Abstract
The mitochondrial peptidasome called presequence protease (PreP) is responsible for the degradation of presequences and other unstructured peptides including the amyloid-β peptide, whose accumulation may have deleterious effects on mitochondrial function. Recent studies showed that PreP activity is reduced in Alzheimer disease (AD) patients and AD mouse models compared to controls, which correlated with an enhanced reactive oxygen species production in mitochondria. In this study, we have investigated the effects of a biologically relevant oxidant, hydrogen peroxide (H2O2), on the activity of recombinant human PreP (hPreP). H2O2 inhibited hPreP activity in a concentration-dependent manner, resulting in oxidation of amino acid residues (detected by carbonylation) and lowered protein stability. Substitution of the evolutionarily conserved methionine 206 for leucine resulted in increased sensitivity of hPreP to oxidation, indicating a possible protective role of M206 as internal antioxidant. The activity of hPreP oxidized at low concentrations of H2O2 could be restored by methionine sulfoxide reductase A (MsrA), an enzyme that localizes to the mitochondrial matrix, suggesting that hPreP constitutes a substrate for MsrA. In summary, our in vitro results suggest a possible redox control of hPreP in the mitochondrial matrix and support the protective role of the conserved methionine 206 residue as an internal antioxidant.
Keywords: Mitochondria, Presequence protease, Peptide degradation, Oxidation, Methionine, Free radicals
Mitochondria have a fundamental role in many cellular processes, including bioenergetics, metabolism of lipids and amino acids, and even coordinated cell death (apoptosis). A recent proteomic analysis estimated that mitochondria contain around 1200 different proteins [1–3], with more than 99% of them being synthesized on cytosolic ribosomes, posttranslationally targeted, and then translocated across the mitochondrial membrane system into one of four compartments: the outer or inner membrane, the intermembrane space, or the mitochondrial matrix (reviewed by [4]).
To ensure correct sorting and targeting after synthesis on cytosolic ribosomes, most of the mitochondrial proteins contain a targeting sequence, designated presequence. This presequence is used for recognition of the translocation machinery by binding to receptor proteins [4]. When the protein reaches its correct destination the presequence is removed by the mitochondrial processing peptidase (reviewed in [5–7]). The free presequence peptides have amphipathic characteristics and have been shown to affect mitochondrial membrane integrity, uncouple respiration, and inhibit enzyme activity [8–10].
Presequence peptides are degraded by a mitochondrial matrix resident peptidasome, the presequence protease (PreP)1 [11–13]. This peptidase has a fundamental role in mitochondrial biogenesis, completing the last step of the protein import process, the degradation of the presequence peptide. PreP was originally identified in Arabidopsis thaliana [14], and homologs were found in yeast [15,16] and also in humans [17]. This important role in mitochondrial biogenesis is supported by the phenotypic analysis of PreP mutants, both in Arabidopsis thaliana and in Saccharomyces cerevisiae: double AtPreP1/AtPreP2 mutants exhibit a growth phenotype, chlorosis, and uncoupling of mitochondria [18], whereas the S. cerevisiae Δmop112 strain has a severe growth defect on nonfermentative carbon sources [15].
In addition to degrading presequences, human PreP (hPreP; UniProt ID: Q5JRX3) was also shown to degrade the amyloid-β peptide (Aβ) [17], suggesting a connection between the activity of hPreP and the progression of Alzheimer disease (AD). This idea is further supported by the observation that hPreP activity is severely diminished in samples of mitochondrial matrix isolated from the temporal lobe of Alzheimer disease patients, compared to age-matched controls [19]. Additionally, this reduction in activity is recapitulated in AD transgenic mice models (overexpressing the human form of amyloid-β precursor protein) [19]. These observations, together with current evidence supporting the localization of Aβ peptide in mitochondria [20–22] and its well-established toxic effects [23–25], point toward an important role of hPreP in Aβ metabolism and in the progression of Alzheimer disease.
Interestingly, even though the activity of hPreP was reduced in AD patients and in transgenic mouse models, this was not a consequence of reduced protein levels [19]. This observation raised the hypothesis that reduced hPreP activity can be caused by oxidation of the enzyme, because an increase in the production of reactive oxygen species (ROS) was observed in AD patients [19].
In this study, we used an in vitro setup with purified hPreP to investigate the effect of oxidation by the biologically relevant oxidant hydrogen peroxide on hPreP activity and established the role of a conserved methionine residue in the process.
Materials and methods
Purification of recombinant wild-type hPreP and variants
Plasmid pGEX6p2 encoding human hPreP lacking its presequence (Δ1–28 aa) was transformed into Escherichia coli Rosetta 2 cells (Novagen). Cell cultures were grown in 1-L batches in Terrific Broth medium (Formedium) containing 100 µg/ml ampicillin (Sigma) and 34 µg/ml chloramphenicol (Sigma) at 37 °C to OD600 ~0.6 and then induced with 0.6 mM isopropyl β-d-1-thiogalactopyranoside (Formedium) for 5 h (with the temperature adjusted to 21 °C). Cells were then harvested and frozen as pellets. The cell pellets were dissolved in GSH-binding buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 pH 7.3, 2.5 mM DTT (dithiothreitol), 10 mM MgCl2, 1 µM Zn acetate) containing 1 mg/ml lysozyme (Sigma) and 10 µg/ml DNase I (Sigma) and then incubated for 1 h at 4 °C for cell lysis. The cell extract was centrifuged at 4000 g to remove unbroken cells and the supernatant further centrifuged at 100,000 g. The soluble fraction was incubated with Glutathione Sepharose (GE Healthcare) for 4 h and then washed four times with GSH-binding buffer and two times with cleavage buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM DTT). Twenty units of HRV3C protease (MoBiTec) were applied and the hPreP protein was eluted after overnight incubation at 4 °C.
hPreP variants were constructed by site-directed mutagenesis on the pGEX_hPreP vector using the QuikChange II kit (Agilent Technologies) and appropriate primers following the manufacturer’s instructions and confirmed by sequencing. hPreP variants were purified using the same procedure.
Oxidation of hPreP with hydrogen peroxide (H2O2)
hPreP and variants (final concentration 0.26 mg ml−1) were incubated with the indicated concentrations of hydrogen peroxide for 4 h at 22 °C, in 50 mM Hepes–KOH, pH 8.2. Hydrogen peroxide (Sigma) concentration was measured at 240 nm (ε = 39.4 ± 0.2 M−1 cm−1). After 4 h, the peroxide was removed by filtration using a 50 k filter (Millipore) or by dialysis against 50 mM Hepes–KOH, pH 8.2, and the hPreP samples were used for activity measurement, SDS–PAGE, and carbonyl Western blot.
hPreP activity measurements
For the analysis of pF1β and Aβ degradation, hPreP samples (previously incubated with peroxide as indicated) were incubated with 0.8 µg pF1β (purified according to [26]) or 1 µg amyloid-β (Sigma) for the indicated times, in degradation buffer (50 mM Hepes, pH 8.2, 10 mM MgCl2), at 37 °C (three independent experiments). The amount of hPreP used was 0.5 µg (per reaction) in the pF1β assay and 1 µg in the amyloid-β assay. After incubation the reactions were stopped with SDS sample buffer and subsequently loaded on NuPAGE 4–12% Bis–Tris gels (Invitrogen) and stained with Coomassie brilliant blue (Sigma). The bands were quantified using the Multi Gauge software.
For the degradation of the C1 peptide (Promega), hPreP fractions (0.8 µg per assay) were incubated with 1 µg C1 peptide in degradation buffer and the reaction allowed to proceed for 20 min at 37 °C. The samples were chilled on ice and subsequently loaded on 0.8% agarose gel and visualized by UV light.
In the substrate V assay, hPreP fractions (0.8 µg per assay) were mixed with 1 µg substrate V (R&D Systems) in degradation buffer, and the increase in fluorescence (excitation 327 nm; emission 395 nm) was immediately recorded in a plate reader (SpectraMax Gemini) during the first 40 s. Results are shown as the substrate V degradation rate and averaged over three independent experiments.
Carbonyl detection
Carbonylation was detected using the OxyBlot kit (Milipore), according to the manufacturer’s instructions.
Limited trypsin proteolysis
hPreP controls (2 µg) and samples treated for 4 h with H2O2 were incubated with 10 ng trypsin at 30 °C, and samples were withdrawn after 1, 10, and 30 min and subsequently analyzed using 10% Tris–glycine gels.
MsrA recovery assays
hPreP was incubated with H2O2 (0.5 or 5 mM) for 4 h at 22 °C. The peroxide was removed by filtration and extensive washes with reaction buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3). Then, hPreP (1.9 µM) was incubated for 3 h at 37 °C with purified recombinant mouse MsrA (as indicated), purified according to [27] in reaction buffer with 10 mM DTT and 5 mM MgCl2. The activity of hPreP was then assayed using the fluorogenic substrate V.
Results
Inactivation of hPreP upon exposure to hydrogen peroxide
The main aim of this study was to investigate the connection between oxidation of hPreP and reduction in activity. To do so, we analyzed the effect of exposure to H2O2 on the activity of recombinant hPreP in an in vitro assay. Nevertheless, hydrogen peroxide is a biologically relevant oxidative agent being produced within the mitochondrial matrix, the cellular compartment where hPreP is localized [28].
To analyze the effect of oxidation on hPreP activity we incubated the purified enzyme with various concentrations of H2O2 (ranging from 0.5 to 5 mM) and assessed the peptidolytic activity of hPreP using four different peptide substrates. The peptides used were previously shown to be substrates for hPreP (or PreP homologs) [19,29–31] and reflect the broad substrate specificity of this peptidase as they vary in length and physicochemical properties. Table 1 summarizes the characteristics of the peptide substrates used: pF1β, a typical mitochondrial presequence peptide (a 54-amino-acid peptide corresponding to the presequence of the Nicotiana plumbaginifolia F1β subunit of ATP synthase); the amyloid-β 1–40 peptide (an endogenous substrate for hPreP); and two broad-range protease substrates, the 11-amino-acid C1 peptide and the fluorogenic bradykinin-derived 9-amino-acid substrate V.
Table 1.
Properties of the peptide substrates used to measure hPreP activity.
| Peptide | No. amino acids | Sequence |
|---|---|---|
| Substrate V | 9 | (7-Methoxycoumarin-4-yl)acetyl–RPPGFSAFK–(2,4-dinitrophenyl) |
| C1 | 11 | PLSRTLSVAAK |
| Amyloid-β | 40 | DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVV |
| pF1β | 53 | ASRRLLASLLRQSAQRGGGLISRSLGNSPKSASRASSRASPKGFLLNRAVQYM |
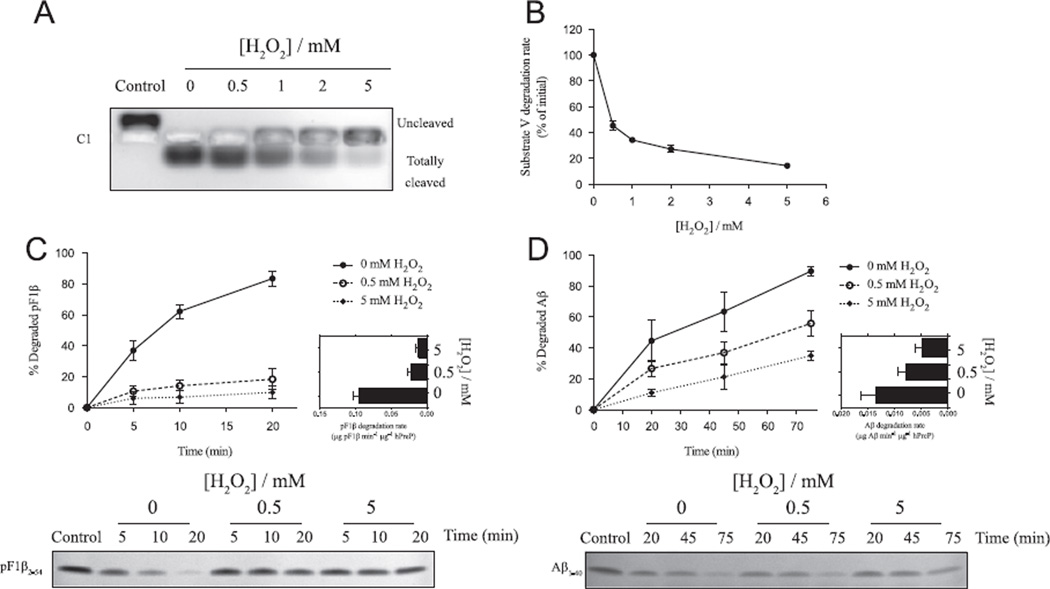
As observed in Fig. 1, exposure of hPreP to H2O2 resulted in a concentration-dependent inhibition of hPreP peptidolytic activity, as analyzed with all four substrates. Fig. 1A shows inhibition of C1 degradation by hPreP exposed to increased concentrations of H2O2. A simple qualitative analysis shows clearly that the cleavage of the C1 peptide, from the slower migrating to the faster migrating form, is reduced when hPreP is exposed to the oxidant.
Fig. 1.
Effect of exposure to hydrogen peroxide on hPreP wild-type activity. After 4 h incubation with H2O2, the hydrogen peroxide was removed and hPreP activity assayed as the cleavage of four substrates. (A) Representative gel showing cleavage of C1 by hPreP, resulting in a change in migration on agarose gel due to the charge profile of the peptide. (B) Effect of oxidation on the rate of substrate V degradation by hPreP (average of three experiments). Time course analysis of (C) pF1β and (D) Aβ degradation by hPreP incubated in the absence or presence (0.5 or 5 mM) of H2O2. Shown are representative gels (corresponding to one of the three experiments) and an estimation of the degradation rate (in the first 10 min for pF1β and in the first 45 min for Aβ).
To provide a more quantitative analysis of the activity inhibition by H2O2 we used the degradation of substrate V to assess hPreP activity. This peptide contains both the fluorescent group 7-methoxycoumarin and the quencher group 2,4-dinitrophenyl, resulting in fluorescence emission upon cleavage of a peptide bond between the two groups. In Fig. 1B we show that there is a concentration-dependent reduction in the degradation rate of substrate V by hPreP upon exposure to increasing concentrations of H2O2. Exposure of hPreP to 0.5 mM H2O2 results in a reduction of activity of about 50–60%, whereas exposure to 5 mM H2O2 results in a reduction of 80–90% of hPreP activity.
We then tested the influence of H2O2 on the hPreP ability to degrade a typical presequence peptide (pF1β) and the endogenous substrate Aβ, using polyacrylamide gel-based assays. Figs. 1C and 1D show time course analyses of pF1β and Aβ degradation, respectively, by hPreP preincubated in the absence or presence of H2O2 (0.5 or 5 mM). In the case of both peptides, it is clear that the rate of hPreP-catalyzed degradation is reduced upon incubation with H2O2. This effect is particularly visible in the case of pF1β degradation, with the hPreP degradation activity reduced by about 70% upon exposure to 0.5 mM H2O2. In the case of Aβ degradation, exposure of hPreP to 0.5 mM H2O2 results in 50% reduction in the cleavage rate.
Even though the extent of hPreP activity inhibition varies between the different assays (probably reflecting the distinct assay conditions and degradation rates), all the data shown in Fig. 1 clearly demonstrate that exposure of hPreP to the oxidative agent H2O2 results in a less active enzyme and consequently in a strong reduction of the degradation activity toward four different substrates.
Considering both the ease of analysis and the reproducibility of results, we selected the substrate V assay to analyze hPreP activity in the remaining experiments reported.
Molecular consequences of hPreP oxidation
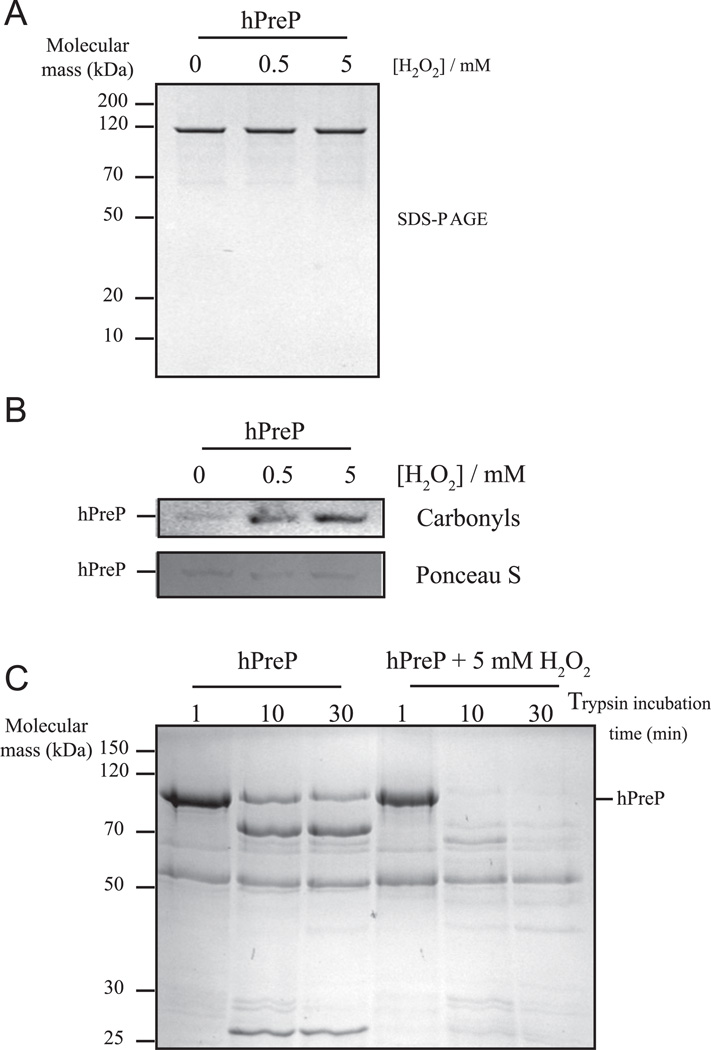
It has been known for long time that incubation with H2O2 can result in protein degradation by chemical hydrolysis of the peptide bond [32]. We evaluated the possibility of hPreP degradation in the presence of H2O2 by simply monitoring hPreP by SDS–PAGE after incubation with H2O2. However, in the case of hPreP oxidation, no degradation was observed (Fig. 2A), indicating that the reduction in activity is rather caused by amino acid oxidation, not by protein degradation. Consistent with this idea, oxidation of hPreP by H2O2 resulted in protein carbonylation, a general hallmark of protein oxidation (Fig. 2B). Interestingly, oxidation probably leads to a change in hPreP conformation resulting in a less stable protein as analyzed by limited trypsin proteolysis. Oxidized hPreP is more sensitive to trypsin proteolysis than the control sample (Fig. 2C), an indicator of altered conformation, possibly reflecting an increased tendency for unfolding.
Fig. 2.
Consequences of hPreP wild-type oxidation. Upon incubation with H2O2 for 4 h, hPreP was analyzed by (A) SDS–PAGE (0.7 µg per lane) and (B) carbonyl immunoblot (4 µg per lane), as described under Materials and methods. Ponceau S staining is shown as a loading control. (C) hPreP samples were incubated in the presence or absence of 5 mM H2O2 for 4 h; subsequently treated with 10 ng trypsin for 1, 10, and 30 min; and then analyzed by SDS–PAGE (2 µg per lane).
It is widely accepted that exposure of proteins to oxidizing agents such as hydrogen peroxide results in the production of oxidized forms of different amino acids [33]. Although it is certain that most amino acid residues can be targets of oxidative agents, those containing sulfur (cysteine and methionine) are among the most readily oxidizable residues. In addition, both oxidation of methionine (to methionine sulfoxide) and oxidation of cysteine (to disulfide) are processes that can be biologically reversed [34–37]. Considering the previous suggestion that hPreP oxidation would lead to formation of a disulfide bridge between C119 and C556 [17] (numbering used throughout this report refers to hPreP preprotein and corresponds to C90 and C527 in the mature portion of the protein as used by Falkevall et al. [17]), we analyzed the effect of H2O2 on hPreP variants in which either C119 or C556 was replaced by serine. In both variants, exposure to H2O2 resulted in a reduction in activity similar to that in wild-type hPreP (not shown). The reduction in activity observed in response to hydrogen peroxide is thus not (or at least not only) due to the formation of an S–S bridge between C119 and C556, as that would be prevented in these substitutions. Additionally, we did not observe any recovery of the activity of oxidized hPreP by adding DTT (see Fig. 5), further supporting the idea that hPreP inactivation by H2O2 is not due to the formation of a disulfide bridge.
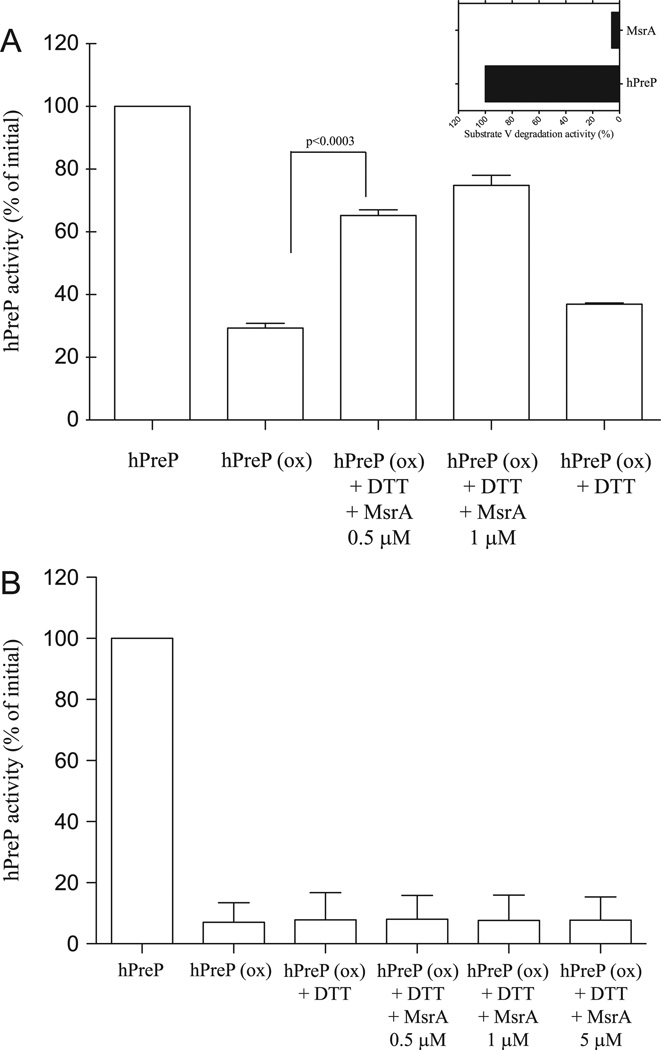
Fig. 5.
Recovery of hPreP wt activity by MsrA. hPreP was exposed to H2O2 for 4 h at the concentrations of 0.5 or 5 mM. The H2O2 was then removed by filtration and hPreP further incubated for 2 h at 37 °C with DTT and MsrA as indicated. After this incubation the hPreP activity was assayed using substrate V. (A) MsrA effect on hPreP oxidized with 0.5 mM H2O2. (B) MsrA effect on hPreP oxidized with 5 mM H2O2. Statistical analysis was made using Student’s t test. Inset in (A) shows that MsrA itself has no degradation activity against substrate V.
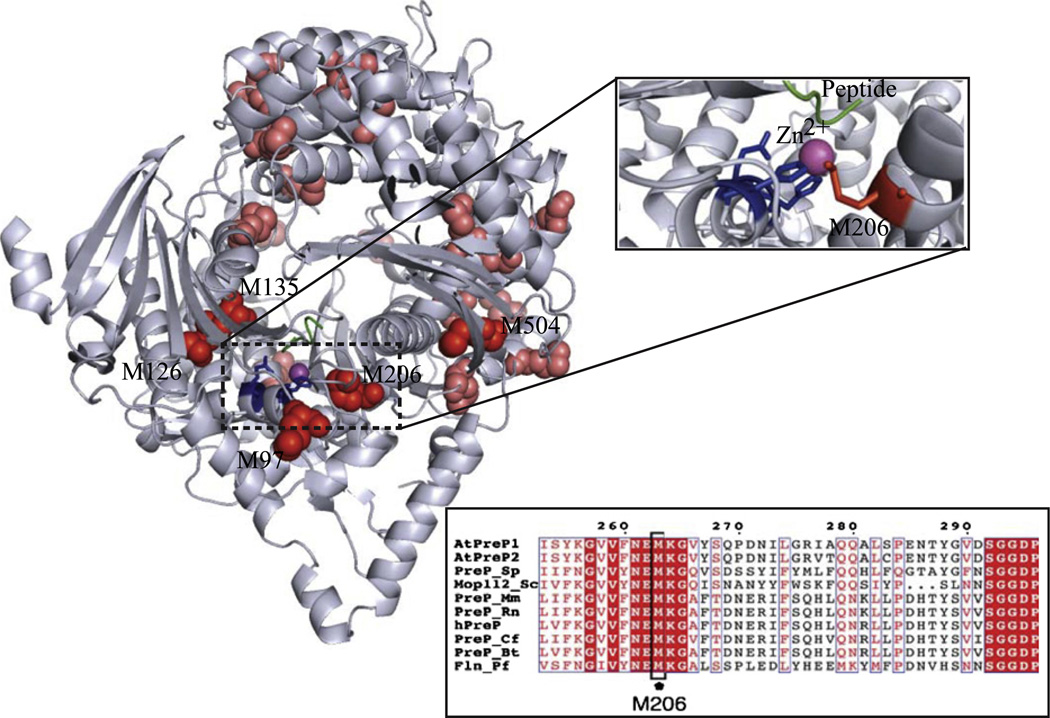
Methionine residues are also readily oxidizable to methionine sulfoxide by reactive oxygen species, including hydrogen peroxide (further oxidation to methionine sulfone occurs only rarely under physiological conditions) [35]. hPreP contains 24 methionine residues in its mature form (residues 29–1037, corresponding to the form processed in mitochondria), with most of them being surface-exposed. However, a few methionine residues are found buried in the structure and close to the active site of the enzyme, according to the structural model of hPreP, built based on the structure of A. thaliana PreP1 (Fig. 3).
Fig. 3.
Localization of the analyzed methionine residues in the hPreP structural model, constructed based on the structure of AtPreP1 (PDB ID: 2FGE) [51]. Methionines are shown in red, the active-site histidines are blue, the substrate peptide is green, and the zinc ion is magenta. The detailed location of M206 within the hPreP active site is highlighted. Inset shows an alignment of PreP sequences from A. thaliana (AtPreP1 and AtPreP2), Plasmodium falciparum (falcilysin, Fln), S. cerevisiae (Mop112), Bos taurus (PreP_Bt), Canis familiaris (PreP_Cf), Mus musculus (PreP_Mm), Rattus norvergicus (PreP_Rn), Schizosaccharomyces pombe (PreP_Sp), and Homo sapiens (hPreP). The alignment is restricted to the region around M206 (full alignment is shown in Supplementary Fig. S2).
An interesting feature of some methionine residues in proteins is their ability to act as internal antioxidants: being easily oxidizable residues, methionines can work as scavengers, preventing further oxidation of other amino acid residues [34,38,39].
To understand the roles of methionine residues and methionine oxidation in hPreP inactivation by H2O2, we analyzed on one hand the possibility of recovering hPreP activity by reduction of methionine sulfoxide and on the other hand the role of methionine residues in hPreP as antioxidants.
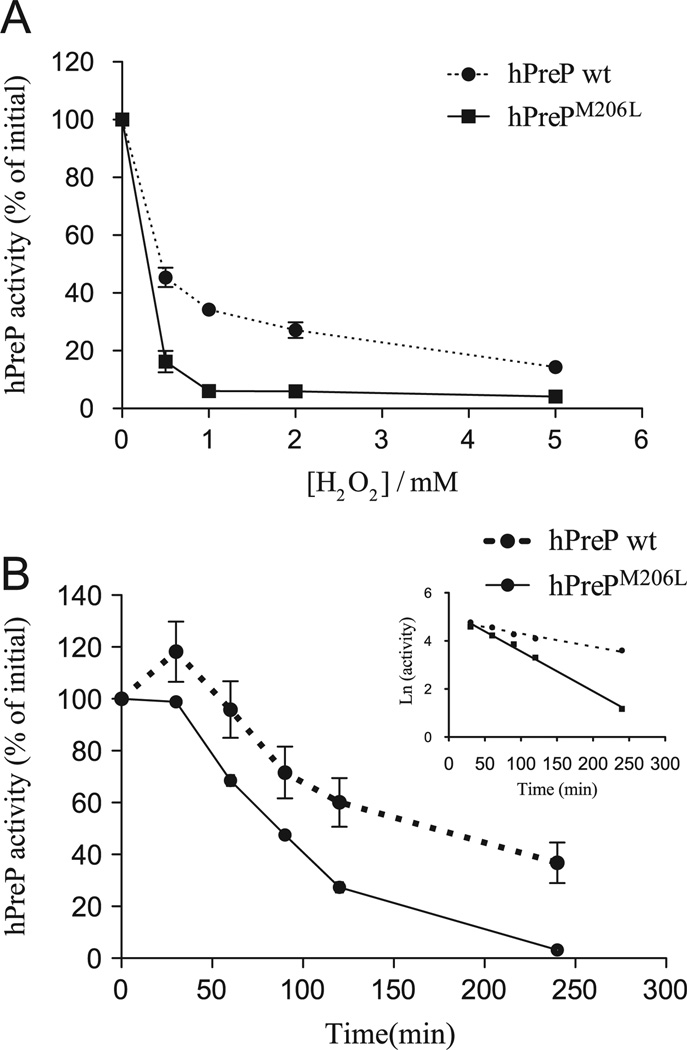
Substitution of methionine 206 for leucine in hPreP results in increased sensitivity to oxidation by hydrogen peroxide
To investigate the role of methionine residues located close to the enzyme active site on hPreP oxidation we produced methionine-to-leucine substitutions of residues M97, M126, M135, M206, and M504. The variants produced resulted in active proteins, with activity comparable to that of wild-type hPreP as measured with the substrate V assay, except for M97L, which showed about half of the activity (not shown). We expected that replacing methionine residues that perform a putative antioxidant function in hPreP with the nonoxidizable amino acid leucine would result in increased sensitivity to H2O2 exposure.
When we measured the peptidolytic activity of the hPreP variants upon exposure to various concentrations of H2O2 (Fig. 4A and Supplementary Fig. S1), we observed that the hPrePM206L variant exhibited a strikingly different inhibition profile compared to wild-type hPreP (Fig. 4A) and the other variants (Supplementary Fig. S1). More specifically, a time course analysis of the oxidative inactivation of hPreP wt and hPrePM206L showed that, upon exposure to 0.5 mM H2O2, the hPrePM206L variant was inactivated three times faster than the wild type (Fig. 4B). Interestingly, in the early time points of the experiment with hPreP wt (Fig. 4B), we consistently observed a slight increase in the activity before observing the activity decrease. Although the reason for this increase is unclear, this phenomenon was observed previously in the oxidative inactivation of glutamine synthetase [40].
Fig. 4.
(A) Effect of H2O2 exposure (4 h) on the activity of hPrePM206L compared to hPreP wild type (wt), as assayed by the degradation of the fluorogenic substrate V. (B) Time course inactivation of hPreP wt and hPrePM206L upon exposure to 0.5 mM H2O2 analyzed by the substrate V assay (results shown are averages of three experiments). The regression lines (inset) were fit for a first-order reaction for time points from 30 to 240 min and gave R2 of 0.96 and 0.99, for the hPreP wt and hPrePM206L data sets, respectively. The ratio between the inactivation rates of hPrePM206L and hPreP wt showed a threefold difference. 100% activity corresponds to a specific activity of 491.6 ± 54.2 ng substrate V degraded min−1 µg protein−1 for wt hPreP and 656.2 ± 32.7 ng substrate V degraded min−1 µg protein−1 for hPrePM206L.
Experiments with hPreP variants revealed that replacing M206 with leucine results in hPreP being more vulnerable to oxidation by H2O2, reflected in a more pronounced effect of this oxidant on the activity, even at low H2O2 concentrations. Considering that in the absence of M206, hPreP activity is more sensitive to H2O2 exposure, it is possible that this methionine residue has a protective role, minimizing further oxidation of other residues at low concentration of the oxidant, although we cannot exclude the possibility that the substitution caused a conformational change that renders residues more susceptible to oxidation.
The importance of the M206 residue is additionally emphasized by its localization within the hPreP active site in the structural model (Fig. 3) and also by the fact that it is the only methionine residue completely conserved in PreP sequences from different organisms, including plants, parasites, yeast, and mammals (Fig. 3 and full sequence alignment in Supplementary Fig. S2). Because of its location, one can speculate that M206 may protect the active-site histidines in the zinc-binding motif (H104ILEH108), avoiding irreparable damage to the enzyme.
Activity of oxidized hPreP can be recovered by MsrA
Methionine oxidation to methionine sulfoxide can be enzymatically reversed by the activity of methionine sulfoxide reductases [41,42]. Msr proteins are divided into two families, MsrA and MsrB, based on the reaction stereospecificity. Oxidation of methionine to methionine sulfoxide results in the formation of two stereoisomers at the sulfur atom (Met-S-SO and Met-R-SO), with MsrA reducing the S isomer and MsrB reducing the R isomer. In mammals, MsrA is dually localized to the cytosol and to the mitochondrial matrix because of alternative translation initiation sites [27,43,44]. In vivo, the regeneration of active MsrA, as part of its catalytic cycle, requires the sequential action of thioredoxin and thioredoxin reductase, whereas in vitro DTT can replace the thioredoxin system as a source of reductant [41].
To evaluate the possibility of recovering hPreP activity upon oxidation we performed incubation with recombinant mouse MsrA [27]. The incubation with MsrA was performed using as substrate hPreP oxidized under either low (0.5 mM H2O2) or high oxidant conditions (5mM H2O2). Fig. 5B shows that when hPreP was oxidized in the presence of 5 mM H2O2, the incubation with MsrA did not increase the activity of hPreP. However, when hPreP was oxidized in the presence of 0.5 mM H2O2 (Fig. 5A) the incubation with MsrA resulted in a significant increase in hPreP activity, whereas incubation with DTT alone had essentially no effect.
Incubation with MsrA resulted in close to the maximal recovery theoretically possible, considering the stereospecificity of MsrA (only one stereoisomer of methionine sulfoxide is reduced and therefore the recovery of activity is about 50% of the activity “lost” because of oxidation).
Our results in an in vitro system with purified proteins suggest that hPreP may be a substrate for mitochondrial MsrA based on the recovery of oxidized hPreP activity by MsrA and also on the known intracellular localization of both hPreP and MsrA in the mitochondrial matrix [17,27,43,44].
Discussion
In this report we show that exposure of purified hPreP to an oxidizing agent, hydrogen peroxide, results in decreased peptidolytic activity. Additionally, we suggest that the evolutionarily conserved methionine 206 plays a protective role against hPreP oxidation, as when this residue is substituted by leucine, hPreP shows increased sensitivity to inactivation by hydrogen peroxide. Our observations show that hPreP is inactivated by H2O2 in a concentration-dependent manner and that oxidation with low concentrations of H2O2 can be reversed by mitochondrial MsrA. Thus, at low concentrations of H2O2, methionine residues in hPreP can be oxidized, resulting in a reduction in activity of about 60%, which can be reversed by the action of MsrA. However, exposure to higher concentrations of H2O2 leads to a reduction in activity of 90–100%, which is not reversed by MsrA. The exposure to 5 mM H2O2 may result in oxidation of residues other than methionines (an increase in protein carbonylation is clearly observed in Fig. 2A, demonstrating further oxidation of other amino acids), reactions that cannot be biologically reversed. Mapping of oxidation sites has been successfully achieved for a number of proteins, especially utilizing HPLC coupled to mass spectrometry [45,46]. However, the large size of hPreP (> 1000 amino acids) and the existence of 24 methionine residues in the mature form make the precise mapping of each methionine oxidation a challenging task. Nevertheless, our current efforts are aimed at tackling this problem.
The present results substantiate the critical role of methionine residues in proteins for protection against oxidative inactivation, in cases in which the proteins are exposed to low concentrations of oxidants. This protective role of methionine residues has been demonstrated in several previous reports [34–36,38,39,47]. For example, it was shown thatmethionine residues in α2-antitrypsin work as internal antioxidants protecting the critical tryptophan residue fromirreversible oxidation and inactivation [47]. Additionally, a recent report showed that partial substitution of methionine residues for the nonoxidizable amino acid nor-leucine in E. coli cells results in increased sensitivity to H2O2 oxidation and in increased protein carbonylation [39]. Our present study further supports the proposal that methionine residues play an important role in protecting proteins from oxidative inactivation by scavenging oxidizing agents and limiting the damage inflicted on catalytically essential residues.
Additionally, the possibility of controlling hPreP activity by reversible methionine oxidation may constitute a novel regulatory mechanism. The activity of hPreP could potentially be controlled within the mitochondrial matrix by localized production of hydrogen peroxide and possibly also by other reactive oxygen species. Although the biological significance of this phenomenon remains to be elucidated, it may provide a new strategy to regulate hPreP activity. Future experiments in human cell lines knocked down for MsrA may provide clues to understanding hPreP regulation.
Although the regulation of protein function by reversible methionine oxidation is still poorly understood, there have been some reports of such a regulatory strategy [48]. One such example is the regulation of Ca2+–calmodulin protein kinase II (CaMKII) [49]. Oxidation of two methionine residues in CaMKII results in activation of the kinase. In CaMKII, a helical segment containing M281/282 is exposed upon binding of Ca2+ and calmodulin, and methionine oxidation to sulfoxide results in retained kinase activity even in the absence of Ca2+–calmodulin. This oxidation can be reversed by MsrA and constitutes an additional regulatory mechanism for CaMKII.
Conclusions
In summary, this report highlights the inhibitory effect of hydrogen peroxide on hPreP activity, describes the role of methionine 206 as an internal antioxidant, and suggests the possibility of hPreP activity being regulated by MsrA. Inactivation by reactive oxygen species was also observed for other amyloid-β-degrading peptidases, such as insulin-degrading enzyme and neprilysin, using in vitro assays [50]. Considering that hPreP activity is reduced in Alzheimer disease patients, our future efforts will be focused on assessing the role of hPreP oxidation in the progression of this pathology.
Supplementary Material
Acknowledgments
This work was supported by research grants from the Swedish Research Council and Alzheimerfonden to E.G.; a doctoral grant from Fundação para a Ciência e a Tecnologia (Portugal) to C.M.P.; a postdoctoral grant from Fundação para a Ciência e a Tecnologia (Portugal) and research grants from the Stohnes and the Sigurd och Elsa Goljes Minne Foundations to P.F.T.; research grants from the Swedish Cancer Society and the Swedish Research Council to J.L.; and the Intramural Research Program of the National Heart, Lung, and Blood Institute to R.L.L. The authors also thank Dr. Therese Eneqvist for the hPreP structural model.
Footnotes
Abbreviations used: hPreP, human presequence protease; MsrA, methionine sulfoxide reductase A; AD, Alzheimer disease; ROS, reactive oxygen species; DTT, dithiothreitol.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.freeradbiomed.2012.09.039.
References
- 1.Meisinger C, Sickmann A, Pfanner N. The mitochondrial proteome: from inventory to function. Cell. 2008;134:22–24. doi: 10.1016/j.cell.2008.06.043. [DOI] [PubMed] [Google Scholar]
- 2.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim. Biophys. Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- 6.Mossmann D, Meisinger C, Vogtle FN. Processing of mitochondrial presequences. Biochim. Biophys. Acta. 2012;1819:1098–1106. doi: 10.1016/j.bbagrm.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Teixeira PF, Glaser E. Processing peptidases in mitochondria and chloroplasts. Biochim. Biophys. Acta. doi: 10.1016/j.bbamcr.2012.03.012. (in press), http://dx.doi.org/10.1016/j.bbamcr.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Hugosson M, Andreu D, Boman HG, Glaser E. Antibacterial peptides and mitochondrial presequences affect mitochondrial coupling, respiration and protein import. Eur. J. Biochem. 1994;223:1027–1033. doi: 10.1111/j.1432-1033.1994.tb19081.x. [DOI] [PubMed] [Google Scholar]
- 9.Nicolay K, Laterveer FD, van Heerde WL. Effects of amphipathic peptides, including presequences, on the functional integrity of rat liver mitochondrial membranes. J. Bioenerg. Biomembr. 1994;26:327–334. doi: 10.1007/BF00763104. [DOI] [PubMed] [Google Scholar]
- 10.Yang MJ, Geli V, Oppliger W, Suda K, James P, Schatz G. The MAS-encoded processing protease of yeast mitochondria: interaction of the purified enzyme with signal peptides and a purified precursor protein. J. Biol. Chem. 1991;266:6416–6423. [PubMed] [Google Scholar]
- 11.Alikhani N, Ankarcrona M, Glaser E. Mitochondria and Alzheimer’s disease: amyloid-beta peptide uptake and degradation by the presequence protease, hPreP. J. Bioenerg. Biomembr. 2009;41:447–451. doi: 10.1007/s10863-009-9244-4. [DOI] [PubMed] [Google Scholar]
- 12.Glaser E, Alikhani N. The organellar peptidasome, PreP: a journey from Arabidopsis to Alzheimer’s disease. Biochim. Biophys. Acta. 2010;1797:1076–1080. doi: 10.1016/j.bbabio.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 13.Kmiec B, Glaser E. A novel mitochondrial and chloroplast peptidasome. PreP. Physiol. Plant. 2012;145:180–186. doi: 10.1111/j.1399-3054.2011.01531.x. [DOI] [PubMed] [Google Scholar]
- 14.Stahl A, Moberg P, Ytterberg J, Panfilov O, Brockenhuus Von Lowenhielm H, Nilsson F, Glaser E. Isolation and identification of a novel mitochondrial metalloprotease (PreP) that degrades targeting presequences in plants. J. Biol. Chem. 2002;277:41931–41939. doi: 10.1074/jbc.M205500200. [DOI] [PubMed] [Google Scholar]
- 15.Kambacheld M, Augustin S, Tatsuta T, Muller S, Langer T. Role of the novel metallopeptidase Mop112 and saccharolysin for the complete degradation of proteins residing in different subcompartments of mitochondria. J. Biol. Chem. 2005;280:20132–20139. doi: 10.1074/jbc.M500398200. [DOI] [PubMed] [Google Scholar]
- 16.Alikhani N, Berglund AK, Engmann T, Spanning E, Vogtle FN, Pavlov P, Meisinger C, Langer T, Glaser E. Targeting capacity and conservation of PreP homologues localization in mitochondria of different species. J. Mol. Biol. 2011;410:400–410. doi: 10.1016/j.jmb.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Falkevall A, Alikhani N, Bhushan S, Pavlov PF, Busch K, Johnson KA, Eneqvist T, Tjernberg L, Ankarcrona M, Glaser E. Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome. PreP. J. Biol. Chem. 2006;281:29096–29104. doi: 10.1074/jbc.M602532200. [DOI] [PubMed] [Google Scholar]
- 18.Nilsson Cederholm S, Backman HG, Pesaresi P, Leister D, Glaser E. Deletion of an organellar peptidasome PreP affects early development in Arabidopsis thaliana. Plant Mol. Biol. 2009;71:497–508. doi: 10.1007/s11103-009-9534-6. [DOI] [PubMed] [Google Scholar]
- 19.Alikhani N, Guo L, Yan S, Du H, Pinho CM, Chen JX, Glaser E, Yan SS. Decreased proteolytic activity of the mitochondrial amyloid-beta degrading enzyme, PreP peptidasome, in Alzheimer’s disease brain mitochondria. J. Alzheimers Dis. 2011;27:75–87. doi: 10.3233/JAD-2011-101716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caspersen, C.;Wang N, Yao J, Sosunov A, Chen X, Lustbader JW, Xu HW, Stern D, McKhann G, Yan SD. Mitochondrial Abeta: a potential focal point for neuronal metabolic dysfunction in Alzheimer’s disease. FASEB J. 2005;19:2040–2041. doi: 10.1096/fj.05-3735fje. [DOI] [PubMed] [Google Scholar]
- 21.Hansson Petersen CA, Alikhani N, Behbahani H, Wiehager B, Pavlov PF, Alafuzoff I, Leinonen V, Ito A, Winblad B, Glaser E, Ankarcrona M. The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc. Natl. Acad. Sci. USA. 2008;105:13145–13150. doi: 10.1073/pnas.0806192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manczak M, Anekonda TS, Henson E, Park BS, Quinn J, Reddy PH. Mitochondria are a direct site of A beta accumulation in Alzheimer’s disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum. Mol. Genet. 2006;15:1437–1449. doi: 10.1093/hmg/ddl066. [DOI] [PubMed] [Google Scholar]
- 23.Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, Wang N, Caspersen C, Chen X, Pollak S, Chaney M, Trinchese F, Liu S, Gunn-Moore F, Lue LF, Walker DG, Kuppusamy P, Zewier ZL, Arancio O, Stern D, Yan SS, Wu HABAD. directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 24.Pagani L, Eckert A. Amyloid-beta interaction with mitochondria. Int. J. Alzheimers Dis. 2011;2011:925050. doi: 10.4061/2011/925050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao J, Irwin RW, Zhao L, Nilsen J, Hamilton RT, Brinton RD. Mitochondrial bioenergetic deficit precedes Alzheimer’s pathology in female mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2009;106:14670–14675. doi: 10.1073/pnas.0903563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavlov PF, Moberg P, Zhang XP, Glaser E. Chemical cleavage of the overexpressed mitochondrial F1beta precursor with CNBr: a new strategy to construct an import-competent preprotein. Biochem. J. 1999;341(Pt 1):95–103. [PMC free article] [PubMed] [Google Scholar]
- 27.Kim G, Cole NB, Lim JC, Zhao H, Levine RL. Dual sites of protein initiation control the localization and myristoylation of methionine sulfoxide reductase A. J. Biol. Chem. 2010;285:18085–18094. doi: 10.1074/jbc.M110.119701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy MP. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Backman HG, Pessoa J, Eneqvist T, Glaser E. Binding of divalent cations is essential for the activity of the organellar peptidasome in Arabidopsis thaliana. AtPreP. FEBS Lett. 2009;583:2727–2733. doi: 10.1016/j.febslet.2009.07.040. [DOI] [PubMed] [Google Scholar]
- 30.Pinho CM, Bjork BF, Alikhani N, Backman HG, Eneqvist T, Fratiglioni L, Glaser E, Graff C. Genetic and biochemical studies of SNPs of the mitochondrial A beta-degrading protease, hPreP. Neurosci. Lett. 2010;469:204–208. doi: 10.1016/j.neulet.2009.11.075. [DOI] [PubMed] [Google Scholar]
- 31.Stahl A, Nilsson S, Lundberg P, Bhushan S, Biverstahl H, Moberg P, Morisset M, Vener A, Maler L, Langel U, Glaser E. Two novel targeting peptide degrading proteases, PrePs, in mitochondria and chloroplasts, so similar and still different. J. Mol. Biol. 2005;349:847–860. doi: 10.1016/j.jmb.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 32.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 33.Toda T, Nakamura M, Morisawa H, Hirota M, Nishigaki R, Yoshimi Y. Proteomic approaches to oxidative protein modifications implicated in the mechanism of aging. Geriatr. Gerontol. Int. 2010;10(Suppl. 1):S25–S31. doi: 10.1111/j.1447-0594.2010.00606.x. [DOI] [PubMed] [Google Scholar]
- 34.Levine RL, Moskovitz J, Stadtman ER. Oxidation of methionine in proteins: roles in antioxidant defense and cellular regulation. IUBMB Life. 2000;50:301–307. doi: 10.1080/713803735. [DOI] [PubMed] [Google Scholar]
- 35.Stadtman ER, Moskovitz J, Levine RL. Oxidation of methionine residues of proteins: biological consequences. Antioxid. Redox Signaling. 2003;5:577–582. doi: 10.1089/152308603770310239. [DOI] [PubMed] [Google Scholar]
- 36.Stadtman ER, Van Remmen H, Richardson A, Wehr NB, Levine RL. Methionine oxidation and aging. Biochim. Biophys. Acta. 2005;1703:135–140. doi: 10.1016/j.bbapap.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Ugarte N, Petropoulos I, Friguet B. Oxidized mitochondrial protein degradation and repair in aging and oxidative stress. Antioxid. Redox Signaling. 2010;13:539–549. doi: 10.1089/ars.2009.2998. [DOI] [PubMed] [Google Scholar]
- 38.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. USA. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo S, Levine RL. Methionine in proteins defends against oxidative stress. FASEB J. 2009;23:464–472. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine RL. Oxidative modification of glutamine synthetase. II. Characterization of the ascorbate model system. J. Biol. Chem. 1983;258:11828–11833. [PubMed] [Google Scholar]
- 41.Boschi-Muller S, Olry A, Antoine M, Branlant G. The enzymology and biochemistry of methionine sulfoxide reductases. Biochim. Biophys. Acta. 2005;1703:231–238. doi: 10.1016/j.bbapap.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 42.Moskovitz J. Methionine sulfoxide reductases: ubiquitous enzymes involved in antioxidant defense, protein regulation, and prevention of aging-associated diseases. Biochim. Biophys. Acta. 2005;1703:213–219. doi: 10.1016/j.bbapap.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Hansel A, Kuschel L, Hehl S, Lemke C, Agricola HJ, Hoshi T, Heinemann SH. Mitochondrial targeting of the human peptide methionine sulfoxide reductase (MSRA), an enzyme involved in the repair of oxidized proteins. FASEB J. 2002;16:911–913. doi: 10.1096/fj.01-0737fje. [DOI] [PubMed] [Google Scholar]
- 44.Vougier S, Mary J, Friguet B. Subcellular localization of methionine sulphoxide reductase A (MsrA): evidence for mitochondrial and cytosolic isoforms in rat liver cells. Biochem. J. 2003;373:531–537. doi: 10.1042/BJ20030443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalle-Donne I, Rossi R, Colombo R, Giustarini D, Milzani A. Biomarkers of oxidative damage in human disease. Clin. Chem. 2006;52:601–623. doi: 10.1373/clinchem.2005.061408. [DOI] [PubMed] [Google Scholar]
- 46.Madian AG, Myracle AD, Diaz-Maldonado N, Rochelle NS, Janle EM, Regnier FE. Differential carbonylation of proteins as a function of in vivo oxidative stress. J. Proteome Res. 2011;10:3959–3972. doi: 10.1021/pr200140x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taggart C, Cervantes-Laurean D, Kim G, McElvaney NG, Wehr N, Moss J, Levine RL. Oxidation of either methionine 351 or methionine 358 in alpha 1-antitrypsin causes loss of anti-neutrophil elastase activity. J. Biol. Chem. 2000;275:27258–27265. doi: 10.1074/jbc.M004850200. [DOI] [PubMed] [Google Scholar]
- 48.Cui ZJ, Han ZQ, Li ZY. Modulating protein activity and cellular function by methionine residue oxidation. Amino Acids. 2012;43:505–517. doi: 10.1007/s00726-011-1175-9. [DOI] [PubMed] [Google Scholar]
- 49.Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O’Donnell SE, Aykin-Burns N, Zimmerman MC, Zimmerman PJ, Ham AJ, Weiss RM, Spitz DR, Shea MA, Colbran RJ, Mohler PJ, Anderson ME. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinall H, Song ES, Hersh LB. Susceptibility of amyloid beta peptide degrading enzymes to oxidative damage: a potential Alzheimer’s disease spiral. Biochemistry. 2005;44:15345–15350. doi: 10.1021/bi050650l. [DOI] [PubMed] [Google Scholar]
- 51.Johnson KA, Bhushan S, Stahl A, Hallberg BM, Frohn A, Glaser E, Eneqvist T. The closed structure of presequence protease PreP forms a unique 10,000 A3 chamber for proteolysis. EMBO J. 2006;25(9):1977–1986. doi: 10.1038/sj.emboj.7601080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.