Abstract
TSF3 encodes one of six (TSF1 to TSF6) recently identified global negative regulators of transcription in Saccharomyces cerevisiae. Mutant tsf3 strains exhibit defects in transcriptional silencing of the GAL1 promoter, allow expression from upstream activation sequence-less promoters, and exhibit pleiotropic defects in cell growth and development. Here we show that TSF3 is involved in transcriptional silencing mediated by the alpha 2 repressor and demonstrate that specific systems of transcriptional silencing may depend on the more global role of TSF3. Cloning and sequencing of TSF3 allowed us to predict a 974-amino-acid gene product identical to SIN4, a negative regulator of transcription of the HO (homothallism) mating type switching endonuclease. TSF3 disruptions are not lethal but result in phenotypes similar to those of the originally isolated alleles. Our results, together with those of Y. W. Jiang and D. J. Stillman (Mol. Cell. Biol. 12:4503-4514, 1992), suggest that TSF3 (SIN4) affects the function of the basal transcription apparatus, and this effect in turn alters the manner in which the latter responds to upstream regulatory proteins.
Full text
PDF



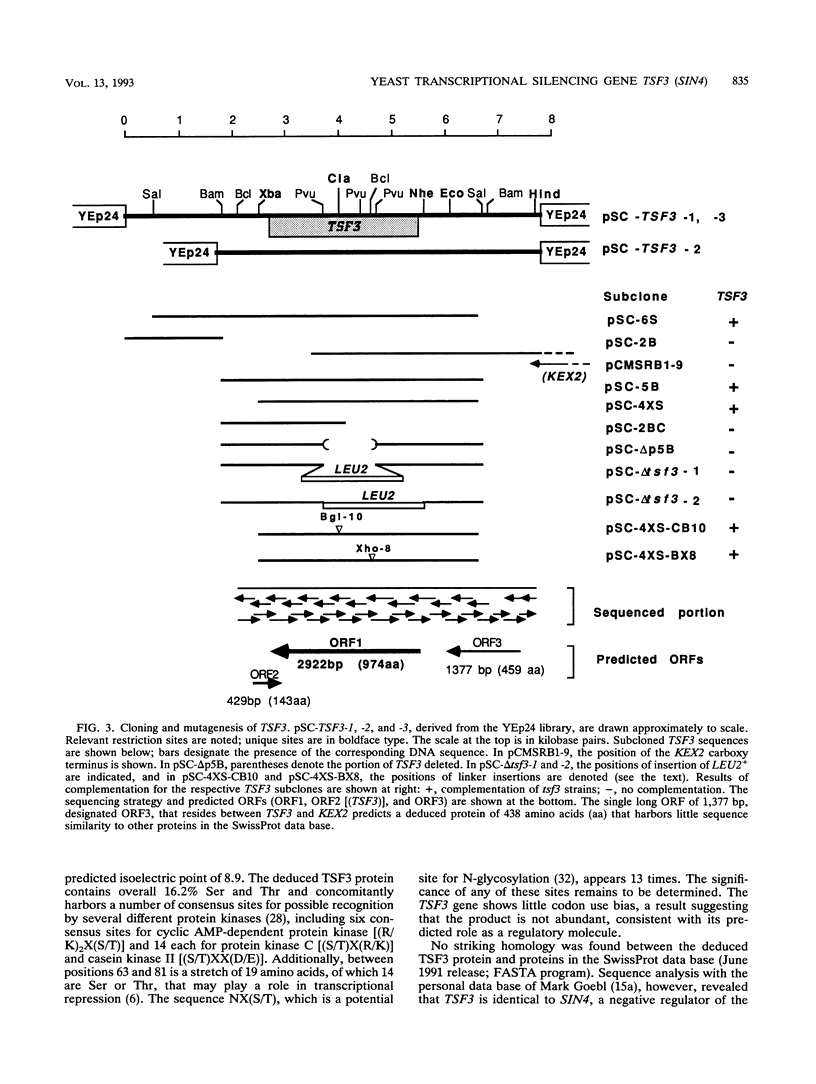


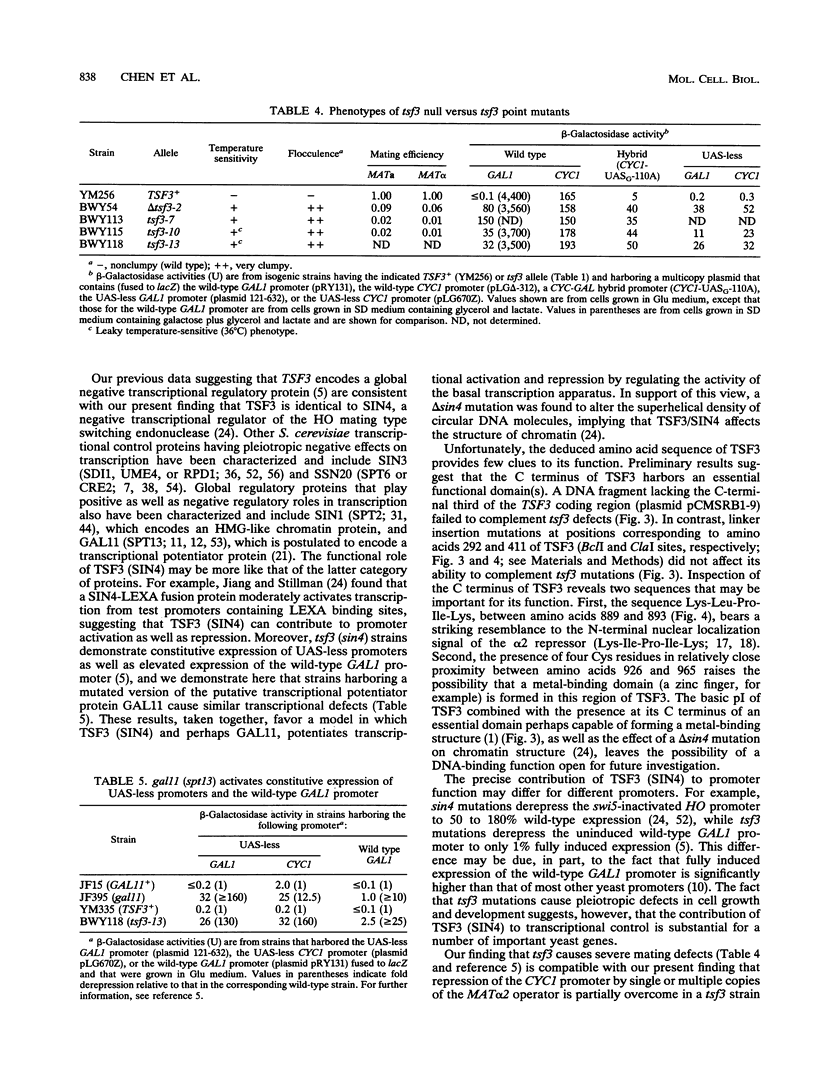


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cavener D. R., Ray S. C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991 Jun 25;19(12):3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran T., Abate C., Cohen D. R., Macgregor P. F., Rauscher F. J., 3rd, Sonnenberg J. L., Connor J. A., Morgan J. I. Inducible proto-oncogene transcription factors: third messengers in the brain? Cold Spring Harb Symp Quant Biol. 1990;55:225–234. doi: 10.1101/sqb.1990.055.01.024. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Malvar T. The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics. 1990 Feb;124(2):283–291. doi: 10.1093/genetics/124.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrin L. K., Mann R. K., Grunstein M. Nucleosome loss activates CUP1 and HIS3 promoters to fully induced levels in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992 Apr;12(4):1621–1629. doi: 10.1128/mcb.12.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassler J. S., Winston F. Isolation and analysis of a novel class of suppressor of Ty insertion mutations in Saccharomyces cerevisiae. Genetics. 1988 Feb;118(2):203–212. doi: 10.1093/genetics/118.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassler J. S., Winston F. The Saccharomyces cerevisiae SPT13/GAL11 gene has both positive and negative regulatory roles in transcription. Mol Cell Biol. 1989 Dec;9(12):5602–5609. doi: 10.1128/mcb.9.12.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley R. L., Jr, Chen S., Ma J., Byrne P., West R. W., Jr Opposing regulatory functions of positive and negative elements in UASG control transcription of the yeast GAL genes. Mol Cell Biol. 1990 Nov;10(11):5663–5670. doi: 10.1128/mcb.10.11.5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley R. L., Jr, West R. W., Jr Differential repression of GAL4 and adjacent transcription activators by operators in the yeast GAL upstream activating sequence. Mol Cell Biol. 1989 Oct;9(10):4282–4290. doi: 10.1128/mcb.9.10.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick J. S., Johnston M. Analysis of URSG-mediated glucose repression of the GAL1 promoter of Saccharomyces cerevisiae. Genetics. 1992 Feb;130(2):295–304. doi: 10.1093/genetics/130.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M., Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991 May;16(5):173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Craik C., Hiraoka Y. Homeodomain of yeast repressor alpha 2 contains a nuclear localization signal. Proc Natl Acad Sci U S A. 1990 Sep;87(18):6954–6958. doi: 10.1073/pnas.87.18.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. N., Hereford L., Herskowitz I. Targeting of E. coli beta-galactosidase to the nucleus in yeast. Cell. 1984 Apr;36(4):1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989 Dec 14;342(6251):749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Himmelfarb H. J., Pearlberg J., Last D. H., Ptashne M. GAL11P: a yeast mutation that potentiates the effect of weak GAL4-derived activators. Cell. 1990 Dec 21;63(6):1299–1309. doi: 10.1016/0092-8674(90)90425-e. [DOI] [PubMed] [Google Scholar]
- Hyman L. E., Seiler S. H., Whoriskey J., Moore C. L. Point mutations upstream of the yeast ADH2 poly(A) site significantly reduce the efficiency of 3'-end formation. Mol Cell Biol. 1991 Apr;11(4):2004–2012. doi: 10.1128/mcb.11.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S., Egli C. M., Braus G. H. Different classes of polyadenylation sites in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3060–3069. doi: 10.1128/mcb.11.6.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. W., Stillman D. J. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae. Mol Cell Biol. 1992 Oct;12(10):4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Keleher C. A., Goutte C., Johnson A. D. The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell. 1988 Jun 17;53(6):927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- Keleher C. A., Passmore S., Johnson A. D. Yeast repressor alpha 2 binds to its operator cooperatively with yeast protein Mcm1. Mol Cell Biol. 1989 Nov;9(11):5228–5230. doi: 10.1128/mcb.9.11.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992 Feb 21;68(4):709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Kemp B. E., Pearson R. B. Protein kinase recognition sequence motifs. Trends Biochem Sci. 1990 Sep;15(9):342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E. A new mapping method employing a meiotic rec-mutant of yeast. Genetics. 1982 Mar;100(3):387–412. doi: 10.1093/genetics/100.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W., Herskowitz I. A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol Cell Biol. 1991 Aug;11(8):4135–4146. doi: 10.1128/mcb.11.8.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukuruzinska M. A., Bergh M. L., Jackson B. J. Protein glycosylation in yeast. Annu Rev Biochem. 1987;56:915–944. doi: 10.1146/annurev.bi.56.070187.004411. [DOI] [PubMed] [Google Scholar]
- Lamphier M. S., Ptashne M. Multiple mechanisms mediate glucose repression of the yeast GAL1 gene. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5922–5926. doi: 10.1073/pnas.89.13.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Young R. A. KEX2 mutations suppress RNA polymerase II mutants and alter the temperature range of yeast cell growth. Mol Cell Biol. 1989 Jun;9(6):2341–2349. doi: 10.1128/mcb.9.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai Y., Harashima S., Oshima Y. AAR1/TUP1 protein, with a structure similar to that of the beta subunit of G proteins, is required for a1-alpha 2 and alpha 2 repression in cell type control of Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jul;11(7):3773–3779. doi: 10.1128/mcb.11.7.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Shore D. Transcriptional regulation in the yeast life cycle. Science. 1987 Sep 4;237(4819):1162–1170. doi: 10.1126/science.3306917. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Stillman D., Kipling D. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell. 1987 Feb 27;48(4):579–587. doi: 10.1016/0092-8674(87)90236-4. [DOI] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991 Nov;10(11):3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L., Celenza J. L., Carlson M. SSN20 is an essential gene with mutant alleles that suppress defects in SUC2 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Feb;7(2):672–678. doi: 10.1128/mcb.7.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M., Suzuki Y., Nogi Y., Matsumoto K., Fukasawa T. Yeast Gal11 protein mediates the transcriptional activation signal of two different transacting factors, Gal4 and general regulatory factor I/repressor/activator site binding protein 1/translation upstream factor. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5373–5377. doi: 10.1073/pnas.87.14.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y. Impact of the Douglas-Hawthorne model as a paradigm for elucidating cellular regulatory mechanisms in fungi. Genetics. 1991 Jun;128(2):195–201. doi: 10.1093/genetics/128.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore S., Maine G. T., Elble R., Christ C., Tye B. K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MAT alpha cells. J Mol Biol. 1988 Dec 5;204(3):593–606. doi: 10.1016/0022-2836(88)90358-0. [DOI] [PubMed] [Google Scholar]
- Perkins D. D. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949 Sep;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Beard C., Smith M., Keranen S. Isolation and characterization of the SPT2 gene, a negative regulator of Ty-controlled yeast gene expression. Mol Cell Biol. 1985 Jul;5(7):1543–1553. doi: 10.1128/mcb.5.7.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Schultz J., Carlson M. Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Oct;7(10):3637–3645. doi: 10.1128/mcb.7.10.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Marshall-Carlson L., Carlson M. The N-terminal TPR region is the functional domain of SSN6, a nuclear phosphoprotein of Saccharomyces cerevisiae. Mol Cell Biol. 1990 Sep;10(9):4744–4756. doi: 10.1128/mcb.10.9.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Roth S. Y., Szent-Gyorgyi C., Simpson R. T. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 1991 Oct;10(10):3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Boguski M. S., Goebl M., Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990 Jan 26;60(2):307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P. W., Stern M. J., Clark I., Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987 Feb 27;48(4):567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Nogi Y., Abe A., Fukasawa T. GAL11 protein, an auxiliary transcription activator for genes encoding galactose-metabolizing enzymes in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4991–4999. doi: 10.1128/mcb.8.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. S., Carlson M., Winston F. SPT6, an essential gene that affects transcription in Saccharomyces cerevisiae, encodes a nuclear protein with an extremely acidic amino terminus. Mol Cell Biol. 1990 Sep;10(9):4935–4941. doi: 10.1128/mcb.10.9.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. S., Malone E. A., Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991 Jun;11(6):3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Clark I., Nicholson P. R., Herskowitz I., Stillman D. J. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol Cell Biol. 1990 Nov;10(11):5927–5936. doi: 10.1128/mcb.10.11.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. W., Jr, Chen S. M., Putz H., Butler G., Banerjee M. GAL1-GAL10 divergent promoter region of Saccharomyces cerevisiae contains negative control elements in addition to functionally separate and possibly overlapping upstream activating sequences. Genes Dev. 1987 Dec;1(10):1118–1131. doi: 10.1101/gad.1.10.1118. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]




