Abstract
Genomic structural variation (SV) is a major determinant for phenotypic variation. Although it has been extensively studied in humans, the nucleotide resolution structure of SVs within the widely used model organism Drosophila remains unknown. We report a highly accurate, densely validated map of unbalanced SVs comprising 8962 deletions and 916 tandem duplications in 39 lines derived from short-read DNA sequencing in a natural population (the “Drosophila melanogaster Genetic Reference Panel,” DGRP). Most SVs (>90%) were inferred at nucleotide resolution, and a large fraction was genotyped across all samples. Comprehensive analyses of SV formation mechanisms using the short-read data revealed an abundance of SVs formed by mobile element and nonhomologous end-joining-mediated rearrangements, and clustering of variants into SV hotspots. We further observed a strong depletion of SVs overlapping genes, which, along with population genetics analyses, suggests that these SVs are often deleterious. We inferred several gene fusion events also highlighting the potential role of SVs in the generation of novel protein products. Expression quantitative trait locus (eQTL) mapping revealed the functional impact of our high-resolution SV map, with quantifiable effects at >100 genic loci. Our map represents a resource for population-level studies of SVs in an important model organism.
SVs, including deletions, insertions, and duplications, are a major contributor to genetic variation, are responsible for the majority of polymorphic nucleotide bases between individuals (Conrad et al. 2010b; The 1000 Genomes Project Consortium 2010; Sudmant et al. 2010; Mills et al. 2011), and have an important influence on phenotypic diversity (Feuk et al. 2006). Owing to inherent difficulties in their ascertainment, however, SVs have remained a relatively poorly understood form of genetic variation in comparison to SNPs.
While there has been a strong recent focus on the characterization of SVs in humans (Conrad et al. 2010b; Sudmant et al. 2010; Mills et al. 2011), mapping common SVs in one of the most widely used model organisms in genetics—the fruit fly Drosophila melanogaster—has lagged further behind. A detailed map of Drosophila SVs would be of immense importance to a large body of genetics studies, by enabling the connection of polymorphic genome rearrangements to systematic phenotypic, functional, and developmental data in a fashion inconceivable in humans or mammalian models. It would also shed light on the frequency of SVs in natural populations, many of which have more resemblance, in terms of genetic diversity, population size, and population substructure, to Drosophila than to human populations. Two studies have recently made initial progress by using microarray-based approaches to provide a first glimpse of D. melanogaster SVs, reporting an abundance of SVs in the fly genome in surveys focusing on five and 15 natural fly isolates, respectively (Dopman and Hartl 2007; Emerson et al. 2008). Owing to constraints of the respective array technologies applied, these studies were limited toward relatively large variants (median SV size of ≥336 bp), were relatively insensitive to mobile element insertions that are difficult to identify by hybridization, and reported SV maps with approximate, rather than nucleotide resolution breakpoint assignments. By comparison, next-generation DNA sequencing (NGS)-based approaches enable mapping SVs across a widened size spectrum (from a few base pairs to Megabases in size), and enable the inference of breakpoints at nucleotide resolution (Mills et al. 2011), a crucial prerequisite for functional analyses (Schlattl et al. 2011) and mechanistic studies of SV formation (Korbel et al. 2007; Conrad et al. 2010a; Kidd et al. 2010; Lam et al. 2010). Based on partial SV maps obtained from low-coverage sequencing (∼0.8×) it was recently concluded that NGS technology is, in principle, suitable for ascertaining SVs in Drosophila (Cridland and Thornton 2010), although deep-sequencing coverage of at least 8× would be required to enable the construction of a more comprehensive, accurate SV map (Cridland and Thornton 2010).
The DGRP project has recently generated deep-sequence coverage (median coverage = 18×) data for a panel of isogenic fly lines. These lines were inbred over 20 generations, yielding a set of isogenic strains distributed across various laboratories—creating an unprecedented community resource for the analysis of population genomics and quantitative traits in Drosophila melanogaster (Ayroles et al. 2009; Mackay et al. 2012). While initial analyses of the DGRP sequencing data have yielded a wealth of SNPs, thus far no SV map has become available for this resource.
Here, we present the first highly accurate NGS-based SV map in the fly, based on analyzing 39 lines from the DGRP. The high resolution of our map enabled us to perform in-depth analyses of SV formation mechanisms and to assess the impact of SVs on functional elements and gene expression variation.
Results
A sequencing-based map of structural variation in a Drosophila melanogaster population
We obtained deep sequencing data for 39 fly lines from the DGRP pilot data set (Table 1; Ayroles et al. 2009; Mackay et al. 2012). These included 38 isogenic lines originating from a wild population, and, in addition, an isolate of the Berkeley strain used to assemble the current version (BDGP R5/dm3) of the fly reference genome (Adams et al. 2000). We applied three complementary approaches for SV mapping (Methods): (1) paired-end mapping, based on identification and analysis of abnormally mapping reads pairs (RP) of size-selected DNA fragments (Tuzun et al. 2005; Korbel et al. 2007; Hormozdiari et al. 2009); (2) read-depth (RD) analysis, which detects SVs by analyzing the depth of sequencing coverage (Alkan et al. 2009; Yoon et al. 2009; Abyzov et al. 2011); and (3) split-read (SR) analysis, based on gapped or clipped alignments of short DNA sequencing reads (Mills et al. 2006; Ye et al. 2009). To capture SVs detectable through these complementary sequence signatures (RP, RD, and SR), and combinations thereof, we integrated the results from four SV discovery tools: Pindel (Ye et al. 2009), CNVnator (Abyzov et al. 2011), Genome STRiP (Handsaker et al. 2011), and DELLY (Rausch et al. 2012b). Our approach, which parallels the SV discovery and genotyping strategy used in the 1000 Genomes Project (Mills et al. 2011), is depicted in Figure 1.
Table 1.
Sample overview
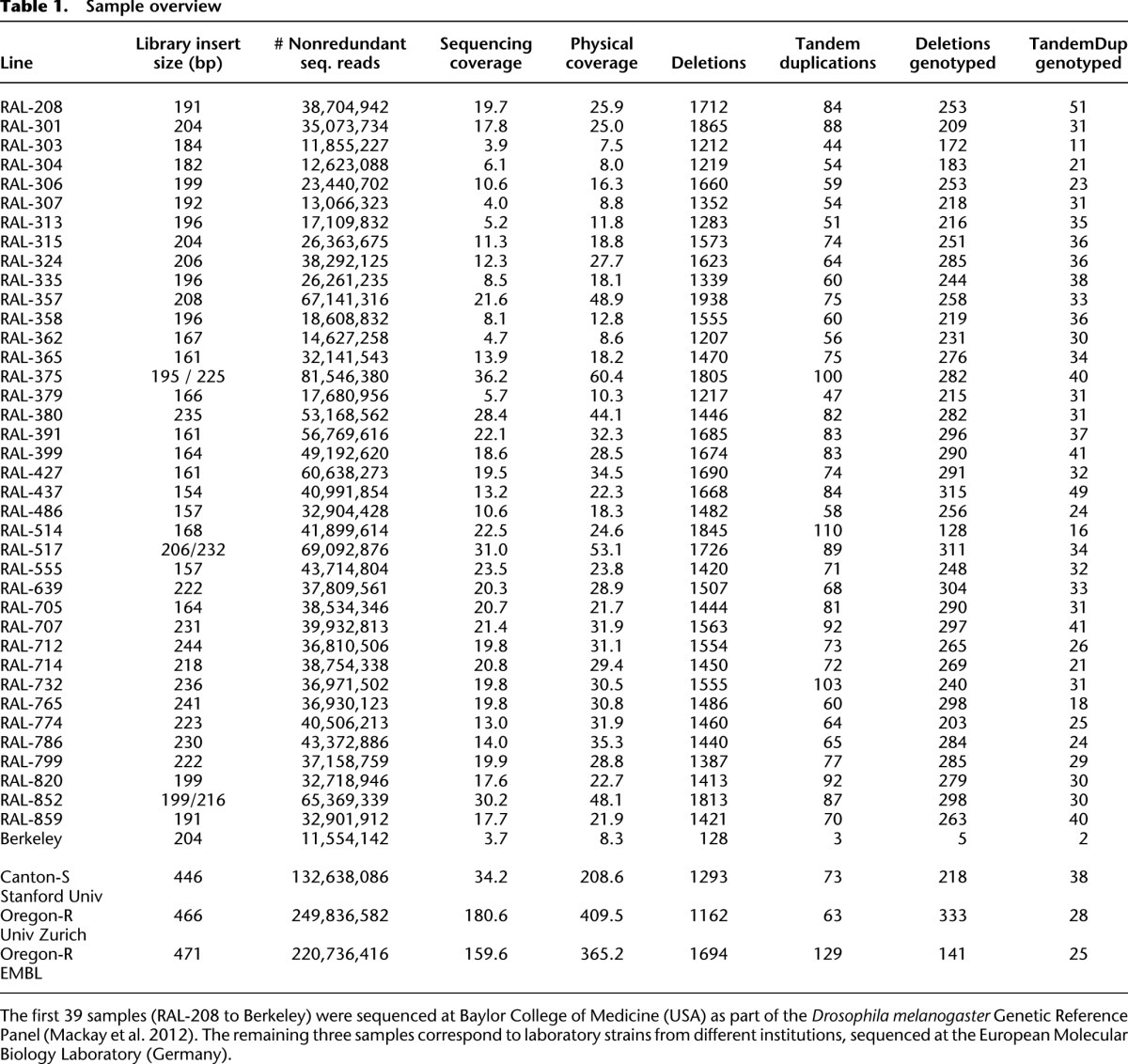
Figure 1.
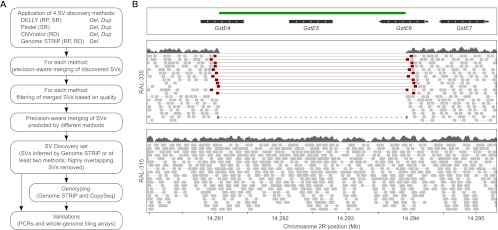
Structural variant (SV) discovery and genotyping. (A) Integrated pipeline for SV discovery, validation, and genotyping. (B) Example of a polymorphic deletion (highlighted in green), supported by discordantly mapping read pairs (red), split reads (blue), and read-depth (dark gray). Light gray boxes indicate individual sequencing reads. Indicated are a sample with the deletion (RAL-335) and one without (RAL-315) that variant.
We applied two steps to merge the SV calls in order to generate a nonredundant variant discovery set of SVs ≥50 bp. First, we merged SV call sets separately for each method (a step not necessary for Genome STRiP, which already provides a merged SV call set). Second, we merged SVs across all four methods based on precision-aware confidence intervals (Methods; Supplemental Text). To assure high confidence, we required each SV to be predicted by at least two methods—with the exception that we kept 283 deletion calls exclusively made by Genome STRiP, a population-based discovery tool that was shown to yield exceptionally high accuracy (false-discovery rate [FDR] < 5%) (Handsaker et al. 2011; Mills et al. 2011). Owing to the nature of duplication calls made by DELLY and Pindel, our SV discovery set contained tandem duplications, but did not comprise dispersed duplications.
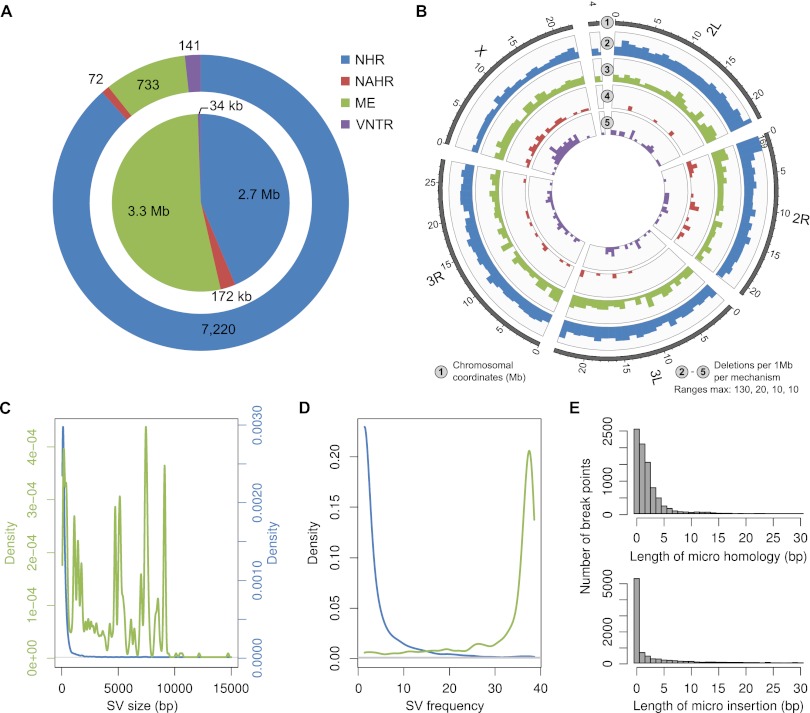
Altogether, we identified 8962 deletions and 916 tandem duplications relative to the reference genome sequence, with sizes ranges of from 50 to 165,327 bp for deletions (median 178 bp), and 78–129,958 bp for duplications (median 2111 bp) (Fig. 2A,B; Supplemental Table 1). For >90% of the SVs (8204/8962 deletions and 903/916 tandem duplications), we inferred breakpoints at nucleotide resolution, yielding the first genome-wide base-pair resolution catalog of these SV forms in D. melanogaster.
Figure 2.
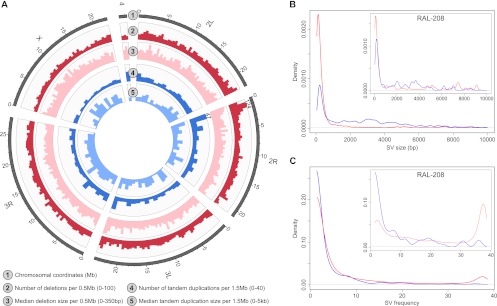
A sequencing-based map of SVs in the Drosophila melanogaster Genetic Reference Panel (DGRP). (A) Spatial and size distribution of deletions and tandem duplications in our SV discovery set. In two of the genomic windows indicated, which includes a SV hotspot region on chromosome 2R, the bars exceeded the displayed range (hence, their absolute height is indicated). (B) Size distribution of deletions (red) and tandem duplications (blue) in our entire discovery set (large plot) and in a single sample (RAL-208, small plot). The differences are owing to different frequency spectra of specific SV classes (see the Results section “Mechanisms of SV formation”). For deletions, several peaks are visible (e.g., 5, 7.5, and 9 kb), corresponding to mobile element insertions into the reference genome or to deletions of DNA transposons, which move by cut-and-paste mechanisms. (C) Frequency spectrum of deletions (red) and tandem duplications (blue) amongst 39 lines, indicated for the entire set of SVs (large plot) and for all SVs discovered in a single sample (RAL-208, small plot). While most SVs were discovered in less than five samples, a subset was present in >35 samples; most of the latter represent mobile element insertion and deletion events.
Significantly fewer SVs per 500 kb genomic window were identified on chromosome X compared with the autosomes (e.g., median 27 vs. 38 for deletions; P = 1.98 × 10−9; Wilcoxon rank-sum test) (Fig. 2A; Supplemental Fig. 1A,B), consistent with previous array-based studies (Dopman and Hartl 2007; Emerson et al. 2008), possibly owing to hemizygosity in males uncovering the effects of recessive mutations (Crow and Kimura 1970; Dopman and Hartl 2007). In addition, lower sequencing coverage on chromosome X may have contributed to this effect.
Altogether, 36% of the deletions and 54% of the tandem duplications were observed in only one line (Fig. 2C). By comparison, 677 SVs were predicted in more than 30 samples, the vast majority of which (671) were inferred as deletions. Further analyses showed that most of these represented insertions of mobile elements into the reference genome, or deletions of DNA transposons present in the reference assembly, both of which we detected as deletions relative to the reference genome.
Genotyping of SVs in Drosophila melanogaster
We further performed SV genotyping to generate a population genotype reference by re-evaluating the occurrence of SVs in all samples using criteria specifically adjusted in an SV locus-specific manner (Waszak et al. 2010; Handsaker et al. 2011). We performed deletion genotyping using Genome STRiP (based on RD and RP analysis) (Handsaker et al. 2011) and genotyped duplications using CopySeq (based on RD analysis) (Waszak et al. 2010). For 3459 of the 8962 deletions, Genome STRiP inferred a high-confidence genotype in at least one sample (Supplemental Table 1). The remaining deletions were either too small, too repetitive, or covered by too few sequencing reads for Genome STRiP to result in a high-confidence genotype call (see Methods). For the majority of the genotyped regions (2834/3459, 82%) Genome STRiP generated homozygous deletion genotypes—as expected for lines undergoing several generations of inbreeding (Supplemental Table 1). We refer to this set as the deletion genotyping reference set. The remaining 18% failing to show homozygous deletions were removed from our genotype set; these comprised to a large extent regions difficult to assess with short read data owing to their high repeat content. Amongst the duplications, 505 (55%) were inferred to have a copy-number genotype of at least 4 (homozygous duplication) in at least one sample forming our tandem duplication genotyping reference set (Supplemental Table 1), with the remaining regions (i.e., predicted to have three copies or less in all samples) comprising an abundance of repeat-rich and relatively small regions (median 580 bp).
Despite those filtering steps, the resulting SV genotyping sets displayed size distributions similar to the respective discovery sets (Supplemental Fig. 2A). We further observed good agreement between the SV frequency spectrum of discovered and genotyped SVs, with the exception that mobile element-associated events were under-represented in the genotyping set (Supplemental Fig. 2B).
Extensive validation of SVs and comparison with a recent microarray-based study
We experimentally assessed the quality of our SV sets by performing extensive PCR and tiling array-based validation experiments. 104/110 PCR validation experiments, carried out in five fly lines, verified the respective SV predictions—with 82/87 of the deletions and 22/23 of the tandem duplications verified by PCR (Supplemental Table 1). We also performed high-density tiling microarray-based validation experiments using arrays densely covering the fly genome with 35 bp median oligonucleotide probe spacing. The arrays enabled us to assess 2588 deletions and 263 tandem duplications in six randomly chosen samples (Fig. 3; Methods). Based on the array data, we estimated FDRs of 3% and 11% for the deletion genotyping set and discovery sets, respectively. For duplications, tiling array-based estimates were FDR = 8% and FDR = 24% for the genotyping and discovery sets, respectively.
Figure 3.
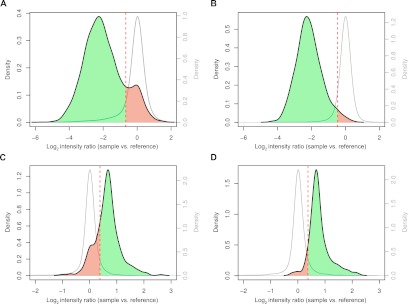
Whole-genome tiling array validation of the DGRP SV map. (A) Deletion discovery set. (Black line) Distribution of median log2 array intensity ratios for deletion loci, recorded between sample and the Berkley reference strain. (Gray line) Distribution for control samples lacking the SV at a given deletion locus of interest, which we used to determine the cutoff for FDR estimation (see Methods for details). (Dashed red line) Cutoff. SVs considered to be validated are highlighted in green, and potential false positive SVs are in red. (B) Deletion genotyping set. (C) Tandem duplication discovery set. (D) Tandem duplication genotyping set. Note that the different nature of homozygous deletions (causing a complete loss of genetic material) compared with homozygous duplications (causing mere duplication of material) likely render array-based FDR-estimations of duplications more error-prone (i.e., compare the better separation of hybridization-based curves for Fig. 3A,B compared with Fig. 3C,D for polymorphic vs. nonpolymorphic regions, and note the aforementioned PCR-based FDR estimates of 4%–6%).
We next compared our predictions with the results of the most comprehensive survey of SVs in Drosophila up to date, i.e., a previously performed array-based survey assessing unbalanced SVs in 15 natural isofemale Drosophila lines (Emerson et al. 2008), identifying 1428 deletions (with an assessed FDR of 47%) (see Emerson et al. 2008) and 2211 duplications (with an FDR of 14%). SVs from our data intersected with 300 (i.e., 175 deletions and 125 duplications) reported by Emerson and colleagues. A possible reason for the small overlap is the distinct origins of the samples used, with Emerson et al. (2008) analyzing samples from sub-Saharan Africa, whereas the DGRP lines originated from Northern America.
Comparison of SVs from wild isolated to that of laboratory fly strains
We further performed high-coverage (>30×) sequencing in three laboratory strains, one Canton-S and two Oregon-R obtained from different laboratories, to examine their diversity and strain genetic relationships, and to compare their SV set with our DGRP-based SV map. Following sequencing, we genotyped deletions as well as tandem duplications and called SNPs in all three laboratory strains. A total of 1242 SVs (1138 deletions and 104 tandem duplications) were genotyped at high confidence in at least one out of three, 1083 of which were inferred in all three samples (Supplemental Table 2)—SV sets that can now be taken into account in research studies based on Canton-S and Oregon-R strains. Somewhat surprisingly, neither at the SV nor at the SNP level did we observe evidence for a higher genetic similarity of the Oregon-R-derived strains compared with the Canton-S-derived strain (Supplemental Table 3A,B), a finding indicating that admixture or contamination events involving these may occur more often than currently appreciated. Furthermore, 770 deletions and 134 tandem duplications discovered in these three strains were not detected in the DGRP set (Supplemental Table 2), suggesting that these are either private to commonly used laboratory strains or display a low population frequency in the DGRP resource.
Analysis of SVs on a population scale—selection and linkage disequilibrium (LD)
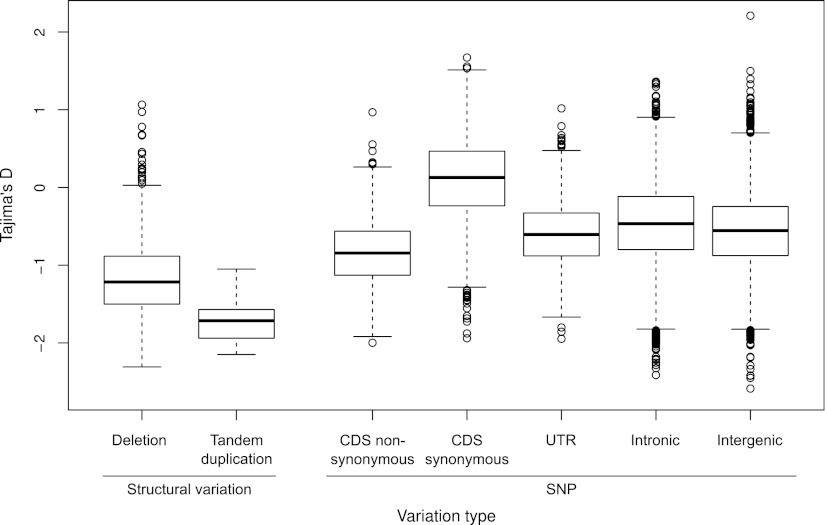
We further performed population genetic analyses in the DGRP strains to analyze selective forces influencing the frequency of SVs in natural populations. These analyses, and other analyses reported in the following, were pursued using our SV discovery set (unless stated otherwise). Using Tajima's D test to assess directional selection (Tajima 1989), we observed strikingly negative values of Tajima's D for both deletions (Tajima's D = −1.15) and tandem duplications (Tajima's D = −1.69). We further compared our summary statistics for SVs to SNPs in different regions of the genome, including both synonymous and nonsynonymous coding SNPs, to examine whether Tajima's D would indicate a stronger tendency for SVs to be selected against than for those SNPS affecting coding regions (Fig. 4) (SNPs were inferred for all samples using JGIL) (Stone 2012; see Methods). Notably, for both deletions and tandem duplications, Tajima's D was significantly more negative than for nonsynonymous SNPs (P < 1 × 10−8), a finding consistent with pervasive negative selection acting against SVs (Fig. 4). Even after accounting for the potential effects of false negatives, Tajima's D continued to show strong evidence for negative selection on SVs within the DGRP population (see Supplemental Figure 3; Supplemental Text).
Figure 4.
Genome-wide values of Tajima's D for SNPs and SVs. Boxplots depict genome-wide distributions of Tajima's D for SNPs in five genomic compartments (intergenic, intronic, nonsynonymous, synonymous, and UTRs), for deletions, and for tandem duplications, as estimated from sliding-window analyses. As classes, both deletions and tandem duplications show Tajima's D values significantly more negative than those of either synonymous or nonsynonymous SNPs (P < 1 × 10−8).
Additional analyses showed a significantly lower Tajima's D value for gene-affecting deletions compared with deletions that were strictly intergenic (Tajima's D = −1.25 vs. −1.01, P = 0.032, t-test). Furthermore, Tajima's D value was less negative on the X chromosome than on the autosomes (see Supplemental Fig. 4; Supplemental Text), an observation that may be explained by selection being less efficient at removing SVs from the X chromosome relative to the autosomes (see Supplemental Text for further discussion).
We also investigated patterns of linkage disequilibrium (LD) between SVs relative to those between SNPs. While relatively few SVs were found within close proximity to one another, relative to SNPs, the average LD between pairs of deletions and pairs of duplications appeared to similarly fall off to an r2 value of ∼0.03 within a few hundred base pairs, remaining at this level for more distantly spaced pairs (Supplemental Fig. 5).
This suggests similar LD properties for both of these distinct classes of genetic variation. Hence, on the basis of LD, selection does not appear to act in profoundly different ways on SVs relative to SNPs.
The impact of SVs on genomic annotation
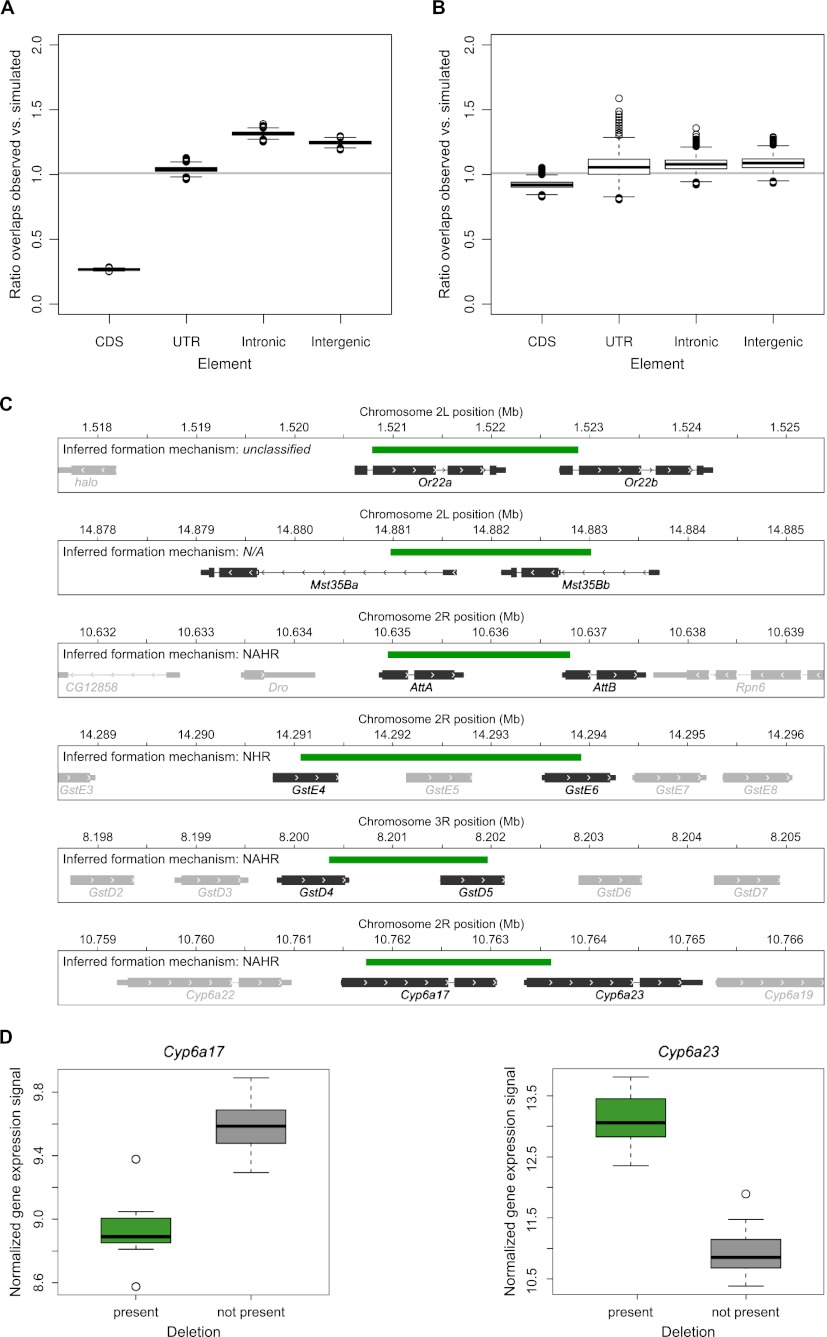
The high resolution of our SV map further enabled us to assess the functional impact of our SVs in detail. To this end, we initially investigated functional impact by relating our SV map to genome annotation. Strikingly, a large number of deletions (562) affected protein-coding sequences, causing gene deletions or partial disruptions (Table 2; Supplemental Table 1). Using simulations, we observed a marked, fourfold depletion of deletions overlapping coding sequences (P < 0.0001; based on permutations), but no depletion for deletions overlapping UTRs or intronic sequences (Fig. 5A; Supplemental Fig. 6A), consistent with pronounced selection acting against gene deletions. Testing for enrichment of gene functional categories amongst the genes affected by SVs through gene deletion (or partial gene disruption) yielded a significant enrichment of genes related to functional categories involved in interactions with the environment, including sensory perception (i.e., chemical stimulus and taste), glutathione transferase activity (involved in detoxification), and others (Supplemental Table 4A). We noticed that SVs affecting genes involved in such functional categories typically involved relatively large deletions, often affecting multiple paralogous genes from one functional category (such as a deletion affecting the glutathione transferase genes GstE4, GstE5, and GstE6) (Fig. 1B). Nonetheless, even when limiting our analysis to those 521 deletions that deleted or disrupted only a single gene, we observed a significant enrichment of genes playing a role in interactions with the environment (i.e., coagulation/hemostasis; see Supplemental Table 4B).
Table 2.
Functional impact of our fine resolution SV set
Figure 5.
Functional impact of SVs. (A) Evidence for purifying selection against deletions overlapping gene coding sequences based on overlaps and expected overlaps with genomic annotation elements. The latter value was determined by randomly moving deletions within a 100-kb region and reanalyzing element overlaps 10,000 times. Intronic and intergenic deletions appeared slightly enriched relative to the randomized set, an effect that can be explained by the appreciable depletion of deletions overlapping coding sequence due to negative selection. (B) Analysis of tandem duplications overlapping annotated functional elements. (C) A collection of discovered putative fusion genes caused by partial gene deletion leading to hybrid genes. (Green) Deletions; (dark gray) fused genes; (light gray) genes in the vicinity of the locus in question. (CDS) Coding sequences; (UTR) upstream and downstream untranslated genic regions. (D) Gene expression analysis in male flies for Cyp6a17 and Cyp6a23 for samples with and without the gene fusing deletion that intersects with both genes. Variant categorization was achieved based on the genotype set (homozygous as well as heterozygous calls were considered as deletion).
Similarly, we observed a depletion of tandem duplications intersecting with coding sequences using simulations (P = 0.0025; based on permutations) (Fig. 5B; Supplemental Fig. 6B). The 227 whole-gene duplications in our discovery set (Supplemental Table 1) comprised several genes involved in environmental response, such as olfactory receptors genes (Supplemental Table 5).
We further identified 78 SVs leading to putative fusion genes. These included well-described gene fusions such as Or22a:Or22b (Turner et al. 2008; Aguade 2009), but also a number of previously undescribed gene fusions of potential functional relevance, for example, the antibacterial protein encoding gene homologues AttA and AttB (Fig. 5C; Supplemental Table 6; Methods). Six out of 24 predicted gene fusions involving deletions led to gene hybrids comprising genes from the same gene family (Fig. 5C), i.e., spanning genomic regions with paralogous sequences of high-sequence similarity that may allow for rearrangements mediated by nonallelic homologous recombination (NAHR). Indeed, further analyses of the SV breakpoint junctions led us to infer that NAHR occurred during SV formation in 3/6 (see below), whereas one SV was formed by rearrangements in the absence of homology stretches occurring directly at the breakpoints, and for two, our analysis was inconclusive. Using PCR, we verified 10/11 tested fusion genes at the DNA level (Supplemental Table 6).
Association of SVs with Drosophila adult gene expression variation
We next assessed how these SVs affect gene expression by performing expression quantitative trait locus (eQTL) mapping (Schlattl et al. 2011) relating mRNA measurements of fly genes, at a genome-wide level, to SVs in the vicinity of those genes. Thereby, we made use of previously published microarray-based adult gene expression data available for 9454 genes across the 38 nonreference fly strains, for which both female and male fly expression data had been independently generated (Ayroles et al. 2009). We pursued eQTL mapping by determining pairwise rank correlation values between the expression value of genes and the presence or absence of SVs located in the surrounding (i.e., such within 50 kb upstream of or downstream from the annotated gene coordinates; see Methods). Using an FDR cutoff of 10%, we identified 79 and 52 deletion-associated eQTLs and 36 and 29 duplication-associated eQTLs in males and females (Table 3; Supplemental Table 7). The majority of SV-associated eQTLs agreed between males and females (Table 3). One notable exception was Ser12, with an established eQTL-association P-value of 3 × 10−7 for females versus 0.99 for males, which we observed to be paralleled by markedly higher Ser12 expression in females—findings that can be attributed to female-specific gene expression (Lawniczak and Begun 2007).
Table 3.
Summary of identified SV-associated eQTLs
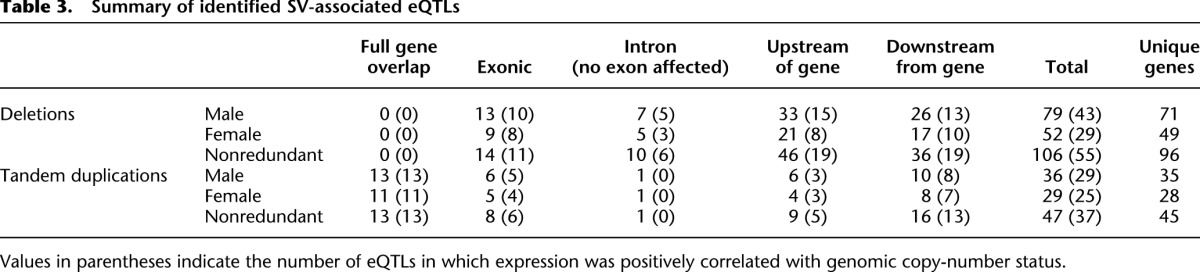
In cases where SVs intersected with protein-coding regions, the relative changes in expression correlated with the gene copy-number alteration in most cases (Table 3). As a notable exception, the expression of Cyp6a23, encoding a cytochrome P450 paralog mapped to a deletion-associated eQTL, displayed a marked negative correlation with the respective copy-number status of its partially intersecting deletion (i.e., samples harboring the deletion showed higher Cyp6a23 expression) (Fig. 5C,D). This deletion led to a gene fusion involving both Cyp6a23 and its paralog Cyp6a17 positioned upstream in tandem orientation (Fig. 5C). Remarkably, the expression of Cyp6a17 was also statistically associated with this SV, showing the intuitively expected positive correlation with deletion status. Further examination of the fusion gene structure revealed that the fusion gene status can account for both the observed negative and positive correlations for Cyp6a23 and Cyp6a17, respectively—with juxtaposition of the active Cyp6a17 promoter into the immediate vicinity of Cyp6a23 serving to explain the observed gene expression variation levels of both genes (Fig. 5C,D).
Despite the observed correlation of copy number and expression for SVs leading to gene disruption or duplication (Table 3), implicating the respective SVs as causally involved, we reasoned that in many cases where SVs do not affect exonic sequence, the actual causal variants may represent SNPs in LD (rather than the respective SVs). To further assess the contribution of SNPs, we extended our eQTL analysis to SNPs (Supplemental Text). We observed SNPs displaying the same or a better association with expression for 91 out of the 129 eQTLs identified with FDR < 5%, which suggests a possible contribution of SNPs in these loci. Conversely, this analysis yielded further strong support for a causal role of those 38 SV-associated eQTLs for which no such correlating SNP was observed (Supplemental Table 7).
Mechanisms of SV formation in the fly genome
We additionally used our SV map to evaluate the relative contribution of different molecular mechanisms leading to SV formation in Drosophila (Hastings et al. 2009; Onishi-Seebacher and Korbel 2011). SV formation mechanisms were identified by scanning DNA sequences surrounding breakpoint junctions for specific diagnostic sequence signatures, using the BreakSeq formation mechanism analysis pipeline (Lam et al. 2010). The mechanisms we were able to distinguish (see Methods) include: (1) NAHR, associated with long sequence similarity stretches around the breakpoints; (2) rearrangements occurring in the absence of homology (here termed nonhomologous rearrangements—NHR), involving nonhomologous end-joining–based DNA double-strand repair (NHEJ) or microhomology-mediated break-induced replication (MMBIR); (3) mobile element (ME) insertion, and deletion events involving retrotransposons or DNA transposons; and (4) the shrinkage or expansion of a variable number of tandem repeats (VNTR) by DNA replication slippage. Owing to our focus on deletions and tandem duplications, our study only detected MEs inserted into the reference genome (and absent in at least one of the DGRP samples), but did not identify MEs that were newly inserted into the DGRP samples.
We initially inferred the formation mechanism for 8204/8962 (91.5%) deletions with base-pair resolution breakpoint information. The vast majority of these SVs (88%) were inferred to be formed by NHR, and 9% corresponded to MEs. The remaining 1% and 2% were attributed to NAHR and DNA replication slippage, respectively (Fig. 6A; Supplemental Table 1). Higher contributions of NAHR were recently reported in the NGS-technology–based SV survey in humans performed by the 1000 Genomes Project Structural Variation Analysis Group, which attributed 15% of the deletions discovered by Illumina sequencing technology to NAHR (Mills et al. 2011). The observed differences may, in part, result from distinct genomic repeat contents in Drosophila vs. humans, with recent bursts of interspersed repeat insertion and segmental duplication (SD) events having shaped the genomes of primates (Bailey et al. 2003; Kim et al. 2008; Marques-Bonet et al. 2009)—since sequence identity-associated SV formation mechanisms are particularly abundant in genomic regions with a high repeat/SD content. It should be kept in mind, however, that limitations of NGS—which biases DNA variant discovery to relatively unique (‘mappable’) genomic regions (The 1000 Genomes Project Consortium 2010) owing the technology's short DNA reads and short paired-end insert sizes (Onishi-Seebacher and Korbel 2011)—also contributed to the picture. In this regard, recent estimates of the fraction of deletions attributable to NAHR in humans were markedly higher (26%) when assessed based on capillary-based paired-end as well as shotgun sequencing of 40-kb long fosmid clones (Kidd et al. 2010)—a result better reflecting the actual contribution of NAHR in humans, since repeat and SD-rich regions were more extensively covered (Kidd et al. 2010). Furthermore, differences in reference genome quality are expected to affect the fraction of NAHR events identified, since repeat/SD-rich regions are typically under-represented in imperfect genome assemblies (Bailey et al. 2004).
Figure 6.
SV formation mechanisms acting in the Drosophila melanogaster genome. (A) Distribution of different SV formation mechanisms inferred amongst deletions for which the breakpoints were discovered at nucleotide resolution (8204 SVs). (Outer circle) Number of deletions per mechanism. (Inner circle) Cumulative size of these events. For 38 events the mechanism was ambiguous (Lam et al. 2010), and hence remained unclassified (data not shown in the plot). (B) Spatial distribution of deletions based on the different formation mechanisms. Colors as in A. In one case the bars exceeded the displayed range (thus, the absolute column height is indicated). (C) Size distribution of deletions related to NHR (blue) and ME (green). The peaks correspond to characteristic size spectra of mobile element classes active in the fruit fly. (D) Frequency distribution of deletions related to NHR (blue) and ME (green). A number of mobile elements were only present in the reference genome and were hence detected as deletions in the majority of samples. (E) Size distribution of micro homologies and micro insertions at deletion breakpoints.
We further related SV formation mechanisms inferred by BreakSeq with SV size, and found that ME-associated events were, on average, significantly larger than SVs formed by any other mechanisms (Fig. 6C; Supplemental Fig. 7A,C,D), an observation explainable by the relatively large size of active transposable elements in D. melanogaster (∼200 bp–10 kb). The ME size spectrum showed several characteristic peaks (Fig. 6C) corresponding to different ME families. MEs that our survey observed most frequently included roo, Doc, 297, BS, copia, and Tirant, all of which were previously identified as active elements (Bergman and Bensasson 2007; Kofler et al. 2012).
Additionally, in contrast to the majority of deletions, ME-associated events were typically inferred in a large number of samples (ME median: 37 samples versus overall median: 2 samples) (Fig. 6D; Supplemental Fig. 7B), suggesting that many may correspond to rare insertions into the D. melanogaster reference genome. Since we observed different SV frequencies for different formation mechanism classes, the distribution of formation mechanisms was different when assessing a single sample instead of the entire sample set (Fig. 6A; Supplemental Fig. 8). For instance, when limiting our analysis to a single sample, about half of the deletion predictions were inferred to be associated with MEs, accounting for over 75% of the affected bases, data in support of the recently reported substantial activity of MEs in Drosophila (Kofler et al. 2012).
We next analyzed the spatial distribution of deletions corresponding to the four different classes of SV formation mechanisms (Fig. 6B). Despite the aforementioned relatively small number of SVs on chromosome X compared with the autosomes, we observed a strong enrichment of VNTR expansion/shrinkage events on chromosome X compared with autosomes (2.1 per Mb [4.3% of all events] vs. 1.0 per Mb [1.3% of all events]; P < 0.0001, based on permutations). This enrichment may be explained by the relatively high fraction of interspersed repeat sequences on chromosome X (10.7%, compared with 8.9% for the autosomes), sequences mediating the shrinkage or expansion of VNTRs. Additionally, most of the NAHR events were inferred on chromosome X and chromosome 2R (enrichment P-values of P < 0.001 for chromosome X, P < 0.01 for chromosome 2R, based on permutations). We further segmented the genome using a recently described statistical approach for SV hotspot detection (Mills et al. 2011), and identified six hotspots of SV formation in the fly genome based on our deletion set, all of which were situated in relatively repeat-dense regions close to centromeres, and three of which were on chromosome 2R, causing a striking abundance of SVs near the chromosome 2R centromere (Fig. 2A; Supplemental Table 8).
We further sought for evidence of NHEJ and MMBIR among SVs classified as NHR based on additional sequence analysis, an analysis suggesting that the vast majority of nonhomology associated SVs involved NHEJ (Supplemental Table 9; Supplemental Text).
We also inferred SV formation mechanisms for 903 tandem duplications for which breakpoints were determined at nucleotide resolution. Out of these, 14 were inferred to be caused by homology-based rearrangements (NAHR), eight likely involved VNTR expansion/shrinkage, five remained unclassified, and all remaining 876 (97%) were inferred to be formed in the absence of sequence homology (NHR). As for deletions, additional sequence analysis suggested that the nonhomology associated tandem duplications involved NHEJ (Supplemental Text).
Discussion
The sequence data resource created by the DGRP provides the basis for advanced genetic studies in one of the most widely applied model genetics organisms. Using sequencing data from the DGRP, we have generated the first high-resolution SV map in Drosophila melanogaster. The nucleotide resolution of most SVs in our set enabled an initial investigation of their likely functional impact, based on intersection with functional regions and based on demonstrating the association of SVs with gene expression variation at >100 genomic loci. Consistent with many SVs having fitness effects, we observed a strong depletion of SVs overlapping coding sequences, a finding corroborated by population genetics analyses and inferred links between SV gene overlap and environmental response functional categories (Dopman and Hartl 2007; Emerson et al. 2008; Mills et al. 2011). Furthermore, we identified several novel fusion genes, including Cyp6a17:Cyp6a23, a possible functional role of which was further substantiated by eQTL analysis making use of substantial gene expression measurements available in male and female flies.
The nucleotide resolution of our data further facilitated the investigation of the relative distribution of mechanisms leading to SVs in the fly. While NHEJ corresponds to the most frequent formation mechanisms, more than half of all variable bases that we inferred relative to the reference genome involved mobile elements, which underscores the importance of transposable and retrotransposable elements in shaping the fly genome. We further demonstrated the utility of our genotyped resource for investigating genetic diversity within widely available laboratory strains, with results that may have implications for the interpretation of results from Drosophila research studies carried out in different laboratories.
Our study focused on the characterization of deletions and tandem duplications. Both are presumed to have a substantial impact on phenotypic diversity (Zhang et al. 2009), and they account for a large fraction of polymorphic genetic differences in flies (Emerson et al. 2008). Due to limitations in read length and the rather low accuracy of existing approaches inferring other SV classes in next-generation sequencing data (Mills et al. 2011), our study did not infer inversions, dispersed duplications, and nonreference transposable elements in the DGRP data. It is further of note that our resource contains a lower number of duplications than previous reports using microarrays (Emerson et al. 2008), possibly owing to our strict focus on tandem duplications and the conservative criteria we used for their ascertainment. We foresee that new DNA sequencing technologies, generating longer reads, and improvements of approaches will increase the genomic regions and SV classes accessible to the analyses that we described here, including regions with high repeat content that are presently difficult to ascertain with short DNA reads.
Despite these remaining limitations, we envision that our SV map will be of great value for the fly research community, further enhancing the utility of the DGRP resource (Mackay et al. 2012). The first dense set of SV genotypes in flies, described in this study, will facilitate genome-wide association studies beyond SV-associated eQTL mapping, performed by imputation or directly through relating SV genotypes to phenotypes (Craddock et al. 2010), to enable the dissection of complex traits in a key genetics model organism (Ayroles et al. 2009). In addition, genetics and genomics-driven analyses using our nucleotide resolution SV map will facilitate further investigations of the interplay of coding and noncoding functional elements in the fruit fly genome, including cis regulatory elements.
Methods
DNA sequencing, sequence data retrieval, and read mapping
Illumina paired-end sequencing data were obtained from the Drosophila melanogaster Genetic Reference Panel (DGRP) (Ayroles et al. 2009; Mackay et al. 2012) (http://www.hgsc.bcm.tmc.edu/projects/dgrp/older_data_releases/lines/). We further performed Illumina GAIIx/HiSeq 2000 paired-end sequencing on three additional fly strains (Canton-S from Stanford University, Oregon-R from the University of Zurich, and Oregon-R from EMBL). The sequencing read length was 36 bp for Canton-S Stanford University and 101 bp for the remaining two samples. DNA library preparation was performed as described recently (Rausch et al. 2012a), with the paired-end libraries showing a median insert size of ∼470 bp. We aligned all reads onto the fly reference genome (BDGP R5/dm3, chromosome Uextra excluded) using Novoalign (v2.06.09s) mapping software (http://www.novocraft.com/); parameter ‘-r Random’. Read pair duplicates were removed using Picard (http://picard.sourceforge.net/).
SV discovery
To discover deletions and tandem duplications we applied the SV detection methods DELLY (Rausch et al. 2012b), Pindel (Ye et al. 2009) v0.2.4d, and CNVnator (Abyzov et al. 2011) v0.2.2 on each sample. We further used Genome STRiP (Handsaker et al. 2011) v1.0.4 to perform simultaneous population-scale deletion discovery on the 39 DGRP samples. For Pindel we set the maximum detectable SV size to 129,472 (parameter ‘-x 6’) and the minimum number of matched bases to 20 (‘-d 20’). For CNVnator we used a bin size of 200 bp. The minimum required mapping quality for Genome STRiP was set to 20.
SV merging and generation of the discovery set
To combine inferred SVs from the different methods, we first merged the predictions of all 39 samples (three samples in the case of the laboratory strains) for each method individually. To do so, we defined confidence intervals around the breakpoints according to the presumed resolution of breakpoint inference for each method as described in the Supplemental Text. To increase the positive predictive value we filtered out SVs that were predicted by a single method only, with the exception that we kept 283 deletion calls solely made by Genome STRiP (filtering steps are described in detail in the Supplemental Text).
SV genotyping
We used the genotyping module of Genome STRiP (Handsaker et al. 2011) for deletion genotyping (genotyping based on pair-end and read-depth information). We required that the genotyping results passed the Genome STRiP internal filtering measures. CopySeq (Waszak et al. 2010) was used for tandem duplication genotyping. Because of the expected homozygosity in the inbreed DGRP lines, we included only those SVs into the genotype set that harbored a homozygous deletion (or duplication) genotype in at least one sample.
PCR-based validations
PCRs were performed in five randomly picked fly lines, both on the selected sample as well as a reference, as previously described (Rausch et al. 2012a).
Whole-genome tiling array-based validations
We prepared whole-genome tiling arrays for six randomly picked DGRP lines as well as the reference strain. DNA was extracted and hybridized to Affymetrix GeneChip Drosophila Tiling 1.0R Arrays (probe annotation was mapped to the current reference genome build dm3). For each probe, the raw intensity was determined. We normalized the intensities of each sample array according to the median intensity. Subsequently, for each of the six lines we obtained all SVs from the discovery set or genotyping set, respectively, that were fully overlapped by at least one oligonucleotide probe position, and computed the median log2 intensity ratio between the sample and the reference for all probes falling into an SV. We excluded SVs for which the median probe intensity on the reference array was among the upper or lower 0.05-quantile of all reference array probe intensities, since outliers in terms of probe response may not be suitable for evaluating SV loci using a hybridization-based technique (those are presumably enriched for regions affected by probe cross-hybridization, or such lacking any response). For estimating the false-discovery rate (FDR) we compared the intensity log2 ratios of each SV locus in question between samples inferred to harbor an SV (e.g., for deletions, mostly negative log2 ratios were observed) and samples inferred not to harbor an SV (i.e., in these cases, a log2 ratio centered at 0 was observed). We computed the median log2 intensity ratio distribution of negative calls (SVs that were not predicted or negatively genotyped in a certain sample; see gray lines in Fig. 3A–D) and used the 0.05-quantile as a cutoff for assuming ‘verification’ of the SV. The 0.05-quantile was inferred based on the opposite right tail of the distribution for deletions (positive log2-ratios), and the left tail for duplications (negative log2-ratios), to avoid possible biases in those “reference” distributions based on false-negative SV calls. For both deletions and tandem duplications we estimated the FDR as the fraction of SVs not reaching the aforementioned cutoff. For the deletion discovery set we also included 3704 inferred deletions that were not assessed by the arrays and for which the formation mechanism analysis inferred a mobile element movement event, assuming an FDR of 5% for these (corresponding to the PCR-based FDR assessment) and weighing the FDR according to the overall number of events falling into this SV class.
SNP calling
To identify SNPs we applied the Joint Genotyper for Inbred Lines (JGIL) (Stone 2012) to the set of DGRP as well as the laboratory strain samples (both sets were analyzed separately). The following parameters were used: Read mapping quality cutoff: 20; number of generation: 20. For this study, we ignored any site for which the JGIL inferred that the SNP quality was less than 20, and set to ‘N’ the genotype of any individual for which the SNP quality score was less than 20 or for which there were fewer than three reads covering the site. For population genetic analyses, we restricted our SNP set to variable sites inferred among the 38 lines of the DGRP.
Population genetic analyses
Tajima's D statistics were obtained with VariScan (Hutter et al. 2006). We classified each mutation (SNP or SV) into distinct genomic compartments (intergenic, intronic, UTR, and coding) using FlyBase annotation version 5.40 (McQuilton et al. 2012). To achieve robust estimates of the summary statistics for each genomic compartment, we used sliding windows covering 100 (in the case of SNPs) or 50 (in the case of SVs) nonoverlapping genomic variants. When analyzing SVs on individual chromosomes we shrank the window size to 30 variants and allowed the windows to overlap. To estimate the size of LD blocks between SNPs and between SVs, we calculated r2 using VCFtools (Danecek et al. 2011).
Analysis of overlap between SVs and functional elements
Analyses of overlap between SVs and functional elements were performed based on protein-coding RefSeq genes obtained from the UCSC Genome Browser on January 9, 2012. Putative fusion genes were determined as pairs of genes where the start of an SV falls into the first gene and its end into the second gene. We furthermore required that both genes were on the same strand and that start and end of an SV overlapped exactly one gene each. The Gene Ontology (GO)-term enrichment analysis was performed using Ontologizer v2.1 software (Bauer et al. 2008).
Expression quantitative trait locus (eQTL) mapping
Mapping of eQTLs was pursued following the same principles used in a recent study relating SVs to gene expression variation in humans (Schlattl et al. 2011). Expression measurements were compared with the SV status in a pairwise fashion, across genic loci, by analysis of their pairwise Spearman correlation (Schlattl et al. 2011), followed by correction for multiple testing by controlling the false discovery rate (FDR) according to Benjamini and Hochberg. We separately mapped eQTLs with male and female gene expression data. The following adjustments relative to Schlattl et al. (2011) were made: First, normalized microarray-based gene expression values obtained from Ayroles et al. (2009), rather than transcriptome sequencing–based data, were used to compute eQTL associations. Second, we limited the search to SVs surrounding a genic locus of interest, thereby defining surrounding regions as segments starting 50 kb upstream and ending 50 kb downstream from each gene of interest.
SV formation mechanism analysis
SV formation mechanism inference was pursued with BreakSeq (Lam et al. 2010). Small template or nontemplate insertions (micro-insertions) were inferred using DELLY and Pindel. To infer their origin, sequences were mapped to the Drosophila melanogaster reference genome using BLAST (Altschul et al. 1990). Only sequences with a unique perfect match were considered for further analysis.
Statistical analyses and figures
Statistical analyses were performed using the software environment R v2.13 (R Development Core Team 2011) unless stated otherwise. Figures were generated using R v2.13, Circos v0.53 (Krzywinski et al. 2009), and the Integrative Genomics Viewer v2.0.17 (Robinson et al. 2011).
Data access
The whole-genome tiling array data of the validation experiments have been submitted to the EBI ArrayExpress Archive (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-1105. The sequencing data of the three laboratory strain samples have been submitted to the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) of the EBI European Nucleotide Archive (ENA) (http://www.ebi.ac.uk/ena/) under accession number ERP001387.
Acknowledgments
We thank Charles Girardot for assistance with the whole-genome tiling array data, Andreas Schlattl for assistance with the eQTL analysis, members of the Korbel group for discussions, and the EMBL GeneCore facility and EMBL's high-performance computing facility for support. We further thank the Drosophila melanogaster Genetic Reference Panel for data access. J.O.K. was funded by an Emmy Noether Fellowship from the German Research Foundation (KO 4037/1-1). D.A.G. was supported by the US National Institutes of Health under a Ruth L. Kirschstein National Research Service Award (1 F32 GM100635-01).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.142646.112.
References
- The 1000 Genomes Project Consortium 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A, Urban AE, Snyder M, Gerstein M 2011. CNVnator: An approach to discover, genotype and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21: 974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Aguade M 2009. Nucleotide and copy-number polymorphism at the odorant receptor genes Or22a and Or22b in Drosophila melanogaster. Mol Biol Evol 26: 61–70 [DOI] [PubMed] [Google Scholar]
- Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, et al. 2009. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet 41: 1061–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan LH, Lawrence F, Anholt RRH, et al. 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 41: 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Liu G, Eichler EE 2003. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet 73: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Church DM, Ventura M, Rocchi M, Eichler EE 2004. Analysis of segmental duplications and genome assembly in the mouse. Genome Res 14: 789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Grossmann S, Vingron M, Robinson PN 2008. Ontologizer 2.0—a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics 24: 1650–1651 [DOI] [PubMed] [Google Scholar]
- Bergman CM, Bensasson D 2007. Recent LTR retrotransposon insertion contrasts with waves of non-LTR insertion since speciation in Drosophila melanogaster. Proc Natl Acad Sci 104: 11340–11345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME 2010a. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet 42: 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. 2010b. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E, et al. 2010. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland JM, Thornton KR 2010. Validation of rearrangement break points identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol Evol 2010: 83–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF, Kimura M 1970. An introduction to population genetics theory. Harper & Row, New York [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman EB, Hartl DL 2007. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci 104: 19920–19925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320: 1629–1631 [DOI] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW 2006. Structural variation in the human genome. Nat Rev Genet 7: 85–97 [DOI] [PubMed] [Google Scholar]
- Handsaker RE, Korn JM, Nemesh J, McCarroll SA 2011. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet 43: 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G 2009. Mechanisms of change in gene copy number. Nat Rev Genet 10: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, Alkan C, Eichler EE, Sahinalp SC 2009. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res 19: 1270–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter S, Vilella AJ, Rozas J 2006. Genome-wide DNA polymorphism analyses using VariScan. BMC Bioinformatics 7: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, Kallicki J, Kaul R, Wilson RK, Eichler EE 2010. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143: 837–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PM, Lam HY, Urban AE, Korbel JO, Affourtit J, Grubert F, Chen X, Weissman S, Snyder M, Gerstein MB 2008. Analysis of copy number variants and segmental duplications in the human genome: Evidence for a change in the process of formation in recent evolutionary history. Genome Res 18: 1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Betancourt AJ, Schlötterer C 2012. Sequencing of pooled DNA samples (Pool-Seq) uncovers complex dynamics of transposable element insertions in Drosophila melanogaster. PLoS Genet 8: e1002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA 2009. Circos: An information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HYK, Mu XJ, Stütz AM, Tanzer A, Cayting PD, Snyder M, Kim PM, Korbel JO, Gerstein MB 2010. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol 28: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak MK, Begun DJ 2007. Molecular population genetics of female-expressed mating-induced serine proteases in Drosophila melanogaster. Mol Biol Evol 24: 1944–1951 [DOI] [PubMed] [Google Scholar]
- Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. 2009. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuilton P, St Pierre SE, Thurmond J 2012. FlyBase 101—the basics of navigating FlyBase. Nucleic Acids Res 40: D706–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE 2006. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res 16: 1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi-Seebacher M, Korbel JO 2011. Challenges in studying genomic structural variant formation mechanisms: The short-read dilemma and beyond. BioEssays 33: 840–850 [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2011. R: A language and environment for statistical computing. R Foundation for Statistical Computing. http://www.R-project.org/
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. 2012a. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148: 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO 2012b. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlattl A, Anders S, Waszak SM, Huber W, Korbel JO 2011. Relating CNVs to transcriptome data at fine resolution: Assessment of the effect of variant size, type, and overlap with functional regions. Genome Res 21: 2004–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EA 2012. Joint genotyping on the fly: Identifying variation among a sequenced panel of inbred lines. Genome Res 22: 966–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J 2010. Diversity of human copy number. Science 330: 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Levine MT, Eckert ML, Begun DJ 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179: 455–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. 2005. Fine-scale structural variation of the human genome. Nat Genet 37: 727–732 [DOI] [PubMed] [Google Scholar]
- Waszak SM, Hasin Y, Zichner T, Olender T, Keydar I, Khen M, Stütz AM, Schlattl A, Lancet D, Korbel JO 2010. Systematic inference of copy-number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Comput Biol 6: e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z 2009. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Xuan Z, Makarov V, Ye K, Sebat J 2009. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res 19: 1586–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR 2009. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 10: 451–481 [DOI] [PMC free article] [PubMed] [Google Scholar]