region for the F gene, we performed genetic linkage analysis using SNP markers (19) and the B. mori genome sequence (22). Using 58 BF1 individuals, the F locus was roughly mapped on chromosome 6, on which the F gene lies (30, 31). The F-linked region was narrowed to an end of chromosome 6, bounded by the SNP marker 06-011 (Fig. 2A).
Next, novel primer sets were designed and fine mapping was performed with 789 BF1 individuals. During the fine mapping, a BAC genomic DNA clone, 055_A23, which bridges the gap between the two scaffolds, Bm_scaf111 and the Bm_scaf78, was sequenced since the F-linked region appeared to occur in this area. Consequently, the F-linked region was narrowed down to a 499 kb region between two SNP markers, Bm_scaf111-1 and F-100222-3-3, both of which were on the same scaffold, Bm_scaf111 (Fig. 2B and supplementary Table III). Twenty-six genes were predicted to lie within the narrowed region of genomic DNA, based on the China gene model of the silkworm genome database, KAIKObase (20) (Fig. 2C). Among these genes, four (i.e., SCRB14, SCRB11, SCRB15, and SCRB12) belong to the CD36 gene family; that is, they are paralogs of Cameo2. The function of the F gene is similar to that of the C gene, despite its unique carotenoid species selectivity. Therefore, we considered these four genes to be possible F gene candidates. No other Cameo2 paralogs, other than SCRB13, were found within the genomic sequence of chromosome 6, including the BAC clone 055_A23 (Fig. 2C).
Expression analysis of Cameo2 paralogs near the F locus
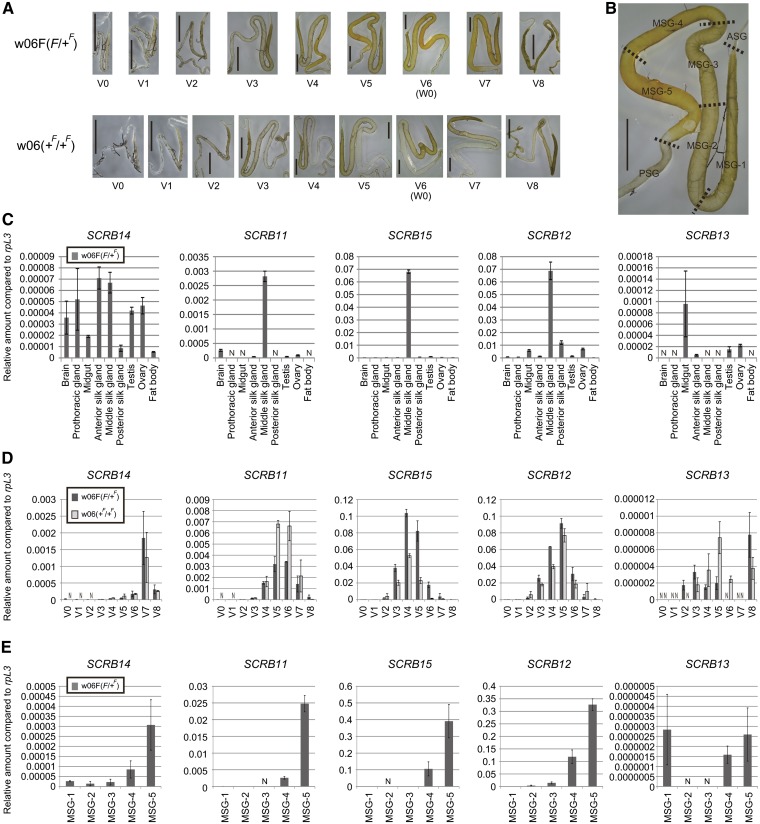
Cellular uptake of β-carotene in the middle silk gland, associated with the F gene, occurs in a spatiotemporally controlled manner (10) (Fig. 3A, B). To examine its relationship to β-carotene uptake, we examined the expression profiles of the F gene candidates with quantitative RT-PCR. SCRB13, which lies outside of the F-linked region, was also included in the analysis for comparison. SCRB11, SCRB15, and SCRB12 exhibited a middle silk gland-specific expression pattern in final instar larvae possessing the F allele, the dominant wild-type allele of the F gene (Fig. 3C). Expression levels of these three genes in the middle silk gland of the strain with the F allele were higher in the mid-stage of the final instar when β-carotene was extensively acquired by the middle silk gland (Fig. 3D). In the posterior portion of the middle silk gland, within which β-carotene mainly accumulates, the highest expression of these three genes was noted (Fig. 3E). In contrast, SCRB14 and SCRB13 expression levels were not specific to the middle silk gland (Fig. 3C) and were not well correlated with developmental changes in β-carotene uptake by the middle silk gland (Fig. 3D).
Fig. 3.
Expression analysis of the F gene candidates by quantitative RT-PCR. (A) Developmental changes in carotenoid pigmentation associated with the F allele in the silk gland during the fifth larval instars. From day 6 of the fifth larval instar [V6 (W0)], larvae spat silk for cocoon formation, resulting in a decrease of carotenoid pigmentation and degradation of the silk gland. The silk gland image from strain w06 (+F/+F) at day 7 of the fifth larval instar (V7) was made by combining two photographs. Scale bar: 0.5 cm. (B) Spatial difference in carotenoid pigmentation of the silk gland. The silk gland depicted from strain w06F (F/+F) at the V5 stage is the same as in (A). Definition of regions in the silk gland is the same as described previously (17): ASG, anterior silk gland; MSG-1–5, middle silk gland-1–5; PSG, posterior silk gland. (C–E) Quantitative RT-PCR analysis of expression levels of the F gene candidates, SCRB14, SCRB11, SCRB15, and SCRB12. SCRB13, which was located outside of the delimited region for the F locus (Fig. 2C), was also included in the analysis. (C) Tissue distribution of the expression of the F gene candidates on day 5 of the fifth larval instar (V5). (D) Developmental expression profile in the middle silk gland of the fifth instar larvae. (E) Spatial expression analysis of the middle silk gland at the V5 stage. The vertical axis shows the transcript levels of each gene normalized to the level of the reference gene ribosomal protein L3 (rpL3) (mean, SE; n = 3). As the efficiency of reverse transcription could differ among target genes, the value of the vertical axis of a particular gene could not necessarily be compared with the value of that of the other genes. Male larvae were used for the data in this figure, except for ovary data. N, not detected.
Genomic insertion in a coding exon severely affects the SCRB15 mRNA structure in the F mutant
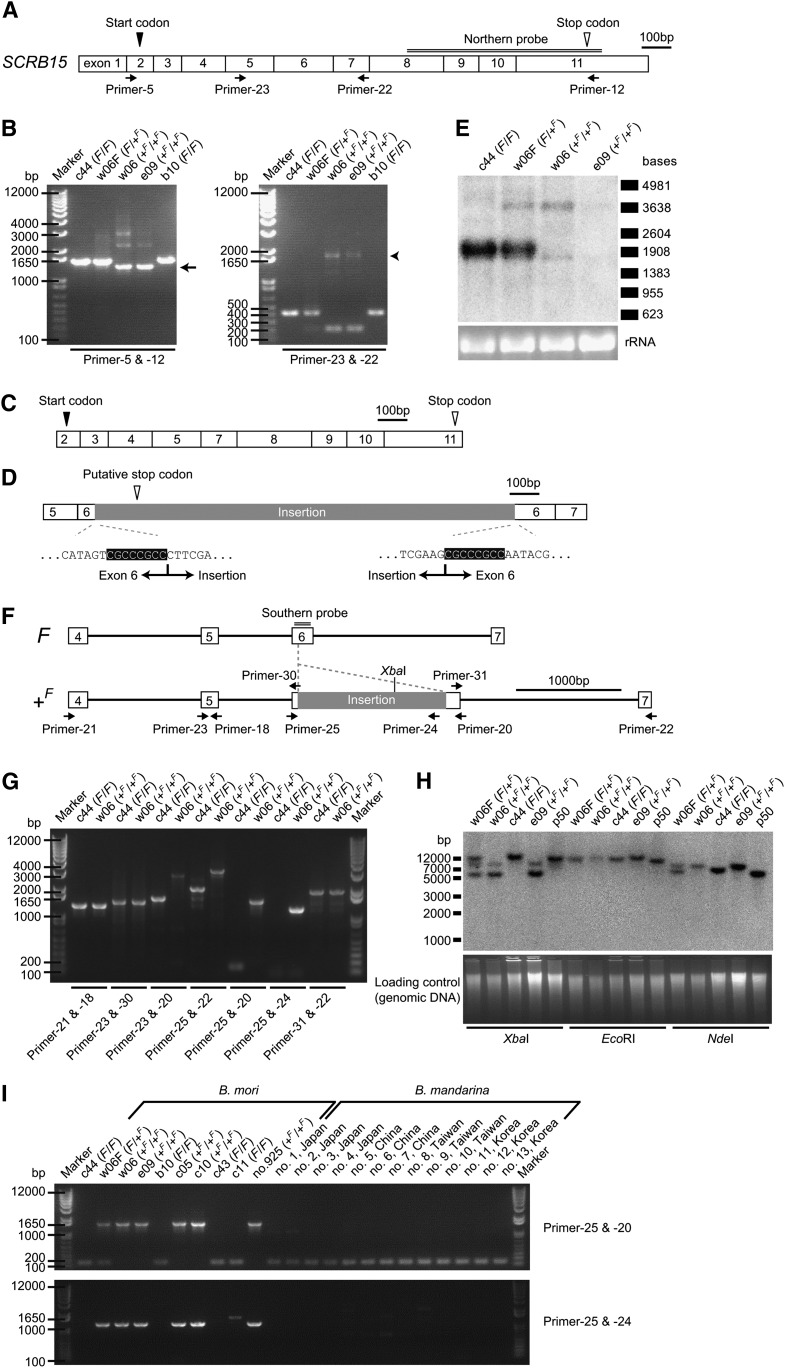
Next, the coding sequences of SCRB11, SCRB15, and SCRB12 were compared between the chromosome containing the F allele and that containing the +F allele by RT-PCR and sequencing. SCRB11 and SCRB12 were well conserved and did not show insertions, deletions, or premature stop codons, but they demonstrated four nonsynonymous substitutions in SCRB11 {i.e., Y19N [from tyrosine at amino acid position 19 in strain c44 (F/F) to asparagine in strain w06 (+F/+F)], I116N, D305E, and V454I}. In contrast, RT-PCR revealed that the SCRB15 coding sequence lengths were significantly different between these chromosomes (Fig. 4A, B). Sequencing of the smaller PCR product from strain w06 (+F/+F) (Fig. 4B, arrow) revealed the absence of 201 base pairs in the coding sequence (Fig. 4C). These 201 base pairs perfectly corresponded to SCRB15 exon 6. Sequencing of the larger PCR product (Fig. 4B, arrowhead) identified an insertion of 1,400 base pairs in exon 6 (Fig. 4D). Although the insertion was not homologous to known transposable elements (32), a duplication of eight nucleotides was found at the insertion site (highlighted in Fig. 4D), which could be a sign of an insertion event. Northern blotting analysis (Fig. 4E) supported the structural difference in SCRB15 between the chromosome bearing the F allele and that bearing the +F allele and revealed a decrease in SCRB15 expression in strains with the +F allele, consistent with the results of quantitative RT-PCR analysis (Fig. 3D).
Fig. 4.
Genomic insertion in an SCRB15 coding exon of the F mutant. (A) Schematic SCRB15 mRNA structure from strain c44, which is homozygous for the F allele. Exon boundaries were determined based on the genomic sequence database (for exons 1–3 and 7–11) (20) and our own genomic sequence data (for exons 4–6). (B) RT-PCR analysis of the SCRB15 mRNA structure from the middle silk gland. PCR primer positions were as depicted in (A). The length of PCR products differed between the F and +F allele. (C) The SCRB15 mRNA structure obtained from a shorter band from strain w06 (+F/+F) [arrow in (B)]; in this case, exon 6, consisting of 201 base pairs, had been spliced out. (D) Another mRNA structure of SCRB15 obtained from a longer band from strain w06 (+F/+F) [arrowhead in (B)]. An insertion of 1.4 kb was present in exon 6. Duplication of eight bases occurred at the insertion site (highlighted). A putative stop codon was present in this insertion. (E) Northern blotting analysis of SCRB15 in the middle silk gland on day 3 of the fifth larval instar [for strains c44 (F/F) and e09 (+F/+F)] or day 4 of the fifth larval instar [for strains w06F (F/+F) and w06 (+F/+F)]. For both strains w06 (+F/+F) and e09 (+F/+F), a longer band, which may correspond to the mRNA containing the 1.4 kb insertion, and a shorter band, which may correspond to the mRNA lacking the 201 base pair exon 6, were detected. The expression of the transcript from strain e09 (+F/+F) was lower than that of the transcript from strain w06 (+F/+F), which was confirmed by quantitative RT-PCR analysis (data not shown). (F) Schematic genomic structure of SCRB15. The structure, except the insertion in exon 6, was based on the data from strain p50. (G) Genotyping of SCRB15 by genomic PCR. Positions of PCR primers are as illustrated in (F). The intron between exons 5 and 6 in both strain c44 and w06 would be longer than that in strain p50. (H) Southern blotting analysis of SCRB15. Genomic DNA completely digested with XbaI, EcoRI, or NdeI was used. The position of the Southern probe is double-lined in (F). The genomic insertion in the +F allele contained one XbaI recognition site, as indicated in (F). No EcoRI or NdeI recognition sites were found within the insertion. Predicted XbaI fragment sizes were 11.6 kb for the F allele and 7.7 kb (weaker) and 5.3 kb (stronger) for the +F allele. Predicted EcoRI fragment sizes were 10.9 kb and 12.3 kb for the F and +F alleles, respectively. Predicted NdeI fragment sizes are 6.4 kb and 7.8 kb for the F and +F alleles, respectively. (I) Genotyping of SCRB15 by genomic PCR in multiple strains of B. mori and multiple individuals of B. mandarina, the putative wild ancestor of B. mori (33). Details of the geographical location of the sampling area of B. mandarina are as described previously (50). The different band from strain c11 obtained with Primer-25 and Primer-24 was an artifact unrelated to SCRB15 that was amplified with Primer-24 as both sense and antisense primers (data not shown).
The genomic sequence of SCRB15 was also compared between the F and +F allele (Fig. 4F–H); this revealed that the same insertion occurred in the genomic sequence of exon 6 of the +F allele. Thus, likely due to this genomic insertion, SCRB15 exon 6 was either spliced out (Fig. 4C) or transcribed with the insertion (Fig. 4D), resulting in the absence of 67 amino acids or an unusual stop codon contained in the insertion sequence, respectively, in the F mutant.
Genotyping of SCRB15 in multiple strains of B. mori and multiple individuals of B. mandarina, the putative wild ancestor of B. mori (33), indicated that the insertion occurred in every +F allele-bearing individual from different strains of B. mori, but it was absent in B. mandarina, suggesting a common and single origin of the +F allele (Fig. 4I). This origin could be associated with the previous categorization of SCRB15 (termed BGIBMGA013438) into the 354 protein-coding genes of the silkworm that represent good candidates for artificial selection during silkworm domestication, identified from a whole-genome analysis of approximately 16 million SNPs (34).
Bioinformatic characterization of the SCRB15 sequence
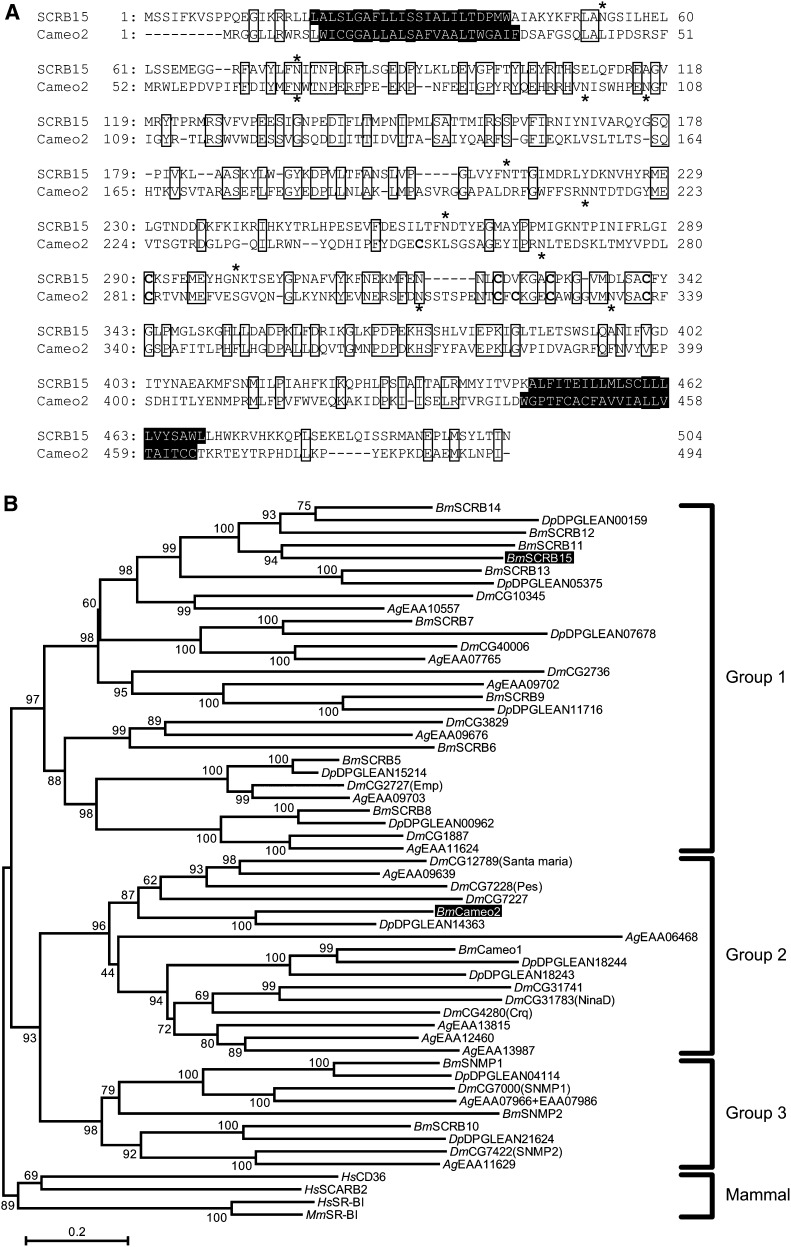
SCRB15 was predicted to encode a 57.6 kDa protein composed of 504 amino acids; glycosylation at five N-glycosylation consensus sites was expected to increase the molecular weight of the protein (Fig. 5A). Its amino acid identity with Cameo2 was 26%. Identical amino acid residues between SCRB15 and Cameo2 were dispersed over the entire sequence, but there were fewer in the central section (residues 215–289 in SCRB15). TMHMM software (35) analysis suggested that SCRB15 comprises a large extracellular loop, anchored to the plasma membrane on each side by transmembrane domains adjacent to short cytoplasmic N-terminal and C-terminal domains. The putative molecular mass, transmembrane topology, and multiple glycosylation sites of SCRB15 are common to members of the CD36 family (36). The theoretical isoelectric point deduced from the primary amino acid sequences was somewhat different for the predicted extracellular loop of SCRB15 (pH 7.8) and that of Cameo2 (pH 5.0). While our previous analysis using the SignalP 3.0 program suggested that the first N-terminal helices of some of CD36 family proteins, including Cameo2, represented a signal peptide (17), analysis using the SignalP 4.0 program (37), a new software version designed to discriminate between signal peptides and transmembrane regions, suggested that the first N-terminal transmembrane helices of SCRB15 and Cameo2 are anchored to the membrane and are not signal peptides.
Fig. 5.
Putative SCRB15 amino acid sequence. (A) Alignment of putative amino acid sequences of SCRB15 and Cameo2. Identical amino acid residues are boxed. Transmembrane helices predicted by TMHMM version 2.0 (35) are highlighted. N-Glycosylation consensus sites (N-X-S/T) and cysteine residues in the putative extracellular region, common features in CD36-related genes (36), are indicated by asterisks and bold type, respectively. (B) A neighbor-joining tree for SCRB15 and other homologs from insects and mammals generated using MEGA5 software (51). SCRB15 and Cameo2 are highlighted. The first two characters of the gene names represent their species: Bm, B. mori; Dm, D. melanogaster; Ag, Anopheles gambiae; Dp, Danaus plexippus; Hs, Homo sapiens; and Mm, Mus musculus. Numbers at nodes indicate the percentage of bootstrap values of 10,000 replicates. Groups are based on previous reports (39, 40). Although 21 homologs were found in the gene database of Danaus plexippus (38), only 11 of those more than 368 amino acids long were included in this tree; the other 10 homologs were shorter than 288 amino acids. DPGLEAN04114 of Danaus plexippus (accession number: EHJ78189) is composed of 1,801 amino acids, while the other CD36 homologs are approximately 500 amino acids. DPGLEAN04114 consists of an aldehyde oxidase-homologous sequence at the N-terminus and a CD36-homologous sequence, closely related to SNMP1, at the C terminus. Both aldehyde oxidase (52) and SNMP1 (53–55) are implicated in pheromone detection in insect olfactory neurons.
A phylogenetic tree of CD36 protein family members from mammals and insects, including the monarch butterfly Danaus plexippus, whose whole genome was recently reported (38), was constructed based on their primary amino acid sequences (Fig. 5B). As shown in previous studies (39, 40), the insect proteins could be divided into three groups, while mammalian proteins formed a single, distinct group. SCRB15 and Cameo2 were somewhat removed from each other and fell into groups 1 and 2, respectively. SCRB11-15 formed a Lepidoptera-specific clade in group 1, suggesting that a small expansion may have occurred in the Lepidopteran lineage.
Enhancement of β-carotene selective uptake into the middle silk gland by transgenic expression of SCRB15
To verify the function of SCRB15 as a product of the F gene, we examined the restoration of β-carotene accumulation in the middle silk gland after transgenic expression of the SCRB15 gene in a strain with the phenotype of the F mutant, using the binary GAL4/UAS system (26). Following transgenic expression of SCRB15 in the middle silk gland by the Ser1-GAL4 driver (29), pigmentation was observed in the middle silk gland (Fig. 6A). Cocoon pigmentation was also restored, although the color was not as intense as those of some nontransgenic strains (Fig. 6B).
Fig. 6.
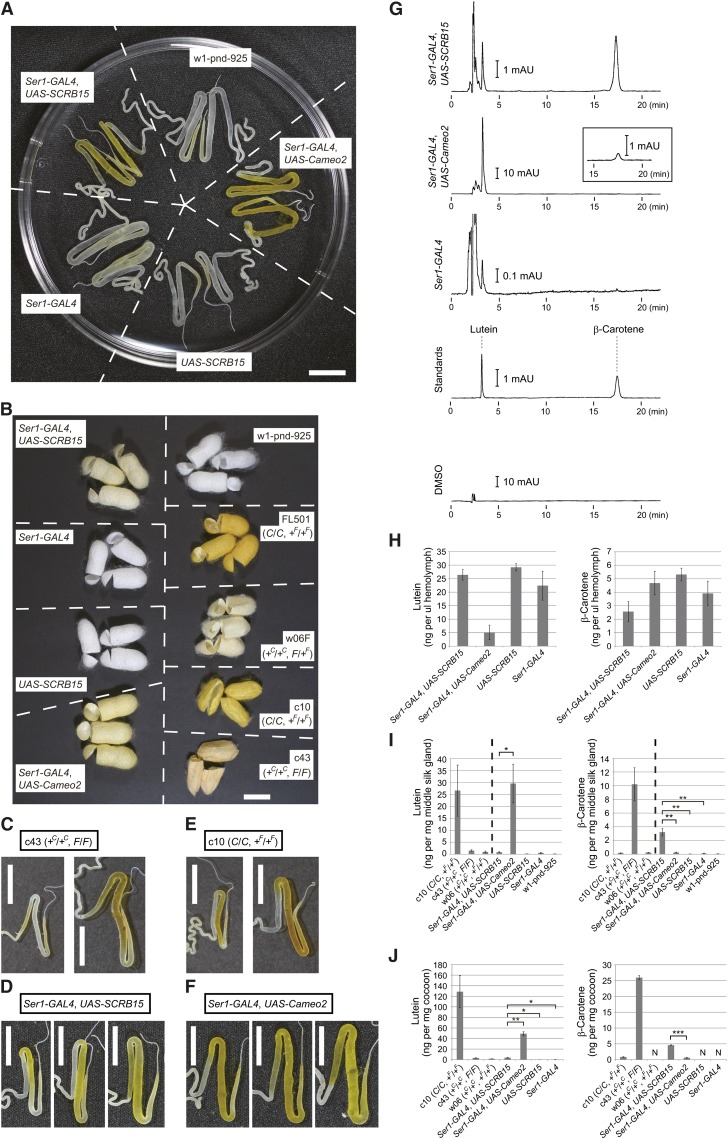
Restoration of selective β-carotene uptake by transgenic expression of SCRB15. (A) Silk glands of the Ser1-GAL4/UAS-SCRB15 line, which expresses SCRB15 in the middle silk gland from the binary GAL4/UAS system (26) with a Ser1-GAL4 driver (29). The Ser1-GAL4 line, the UAS-SCRB15 line, and the w1-pnd-925 line are control lines that do not express SCRB15. The Ser1-GAL4/UAS-Cameo2 line (17) expresses Cameo2 and selectively accumulates lutein in the middle silk gland. The stage was at one or two days prior to W0. (B) Cocoon colors of the transgenic lines and nontransgenic strains. For nontransgenic strains, the genotypes of the C and the F genes are shown in parentheses. (C–F) Distribution of carotenoid pigmentation in the middle silk gland of the transgenic lines and nontransgenic strains during the last larval instar. The later and larger silk gland is shown on the right in each panel. (G) Representative charts of the reverse-phase HPLC analysis of carotenoid composition of the middle silk gland in the transgenic larvae, at one or two days prior to W0. Detection was performed at 445 nm. The inset in the Ser1-GAL4/UAS-Cameo2 line is a magnification near the β-carotene peak. Most peaks eluting before lutein were considered to be due to the DMSO used for carotenoid extraction. (H–J) Lutein and β-carotene concentration in the hemolymph (H), the middle silk gland (I), and cocoons (J) (mean, SE; n = 3–5 individuals). The stage of the hemolymph and the middle silk gland was at one or two days prior to W0, except for strains c10 (W0), c43 (W0), and w06 (W0). Statistical significance (* P < 0.05; ** P < 0.01; *** P < 0.00005) was analyzed using Student t-test. The cocoon color of strain w06 (+C/+C, +F/+F) is shown in Fig. 1C. The late and bigger silk gland images in (C) and (E) are the same as those in Fig. 1C. Scale bar: 1 cm.
We next focused on the region of pigmentation in the middle silk gland of the last larval instar. As mentioned before, pigmentation in the larvae bearing the F allele commences in the posterior part and spreads into the middle of the gland (Figs. 3A and 6C) (10), which likely reflects the expression pattern of SCRB15 (Fig. 3E) and migration of liquid silk toward the anterior part of the silk gland. In contrast, pigmentation of the transgenic larvae, in which the SCRB15 expression was driven by Ser1-GAL4, commenced from the middle part and spread into the posterior part of the gland (Fig. 6D). This pattern was concordant with the property of the Ser1-GAL4 driver as monitored using the UAS-EGFP gene (29). On the other hand, the pigmentation pattern in larvae bearing the C allele commences from the middle part of the gland but spreads into the posterior region only minimally (Fig. 6E) (10), which likely reflects the middle-specific expression pattern of Cameo2 (17). In the transgenic larvae, in which Cameo2 expression was driven by Ser1-GAL4, pigmentation commenced from the middle and spread into the posterior part (Fig. 6F) (17), which is similar to what was observed in the SCRB15 transgenic larvae but not larvae bearing the F or C allele. Overall, the pigmentation seemed to occur where SCRB15 or Cameo2 were expressed. These data suggest that carotenoid uptake by SCRB15 or Cameo2 do not require cofactors of which the expression is restricted to limited regions in the middle and posterior parts of the middle silk gland. It should be noted that the carotenoid-binding protein (CBP), which is an obligate cofactor of the products of both the C gene and the F gene for carotenoid transport into the middle silk gland (10, 12), is expressed from the middle to the posterior part of the middle silk gland (17, 41).
Carotenoid content of larval hemolymph, middle silk gland, and cocoons of the transgenic lines were analyzed by HPLC with a reverse-phase column (Fig. 6GndashJ). A decrease in β-carotene, but not lutein content, was observed in hemolymph of transgenic larvae expressing SCRB15 (Fig. 6H). This may be due to selective uptake of β-carotene by the middle silk gland (Fig. 6I). Cocoons produced by the transgenic larvae expressing SCRB15 selectively accumulated β-carotene (Fig. 6J). The β-carotene quantities did not reach the same level as in a strain bearing the F allele, which is consistent with observations regarding the color intensity of the cocoons (Fig. 6B).
From the enhancement of selective β-carotene uptake into the middle silk gland and cocoons, we conclude that the F allele encodes SCRB15.
DISCUSSION
Regardless of the essential roles of carotenoids in animals, handling of carotenoids in the body requires special processes due to its water insolubility. The molecular events involved in the transport of any carotenoid from the gut lumen to the gut epithelial cells, to the blood, and to its target cells are not yet understood. In B. mori, several mutants with altered cocoon colors are defective in one of the steps involved in the transport of carotenoids from the midgut lumen to its target tissue (the middle silk gland). We have been characterizing carotenoid transport at the molecular level using the B. mori cocoon color mutants (12, 17).
In this study, we identified the F gene responsible for control of the selective cellular uptake of β-carotene as SCRB15, following the identification of the C gene that controls selective cellular uptake of lutein (17). A homology search revealed that SCRB15 belongs to the CD36 gene family, which includes Cameo2, encoded by the C gene. Functional differentiation after gene duplication therefore likely facilitated differences in selectivity for carotenoid species. To date, it has been shown that members of the CD36 family are critical for a large variety of biological activities in species from mammals to insects: these activities include angiogenesis, viral and bacterial recognition by the immune system, transport of lipids from cholesteryl ester to fatty acids, and taste and olfactory perception (42). Our study indicates that this family is also critical for discrimination of chemical structures that differ only marginally, such as carotenoids in the silkworm (Fig. 1).
A recent report indicated that the mammalian CD36 was involved in both lycopene and lutein uptake by adipocytes (43). CD36 homologs may not always discriminate among carotenoid species strictly.
Carotenoids are carried in the hemolymph by lipophorin, a high-density lipoprotein in insects, which contains both β-carotene and lutein (11). As lipophorin contains both carotenoids in its hydrophobic core, transfer of β-carotene or lutein from lipophorin to the middle silk gland occurs at the interface between hemolymph and cell membrane of the middle silk gland. Consistent with the emerging view, SCRB15 and Cameo2 are transmembrane proteins expressed in the middle silk gland. We expect that SCRB15 and Cameo2 function as noninternalizing lipophorin receptors that facilitate selective uptake of carotenoids, as shown for SR-BI for cellular uptake of cholesteryl ester from high-density lipoproteins (18).
Expression profiles of SCRB11 and SCRB12 were similar to that of SCRB15 (Fig. 3C). Although the SCRB15 mutation is apparently a main cause of β-carotene deficiency of the F mutant, it could be possible that SCRB11 and SCRB12 are involved in the cellular uptake of β-carotene. Furthermore, this study does not exclude the possibility that the four nonsynonymous substitutions in SCRB11, observed between strain c44 (F/F) and strain w06 (+F/+F), were a cause of the modest rescue in β-carotene accumulation by transgenic expression of SCRB15 (Fig. 6).
β-carotene is absorbed into the midgut and transferred into the hemolymph irrespective of which allele of the F gene is present. Considering the genomic disruption of a coding exon in the +F allele (Fig. 4) and the middle silk gland-specific expression pattern (Fig. 3C), SCRB15 may not be essential for absorption of β-carotene in the midgut. Thus, the β-carotene transport system in the midgut is likely to be different from that in the middle silk gland.
Selective transport of lipids other than carotenoids also occurs in insects. For example, diacylglycerol is transported by lipophorin to the flight muscle to provide an energy source for flight (44). As CD36 family members are engaged in cellular uptake of lipids other than carotenoids in mammals, such as fatty acids or cholesteryl ester (42), it is interesting to consider the possibility that these molecules in silkworm are also involved in the transport of lipids other than carotenoids. Identification of SCRB15 as the F gene casts a spotlight on this possibility for group 1 (Fig. 5B), since the function of the group 1 insect proteins remains largely unknown. It should be noted that although the SCRB13 expression was barely detected in the tissues used in this study (Fig. 3CndashE), an SCRB13 EST clone (accession number: AK385087) was found in the corpora allata in the larvae, where juvenile hormone, a sesquiterpenoid hormone that is critical for insect development, is synthesized and secreted (45).
The molecular mechanism underlying the specific carotenoid selectivity of SCRB15 and Cameo2 is a subject for further study. While it is unknown whether the interaction between carotenoids and these proteins is direct or indirect (i.e., needing unidentified cofactors), the coding sequence of SCRB15 should contain key residues facilitating the selectivity for β-carotene that would not be present in Cameo2, as the selectivity for carotenoids was observed by transgenic expression using the same Ser1-GAL4 driver (Figs. 6G–J). Because production of transgenic larvae is not suitable for high-throughput studies, in vitro reconstitution of selective carotenoid uptake by SCRB15 and Cameo2, such as has been achieved for cellular cholesteryl ester uptake by SR-BI (46–48), is required to examine the selectivity mechanism by point mutagenesis or analysis of chimeric genes of SCRB15 and Cameo2.
Supplementary Material
Acknowledgments
The authors thank the members of the Insect Genome Research Unit at the National Institute of Agrobiological Sciences (Japan) for technical assistance in the sampling of BF1 individuals for mapping; Akitoshi Kitamura and Yuri Ooi (Fuji Chemical Industry Co., Ltd.) for technical support and advice on the HPLC analysis of carotenoids; Yumiko Nakajima for providing samples of genomic DNA from B. mandarina; Robert O. Ryan for critical reading of the manuscript; and Atsushi Kato, Hirofumi Fujimoto, and Naoko Honda for support.
Footnotes
Abbreviations:
- BAC
- bacterial artificial chromosome
- C
- Yellow cocoon
- Cameo2
- C locus-associated membrane protein homologous to a mammalian HDL receptor 2
- EGFP
- enhanced green fluorescent protein
- F
- Flesh
- rpL3
- ribosomal protein L3
- RT
- reverse transcription
- SNP
- single nucleotide polymorphism
- SR-BI
- scavenger receptor class B type I
- UAS
- upstream activating sequence
- V0
- day 0 of the fifth instar
- W0
- day 0 of the wandering stage
This work was supported by the Teimei Empress Memorial Foundation (Japan), the Futaba Electronics Memorial Foundation (Japan), and JSPS KAKENHI Grant 21380045, 22770138 (Japan). Funding was also received from the Insect Technology Project of the Ministry of Agriculture, Forestry, and Fisheries (Japan) and the National Bioresource Project (Silkworm) of the Ministry of Education, Culture, Sports, Science, and Technology (Japan). The authors declare no competing financial interests.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure and three tables.
REFERENCES
- 1.Landrum J. T., editor. 2009. Carotenoids: Physical, Chemical, and Biological Functions and Properties. CRC Press, Boca Raton, FL. [Google Scholar]
- 2.Moran N. A., Jarvik T. 2010. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science. 328: 624–627 [DOI] [PubMed] [Google Scholar]
- 3.Reboul E., Borel P. 2011. Proteins involved in uptake, intracellular transport and basolateral secretion of fat-soluble vitamins and carotenoids by mammalian enterocytes. Prog. Lipid Res. 50: 388–402 [DOI] [PubMed] [Google Scholar]
- 4.Borel P. 2012. Genetic variations involved in interindividual variability in carotenoid status. Mol. Nutr. Food Res. 56: 228–240 [DOI] [PubMed] [Google Scholar]
- 5.Harrison E. H. 2012. Mechanisms involved in the intestinal absorption of dietary vitamin A and provitamin A carotenoids. Biochim. Biophys. Acta. 1821: 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Lintig J. 2010. Colors with functions: elucidating the biochemical and molecular basis of carotenoid metabolism. Annu. Rev. Nutr. 30: 35–56 [DOI] [PubMed] [Google Scholar]
- 7.von Lintig J. 2012. Metabolism of carotenoids and retinoids related to vision. J. Biol. Chem. 287: 1627–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landrum J. T., Bone R. A. 2001. Lutein, zeaxanthin, and the macular pigment. Arch. Biochem. Biophys. 385: 28–40 [DOI] [PubMed] [Google Scholar]
- 9.Vachali P., Li B., Nelson K., Bernstein P. S. 2012. Surface plasmon resonance (SPR) studies on the interactions of carotenoids and their binding proteins. Arch. Biochem. Biophys. 519: 32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakajima M. 1963. Physiological studies on the function of genes concerning carotenoid permeability in the silkworm. Bull. Fac. Agric. Tokyo Univ. Agric. Techol. 8: 1–80 [Google Scholar]
- 11.Tsuchida K., Arai M., Tanaka Y., Ishihara R., Ryan R. O., Maekawa H. 1998. Lipid transfer particle catalyzes transfer of carotenoids between lipophorins of Bombyx mori. Insect Biochem. Mol. Biol. 28: 927–934 [DOI] [PubMed] [Google Scholar]
- 12.Sakudoh T., Tsuchida K.2009. Transport of carotenoids by a carotenoid-binding protein in the silkworm. In Carotenoids: Physical, Chemical, and Biological Functions and Properties. J. T. Landrum, editor. CRC Press, Boca Raton, FL. 511–523.
- 13.Tazima Y. 1964. The Genetics of the Silkworm. Logos Press, UK [Google Scholar]
- 14.Ishii K. 1917. On a yellow blood and white cocoon mutant of the silkworm. Sakurakai Zasshi. 1: 113–115 [Google Scholar]
- 15.Uda H. 1919. On the relation between blood colour and cocoon colour in the silkworm,with special reference to Mendel's law of heredity. Genetics. 4: 395–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleghorn M. L. 1918. First report on the inheritance of visible and invisible characters in silkworms. Proc. Zool. Soc. Lond. 88: 133–146 [Google Scholar]
- 17.Sakudoh T., Iizuka T., Narukawa J., Sezutsu H., Kobayashi I., Kuwazaki S., Banno Y., Kitamura A., Sugiyama H., Takada N., et al. 2010. A CD36-related transmembrane protein is coordinated with an intracellular lipid-binding protein in selective carotenoid transport for cocoon coloration. J. Biol. Chem. 285: 7739–7751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rigotti A., Krieger M.2010. The HDL receptor SR-BI. In High Density Lipoproteins, Dyslipidemia, and Coronary Heart Disease. E. J. Schaefer, editor. Springer, New York. 103–109.
- 19.Yamamoto K., Nohata J., Kadono-Okuda K., Narukawa J., Sasanuma M., Sasanuma S. I., Minami H., Shimomura M., Suetsugu Y., Banno Y., et al. 2008. A BAC-based integrated linkage map of the silkworm Bombyx mori. Genome Biol. 9: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimomura M., Minami H., Suetsugu Y., Ohyanagi H., Satoh C., Antonio B., Nagamura Y., Kadono-Okuda K., Kajiwara H., Sezutsu H., et al. 2009. KAIKObase: an integrated silkworm genome database and data mining tool. BMC Genomics. 10: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niwa R., Sakudoh T., Namiki T., Saida K., Fujimoto Y., Kataoka H. 2005. The ecdysteroidogenic P450 Cyp302a1/disembodied from the silkworm, Bombyx mori, is transcriptionally regulated by prothoracicotropic hormone. Insect Mol. Biol. 14: 563–571 [DOI] [PubMed] [Google Scholar]
- 22.The International Silkworm Genome Consortium 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38: 1036–1045 [DOI] [PubMed] [Google Scholar]
- 23.Koike Y., Mita K., Suzuki M. G., Maeda S., Abe H., Osoegawa K., deJong P. J., Shimada T. 2003. Genomic sequence of a 320-kb segment of the Z chromosome of Bombyx mori containing a kettin ortholog. Mol. Genet. Genomics. 269: 137–149 [DOI] [PubMed] [Google Scholar]
- 24.Mita K., Kasahara M., Sasaki S., Nagayasu Y., Yamada T., Kanamori H., Namiki N., Kitagawa M., Yamashita H., Yasukochi Y., et al. 2004. The genome sequence of silkworm, Bombyx mori. DNA Res. 11: 27–35 [DOI] [PubMed] [Google Scholar]
- 25.Sakudoh T., Tsuchida K., Kataoka H. 2005. BmStart1, a novel carotenoid-binding protein isoform from Bombyx mori, is orthologous to MLN64, a mammalian cholesterol transporter. Biochem. Biophys. Res. Commun. 336: 1125–1135 [DOI] [PubMed] [Google Scholar]
- 26.Imamura M., Nakai J., Inoue S., Quan G. X., Kanda T., Tamura T. 2003. Targeted gene expression using the GAL4/UAS system in the silkworm Bombyx mori. Genetics. 165: 1329–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakudoh T., Sezutsu H., Nakashima T., Kobayashi I., Fujimoto H., Uchino K., Banno Y., Iwano H., Maekawa H., Tamura T., et al. 2007. Carotenoid silk coloration is controlled by a carotenoid-binding protein, a product of the Yellow blood gene. Proc. Natl. Acad. Sci. USA. 104: 8941–8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura T., Thibert C., Royer C., Kanda T., Abraham E., Kamba M., Komoto N., Thomas J. L., Mauchamp B., Chavancy G., et al. 2000. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat. Biotechnol. 18: 81–84 [DOI] [PubMed] [Google Scholar]
- 29.Tatematsu K., Kobayashi I., Uchino K., Sezutsu H., Iizuka T., Yonemura N., Tamura T. 2010. Construction of a binary transgenic gene expression system for recombinant protein production in the middle silk gland of the silkworm Bombyx mori. Transgenic Res. 19: 473–487 [DOI] [PubMed] [Google Scholar]
- 30.Simodaira M. 1947. Studies of linkage in the silkworm. I. Relation between VI and VIII linkage group. Jpn. J. Genet. 22: 82–84 [Google Scholar]
- 31.Banno Y., Sakaida K., Nakamura T., Tsuchida K., Kawaguchi Y., Koga K., Doira H. 1997. Reassessment of mapping of the E homeotic gene complex of Bombyx mori by linkage analysis and in situ hybridization with an Antennapedia clone as a probe. J. Seric. Sci. Jap. 66: 151–155 [Google Scholar]
- 32.Osanai-Futahashi M., Suetsugu Y., Mita K., Fujiwara H. 2008. Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 38: 1046–1057 [DOI] [PubMed] [Google Scholar]
- 33.Goldsmith M. R.2009. Recent progress in silkworm genetics and genomics. In Molecular Biology and Genetics of the Lepidoptera. M. R. Goldsmith and F. Marec, editors. CRC Press, Boca Raton, FL. 25–48.
- 34.Xia Q., Guo Y., Zhang Z., Li D., Xuan Z., Li Z., Dai F., Li Y., Cheng D., Li R., et al. 2009. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science. 326: 433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonnhammer E. L., von Heijne G., Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6: 175–182 [PubMed] [Google Scholar]
- 36.Hoosdally S. J., Andress E. J., Wooding C., Martin C. A., Linton K. J. 2009. The Human Scavenger Receptor CD36: glycosylation status and its role in trafficking and function. J. Biol. Chem. 284: 16277–16288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petersen T. N., Brunak S., von Heijne G., Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 8: 785–786 [DOI] [PubMed] [Google Scholar]
- 38.Zhan S., Merlin C., Boore J. L., Reppert S. M. 2011. The monarch butterfly genome yields insights into long-distance migration. Cell. 147: 1171–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichols Z., Vogt R. G. 2008. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 38: 398–415 [DOI] [PubMed] [Google Scholar]
- 40.Vogt R. G., Miller N. E., Litvack R., Fandino R. A., Sparks J., Staples J., Friedman R., Dickens J. C. 2009. The insect SNMP gene family. Insect Biochem. Mol. Biol. 39: 448–456 [DOI] [PubMed] [Google Scholar]
- 41.Tsuchida K., Jouni Z. E., Gardetto J., Kobayashi Y., Tabunoki H., Azuma M., Sugiyama H., Takada N., Maekawa H., Banno Y., et al. 2004. Characterization of the carotenoid-binding protein of the Y-gene dominant mutants of Bombyx mori. J. Insect Physiol. 50: 363–372 [DOI] [PubMed] [Google Scholar]
- 42.Silverstein R. L., Febbraio M. 2009. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2: re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moussa M., Gouranton E., Gleize B., Yazidi C. E., Niot I., Besnard P., Borel P., Landrier J. F. 2011. CD36 is involved in lycopene and lutein uptake by adipocytes and adipose tissue cultures. Mol. Nutr. Food Res. 55: 578–584 [DOI] [PubMed] [Google Scholar]
- 44.Ryan R. O., Van der Horst D. J.2011. Lipid transport. In Insect Molecular Biology and Biochemistry. L. I. Gilbert, editor. Academic Press, Waltham, MA. 317–345.
- 45.Daimon T., Kozaki T., Niwa R., Kobayashi I., Furuta K., Namiki T., Uchino K., Banno Y., Katsuma S., Tamura T., et al. 2012. Precocious metamorphosis in the juvenile hormone-deficient mutant of the silkworm, Bombyx mori. PLoS Genet. 8: e1002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papale G. A., Nicholson K., Hanson P. J., Pavlovic M., Drover V. A., Sahoo D. 2010. Extracellular hydrophobic regions in scavenger receptor BI play a key role in mediating HDL-cholesterol transport. Arch. Biochem. Biophys. 496: 132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu J., Zhang Z., Shen W. J., Nomoto A., Azhar S. 2011. Differential roles of cysteine residues in the cellular trafficking, dimerization, and function of the high-density lipoprotein receptor, SR-BI. Biochemistry. 50: 10860–10875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nieland T. J., Xu S., Penman M., Krieger M. 2011. Negatively cooperative binding of high-density lipoprotein to the HDL receptor SR-BI. Biochemistry. 50: 1818–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka H., Ishibashi J., Fujita K., Nakajima Y., Sagisaka A., Tomimoto K., Suzuki N., Yoshiyama M., Kaneko Y., Iwasaki T., et al. 2008. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem. Mol. Biol. 38: 1087–1110 [DOI] [PubMed] [Google Scholar]
- 50.Sakudoh T., Nakashima T., Kuroki Y., Fujiyama A., Kohara Y., Honda N., Fujimoto H., Shimada T., Nakagaki M., Banno Y., et al. 2011. Diversity in copy number and structure of a silkworm morphogenetic gene as a result of domestication. Genetics. 187: 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pelletier J., Bozzolan F., Solvar M., Francois M. C., Jacquin-Joly E., Maibeche-Coisne M. 2007. Identification of candidate aldehyde oxidases from the silkworm Bombyx mori potentially involved in antennal pheromone degradation. Gene. 404: 31–40 [DOI] [PubMed] [Google Scholar]
- 53.Rogers M. E., Sun M., Lerner M. R., Vogt R. G. 1997. Snmp-1, a novel membrane protein of olfactory neurons of the silk moth Antheraea polyphemus with homology to the CD36 family of membrane proteins. J. Biol. Chem. 272: 14792–14799 [DOI] [PubMed] [Google Scholar]
- 54.Benton R., Vannice K. S., Vosshall L. B. 2007. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 450: 289–293 [DOI] [PubMed] [Google Scholar]
- 55.Jin X., Ha T. S., Smith D. P. 2008. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc. Natl. Acad. Sci. USA. 105: 10996–11001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.