Abstract
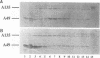
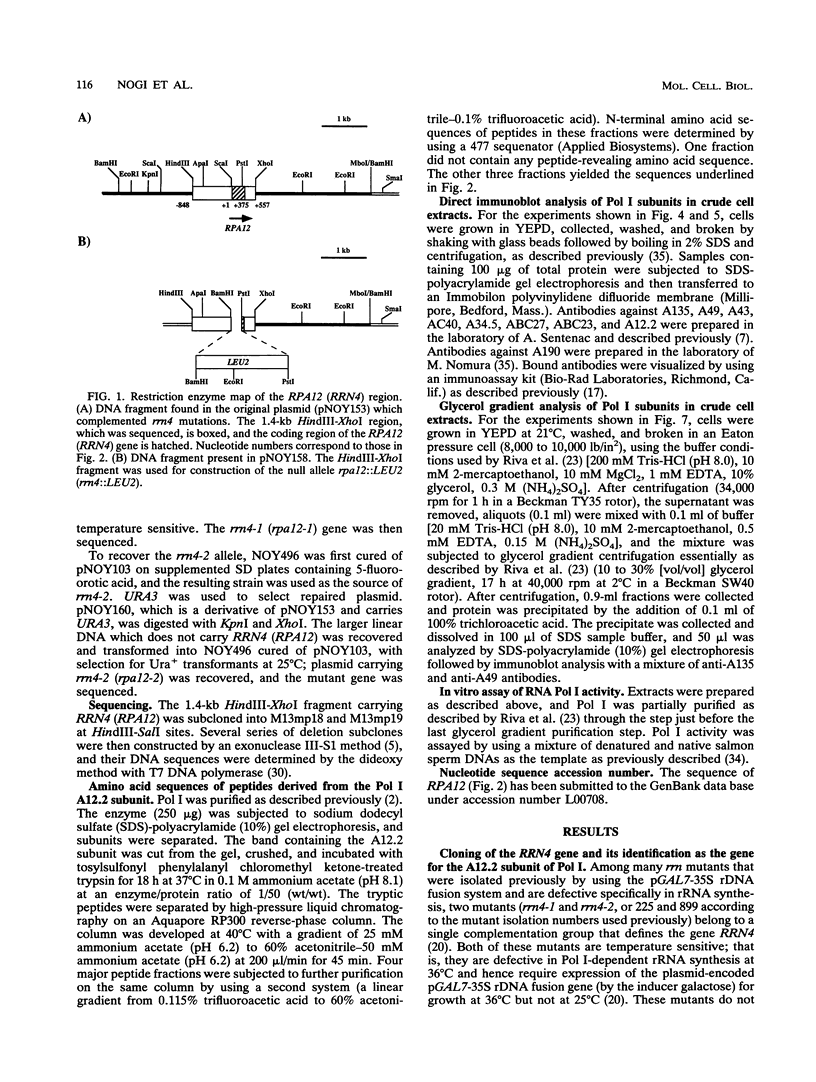
We have previously isolated mutants of Saccharomyces cerevisiae that are primarily defective in transcription of 35S rRNA genes by RNA polymerase I and have identified genes (RRN1 to RRN9) involved in this process. We have now cloned the RRN4 gene by complementation of the temperature-sensitive phenotype of the rrn4-1 mutant and have determined its complete nucleotide sequence. The following results demonstrate that the RRN4 gene encodes the A12.2 subunit of RNA polymerase I. First, RRN4 protein expressed in Escherichia coli reacted with a specific antiserum against A12.2. Second, amino acid sequences of three tryptic peptides obtained from A12.2 were determined, and these sequences are found in the deduced amino acid sequence of the RRN4 protein. The amino acid sequence of the RRN4 protein (A12.2) is similar to that of the RPB9 (B12.6) subunit of yeast RNA polymerase II; the similarity includes the presence of two putative zinc-binding domains. Thus, A12.2 is a homolog of B12.6. We propose to rename the RRN4 gene RPA12. Deletion of RPA12 produces cells that are heat but not cold sensitive for growth. We have found that in such null mutants growing at permissive temperatures, the cellular concentration of A190, the largest subunit of RNA polymerase I, is lower than in the wild type. In addition, the temperature-sensitive phenotype of the rpa12 null mutants can be partially suppressed by RPA190 (the gene for A190) on multicopy plasmids. These results suggest that A12.2 plays a role in the assembly of A190 into a stable polymerase I structure.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Buhler J. M., Sentenac A., Fromageot P. Isolation, structure, and general properties of yeast ribonucleic acid polymerase A (or I). J Biol Chem. 1974 Sep 25;249(18):5963–5970. [PubMed] [Google Scholar]
- Dequard-Chablat M., Riva M., Carles C., Sentenac A. RPC19, the gene for a subunit common to yeast RNA polymerases A (I) and C (III). J Biol Chem. 1991 Aug 15;266(23):15300–15307. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Huet J., Sentenac A., Fromageot P. Spot-immunodetection of conserved determinants in eukaryotic RNA polymerases. Study with antibodies to yeast RNA polymerases subunits. J Biol Chem. 1982 Mar 10;257(5):2613–2618. [PubMed] [Google Scholar]
- Iborra F., Huet J., Breant B., Sentenac A., Fromageot P. Identification of two different RNase H activities associated with yeast RNA polymerase A. J Biol Chem. 1979 Nov 10;254(21):10920–10924. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner T. J., Hill J. E., Myers A. M., Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- Kolodziej P. A., Young R. A. Mutations in the three largest subunits of yeast RNA polymerase II that affect enzyme assembly. Mol Cell Biol. 1991 Sep;11(9):4669–4678. doi: 10.1128/mcb.11.9.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P., Young R. A. RNA polymerase II subunit RPB3 is an essential component of the mRNA transcription apparatus. Mol Cell Biol. 1989 Dec;9(12):5387–5394. doi: 10.1128/mcb.9.12.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A. J., Olson M. V. Physical map of the Saccharomyces cerevisiae genome at 110-kilobase resolution. Genetics. 1991 Apr;127(4):681–698. doi: 10.1093/genetics/127.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Martindale D. W. A conjugation-specific gene (cnjC) from Tetrahymena encodes a protein homologous to yeast RNA polymerase subunits (RPB3, RPC40) and similar to a portion of the prokaryotic RNA polymerase alpha subunit (rpoA). Nucleic Acids Res. 1990 May 25;18(10):2953–2960. doi: 10.1093/nar/18.10.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker J. H., Yamagishi M., Kolb J. M., Nomura M. Suppressor analysis of temperature-sensitive RNA polymerase I mutations in Saccharomyces cerevisiae: suppression of mutations in a zinc-binding motif by transposed mutant genes. Mol Cell Biol. 1991 Feb;11(2):746–753. doi: 10.1128/mcb.11.2.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mémet S., Gouy M., Marck C., Sentenac A., Buhler J. M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988 Feb 25;263(6):2830–2839. [PubMed] [Google Scholar]
- Nakayama N., Miyajima A., Arai K. Nucleotide sequences of STE2 and STE3, cell type-specific sterile genes from Saccharomyces cerevisiae. EMBO J. 1985 Oct;4(10):2643–2648. doi: 10.1002/j.1460-2075.1985.tb03982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Vu L., Nomura M. An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7026–7030. doi: 10.1073/pnas.88.16.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano R., Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H. rRNA synthesis in the nucleolus. Trends Genet. 1990 Dec;6(12):390–395. doi: 10.1016/0168-9525(90)90298-k. [DOI] [PubMed] [Google Scholar]
- Riva M., Buhler J. M., Sentenac A., Fromageot P., Hawthorne D. C. Natural variation in yeast RNA polymerase A. Formation of a mosaic RNA polymerase A in a meiotic segregant from an interspecific hybrid. J Biol Chem. 1982 Apr 25;257(8):4570–4577. [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Mougey E. B. News from the nucleolus: rRNA gene expression. Trends Biochem Sci. 1991 Feb;16(2):58–62. doi: 10.1016/0968-0004(91)90025-q. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treich I., Carles C., Riva M., Sentenac A. RPC10 encodes a new mini subunit shared by yeast nuclear RNA polymerases. Gene Expr. 1992;2(1):31–37. [PMC free article] [PubMed] [Google Scholar]
- Treich I., Riva M., Sentenac A. Zinc-binding subunits of yeast RNA polymerases. J Biol Chem. 1991 Nov 15;266(32):21971–21976. [PubMed] [Google Scholar]
- Wittekind M., Dodd J., Vu L., Kolb J. M., Buhler J. M., Sentenac A., Nomura M. Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae. Mol Cell Biol. 1988 Oct;8(10):3997–4008. doi: 10.1128/mcb.8.10.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekind M., Kolb J. M., Dodd J., Yamagishi M., Mémet S., Buhler J. M., Nomura M. Conditional expression of RPA190, the gene encoding the largest subunit of yeast RNA polymerase I: effects of decreased rRNA synthesis on ribosomal protein synthesis. Mol Cell Biol. 1990 May;10(5):2049–2059. doi: 10.1128/mcb.10.5.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N. A., Lane W. S., Young R. A. Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. J Biol Chem. 1991 Oct 5;266(28):19053–19055. [PubMed] [Google Scholar]
- Woychik N. A., Liao S. M., Kolodziej P. A., Young R. A. Subunits shared by eukaryotic nuclear RNA polymerases. Genes Dev. 1990 Mar;4(3):313–323. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]
- Yano R., Nomura M. Suppressor analysis of temperature-sensitive mutations of the largest subunit of RNA polymerase I in Saccharomyces cerevisiae: a suppressor gene encodes the second-largest subunit of RNA polymerase I. Mol Cell Biol. 1991 Feb;11(2):754–764. doi: 10.1128/mcb.11.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T., Ishihama A. Genetics of bacterial RNA polymerases. Annu Rev Genet. 1979;13:59–97. doi: 10.1146/annurev.ge.13.120179.000423. [DOI] [PubMed] [Google Scholar]