Abstract
The rhizarian amoeba Paulinella chromatophora harbors two photosynthetically active organelles of cyanobacterial origin that have been acquired independently of classic primary plastids. Because their acquisition did take place relatively recently, they are expected to provide new insight into the ancient cyanobacterial primary endosymbiosis. During the process of Paulinella endosymbiont-to-organelle transformation, more than 30 genes have been transferred from the organelle to the host nuclear genome via endosymbiotic gene transfer (EGT). The article discusses step-by-step protein import of EGT-derived proteins into Paulinella photosynthetic organelles with the emphasis on the nature of their targeting signals and the final passage of proteins through the inner organelle membrane. The latter most probably involves a simplified Tic translocon composed of Tic21- and Tic32-like proteins as well as a Hsp70-based motor responsible for pulling of imported proteins into the organelle matrix. Our results indicate that although protein translocation across the inner membrane of Paulinella photosynthetic organelles seems to resemble the one in classic primary plastids, the transport through the outer membrane does not. The differences could result from distinct integration pathways of Paulinella photosynthetic organelles and primary plastids with their respective host cells.
Keywords: Endosymbiosis, Molecular motor, Plastid, Protein import, Signal peptide, Vesicular trafficking
Introduction
The acquisition of a cyanobacterium by a eukaryotic cell and its subsequent transformation into a primary plastid, called primary endosymbiosis, turned out to be one of the most important transitions in the evolution of life on our planet. It triggered the emergence of new photosynthetic eukaryotic lineages: glaucophytes, red algae, and green plants, united under the name of Archaeplastida/Plantae (Rodriguez-Ezpeleta et al. 2005; Adl et al. 2012). The primary plastid endosymbiosis happened more than 1,500 million years ago (Yoon et al. 2004) and it is still obscure in many details although intensively studied (e.g.: Bhattacharya et al. 2007; Bodył et al. 2009; Gross and Bhattacharya 2011).
Interestingly, primary photosynthetic endosymbiosis took place once again in the history of our biosphere. The new photosynthetic consortium was created between the testate filose amoeba Paulinella chromatophora, a member of the supergroup Rhizaria, and an α-cyanobacterium (Fig. 1) (Marin et al. 2005; Yoon et al. 2006, 2009; see for reviews: Bodył et al. 2012; Nakayama and Archibald 2012). Interestingly, this endosymbiosis happened about 60 million years ago (Nowack et al. 2008). Because Paulinella endosymbionts/organelles were acquired much more recently than the primary plastids of Archaeplastida, they offer a new insight into the process of endosymbiont-to-organelle transformation and, in particular, into the evolution of protein import mechanisms into prokaryote-derived organelles at their early stage.
Fig. 1.
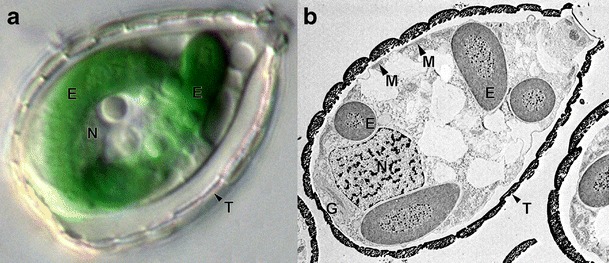
Paulinella chromatophora viewed under the optical (a) and electron (b) microscope. This testate filose amoeba is surrounded by the cell wall called theca (T), which is composed of silica scales. Apart from typical eukaryotic organelles such as nucleus (N) and mitochondria (M), it harbors two cyanobacterium-derived endosymbionts (E). The endosymbionts are photosynthetically active, deeply integrated with the host cell, and their genome have been reduced to one third in comparison to their cyanobacterial ancestors. Moreover, the endosymbionts import proteins encoded by genes that were transferred from the endosymbiont to the host nuclear genome. This import proceeds co-translationally via the host endomembrane system, involving the Golgi apparatus (G). All these features justify to call Paulinella photosynthetic endosymbionts true cell organelles. The Paulinella images were kindly supplied by Dr Eva Nowack
For many years, Paulinella endosymbionts have been the subject of a hot debate whether they really represent true cell organelles (Bhattacharya and Archibald 2006; Theissen and Martin 2006; Bodył et al. 2007). They divide synchronously with the Paulinella host, exchange metabolites with its cytosol, and are incapable of growing independently of the host cell (Kies and Kremer 1979; Yoon et al. 2006; Marin et al. 2005). Moreover, the genome of Paulinella endosymbionts was reduced by a factor of three compared to their closest free-living cyanobacterial relatives (Nowack et al. 2008; Reyes-Prieto et al. 2010) and lost many genes, including those involved in essential biosynthetic pathways (Nowack et al. 2008; Reyes-Prieto et al. 2010). The genome-size reduction was accompanied by movement of genes from the endosymbiont to the host nuclear genome, a process known as endosymbiotic gene transfer (EGT). The genome and transcriptome analyses identified more than 30 such EGT-derived genes in the Paulinella nuclear genome (Nakayama and Ishida 2009; Reyes-Prieto et al. 2010; Nowack et al. 2011).
The debate about the organellar status of Paulinella photosynthetic endosymbionts was finally settled by the recent studies of Nowack and Grossman (2012). The authors showed that the endosymbionts import nucleus-encoded and EGT-derived photosynthetic proteins, such as PsaE, PsaK1, and PsaK2, and thus should be called true cell organelles. They also demonstrated that one of these proteins, PsaE, is targeted via the endomembrane system to these organelles but, unexpectedly, not by means of a classic signal peptide, which is commonly used by proteins imported to this system. The co-translational import of PsaE clearly contrasts with the post-translational transit peptide- and Toc-Tic-based mechanism that is characteristic of the Archaeplastida primary plastids (Fig. 2) (see for reviews: Li and Chiu 2010; Shi and Theg 2012).
Fig. 2.
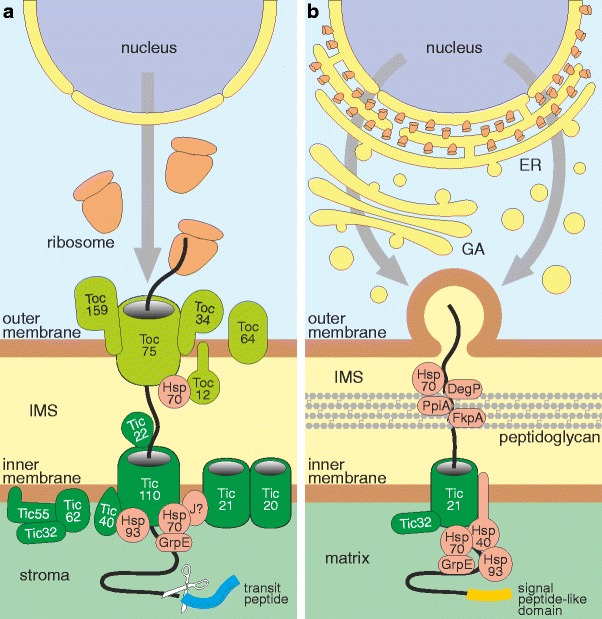
Protein import into classic primary plastids (a) and Paulinella photosynthetic organelles (b). (a) The majority of nucleus-encoded plastid proteins are imported into classic primary plastids post-translationally using N-terminal transit peptide and Toc-Tic supercomplex. The latter consists of many specialized protein subunits that function as transit peptide receptors (Toc34, Toc64, and Toc159), protein-conducting channels (Toc75, Tic20, Tic21, and Tic110), regulatory elements (Tic55, Tic62, and Tic32), scaffold proteins (Tic110), Toc-Tic translocons connecting subunits (Toc12, Tic22), chaperones (Hsp70, Hsp93), and co-chaperones (Tic40). There are two independent molecular motors pulling imported proteins into the stroma: one consists of Tic110, Tic40, and Hsp93, whereas the second engages Hsp70, possibly GrpE, and some unknown J-domain protein (J?). (b) In contrast to classic primary plastids, nucleus-encoded photosynthetic proteins of Paulinella chromatophora are usually equipped with a signal peptide-like domain and are targeted to its photosynthetic organelles via the endomembrane system involving the endoplasmic reticulum (ER) and Golgi apparatus (GA). After the release of imported proteins into the intermembrane space (IMS), their further trafficking through the peptidoglycan wall could be facilitated by molecular chaperones, such as DegP, FkpA, PpiA, and Hsp70. The final import step, translocation across the inner membrane, may proceed via the Tic21 channel homolog with the help of Tic32-like protein, a calcium- and redox-sensing regulatory subunit. The imported proteins are hypothesized to be finally pulled into the organelle matrix by a molecular motor composed of Hsp93, Hsp70, Hsp40, and GrpE
Although the findings of Nowack and Grossman (2012) are invaluable contributions to the subject of Paulinella endosymbiosis, and thus endosymbiosis in general, there are still two critical issues unsolved: (i) the nature of targeting signals in proteins imported into Paulinella photosynthetic organelles and (ii) mechanisms of further protein transport through the subsequent organelle envelope barriers, including the peptidoglycan wall and the inner plastid membrane. Therefore, we have decided to raise these important questions here. We also discuss implications of the new findings for understanding of the classic primary plastid endosymbiosis, especially early stages in the evolution of photosynthetic organelles derived from cyanobacterial endosymbionts.
Targeting signals of proteins imported into Paulinella photosynthetic organelles
The PsaE protein from Paulinella CCAC0185 strain that Nowack and Grossman (2012) analyzed is indeed devoid of a hydrophobic domain that could function as a signal peptide. This strongly contrasts with its homolog from Paulinella FK01 strain, where a signal peptide was predicted with high confidence (Mackiewicz and Bodył 2010). Thus, although both PsaE homologs are imported into the same cellular compartment via the endomembrane system, they appear to have evolved distinct targeting mechanisms in the two Paulinella strains. Notably, their mature sequences differ by one indel and 21 substitutions per only 69 amino acid residues (Fig. 3). Eight of these substitutions are non-conserved (according to Blosum62 substitution matrix) and lead to changes in biochemical properties of amino acid residues. Consequently, these substitutions could reflect the evolution of an internal PsaE targeting signal in the Paulinella CCAC0185 strain investigated by Nowack and Grossman (2012).
Fig. 3.

Alignment of PsaE proteins from two Paulinella strains. PsaE from FK01 strain is equipped with an N-terminal extension (yellow box) that was predicted with high confidence as a signal peptide by 27 from 30 algorithms with an unambiguous cleavage site (Mackiewicz and Bodył 2010). Eight non-conserved substitutions according to Blosum62 matrix (in red), which change biochemical properties of residues, may constitute some internal PsaE targeting signal in the CCAC0185 strain
Although targeting mechanisms for the Paulinella PsaK proteins were not determined by Nowack and Grossman (2012), it is possible that they are also imported into Paulinella photosynthetic organelles via the endomembrane system. According to these authors, the C-terminal hydrophobic domain of Paulinella PsaK proteins could function as an internal targeting signal; however, it seems more likely that their N-terminal hydrophobic domains represent signal peptides (Mackiewicz et al. 2012). Compared to their cyanobacterial homologs, these domains have lost several hydrophilic residues and acquired some hydrophobic ones, thereby gaining features of signal peptides (Mackiewicz et al. 2012). In support of this, experimental studies showed that similar changes in mitochondrial and plastid membrane proteins caused their targeting to the endomembrane system (Lee et al. 2004; Kanaji et al. 2000; Horie et al. 2003; Waizenegger et al. 2003).
N-terminal hydrophobic domains in two other nucleus-encoded and endosymbiont-derived Paulinella proteins, homologs of PsbN and Synechococcus WH5701_13415, respectively (Nowack et al. 2011), could also be adapted to signal peptide functions because they show similar compositional changes to the PsaK proteins (Mackiewicz et al. 2012). These data suggest that not only PsaE but also other Paulinella endosymbiont-derived proteins encoded by the host nuclear genome can utilize vesicular trafficking to reach the photosynthetic organelles.
Import of Paulinella proteins across the intermembrane space and the inner envelope membrane of its photosynthetic organelles
Crossing the peptidoglycan wall
After fusion of endomembrane system-derived transport vesicles with the outer membrane of Paulinella photosynthetic organelles, imported proteins are released into the intermembrane space and then must cross the peptidoglycan wall (Fig. 2b). Unfortunately, in contrast to the glaucophyte muroplast (Pfanzagl et al. 1996a, b; Pfanzagl and Löffelhardt 1999), nothing is known about the thickness and crosslinkage as well as eventual modification of the Paulinella organelle peptidoglycan. Nevertheless, adaptations required for this translocation appear to be the low molecular weight and nearly neutral charge. These features are common to Paulinella EGT-derived proteins and fit well with the properties characteristic of proteins known to cross dense and negatively charged peptidoglycan walls (Mackiewicz et al. 2012). Moreover, the process could be assisted by molecular chaperones, homologous to bacterial periplasmic space proteins, such as DegP, FkpA, and PpiA, which are encoded in the Paulinella organelle genome (Table 1) (Mackiewicz and Bodył 2010).
Table 1.
List of Paulinella chromatophora proteins that can be involved in protein import into its photosynthetic organelles
| Accession number | Description | Putative role in protein import |
|---|---|---|
| YP_002049046 | homolog to Hsp70 (DnaK), molecular chaperone | passage of proteins through the intermembrane space |
| YP_002048762 | homolog to bacterial DegP, serine endoprotease | passage of proteins through the intermembrane space |
| YP_002048801 | homolog to bacterial FkpA, FKBP-type peptidyl-prolyl cis-trans isomerase (rotamase) | passage of proteins through the intermembrane space |
| YP_002048701 | homolog to bacterial PipA, peptidyl-prolyl cis-trans isomerase A (rotamase A) | passage of proteins through the intermembrane space |
| YP_002048795 | homolog to plant Tic21, protein-conducting channel in the inner organelle membrane | protein translocation across the inner organelle membrane |
| YP_002049520 | homolog to plant Tic32, short-chain dehydrogenase | redox regulation of the protein import |
| YP_002049484 | homolog to Hsp93, ATPase | pulling of imported proteins into the organelle matrix |
| YP_002049416 | homolog to Hsp93, ATPase | pulling of imported proteins into the organelle matrix |
| YP_002048846 | homolog to Hsp40 (DnaJ), heat shock protein | pulling of imported proteins into the organelle matrix; stimulates ATPase activity of Hsp70 |
| YP_002048821 | homolog to Hsp70 (DnaK), molecular chaperone | pulling of imported proteins into the organelle matrix; interacts with Hsp40 and GrpE |
| YP_002048880 | homolog to Hsp70 (DnaK), molecular chaperone | pulling of imported proteins into the organelle matrix; interacts with Hsp40 and GrpE |
| YP_002049062 | homolog to GrpE, heat shock protein | pulling of imported proteins into the organelle matrix; nucleotide exchange factor for Hsp70 |
One of three potential Paulinella Hsp70 (DnaK) chaperones, YP_002049046, could also be involved in protein passage across the intermembrane space and the peptidoglycan wall. YP_002049046 was predicted by SecretomeP 2.0 Server to be a non-classic secreted protein targeted by a signal peptide-independent pathway (Bendtsen et al. 2005). In support of this, some studies identified Hsp70 in the intermembrane space of primary plastids indicating its role in protein translocation across this space (Schnell et al. 1994; Kessler and Blobel 1996; Becker et al. 2004).
Crossing the inner organelle membrane with Tic21-like protein
The passage through the inner organelle membrane could be mediated by a homolog of the plastid Tic21 protein, encoded by the Paulinella organelle genome (Fig. 2b, Table 1) (Bodył et al. 2010). The Paulinella Tic21-like protein contains four well-predicted α-helical transmembrane domains, similar to its higher plant counterpart, which suggests its capability to form a protein translocation pore. Interestingly, Tic21 molecular weight and number of transmembrane domains are characteristic of other protein-conducting channels, e.g. Tic20 – the classic Tic translocon (Chen et al. 2002; Hirabayashi et al. 2011; Kovacs-Bogdan et al. 2011) and Tim23/Tim17 – a mitochondrial inner membrane translocon (Marom et al. 2011). These common features imply parallel evolution of import functions in Tic21, Tic20, and Tim23/Tim17 channel proteins (Kikuchi et al. 2009).
Although plant Tic21 was claimed to be an iron transporter (Duy et al. 2007), recent studies by Kikuchi et al. (2009) and Hirabayashi et al. (2011) have confirmed previous suggestions on its role in the plastid protein import (Teng et al. 2006). It is probable that the Paulinella Tic21 homolog fulfills much the same functions in its photosynthetic organelles because the genetic complementation of Arabidopsis tic21 knockout mutant with its cyanobacterial Synechocystis ortholog turned out to be successful (Lv et al. 2009).
The protein import into Paulinella photosynthetic organelles may also involve a confident, encoded by their genome, homolog to plant Tic32 (Table 1). This protein, together with Tic55 and Tic62, is responsible for regulation of protein import by redox-sensing and calcium-signaling in the plastids of green plants (or chloroplasts) (Li and Chiu 2010; Shi and Theg 2012; Stengel et al. 2010). The Paulinella Tic32 possesses all the essential domains: dehydrogenase⁄reductase (SDR), NAD(P)-binding, and calmodulin (CaM)-binding domain and all the conserved motifs and residues associated with dehydrogenase⁄reductase activity (Bodył et al. 2010). Although homologs to Tic55 and Tic62 were not identified in the genome of Paulinella photosynthetic organelles (Bodył et al. 2010), they may be encoded by endosymbiont genes transferred to the host nuclear genome. All these three proteins would then constitute a redox-sensing complex in the Paulinella organelle inner membrane, but it cannot be excluded that Tic32 plays the regulatory function alone.
A molecular motor associated with protein import across the inner envelope membrane of Paulinella photosynthetic organelles
Two protein import motors in green plant plastids
The final passage of proteins through the inner envelope membrane of plastids is driven by a molecular motor pulling imported proteins into the stroma. There are two such Tic-associated motor complexes that act in parallel (Fig. 2) (see for reviews: Shi and Theg 2011; Flores-Perez and Jarvis 2012). In the first motor, Tic110, Hsp93 (the Hsp100-type) chaperone, and Tic40 co-chaperone cooperate with each other (Kovacheva et al. 2005). The Tic40 is anchored in the inner membrane with its N-terminal transmembrane region (Chou et al. 2003; Kao et al. 2012). Moreover, the protein is associated with Tic110 by the TPR domain, whereas its C-terminal Sti1 (Hip/Hop) domain engages ATPase domains of Hsp93 to ATP hydrolysis. The ATP hydrolysis generates energy necessary for conformational changes of Hsp93 that enable pulling of proteins into the stroma.
The second, recently discovered, import motor involves stromal Hsp70 chaperone as the driving force for protein import and was identified in both the moss Physcomitrella patens (Shi and Theg 2010) and the higher plant Arabidopsis thaliana (Su and Li 2010). Because Hsp70 requires proteins with J domain and nucleotide exchange factors (e.g. GrpE) for effective functioning (Kampinga and Craig 2010), such partners are expected in the Hsp70-based molecular motor. The J-domain proteins play a crucial role in stimulation of ATP hydrolysis, whereas GrpE facilitates the release of ADP from the chaperone. In agreement with that, it was found that one of the stromal co-chaperones, CGE (Chloroplast GrpE homolog), has an essential role in plastid protein import (Shi and Theg 2010). Although the involvement of J-domain proteins as a molecular motor component has not yet been confirmed, their role cannot be excluded on the basis of dnaJ-J8 mutants, which still at low level could express truncated J proteins masking expected defects in protein import (Chiu et al. 2010).
A hypothetical Hsp70-dependent import motor in Paulinella photosynthetic organelles
The genome of Paulinella photosynthetic organelles encodes two homologs to the plant Hsp93 (Table 1), but it lacks genes for Tic110- and Tic40-like proteins (Bodył et al. 2010). Their absence from the Paulinella organelle genome should be expected because the plant tic110 and tic40 genes are of eukaryotic origin (Shi and Theg 2012). The preliminary analyses by Nowack et al. (2011) also indicate that they, most probably, do not reside in the host nuclear genome. Consequently, Paulinella photosynthetic organelles are probably devoid of a Tic110-Hsp93-Tic40-based molecular motor typical of higher plant plastids.
Interestingly, we found at least two putative stromal Hsp70 (DnaK) homologs, encoded by the Paulinella photosynthetic organelle (Table 1), implying that an Hsp70-dependent import motor similar to the alternative one in plant primary plastids operates in the protist (Fig. 2b). The other potential component of this motor is YP_002048846, one of four Hsp40 (DnaJ) proteins encoded in the Paulinella organelle genome (Bodył et al. 2010). This protein is equipped with clear DnaJ and α-helical membrane-anchoring domains, and thus may function as a co-chaperone for Hsp70 by stimulation of its ATPase activity. The majority of programs used to predict the topology of this transmembrane helix showed internal orientation of the N-terminal part of YP_002048846 indicating that its J domain is exposed to the organelle matrix (Bodył et al. 2010). Such organization of the Paulinella Hsp40 suggests that it could indeed represent a component of the Hsp70-based molecular motor. The third key element of the pulling machinery is probably a clear Paulinella homolog to GrpE (Table 1). The GrpE-like protein, encoded by the Paulinella organelle genome, could function as a nucleotide exchange factor for Hsp70.
In support of the presence of Hsp70-based import motor in Paulinella photosynthetic organelles, mitochondrial protein import motor is also driven by matrix Hsp70 chaperones (Schmidt et al. 2010). Tim14 (Pam18) is anchored in the inner mitochondrial membrane by its transmembrane domain and functions as a J-domain protein that in co-operation with Tim16 (Pam16) stimulates the ATPase activity of Hsp70. The nucleotide exchange factor called Mge1 (Mitochondrial GrpE-related protein 1) participates in this process as well. These data indicate that the functionally similar molecular motors evolved three times independently: in mitochondria, in classic primary plastids, and very likely in Paulinella photosynthetic organelles.
At present, it cannot be excluded that Paulinella Hsp93 and Hsp70 proteins do not cooperate with their co-chaperones and act according to the Brownian ratchet (or trapping) model proposed for mitochondrial Hsp70 (Yamano et al. 2008). According to this model, random motion (Brownian oscillations) causes a short segment of an imported polypeptide to become temporarily exposed from the translocation pore to the matrix. Then, Hsp70 chaperones bind to this segment and prevent the reverse movement of the polypeptide chain. In this way, the protein has no alternative but to enter the matrix.
Does Paulinella recapitulate early stages of primary plastid evolution?
The findings that most of the EGT-derived proteins are imported co-translationally into Paulinella photosynthetic organelles suggest that proteins could have been initially targeted to classic primary plastids also via the endomembrane system and the current post-translational protein import based on both transit peptides and Toc and Tic translocons evolved later (Bhattacharya et al. 2007). In fact, the precursors of some higher plant plastid proteins, such as α-carbonic anhydrase, nucleotide pyrophosphatase/phosphodiesterase, and α-amylases, do carry signal peptides (Chen et al. 2004; Villarejo et al. 2005; Nanjo et al. 2006; Kitajima et al. 2009). However, they represent only a small group of non-photosynthetic proteins for which endomembrane system-mediated trafficking probably evolved to permit glycosylation in the Golgi apparatus or for dual trafficking to both the plastid and the cell wall (see for discussion: Bodył et al. 2009). Moreover, it is difficult to imagine how selection could have favored a stepwise modification of signal peptides into transit peptides in hundreds of plastid proteins. This would have caused substantial mistargeting and disruption of plastid functions (Bodył et al. 2009).
Paulinella photosynthetic organelles and classic primary plastids were acquired independently (the latter much earlier) from distinct cyanobacteria: α and β, respectively (Marin et al. 2005, 2007), which probably resulted in their distinct way of integration into the host cells. This is emphasized by the fact that, in contrast to Paulinella, no photosynthetic proteins are imported into primary plastids via the endomembrane system. The reason why Paulinella could not evolve classic Toc-Tic import machinery is lack of genes encoding many Toc and Tic components, especially Toc75 – the major protein-conducting channel in the outer plastid membrane (Li and Chiu 2010; Shi and Theg 2012). These proteins were most probably lost from the photosynthetic organelle genome (Bodył et al. 2010) without their prior successful transfer to the host nucleus (Nowack et al. 2011). Therefore, although Paulinella photosynthetic organelles provide exciting new insights into mechanisms of organellogenesis, it is difficult to use them as a model for the evolution of early stages of protein targeting to classic primary plastids. Nevertheless, protein import across the inner envelope membrane, such as the previously discussed molecular import motor, seems to be an example of parallel evolution in the case of classic primary plastids and Paulinella photosynthetic organelles.
Perspectives
Apart from Paulinella, there are many other eukaryotes in endosymbiotic relationships with cyanobacteria, such as euglenids, cercozoans, dinoflagellates, diatoms, land plants, fungi, sponges, corals, and tunicates (Carpenter and Foster 2003; Rasmussen and Nilsson 2003; Raven 2003; Kneip et al. 2007). Especially notable are the protozoans Auranticordis quadriverberis (Chantangsi et al. 2008), Ebria tripartite, and Hermesinum adriaticum (Hargraves 2002) because they are, like Paulinella, classified to the phylum Cercozoa (Hoppenrath and Leander 2006; Chantangsi et al. 2008; Heger et al. 2010; Adl et al. 2012). It will be interesting to learn how advanced these relationships are, and if parallel or different mechanisms of endosymbiont establishment evolved in these species.
Well documented cases of deeply integrated cyanobacterial endosymbionts in non-cercozoan hosts are also worth noting. Good examples are the diatoms Climacodium frauenfeldianum (Carpenter and Foster 2003) and Rhopalodia gibba (Prechtl et al. 2004) whose intracellular permanent symbionts are related to Cyanothece-like cyanobacteria (Prechtl et al. 2004). Studies of R. gibba revealed that its endosymbionts are vertically transmitted (Prechtl et al. 2004) and show many modifications in their genome typical of other endosymbionts, such as numerous deletions, elevated AT content, loss/fusion/truncation of genes, pseudogenization, and expansion of non-coding regions (Prechtl et al. 2004; Kneip et al. 2008). A stable long-term association involving endosymbiotic cyanobacteria were also found in the scleractinian coral Montastraea cavernosa (Lesser et al. 2004). In the case of M. cavernosa, the cyanobacteria are located within the epithelial host cells coexisting with the symbiotic dinoflagellates (zooxanthellae) (Lesser et al. 2004).
One of the paradigms of contemporary evolutionary cell biology is the assumption that transformations of endosymbionts into cell organelles are a rarity (Cavalier-Smith and Lee 1985; Cavalier-Smith 2000). The paradigm is based on presumptive difficulties with the origin of an import apparatus for nucleus-encoded organelle proteins, but such machinery could evolve via evolutionary tinkering from components pre-existing in the endosymbiont and/or the host (Gross and Bhattacharya 2011; Bodył et al. 2012; Shi and Theg 2012). Thus, when considering these data and the case of Paulinella photosynthetic organelles, it is possible that also other cyanobacterial endosymbionts are undergoing transformation into true cell organelles.
Acknowledgments
We are very grateful to Dr E. Nowack for providing Paulinella images, and to two anonymous Reviewers for their excellent comments and remarks that significantly improved the paper. This work was supported by grant UMO-2011/01/N/NZ8/00150 to P.G. and P.M. and Wrocław University grant 1069/S/KBEE/2012 to A.B.
Contributor Information
Paweł Mackiewicz, Phone: +48-713756303, FAX: +48-713756234, Email: pamac@smorfland.uni.wroc.pl.
Andrzej Bodył, Email: bodyl@biol.uni.wroc.pl.
Przemysław Gagat, Email: przemyslaw.gagat@smorfland.uni.wroc.pl.
References
- Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, le Gall L, Lynn DH, McManus H, Mitchell EA, Mozley-Stanridge SE, Parfrey LW, Pawlowski J, Rueckert S, Shadwick L, Schoch CL, Smirnov A, Spiegel FW. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–514. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Hritz J, Vogel M, Caliebe A, Bukau B, Soll J, Schleiff E. Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts. Mol Biol Cell. 2004;15:5130–5144. doi: 10.1091/mbc.E04-05-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Kiemer L, Fausboll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Archibald J. The difference between organelles and endosymbionts. Response to Theissen and Martin. Curr Biol. 2006;16:R1017–R1018. doi: 10.1016/j.cub.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Archibald JM, Weber AP, Reyes-Prieto A. How do endosymbionts become organelles? Understanding early events in plastid evolution. Bioessays. 2007;29:1239–1246. doi: 10.1002/bies.20671. [DOI] [PubMed] [Google Scholar]
- Bodył A, Mackiewicz P, Stiller JW. The intracellular cyanobacteria of Paulinella chromatophora: endosymbionts or organelles? Trends Microbiol. 2007;15:295–296. doi: 10.1016/j.tim.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Bodył A, Mackiewicz P, Stiller JW. Early steps in plastid evolution: current ideas and controversies. Bioessays. 2009;31:1219–1232. doi: 10.1002/bies.200900073. [DOI] [PubMed] [Google Scholar]
- Bodył A, Mackiewicz P, Stiller JW. Comparative genomic studies suggest that the cyanobacterial endosymbionts of the amoeba Paulinella chromatophora possess an import apparatus for nuclear-encoded proteins. Plant Biol (Stuttg) 2010;12:639–649. doi: 10.1111/j.1438-8677.2009.00264.x. [DOI] [PubMed] [Google Scholar]
- Bodył A, Mackiewicz P, Gagat P. Organelle evolution: Paulinella breaks a paradigm. Curr Biol. 2012;22:R304–R306. doi: 10.1016/j.cub.2012.03.020. [DOI] [PubMed] [Google Scholar]
- Carpenter EJ, Foster RA. Marine cyanobacterial symbioses. In: Rai AN, Bergman B, Rasmussen U, editors. Cyanobacteria in symbiosis. Netherlands: Springer; 2003. pp. 11–17. [Google Scholar]
- Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000;5:174–182. doi: 10.1016/S1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Lee JJ. Protozoa as hosts for endosymbioses and the conversion of symbionts into organelles. J Eukaryot Microbiol. 1985;32:376–379. doi: 10.1111/j.1550-7408.1985.tb04031.x. [DOI] [Google Scholar]
- Chantangsi C, Esson HJ, Leander BS. Morphology and molecular phylogeny of a marine interstitial tetraflagellate with putative endosymbionts: Auranticordis quadriverberis n. gen. et sp. (Cercozoa) BMC Microbiol. 2008;8:123. doi: 10.1186/1471-2180-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Smith MD, Fitzpatrick L, Schnell DJ (2002) In vivo analysis of the role of atTic20 in protein import into chloroplasts. Plant Cell 14:641–654 [DOI] [PMC free article] [PubMed]
- Chen MH, Huang LF, Li HM, Chen YR, Yu SM. Signal peptide-dependent targeting of a rice alpha-amylase and cargo proteins to plastids and extracellular compartments of plant cells. Plant Physiol. 2004;135:1367–1377. doi: 10.1104/pp.104.042184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CC, Chen LJ, Li HM. Pea chloroplast DnaJ-J8 and Toc12 are encoded by the same gene and localized in the stroma. Plant Physiol. 2010;154:1172–1182. doi: 10.1104/pp.110.161224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Fitzpatrick LM, Tu SL, Budziszewski G, Potter-Lewis S, Akita M, Levin JZ, Keegstra K, Li HM. Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 2003;22:2970–2980. doi: 10.1093/emboj/cdg281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy D, Wanner G, Meda AR, von Wiren N, Soll J, Philippar K. PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell. 2007;19:986–1006. doi: 10.1105/tpc.106.047407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Perez U, Jarvis P (2012) Molecular chaperone involvement in chloroplast protein import. Biochim Biophys Acta in press. doi:10.1016/j.bbamcr.2012.1003.1019 [DOI] [PubMed]
- Gross J, Bhattacharya D. Endosymbiont or host: who drove mitochondrial and plastid evolution? Biol Direct. 2011;6:12. doi: 10.1186/1745-6150-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargraves PE. The ebridian flagellates Ebria and Hermesinum. Plankton Biol Ecol. 2002;49:9–16. [Google Scholar]
- Heger TJ, Mitchell EAD, Todorov M, Golemansky V, Lara E, Leander BS, Pawlowski J. Molecular phylogeny of euglyphid testate amoebae (Cercozoa: Euglyphida) suggests transitions between marine supralittoral and freshwater/terrestrial environments are infrequent. Mol Phylogenet Evol. 2010;55:113–122. doi: 10.1016/j.ympev.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Kikuchi S, Oishi M, Nakai M (2011) In vivo studies on the roles of two closely related Arabidopsis Tic20 proteins, AtTic20-I and AtTic20-IV. Plant Cell Physiol 52:469–478 [DOI] [PubMed]
- Hoppenrath M, Leander BS. Ebriid phylogeny and the expansion of the Cercozoa. Protist. 2006;157:279–290. doi: 10.1016/j.protis.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Horie C, Suzuki H, Sakaguchi M, Mihara K. Targeting and assembly of mitochondrial tail-anchored protein Tom5 to the TOM complex depend on a signal distinct from that of tail-anchored proteins dispersed in the membrane. J Biol Chem. 2003;278:41462–41471. doi: 10.1074/jbc.M307047200. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaji S, Iwahashi J, Kida Y, Sakaguchi M, Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J Cell Biol. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y-F, Lou Y-C, Yeh Y-H, Hsiao C-D, Chen C. Solution structure of the C-terminal NP-repeat domain of Tic40, a co-chaperone during protein import into chloroplasts. J Biochem. 2012;152:443–451. doi: 10.1093/jb/mvs086. [DOI] [PubMed] [Google Scholar]
- Kessler F, Blobel G. Interaction of the protein import and folding machineries of the chloroplast. Proc Natl Acad Sci U S A. 1996;93:7684–7689. doi: 10.1073/pnas.93.15.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kies L, Kremer BP. Function of cyanelles in the tecamoeba Paulinella chromatophora. Naturewissenschaften. 1979;66:578–579. doi: 10.1007/BF00368819. [DOI] [Google Scholar]
- Kikuchi S, Oishi M, Hirabayashi Y, Lee DW, Hwang I, Nakai M. A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell. 2009;21:1781–1797. doi: 10.1105/tpc.108.063552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima A, Asatsuma S, Okada H, Hamada Y, Kaneko K, Nanjo Y, Kawagoe Y, Toyooka K, Matsuoka K, Takeuchi M, Nakano A, Mitsui T. The rice alpha-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell. 2009;21:2844–2858. doi: 10.1105/tpc.109.068288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneip C, Lockhart P, Voss C, Maier UG. Nitrogen fixation in eukaryotes-new models for symbiosis. BMC Evol Biol. 2007;7:55. doi: 10.1186/1471-2148-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneip C, Voss C, Lockhart PJ, Maier UG. The cyanobacterial endosymbiont of the unicellular algae Rhopalodia gibba shows reductive genome evolution. BMC Evol Biol. 2008;8:30. doi: 10.1186/1471-2148-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva S, Bedard J, Patel R, Dudley P, Twell D, Rios G, Koncz C, Jarvis P (2005) In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J 41:412–428 [DOI] [PubMed]
- Kovacs-Bogdan E, Benz JP, Soll J, Bolter B. Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol. 2011;11:133. doi: 10.1186/1471-2229-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Sohn EJ, Lee KH, Lee DW, Hwang I. The transmembrane domain of AtToc64 and its C-terminal lysine-rich flanking region are targeting signals to the chloroplast outer envelope membrane. Mol Cells. 2004;17:281–291. [PubMed] [Google Scholar]
- Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science. 2004;305:997–1000. doi: 10.1126/science.1099128. [DOI] [PubMed] [Google Scholar]
- Li HM, Chiu CC. Protein transport into chloroplasts. Annu Rev Plant Biol. 2010;61:157–180. doi: 10.1146/annurev-arplant-042809-112222. [DOI] [PubMed] [Google Scholar]
- Lv HX, Guo GQ, Yang ZN. Translocons on the inner and outer envelopes of chloroplasts share similar evolutionary origin in Arabidopsis thaliana. J Evol Biol. 2009;22:1418–1428. doi: 10.1111/j.1420-9101.2009.01755.x. [DOI] [PubMed] [Google Scholar]
- Mackiewicz P, Bodył A. A hypothesis for import of the nuclear-encoded PsaE protein of Paulinella chromatophora (Cercozoa, Rhizaria) into its cyanobacterial endosymbionts/plastids via the endomembrane system. J Phycol. 2010;46:847–859. doi: 10.1111/j.1529-8817.2010.00876.x. [DOI] [Google Scholar]
- Mackiewicz P, Bodył A, Gagat P. Possible import routes of proteins into the cyanobacterial endosymbionts/plastids of Paulinella chromatophora. Theory Biosci. 2012;131:1–18. doi: 10.1007/s12064-011-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin B, Nowack EC, Melkonian M. A plastid in the making: evidence for a second primary endosymbiosis. Protist. 2005;156:425–432. doi: 10.1016/j.protis.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Marin B, Nowack EC, Glockner G, Melkonian M. The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium. BMC Evol Biol. 2007;7:85. doi: 10.1186/1471-2148-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marom M, Azem A, Mokranjac D. Understanding the molecular mechanism of protein translocation across the mitochondrial inner membrane: still a long way to go. Biochim Biophys Acta. 2011;1808:990–1001. doi: 10.1016/j.bbamem.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Archibald JM. Evolving a photosynthetic organelle. BMC Biol. 2012;10:35. doi: 10.1186/1741-7007-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Ishida K-I. Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr Biol. 2009;19:R284–R285. doi: 10.1016/j.cub.2009.02.043. [DOI] [PubMed] [Google Scholar]
- Nanjo Y, Oka H, Ikarashi N, Kaneko K, Kitajima A, Mitsui T, Munoz FJ, Rodriguez-Lopez M, Baroja-Fernandez E, Pozueta-Romero J (2006) Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. Plant Cell 18:2582–2592 [DOI] [PMC free article] [PubMed]
- Nowack EC, Grossman AR. Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora. Proc Natl Acad Sci U S A. 2012;109:5340–5345. doi: 10.1073/pnas.1118800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack EC, Melkonian M, Glockner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 2008;18:410–418. doi: 10.1016/j.cub.2008.02.051. [DOI] [PubMed] [Google Scholar]
- Nowack EC, Vogel H, Groth M, Grossman AR, Melkonian M, Glockner G. Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora. Mol Biol Evol. 2011;28:407–422. doi: 10.1093/molbev/msq209. [DOI] [PubMed] [Google Scholar]
- Pfanzagl B, Löffelhardt W (1999) In vitro synthesis of peptidoglycan precursors modified with N-acetylputrescine by Cyanophora paradoxa cyanelle envelope membranes. J Bacteriol 181:2643–2647 [DOI] [PMC free article] [PubMed]
- Pfanzagl B, Allmaier G, Schmid ER, de Pedro MA, Loffelhardt W. N-acetylputrescine as a characteristic constituent of cyanelle peptidoglycan in glaucocystophyte algae. J Bacteriol. 1996;178:6994–6997. doi: 10.1128/jb.178.23.6994-6997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanzagl B, Zenker A, Pittenauer E, Allmaier G, Martinez-Torrecuadrada J, Schmid ER, De Pedro MA, Loffelhardt W. Primary structure of cyanelle peptidoglycan of Cyanophora paradoxa: a prokaryotic cell wall as part of an organelle envelope. J Bacteriol. 1996;178:332–339. doi: 10.1128/jb.178.2.332-339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prechtl J, Kneip C, Lockhart P, Wenderoth K, Maier UG. Intracellular spheroid bodies of Rhopalodia gibba have nitrogen-fixing apparatus of cyanobacterial origin. Mol Biol Evol. 2004;21:1477–1481. doi: 10.1093/molbev/msh086. [DOI] [PubMed] [Google Scholar]
- Rasmussen U, Nilsson M. Cyanobacterial diversity and specificity in plant symbioses. In: Rai AN, Bergman B, Rasmussen U, editors. Cyanobacteria in symbiosis. Netherlands: Springer; 2003. pp. 313–328. [Google Scholar]
- Raven JA. Evolution of cyanobacterial symbioses. In: Rai AN, Bergman B, Rasmussen U, editors. Cyanobacteria in symbiosis. Netherlands: Springer; 2003. pp. 329–346. [Google Scholar]
- Reyes-Prieto A, Yoon HS, Moustafa A, Yang EC, Andersen RA, Boo SM, Nakayama T, Ishida K, Bhattacharya D. Differential gene retention in plastids of common recent origin. Mol Biol Evol. 2010;27:1530–1537. doi: 10.1093/molbev/msq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Loffelhardt W, Bohnert HJ, Philippe H, Lang BF. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G. Isolation of components of the chloroplast protein import machinery. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- Shi LX, Theg SM. A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell. 2010;22:205–220. doi: 10.1105/tpc.109.071464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LX, Theg SM. The motors of protein import into chloroplasts. Plant Signal Behav. 2011;6:1397–1401. doi: 10.4161/psb.6.9.16916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LX, Theg SM (2012) The chloroplast protein import system: from algae to trees. Biochim Biophys Acta in press. doi:10.1016/j.bbamcr.2012.1010.1002 [DOI] [PubMed]
- Stengel A, Benz JP, Soll J, Bolter B. Redox-regulation of protein import into chloroplasts and mitochondria: similarities and differences. Plant Signal Behav. 2010;5:105–109. doi: 10.4161/psb.5.2.10525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li HM. Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell. 2010;22:1516–1531. doi: 10.1105/tpc.109.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng YS, Su YS, Chen LJ, Lee YJ, Hwang I, Li HM. Tic21 is an essential translocon component for protein translocation across the chloroplast inner envelope membrane. Plant Cell. 2006;18:2247–2257. doi: 10.1105/tpc.106.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen U, Martin W. The difference between organelles and endosymbionts. Curr Biol. 2006;16:R1016–R1017. doi: 10.1016/j.cub.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Villarejo A, Buren S, Larsson S, Dejardin A, Monne M, Rudhe C, Karlsson J, Jansson S, Lerouge P, Rolland N, von Heijne G, Grebe M, Bako L, Samuelsson G. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat Cell Biol. 2005;7:1224–1231. doi: 10.1038/ncb1330. [DOI] [PubMed] [Google Scholar]
- Waizenegger T, Stan T, Neupert W, Rapaport D. Signal-anchor domains of proteins of the outer membrane of mitochondria: structural and functional characteristics. J Biol Chem. 2003;278:42064–42071. doi: 10.1074/jbc.M305736200. [DOI] [PubMed] [Google Scholar]
- Yamano K, Kuroyanagi-Hasegawa M, Esaki M, Yokota M, Endo T. Step-size analyses of the mitochondrial Hsp70 import motor reveal the Brownian ratchet in operation. J Biol Chem. 2008;283:27325–27332. doi: 10.1074/jbc.M805249200. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Reyes-Prieto A, Melkonian M, Bhattacharya D. Minimal plastid genome evolution in the Paulinella endosymbiont. Curr Biol. 2006;16:R670–R672. doi: 10.1016/j.cub.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Nakayama T, Reyes-Prieto A, Andersen RA, Boo SM, Ishida K, Bhattacharya D. A single origin of the photosynthetic organelle in different Paulinella lineages. BMC Evol Biol. 2009;9:98. doi: 10.1186/1471-2148-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]


